Abstract
Purpose:
To study the susceptibilities of Aspergillus species against amphotericin B in infectious keratitis and to find out if drug resistance had any association with the molecular characteristics of the fungi.
Materials and Methods:
One hundred and sixty Aspergillus isolates from the corneal scrapings of patients with keratitis were tested for susceptibilities to amphotericin B by broth microdilution method. These included Aspergillus flavus (64 isolates), A. fumigatus (43) and A. niger (53). Fungal DNA was extracted by glass bead vertexing technique. Polymerase chain reaction (PCR) assay was standardized and used to amplify the 28S rRNA gene. Single-stranded conformational polymorphism (SSCP) of the PCR product was performed by the standard protocol.
Results:
Of the 160 isolates, 84 (52.5%) showed low minimum inhibitory concentration (MIC) values (≤ 1.56 μg/ml) and were designated as amphotercin B-sensitive. Similarly, 76 (47.5%) had high MICs (≥ 3.12 μg/ml) and were categorized as amphotericin B-resistant. MIC50 and MIC90 values ranged between 3.12-6.25 μg/ml and 3.12-12.5 μg/ml respectively. A. flavus and A. niger showed higher MIC50 and MIC90 values than A. fumigatus. The SSCP pattern exhibited three extra bands (150 bp, 200 bp and 250 bp each) in addition to the 260 bp amplicon. Strains (lanes 1 and 7) lacking the 150 bp band showed low MIC values (≤ 1.56 μg/ml).
Conclusion:
A. niger and A. flavus isolates had higher MICs compared to A. fumigatus, suggesting a high index of suspicion for amphotericin B resistance. PCR-SSCP was a good molecular tool to characterize Aspergillus phenotypes in fungal keratitis.
Keywords: Antifungal sensitivity testing, amphotericin B, Aspergillus, fungal keratitis, polymerase chain reaction--single-stranded conformational polymorphism.
Fungal keratitis, if not diagnosed and treated early, can lead to corneal opacity and blindness.[1] Empiric therapy, without knowing the antifungal susceptibility pattern, often gives rise to complications, if the causative fungal agent is unresponsive to the drug.[2] In view of the increasing resistance amongst mycelial fungi to one or more antifungal agents,[3] it is often advisable to conduct the antifungal drug susceptibility testing before administering the drug. Not infrequently, amphotericin B in its topical form (0.15% drops) is used for the treatment of suspected cases of fungal keratitis in our setup,though natamycin is the preferred first line polyene antifungal. Since there are occasional reports of natamycin resistance amongst the clinical isolates of Aspergillus[4] as well as increasing reports of natamycin unresponsiveness in patients with mycotic keratitis,[5] it was felt imperative to study the response of the Aspergillus isolates towards the other commonly used polyene i.e. amphotericin B . However, antifungal sensitivity testing for filamentous fungi like Aspergillus species has certain limitations because the process is often cumbersome, time-consuming and requires standardization of many variables.[4,6] In view of the above , there is always a need for a rapid and confirmatory tool, which would be a reliable adjunct to the routine antifungal tests.
Recently, universal primers common to all fungi were used as a promising approach for clinical microbiological diagnosis.[7] Thus far, a few techniques were utilized in the past to classify and recognize medically important fungi.[8–10] In the same context, other researchers[11] put forth a novel approach for distinguishing opportunistic fungal pathogens by employing polymerase chain reaction (PCR)-based amplification of the conserved region of the 18S rRNA gene, followed by single-stranded conformational polymorphism (SSCP) assay. However, the applicability of this technique has never been sought in ocular infections. Since SSCP was used in the past to discriminate between and to characterize medically important opportunistic fungi, we decided to look for an alternative application of the above strategy[11] to elucidate if PCR-SSCP analysis could help in characterizing the Aspergillus species isolated from the corneas of patients with fungal kertitis, in terms of their drug susceptibility patterns.
Materials and Methods
One hundred and sixty Aspergillus species isolated from the corneal scrapings of patients with infective keratitis, comprising 64 isolates of Aspergillus flavus, 43 of A. fumigatus, and 53 of A. niger were subjected to testing for their susceptibilities to amphotericin B (Himedia, Mumbai, India) by the recommended protocol.[12] Candida parapsilosis ATCC 90018 and Candida krusei ATCC 6258, the standard reference strains were tested simultaneously with each batch of sensitivity testing as control experiments. MIC50 (50th percentile) and MIC90 (90th percentile) values of the drug against all the isolates were calculated using the SATA 10.0 software.
