Abstract
In Sindbis, Venezuelan equine encephalitis and related alphaviruses, the polymerase is translated as a fusion with other non-structural proteins via readthrough of a UGA stop codon. Surprisingly, earlier work reported that the signal for efficient readthrough comprises a single cytidine residue 3′-adjacent to the UGA. However, analysis of variability at synonymous sites revealed strikingly enhanced conservation within the ∼150 nt 3′-adjacent to the UGA, and RNA folding algorithms revealed the potential for a phylogenetically conserved stem–loop structure in the same region. Mutational analysis of the predicted structure demonstrated that the stem–loop increases readthrough by up to 10-fold. The same computational analysis indicated that similar RNA structures are likely to be relevant to readthrough in certain plant virus genera, notably Furovirus, Pomovirus, Tobravirus, Pecluvirus and Benyvirus, as well as the Drosophilia gene kelch. These results suggest that 3′ RNA stimulatory structures feature in a much larger proportion of readthrough cases than previously anticipated, and provide a new criterion for assessing the large number of cellular readthrough candidates that are currently being revealed by comparative sequence analysis.
INTRODUCTION
There are two types of exceptions to universality of the genetic code. In one, the meaning of a codon is globally reassigned in a context independent manner (1). In the other, codon redefinition is in competition with standard decoding and it is codon context dependent (2). Though there is an example where the meaning of a sense codon is redefined (3), most cases of codon redefinition involve one of the three stop codons of the standard code (UGA, UAG or UAA) specifying an amino acid at least a proportion of the time that it is decoded. Where the significant feature of stop codon redefinition is to allow ribosomes to continue translation into a downstream open reading frame (ORF), rather than the identity of the amino acid specified, then it is generally termed stop codon readthrough (RT) (4). In contrast, when selenocysteine or pyrrolysine are specified by UGA or UAG, respectively, then the important features are the special properties of these non-universal amino acids (5–7). Both types of non-global codon redefinition are just one aspect of the variety of ways (collectively referred to as ‘recoding’) in which genetic readout can be dynamically altered in a site- or mRNA-specific manner (8,9).
Numerous studies have shown that the identity of the 3′-adjacent nucleotide influences stop codon leakiness in both prokaryotes and eukaryotes and, correspondingly, there is considerable bias in the identity of the nucleotide at this position for natural gene terminators (10–16). Of great interest was the discovery that RT of the coat protein (CP) gene terminator of the phage Qβ yields a greatly extended protein that is important for viral propagation (17,18). Shortly afterwards, studies that utilized purified yeast suppressor tRNAs in in vitro experiments found that several plant viruses, including tobacco mosaic tobamovirus (TMV), also utilize RT to express their replicase proteins (19–21). Similarly murine leukemia gammaretrovirus (MuLV), whose relevant sequence is identical to that in xenotropic MuLV-related virus (XMRV), utilizes RT of the gag gene terminator to allow ribosomes to enter the pol gene and synthesize the Gag-Pol polyprotein that is the source of viral reverse transcriptase (22,23). MuLV Pol binds to the translation release factor, eRF1, and non-interacting mutants of Pol failed to synthesize adequate levels of Gag-Pol to permit replication (24). This raises the possibility of temporal control of RT (25). The efficiency of RT in the Drosophila gene kelch also appears to be developmentally regulated (26). Two other Drosophila genes are known to employ RT—headcase and out-at-first—though another approximately 150 candidate cases have recently been identified via comparative genomic approaches utilizing sequences from 12 Drosophila species (27–29). Although some of these candidates may actually be cases of alternative splicing or RNA editing, the indication is that utilized RT may be significantly more common in cellular organisms than previously supposed.
Several alphaviruses, including Sindbis virus (SINV), utilize RT of a UGA stop codon in their replicase gene (30,31). For SINV, primarily on the basis of in vitro translation studies, the only contextual feature reported to be important for RT was the identity of the cytidine nucleotide immediately 3′ of the stop codon, directly analogous to the results of the early stop codon leakiness studies (32). Similarly, in the tobraviruses (specifically tobacco rattle virus) and, by implication, the pecluvirus, furovirus and pomovirus replicase gene, and the furovirus CP-extension gene, it has been reported that RT of the UGA stop codon might depend on just the three 3′-adjacent nucleotides (33). For these plant viruses, and alphaviruses that utilize RT, the consensus motif in wild-type (WT) viruses is UGA-CUA or UGA-CGG (34). In contrast, for TMV (where the RT codon is UAG), plant tissue culture experiments showed that the 6 nt immediately 3′ of the stop codon are relevant, with the consensus motif for efficient RT being UAG-CAR-YYA (35,36). The same motif is utilized by a number of other plant viruses, while the motif UAG-CAR-NBA stimulates RT in yeast (37). In terms of 5′ stimulatory motifs, adenines at the −1 and −2 nucleotide positions have been shown to positively modulate RT in yeast and are a feature common to many virus RT sites, notably in the tobamoviruses, poleroviruses and luteoviruses (38).