Glass bead vertexing method[13] was adopted and standardized according to our laboratory conditions. Fungal mycelia from a young culture were transferred to a locking microfuge tube using a sterile spatula and suspended in 400 μl extraction buffer [2% Triton X-100, 1% Sodium dodecyl sulfate (SDS), 100mM Tris-Cl (pH 8.0), and 1mM Ethylenediamine tetra-acetic acid (EDTA)]. Glass beads 0.6 mm in diameter (Beads and Ceramics, Plot No.3, UP State Industrial Development Corporation, Site No.2, Firozabad, UP, India) which were previously acid-washed and dried were added (500 mg per tube). This was followed by the addition of 400 μl phenol/chloroform/iso-amyl alcohol (Phe/Chl/IAA in the ratio of 24:24:1), and continuous vertexing for 30 min. The aqueous layer was then removed and re-extracted with an equal volume of Phe/Chl/IAA (24:24:1) twice, an equal volume of Chl/IAA (24:1) once, and precipitated with 0:1 volume of 10M ammonium acetate followed by 2.0 volume 100% ethanol. The resulting fungal DNA pellet was resuspended in 100 μl 10mM Tris (pH 8.0), 1mM EDTA and 1 μl 500 μg/ml RNAse (Boehringer Mannheim, Indianapolis, IN, USA). The final digest after RNAse treatment was incubated at 37°C for 1 h. To remove residual cellular debris, the tube was spun at high speed in a micro centrifuge for 10 min (10 000 g), and the supernatant was then transferred to a new tube.
The PCR was carried out using the following panfungal primers for the amplification of the 28S rRNA gene, common to all medically important fungi.[14]
5’ GTG AAA TTG T TG AAA GGG AA 3’
5’ GAC TCC TTG GTC CGT GTT 3’
All PCR reactions were carried out in a 25-μl reaction volume. The reagents added to the tube were 2.5 μl of 10 × PCR buffer, 2 μl of 10mM deoxynucleotidetriphosphate (dNTP) mix, 1 μl each of nucleic acid primer (10 pM), 1 unit Taq polymerase, 3 μl of DNA template and sterile water to make up volume to 25 μl.
All the above quantitative parameters were standardized one by one to give a reproducible result. PCR amplification conditions were as follows: an initial step at 94°C for 15 min, followed by 30 cycles carried out at 94°C for 30 sec, 50°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 15 min.
The above PCR protocol was standardized by varying different temperature conditions and the duration of the steps in the cycles of denaturation, annealing and extension. PCR product was visualized by subjecting a 10-μl volume of the amplified reaction mixture to electrophoresis in Tris-borate buffer on a 2% agarose gel incorporating 0.5 μg/ml ethidium bromide. The gel was examined on an ultraviolet transilluminator and photographed. A 50 bp ladder was run alongside the tests. The criterion for positivity was the presence of 260 bp amplicon.
The SSCP analysis was conducted according to the earlier improvised technique of Walsh et al.,[11] with minor modifications. Briefly, an acrylamide-bis-acrylamide-glycerol gel (9% acrylamide, 1.2% bis-acrylamide, 5% glycerol) was used to analyze the PCR product. The gel consisted of 6 ml of acrylamide (Cisco Research Laboratories Pvt. Ltd., Mumbai, India), 0.9114 ml of bis-acrylamide (Cisco Research Laboratories Pvt. Ltd., Mumbai, India), 1 ml of 5% glycerol, 1.92 ml of 10X Tris-borate-EDTA, and 10 ml of double-distilled water. After this, 100 μl of ammonium persulfate, and 20 μl of NNNN-tetramethylethylenediamine (TEMED), were added and mixed to induce gel polymerization, prior to the gels being poured. The Thermo EC 120 Mini Vertical Gel System was used for electrophoresis.
Gels were allowed to solidify for 1 h at room temperature. Solidified gels were set into the buffer chamber containing 0.5X Tris-borate EDTA running buffer. Samples were heated to 95°C for 5 min and then loaded directly onto the gels after mixing with 5 ul of loading dye (95% formamide, 0.5% xylene cyanol, 0.5% bromophenol blue).
Gels were run at room temperature at 50 V for 2 h along with a molecular weight marker (50 bp ladder). Gels were then silver-stained using AgNo3 and formaldehyde in appropriate concentrations for 15 min and developed by adding 0.76N NaOH and subsequently 750 μl of formaldehyde drop by drop. Lastly, the gels were photographed.
The above protocol was standardized previously so far as the acrylamide bis-acrylamide concentration, ammonium persulfate and TEMED concentration etc. were concerned.
Results
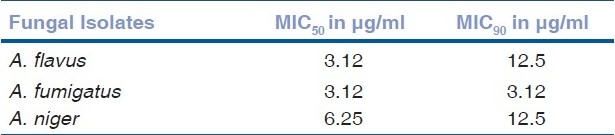
In accordance with the standardized criteria laid down in our previous study,[6] isolates showing MIC values of ≥3.12 μg/ml were designated as amphotericin B-resistant and those having MIC values of ≤1.56 μg/ml were labeled as sensitive. Therefore considering the cutoff value of 1.56 μg/ml, 84 (52.5%) of our isolates showed low MIC values (MICs ≤ 1.56 μg/ml) and 76 (47.5%) had high MIC values (MICs ≥ 3.12 μg/ml) as depicted in Table 1. Table 2 shows the MIC50 and MIC90 values of all the three groups of fungi. A. flavus, and A. niger had higher MIC50 and MIC90 values as compared to A. fumigatus.
Table 1.
Minimum inhibitory concentrations of amphotericin B for ocular isolates of Aspergillus species
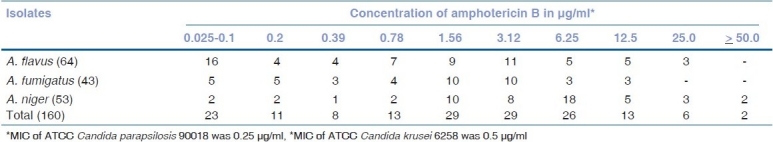
Table 2.
MIC50 and MIC90 of amphoterticin B against ocular isolates of Aspergillus species
The glass bead vertexing technique of fungal DNA extraction[13] worked perfectly well and yielded reproducible results according to our own laboratory conditions [Fig. 1]. PCR results were reproducible and we obtained 260 base pair (bp) bands from all the isolates using the panfungal primer for the amplification of the 28S rRNA genes [Fig. 2].
Figure 1.

Agarose gel electrophoresis showing the DNA bands after glass bead vertexing extraction method. Lane numbers 1-5 represent extracted DNA from five different fungi, all showing bands
Figure 2.
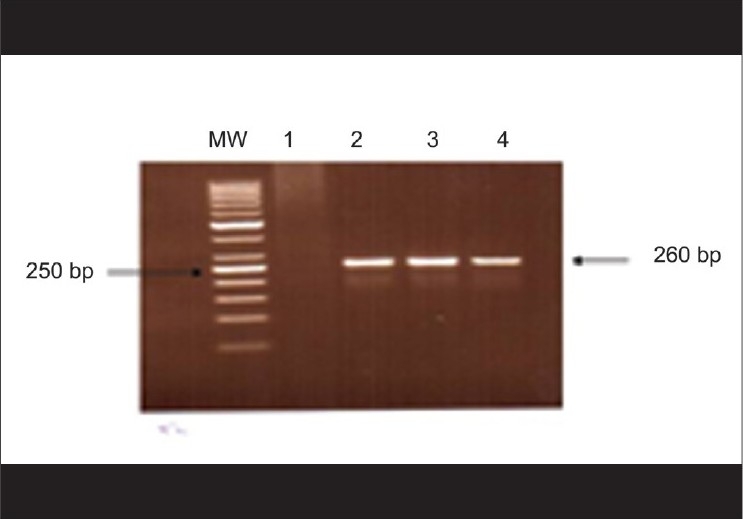
Amplified products of fungal PCR showing 260 bp fragments after agarose gel electrophoresis. Lane on the extreme left (MW) shows 50 bp ladder (Molecular Weight Marker)
SSCP patterns of the PCR products are depicted in Fig. 3. In addition to the 260 bp (PCR amplicon) band, we observed three more bands of 150 bp, 200 bp and 250 bp each. Whereas Lanes 2-6 had three bands (150, 250 and 260 bp) each, Lanes 1 and 7 were lacking the 150 bp band. The strain on Lane 7, however, was totally different from the rest, as it showed two clear bands of 200 and 250 bp in addition to the 260 bp band which was common to all the strains. Strains on lane numbers 1 and 7 as well as that on Lane 6 had lower MIC values (≤1.56 μg/ml; amphotericin B-sensitive) as compared to the rest. After analyzing four sets of experiments (each comprising seven test strains, and thus studying randomly selected 28 strains), it was our observation that all strains (excepting the one on Lane 6) lacking the 150 bp fragment were found to be sensitive to amphotericin B. In order to check the reproducibility of our results, all the four batches of experiments were repeated thrice. As the procedure was laborious and needed stringent parameters at every step, we could not afford to test more number of strains. However, of the total of 28 isolates studied, 15 (53.6%) were lacking the 150 bp band and had low MIC values (≤1.56 μg/ml; amphotericin B-sensitive); whereas 12 (43%) having the 150 bp band had high MIC values (≥ 3.12 μg/ml; amphotericin B-resistant). The only anonymous result was obtained with one strain (Lane 6, as stated above) that possessed the 150 bp band, but was amphotericin B-sensitive (MIC < 1.56 μg/ml).