For a relatively small number of cases of utilized RT, the known stimulatory signals involve an mRNA structure 3′ of the stop codon. In MuLV, in vitro translation studies showed that a compact pseudoknot structure 3′ of the gag terminator, UAG, is essential for meaningful levels of RT, with the identity of certain nucleotides in the 8 nt ‘spacer’ region between the stop codon and the pseudoknot, as well as some of the nucleotides in loop 2 of the pseudoknot, being important (39–42). The location of the pseudoknot (8 nt 3′ of the stop codon) may permit it to act at the mRNA unwinding site half-way through the mRNA entrance channel of the ribosome (43). A very different stimulatory element is present in the plant luteoviruses, where RT at the end of the CP gene produces a much larger CP-extension protein that is important for aphid transmission (44). In the best-studied of these viruses, barley yellow dwarf, both 3′-adjacent sequences and an element ∼700–750 nt 3′ of the UAG stop codon have been identified as important for RT and long-range RNA base pairing between the 3′-proximal and 3′-distal elements has been suggested as a possible mechanism (45). Similar results were found for beet western yellows virus (46).
Although cytidine residues are under-represented at the position immediately 3′-adjacent to UGA (and other) terminators in eukaryotes, they are by no means absent (11,12). Thus we hypothesized that, at least in vivo, RT in SINV and other alphaviruses might be modulated by additional sequence elements. To test for the existence of such elements, we investigated the degree of phylogenetic conservation at synonymous sites downstream of known RT stop codons in alphavirus genomes, and then extended the analyses to other RNA viruses and selected cellular RT genes. Regions of enhanced conservation at synonymous sites are indicative of overlapping functional elements such as RNA secondary structures or primary nucleotide sequences with functions in addition to amino acid coding. In many cases, and in particular those cases where RT of a UGA codon had been previously assumed to be stimulated simply by the 3′-adjacent nucleotides CUA or CGG, we found considerably enhanced conservation at synonymous sites in the 3′-adjacent sequence, typically extending over a region of 100–200 nt 3′-adjacent to the stop codon. Here, we computationally and experimentally explore these conserved regions and their significance for RT.
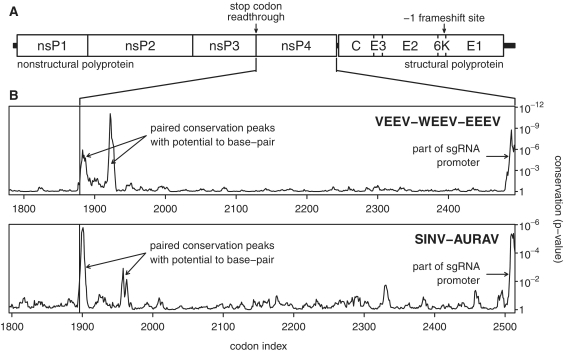
The genus Alphavirus encompasses approximately 30 described species, many of which infect humans and livestock, causing rashes, painful arthritis, fever and potentially fatal encephalitis (reviewed in reference 47; see reference 48 for a phylogeny). Transmission is generally via arthropods such as mosquitoes. The single-stranded positive sense genomic RNA is about 11–12 kb long and contains two long ORFs separated by a short non-coding sequence (Figure 1A). The 5′-proximal ORF codes for the non-structural proteins nsP1–nsP2–nsP3–nsP4 while the 3′-proximal ORF, which is translated from a subgenomic RNA, codes for the structural polyprotein C-E3-E2-6K-E1 and, via programmed ribosomal frameshifting, C-E3-E2-TF (49). In SINV, Venezuelan equine encephalitis (VEEV), eastern equine encephalitis (EEEV), western equine encephalitis (WEEV) and related alphaviruses, a UGA stop codon separates the coding sequence for nsP4 (RNA-dependent RNA polymerase, RdRp) from the coding sequence for nsP123 (30,50). In contrast, the salmonid alphaviruses lack the UGA stop codon while, for alphaviruses in the Semliki Forest complex, the stop codon tends to be present in some but not all strains even within a single species, possibly as a result of conflicting selective forces in alternating arthropod and vertebrate hosts (passaging in cell culture may also drive selection for or against a stop codon at this location; see ref. 51 and references therein).