Figure 3.
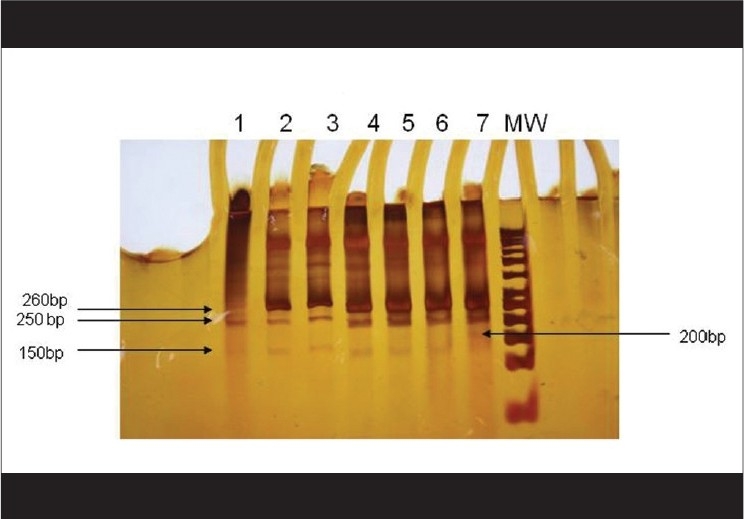
SSCP pattern of the PCR products of drug-resistant and drug-sensitive fungal isolates {Starting from the left; Lanes 2,3,4,5 and 6 show three bands each that are different from the Lanes1 and 7 which lack the 150 bp band. However, the pattern on Lane 7 is totally different from the rest as it shows three clear bands of 250 bp, 200 bp, in addition to the 260 bp band which is common to all. Products on Lanes 1 and 7 belonged to strains which had shown low MIC values (≤ 1.56 μg/ml)} Lane on the extreme right (MW) shows 50 bp ladder (Molecular Weight Marker)
Discussion
In the present study, the MIC90 values of all the three Aspergillus species were found to be higher than those reported for amphotericin B by others,[15,16] though the MIC50 values were comparable. This might be due to prolonged exposure to empiric therapy with amphotericin B topical by some of our patients, after acquiring the infection in the community, as it happens quite often in the semi-urban setup in India, much before they report to the tertiary care hospitals.[1] Aspergillus species being the commonest agents responsible for fungal keratitis in this part of the country,[2] some of the strains exhibiting such in vitro resistance would not be unusual. However, the overall picture was that corneal ulcer isolates of A. niger and A. flavus showed much higher MIC50 and MIC90s as compared to A. fumigatus. Thus, there should always be a high index of suspicion regarding amphotericin B resistance amongst A. niger and A. flavus isolates in infectious keratitis. In such a clinical situation the therapeutic modalities may need to be changed.