Figure 1.
Synonymous site conservation in alphaviruses. (A) Map of the alphavirus genome. (B) Synonymous site conservation in VEEV-EEEV-WEEV and SINV-AURAV sequence alignments. The plot depicts the probability that the degree of conservation within a 9-codon sliding window could be obtained under a null model of neutral evolution at synonymous sites. Note that the RT stop codon itself has been excluded from the conservation statistics. In order to map the conservation statistic onto the coordinates of a specific sequence in each alignment, all alignment columns with gaps in a chosen reference sequence were removed prior to calculation of conservation. The following reference sequences (GenBank accession numbers) were used: VEEV NC_001449, SINV NC_001547.
MATERIALS AND METHODS
Computational analysis
Virus sequences were obtained from GenBank in May 2009, updated in October 2010, and processed using BLAST, EMBOSS and ClustalW (52–54). The accession numbers of all sequences used are given in the Supplementary Data. Coding sequences were extracted, translated, aligned with ClustalW and back-translated to nucleotide sequence alignments, and manually adjusted in a few cases. For the synonymous site conservation plots, alignment columns in which the reference sequence (Figure 4) contained gap characters were removed so that the plots are in reference sequence coordinates. RNA structures were predicted using a combination of Vienna RNA RNAfold and alidot, pknotsRG and manual inspection (55,56).
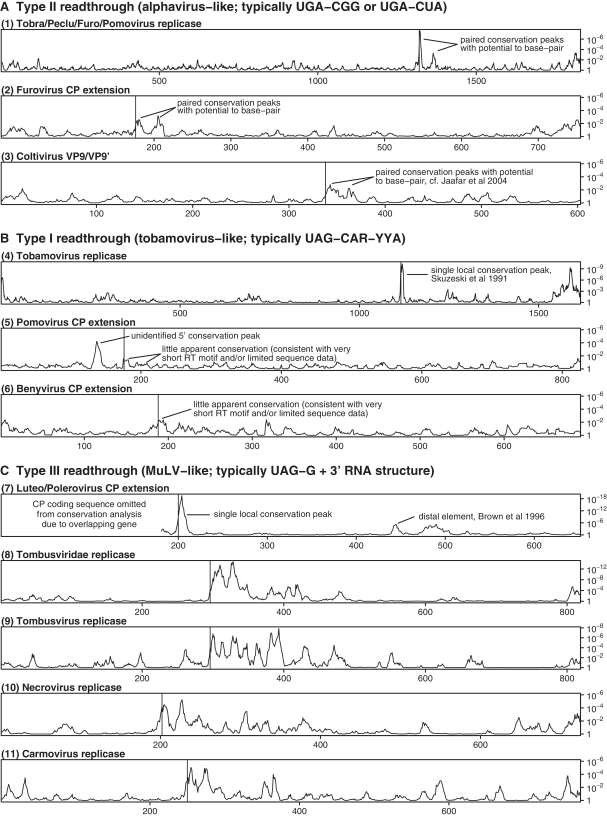
Figure 4.
Synonymous site conservation for RT coding sequences in various plant viruses and also coltiviruses. RT Types II (A), I (B) and III (C) are according to the classification of Beier and Grimm (34). Codon index within the RT coding sequence is plotted on the horizontal axis and the conservation P-value is plotted on the vertical axis (see caption to Figure 1B for details). In each case, the RT site is indicated with a vertical line. The following reference sequences (GenBank accession numbers) were used: (1) NC_002041, (2) NC_002042, (3) NC_004180, (4) NC_001367, (5) NC_003724, (6) NC_003515, (7) NC_004750, (8) NC_001554, (9) NC_001554, (10) NC_001777 and (11) NC_003821. The Tombusviridae plot excludes viruses from the genus Dianthovirus, in which the replicase is expressed via ribosomal frameshifting instead of RT.
Conservation at synonymous sites was analyzed as described in ref. (57; a procedure inspired by the SSSV statistic of ref. 58). The procedure takes into account whether synonymous site codons are 1-, 2-, 3-, 4- or 6-fold degenerate and the differing probabilities of transitions and transversions. Briefly, for a given pair of sequences within an alignment, a codon position was defined as a synonymous site if the same amino acid was encoded in both sequences. A ‘null’ substitution model was defined such that the relative probability of each possible synonymous codon substitution (including substitution with itself) at such sites may be calculated by assuming that the component nucleotides evolve neutrally. Neutral evolution was modelled using a Kimura nucleotide substitution matrix with κ = 3 (59). For each sequence pair, the divergence parameter t was set so that the total expected number of nucleotide substitutions at synonymous sites under the null model was equal to the total observed number. Next, the difference between the expected number and observed number of nucleotide substitutions was calculated at each synonymous site in the pairwise comparison. The variance at each site was calculated from the expected probabilities of each possible synonymous codon substitution, assuming a multinomial distribution. Statistics were summed, at each alignment codon position, over a phylogenetic tree as described in ref. (60). Finally the statistics were averaged over a sliding window. An approximate P-value (probability that the mean conservation in the sliding window would be as high as observed if the null model were true) was also calculated, under the assumption of a normal distribution as an approximation to the sum of many independent multinomial distributions.
Dual luciferase reporter constructs
The sequences encompassing the RT site and the predicted 3′ stem–loop structure for VEEV and SINV were synthesized by GenScript and cloned into the XhoI and BglII sites of pDluc, a derivative of the p2luc vector (61,62). The firefly luciferase gene is in the same reading frame relative to the upstream renilla luciferase gene such that RT of the stop codon results in a renilla-firefly luciferase fusion product. Derivative constructs were generated by PCR using appropriate primers and recloning into pDluc. All plasmids were verified by DNA sequencing.
In vitro RT assays
Plasmid DNAs (0.2 μg) were used as templates in 10 μl reactions of the rabbit reticulocyte lysate TNT® T7 Quick Coupled Transcription/Translation system (Promega). 35S-methionine (Perkin Elmer) was included in the reactions and protein products were separated by SDS–PAGE. Dried gels were analyzed using a Typhoon PhosphorImager (GE Healthcare) and the amount of radioactivity in each product was determined using the ImageQuant 5.2 program (Molecular Dynamics). After normalization for the number of methionine residues in termination and RT products (9 and 22, respectively), the RT efficiencies were calculated as [RT/(RT + termination)].
Tissue culture RT assays
RT assays were performed using the dual luciferase reporter constructs, as previously described (62,63). To control for possible differences in stability of specific mRNA sequences, each RT construct was compared with a control construct that was identical except that the TGA stop codon was replaced with a TGG codon. RT efficiencies were calculated as (firefly activity/renilla activity) for the RT sequence normalized by (firefly activity/renilla activity) for the corresponding TGG control sequence. Standard deviations were calculated based on six independent transfections.
RESULTS
Computational analysis of alphavirus sequences
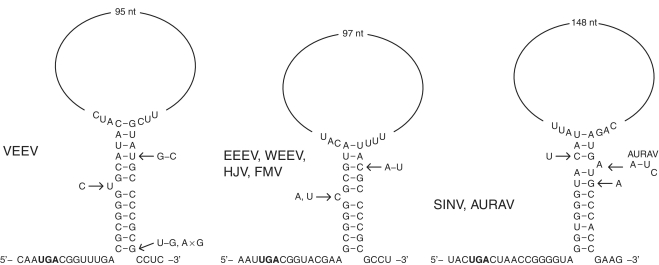
Sequence alignments of coding sequences containing RT stop codons were generated for a number of RNA virus taxa and the degree of conservation at synonymous sites was analyzed as described in the ‘Materials and Methods’ section. For an alignment of 63 VEEV, EEEV and WEEV sequences, this analysis revealed significantly enhanced conservation in a region comprising the ∼140 nt 3′-adjacent to the RT stop codon and a 9-codon sliding window size clearly resolved the conservation into two distinct peaks (Figure 1B). Inspection of the sequence alignment demonstrated the potential for base pairing between the sequences corresponding to these two peaks to form a stem–loop structure. In VEEV, the 5′-end of the 5′ component of the stem is separated from the stop codon by an 8–9 nt ‘spacer’ and the 5′ and 3′ components of the stem are separated by a less-conserved ‘loop’ region (which may nonetheless contain structured elements) of 101 nt (Figure 2). The predicted stem has 11–12 bp with a 1 nt asymmetric bulge in the centre of the 5′ component and, despite the enhanced conservation, is further supported by a compensatory A:U to G:C substitution that occurs in some strains at the fourth base pair from the ‘top’ of the stem. In EEEV and WEEV, the predicted stem has 10 bp with a 1 nt asymmetric 5′ bulge, and is separated from the stop codon by a 9 nt ‘spacer’ (Figure 2). Again, the predicted stem is supported by a compensatory G:C to A:U substitution in the related Fort Morgan virus (FMV). High conservation was also noted for the 1–2 codons immediately 3′-adjacent to the 3′ component of the predicted stem in VEEV, EEEV and WEEV.