In our earlier study,[6] we noticed proteinase production as a phenotypic marker amongst the majority of the fungal isolates showing a high level of amphotericin B resistance. Clinical significance of proteinase was previously shown in Candida species isolated from vulvovaginitis cases and from cases with other deep-seated Candida infections.[17,18] Besides the above mentioned phenotypic characteristics, there was no molecular marker of virulence which could be ascribed to any of the filamentous fungi causing keratitis. The present study, however, documented the evidence that there were distinct SSCP patterns amongst Aspergillus species presenting with high MICs and low MICs against amphotericin B [Fig. 3]. Thus, a particular SSCP pattern could well be denoted as a molecular characteristic for the drug-resistant phenotype. As was evident from our observations, some of the isolates lacked the 150 bp b - Lanes 1, 7 [Fig. 3]. Truly, these were the strains which had lower MIC values as compared to the others. Thus, further studies on the molecular epidemiology of ocular fungal pathogens involving a large number of isolates will be required to predict the role, if any, of this 150 bp fragment in amphotericin B resistance. However, the fact that the presence of this fragment could be a predictor of such resistance, cannot be ruled out altogether, because 15 out of 16 strains that were sensitive to the drug (MIC ≤1.56 μg/ml) lacked this component. It is noteworthy that the utility of PCR-SSCP has been documented in the past for the study of molecular taxonomy, epidemiologic investigations and for the detection of drug resistance.[11,19] SSCP analysis was earlier used as a valuable tool in understanding the mechanism of drug resistance in Mycobacterium tuberculosis.[20]
PCR-SSCP has widely been used earlier as a screening method for the detection of mutations, by amplifying the target sequence of interest and separating this as single-stranded molecules by electrophoresis in a nondenaturating polyacrylamide gel.[21] Many workers used this technique to identify sequence variation in a single strand of DNA due to the adaptability of the single strand to unique conformation under nondenaturating condition.[21,22] As the sensitivity and resolution of PCR-SSCP can be influenced by many parameters, including size and guanine-cytosine (GC) content of the PCR product; gel temperature during electrophoresis; buffer composition (e.g. ionic strength, pH etc.); buffer additives, mainly glycerol, formamide, polyethylene glycol; gel matrix composition and concentration; as well as primer concentration in the PCR product, we, therefore, standardized all these variables to suit to our own laboratory conditions as mentioned in the methods. Thus we utilized this technique to analyze the amphotericin B susceptible and resistant phenotypes of Aspergillus species in our study.
As resistance to antifungal drugs continues to expand, and antifungal sensitivity testing of filamentous fungi is time-consuming and needs standardization of many variables,[3,12] elucidation of a good marker for resistance becomes increasingly important. In our previous study we hypothesized that proteinase production was a good phenotypic characteristic of amphotericin B resistance amongst fungi in mycotic keratitis.[6] Although extrapolations from our present data did not exactly support that PCR-SSCP was able to unfold the mechanism of drug resistance, it would not be inappropriate to infer that this technique had the potential for further distinctive characterization (beyond proteinase activity) of amphotericin B-resistant phenotypes of Aspergillus species in fungal keratitis.
However, the future novel applicability of the SSCP technique in fungal infections, particularly so in the fields of molecular epidemiology, pathogenesis, and drug resistance needs further attention with much more extensive study and diligent analysis.
Footnotes
Source of Support: Indian Council of Medical Research.
Conflict of Interest: None declared.
References
- 1.Agarwal V, Biswas J, Madhwan HN, Mangat G, Reddy MK, Saini JS, et al. Current perspectives in infectious keratitis. Indian J Ophthalmol. 1994;42:171–91. [PubMed] [Google Scholar]
- 2.Satpathy G, Vishalkashi P. Ulcerative keratitis: Microbial profile and sensitivity pattern: A five year study. Ann Ophthalmol. 1995;27:301–6. [Google Scholar]
- 3.Pujol I, Guarro J, Llop C, Soler L, Fernandez BJ. Comparison study of broth macrodilution and microdilution antifungal susceptiblity tests for the filamentous fungi. Antimicrob Agents Chemother. 1996;40:2106–10. doi: 10.1128/aac.40.9.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day S, Lalitha P, Haug S, Fothergill AW, Cevallos V, Vijayakumar R, et al. Activity of antibiotics against Fusarium and Aspergillus. Br J Ophthalmol. 2009;93:116–9. doi: 10.1136/bjo.2008.142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahman MR, Johnson GJ, Husain R, Howlader SA, Minassian DC. Randomised trial of 0.2% chlorhexidine gluconate and 2.5%% natamycin for fungal keratitis in Bangladesh. Br J Ophthalmol. 1998;82:919–25. doi: 10.1136/bjo.82.8.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayak N, Satpathy G, Prasad S, Vajpayee RB, Pandey RM. Correlation of proteinase production with amphotericin B resistance in fungi from mycotic keratitis. Ophthal Res. 2010;44:113–8. doi: 10.1159/000315360. [DOI] [PubMed] [Google Scholar]
- 7.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. San Diego: Academic Press Inc; 1990. pp. 315–24. [Google Scholar]
- 8.Hopfer RL, Walden P, Setterquist S, Highsmith WE. Detection and differentiation of fungi in clinical specimens using polymerase chain reaction (PCR) amplification and restriction enzyme analysis. J Med Vet Mycol. 1993;31:65–75. doi: 10.1080/02681219380000071. [DOI] [PubMed] [Google Scholar]
- 9.Kan VL. Polymerase chain reaction for the diagnosis of candidemia. J Infect Dis. 1993;168:779–83. doi: 10.1093/infdis/168.3.779. [DOI] [PubMed] [Google Scholar]
- 10.Maiwald M, Kappe R, Sonntag HG. Rapid presumptive identification of medically relevant yeasts to the species level by polymerase chain reaction and restriction enzyme analysis. J Med Vet Mycol. 1994;32:115–22. doi: 10.1080/02681219480000161. [DOI] [PubMed] [Google Scholar]
- 11.Walsh TJ, Francesconi A, Kasai M, Chanock SJ. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J Clin Microbiol. 1995;33:3216–20. doi: 10.1128/jcm.33.12.3216-3220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pujol I, Guarro J, Sala J, Riba MD. Effects of incubation temperature, inoculum size and time of reading on broth microdilution susceptibility test results on amphotericin B against Fusarium. Antimicrob Agents Chemother. 1997;41:808–11. doi: 10.1128/aac.41.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burik AHV, Schreckhise RW, White TC, Bowden RA, Myerson D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med Mycol. 1998;36:299–303. [PubMed] [Google Scholar]
- 14.Anand AR, Madhavan HN, Neelam V, Therese LK. Use of polymerase chain reaction in the diagnosis of endophthalmitis. Ophthalmology. 2001;108:326–30. doi: 10.1016/s0161-6420(00)00517-0. [DOI] [PubMed] [Google Scholar]
- 15.Lalitha P, Shapiro BL, Srinivasan M, Venkatesh PN, Acharya NR, Fothergill AW, et al. Antimicrobial susceptibility of Fusarium, Aspergillus and other filamentous fungi isolated from keratitis. Arch Ophthalmol. 2007;125:789–93. doi: 10.1001/archopht.125.6.789. [DOI] [PubMed] [Google Scholar]
- 16.Therese KL, Bagyalakshmi R, Madhavan HN, Deepa P. In-vitro susceptibility testing by agar dilution method to determine the minimum inhibitory concentrations of amphotericin B, fluconazole and ketoconazole against ocular fungal isolates. Indian J Med Microbiol. 2006;24:273–9. doi: 10.4103/0255-0857.29386. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti A, Nayak N, Talwar P. In vitro proteinase production by Candida species. Mycopathologia. 1991;114:163–8. doi: 10.1007/BF00437209. [DOI] [PubMed] [Google Scholar]
- 18.Cassone A, Bernadis FD, Medello F, Ceddia T. Evidence for a correlation between proteinase secretion and vulvovaginal candidiasis. J Infect Dis. 1987;156:777–83. doi: 10.1093/infdis/156.5.777. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell TG, Saudin RL, Bowman BH, Meyer W, Merz WG. Molecular mycology: DNA probes and applications of PCR technology. J Med Vet Mycol. 1994;32:3351–66. doi: 10.1080/02681219480000961. [DOI] [PubMed] [Google Scholar]
- 20.Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs WR, Jr, et al. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: A molecular study. Lancet. 1994;344:293–8. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 21.Sheffield VC, Beck JS, Kwitek AE, Sandstrom DW, Stone EM. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics. 1993;16:325–32. doi: 10.1006/geno.1993.1193. [DOI] [PubMed] [Google Scholar]
- 22.Lugovtsev VY, Vodeiko GM, Strupczewski CM, Levandowski RA. Simple and rapid strategy for genetic characterization of influenza B virus reassortants. J Virol Met. 2005;124:203–10. doi: 10.1016/j.jviromet.2004.11.024. [DOI] [PubMed] [Google Scholar]