Figure 2.
Predicted RNA structures for selected alphaviruses. Reference sequences shown are for NC_001449 (VEEV), NC_003899 (EEEV) and NC_001547 (SINV). Variations within the predicted stems are indicated but variations in single-stranded regions are omitted. AURAV is similar to SINV but the unpaired bulge nucleotide is positioned 1 nt further 3′.
With respect to the non-structural polyprotein, SINV and Aura virus (AURAV) form a separate clade from VEEV, WEEV and EEEV but, again, the conservation analysis revealed striking tandem conservation peaks 3′ of the RT site (Figure 1B) and, again, the conservation peaks corresponded to sequences with the potential to base pair to form an RNA structure—this time comprising an 11 bp stem with a 1 nt asymmetric 3′ bulge, a 12 nt ‘spacer’ from the RT stop codon, and a 154 nt ‘loop’ region (Figure 2). For those alphavirus species where there appears to be a constant flux between presence and absence of the RT stop codon, it is not unreasonable to suspect that the 3′ structure, if any, will be present whether or not the stop codon is present in any particular sequence. However, although we found the potential for conserved RNA stems to form in a number of these species (e.g. Ross River, getah, Semliki Forest and chikungunya viruses; Figure 3 and Supplementary Data), the range of divergences in the available sequence data proved inadequate to obtain supporting evidence from an analysis of conservation at synonymous sites.
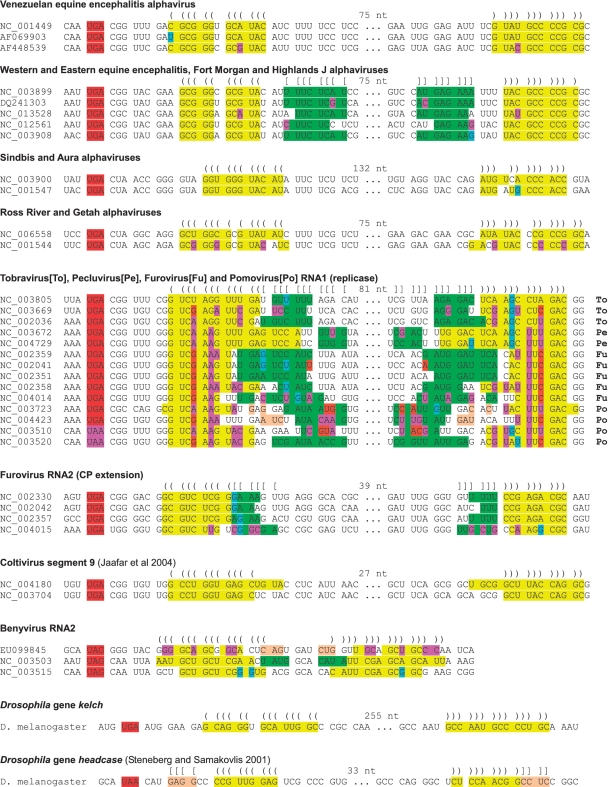
Figure 3.
Predicted RNA structures for a selection of RT cassettes (see Supplementary Data for considerably more annotated sequences). The RT stop codon is indicated in red or pink. Predicted stems are indicated in yellow (for the main, or basal, stem) and green (stem extension), and predicted base pairings are indicated with ‘()’s and ‘[]’s. In general, the large loop regions are also predicted to fold but the fold is less highly conserved. Single substitutions that preserve the predicted base pairings (e.g. G-C to G-U and G-U to A-U) are indicated with pale blue. Compensatory substitutions (paired substitutions that preserve the predicted base pairings) are indicated with pink or orange. For the tobravirus, pecluvirus, furovirus and pomovirus RNA1 sequences, cross-family variations are indicated in the lower (yellow) stem but, within the upper (green) stem (whose position varies between different genera), only cross-genus variations are indicated. GenBank accession numbers of representative virus sequences are indicated; see Supplementary Data for the individual species names.
Tobraviruses, pecluviruses, furoviruses, pomoviruses and coltiviruses
Curiously this phenomenon was not just limited to the alphaviruses. The potential to form an extended stem–loop structure 3′-adjacent to a RT stop codon—phylogenetically conserved and supported by a pair of peaks in synonymous site conservation—was also found in a number of plant virus RT cases, for example, in the replicase gene in the genera Tobravirus, Pecluvirus, Furovirus and Pomovirus (family Virgaviridae; ref. 64). Here, with a few exceptions in certain species, RT occurs at a UGA stop codon followed by the triplet CGG (pomoviruses beet virus Q and beet soil-borne virus utilize a UAA stop codon instead of UGA, and potato mop–top pomovirus has 3′-adjacent CGC instead of CGG). The motif xxA-stop-CGx-xxx-xxG, however, is completely conserved in the 35 sequences analyzed (despite lack of amino acid conservation at the −1 and +3 codons). Our analysis supported the existence of an RNA stem with typically 12+ bp, separated from the stop codon by a ‘spacer’ of 8 nt, together spanning a region of 138 nt 3′-adjacent to the stop codon and corresponding to two striking peaks in synonymous site conservation 3′-adjacent to the RT site (Figures 3 and 4). Further, the predicted stem is well-supported by a large number of compensatory substitutions—i.e. paired substitutions that preserve the predicted base pairings—between the different species (Figure 3 and Supplementary Data).
The furoviruses and pomoviruses have a second RT site in the CP gene. Here, however, there is a marked dichotomy between the two genera in the RT context. In the furoviruses, the RT context is generally UGA-CGG (UGA-UGG in the highly divergent sorghum chlorotic spot virus, AB033692) and there was evidence for tandem synonymous site conservation peaks and an associated stem–loop structure that, together with a 7 nt ‘spacer’, covered 96 nt 3′-adjacent to the stop codon (Figures 3 and 4). In the pomoviruses, however, the RT context is generally A-UAG-CAA-UYA (A-UAA-CAA-UUA in the highly divergent broad bean necrosis virus, D86637) and the synonymous site conservation analysis failed to reveal extended conservation in the vicinity of the RT site (Figure 4). Thus the furovirus context and predicted structure is alphavirus-like while the pomovirus context and lack of predicted structure is tobamovirus-like (see below). The animal-infecting coltiviruses also have an alphavirus-like RT site (UGA-CGG) in the VP9/VP9′-coding sequence and, again, there is potential to form a 3′-adjacent RNA stem–loop structure (Figure 3; as noted previously in ref. 65), which is tentatively supported by our conservation analysis (Figure 4).
Tobamoviruses, luteoviruses, benyviruses, tombusviruses and other viruses
Stop codon RT is also utilized by members of the plant virus taxa Tombusviridae, Luteoviridae, Benyvirus and Tobamovirus but the RT signals for these viruses had previously been grouped separately from those utilized by the alphaviruses, coltiviruses, tobraviruses, pecluviruses, furoviruses and pomoviruses (excepting pomovirus RNA2), and our analysis likewise supported this distinction at the level of extended 3′-adjacent synonymous site conservation (34). In the case of the tobamoviruses, greatly enhanced synonymous site conservation is seen from codons −1 to +3 relative to the UAG stop codon, and the motif xxA-UAG-CAA-UUA-xxG is completely conserved in the 105 sequences analyzed (despite lack of amino acid conservation at the −1 and +3 codons). However, more extended conservation of the type seen in the alphaviruses was not observed (Figure 4). In the luteoviruses and poleroviruses (family Luteoviridae), the stop codon context AAA-UAG-GUA is completely conserved in all except one of 247 sequences analyzed (rose spring dwarf-associated virus, EU024678, has GAA-UGA-CGG), and enhanced synonymous site conservation was also observed over several further codons, especially codons −1 to +5. However, while this region may well interact with distal elements as discussed in ref. (45), the extended 3′-adjacent conservation of the type seen in the alphaviruses was not observed in the Luteoviridae (Figure 4). The highly conserved local nucleotide contexts of the different RT sites mentioned here have been noted, discussed and characterized in detail in a number of previous works (34 and references therein). A compilation of our own sequence analysis is given in the Supplementary Data and, to our knowledge, represents the largest such compilation to date.
In the benyviruses—which generally have a tobamovirus-like stop codon context (i.e. UAG-CAA-UUA; however, highly divergent rice stripe necrosis virus, EU099845, has UAG-GGG-UAC), the potential was observed for a local stem–loop structure (e.g. 9 nt spacer, 12 nt stem, 8 nt loop in beet necrotic yellow vein virus, D84411; Figure 3), but there was insufficient sequence data to obtain strong support from the synonymous site conservation analysis (Figure 4). Previous deletion experiments in beet necrotic yellow vein benyvirus have shown, incidentally, that RT efficiency is considerably reduced when sequence corresponding to codons +3 to +34 from the RT codon is deleted, even though the immediately 3′-adjacent nucleotide context UAG-CAA-UUA is left intact (66). In contrast, deletion of codons +28 to +61 had little effect on RT. These results indicate that there is an additional stimulatory element within the region defined by codons +3 to +27, consistent with the predicted stem–loop structure (codons +4 to +14).
In the Tombusviridae family (including genera Tombusvirus, Carmovirus, Necrovirus and others), RT occurs at a UAG stop codon followed by GGR, but enhanced synonymous site conservation was observed for approximately 200 codons 3′-adjacent to the UAG (Figure 4). Some of this conservation, however, may be explained by other conserved elements in the region (see ref. 67 and references therein; see also refs 68, 69). RNA folding software predicted alphavirus-like 3′-adjacent stem–loops in some species and more complex structures in other species, a detailed analysis of which is beyond the scope of this article. The RT site in gammaretroviruses has been studied in depth and our computational analysis supported the known stimulatory spacer sequence and pseudoknot structure but did not reveal further conservation in the vicinity (39–42). RT sites in enamoviruses, carrot red leaf luteovirus-associated RNA, Middelburg and Barmah Forest alphaviruses, Providence tetravirus and others, were not analyzed in detail due to lack of sequence data for useful comparative computational analysis (70–72).
Non-selenocysteine RT in cellular genes
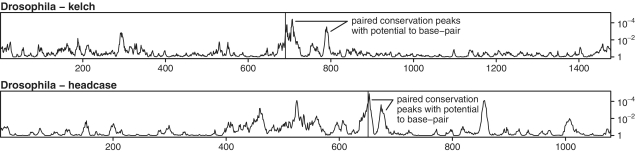
A 3′-proximal RNA structure has been previously implicated in UAA RT in the Drosophila gene headcase, and our computational analysis demonstrated that at least the basal part of the predicted stem is conserved in 12 sequenced Drosophila species, while a 52 nt (in Drosophila melanogaster; alignment gaps in other species) ‘loop’ region is far less conserved (27, 73; Figures 3 and 5 and Supplementary Data). In contrast, to our knowledge, no stimulatory RNA structure has been previously proposed for UGA RT in kelch. However, when we applied our computational analysis to kelch, we found tandem synonymous site conservation peaks 3′ of the RT codon and the corresponding sequences were predicted to form an RNA stem (268 nt loop in D. melanogaster) that is conserved in all 12 Drosophila species (Figures 3 and 5; Supplementary Data). The predicted stem has 14 bp with a 1 nt asymmetric bulge near the center of the 5′ component, and is separated from the RT codon by an 8 nt ‘spacer’ sequence that is completely conserved in all 12 Drosophila species but, perhaps unusually, the RT codon context is UGA-AUG (UGA-AGC in Anopheles, Culex and Aedes mosquitoes).
Figure 5.
Synonymous site conservation for the Drosophila genes kelch and headcase based on alignments of sequences from 12 Drosophila species. The codon index (relative to the D. melanogaster sequence) is plotted on the horizontal axis and the conservation P-value is plotted on the vertical axis (see caption to Figure 1B for details). In each case, the RT site is indicated with a vertical line.
Experimental analysis of the predicted structure in VEEV and SINV
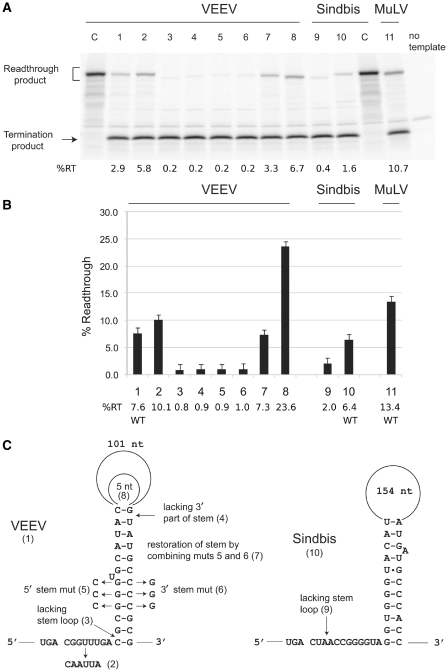
In order to verify and investigate the functionality of the predicted RNA structures in VEEV and SINV, local sequences (15 nt 5′ of the UGA stop codon and 156 nt 3′ for VEEV or 204 nt 3′ for SINV) were cloned in-frame between the renilla luciferase and firefly luciferase genes in vector pDluc. The firefly luciferase gene lacks an initiation codon and its expression is dependent on RT of the UGA codon. RT efficiencies were determined both in vitro using rabbit reticulocyte lysate, and in HEK293 tissue culture cell lysates. A positive control for RT, the MuLV gag-pol RT site and 3′-adjacent pseudoknot, was included in all assays (Figure 6A and B, lane 11).
Figure 6.
RT assays of dual luciferase reporter constructs. (A) In vitro assays in the rabbit reticulocyte lysate system. The WT and mutant constructs assayed are as follows: Left-hand C lane, VEEV RT cassette with UGA replaced by UGG; lane 1, WT VEEV RT stem–loop cassette; lane 2, VEEV RT stem–loop cassette with 6 nt immediately 3′ of the stop codon replaced by CAAUUA; lane 3, VEEV RT site with just 9 nt 3′ (i.e. lacking the stem–loop structure); lane 4, VEEV cassette lacking 3′ part of the stem; lane 5, VEEV with 3C substitution in the 5′ part of the stem; lane 6, VEEV with 3G substitution in the 3′ part of the stem; lane 7, VEEV with 3C and 3G substitutions in the 5′ and 3′ parts of the stem, respectively; lane 8, VEEV with only 5 nt in the loop of the stem–loop structure; lane 9, SINV RT site with just 3 nt 3′; lane 10, WT SINV RT stem–loop cassette; right-hand C lane, SINV stem–loop cassette with UGA replaced by UGG; lane 11, MuLV RT cassette. The positions of the RT and termination products are indicated. The sizes of the RT products vary due to differences in the extent of sequence in the RT cassettes. (B) RT assays performed in tissue culture cells. The y-axis indicates the RT efficiency. The various constructs are listed on the x-axis and correspond to the numbering in (A). Error bars indicate the standard deviation of six independent transfections for each construct. (C) Schematic of the constructs analyzed in (A) and (B). Mutants numbered (2–9) correspond to the lanes/bars in (A)/(B).
The WT VEEV and SINV constructs promoted RT in vitro at 2.9% and 1.6%, respectively (Figure 6A, lanes 1 and 10). The RT efficiencies in tissue culture cells were much higher: 7.6% for VEEV and 6.4% for SINV (Figure 6B, lanes 1 and 10). Substitution of the 6 nt immediately 3′ of the UGA codon in VEEV with the tobamovirus-like RT stimulator, CAA-UUA, increased RT both in vitro (5.8%) and in tissue culture cells (10.1%; Figure 6A and B, lane 2). Derivative constructs lacking the sequences for the predicted structures were generated (Figure 6C). The VEEV derivative containing only 9 nt 3′ of the UGA codon directed just 0.2% RT in vitro and 0.8% in tissue culture cells (Figure 6A and B, lane 3), while the SINV derivative containing only 3 nt 3′ of the UGA codon directed just 0.4% RT in vitro and 2.0% in tissue culture cells (Figure 6A and B, lane 9). Thus the stimulatory effect of the stem–loop sequence is ∼10- to 14-fold for VEEV and 3- to 4-fold for SINV, depending on the assay system. This is in direct contrast to ref. 32 where no difference was found in vitro between an insert comprising just SINV UGA-CUA and an insert comprising the entire SINV nsP3+nsP4-coding sequences.
The VEEV stem–loop sequence was chosen for further analysis due to the higher RT efficiency and its greater stimulatory effect. When the 3′ part of the stem was deleted, RT was reduced to 0.2% in vitro and 0.9% in tissue culture cells (Figure 6A and B, lane 4), thus demonstrating the importance of the sequence corresponding to the 3′ component of the predicted stem, >120 nt downstream of the RT codon. To address base pairing within the predicted stem, two mutations were constructed that were predicted to disrupt Watson–Crick interactions: 3 G residues in the 5′ part of the stem were changed to Cs or 3 C residues in the 3′ part of the stem were changed to Gs (Figure 6C). In both cases, RT was drastically reduced in both in vitro and tissue culture cell assays (Figure 6A and B, lanes 5 and 6). However, when the two mutations were combined such that the predicted base pairings were restored, RT recovered to near the WT level (Figure 6A and B, lane 7). The importance of the sequence between the two halves of the stem (referred to here as the ‘loop’ region but without implication about internal structure) was tested by deleting all but 5 nt of its sequence. Interestingly, this resulted in substantially higher RT than the WT level in both assay systems (Figure 6A and B, lane 8).
DISCUSSION
We have shown that the stimulatory elements for efficient RT in VEEV and probably also SINV include not just the immediately 3′-adjacent nucleotides, but also a stem–loop structure that spans ∼140 nt 3′ of the stop codon. Computational analyses provide strong evidence that similar structures are relevant for RT in several other alphaviruses, and in plant viruses where RT occurs at a UGA codon. Although this RNA structure is clearly not essential for some level of RT to occur in some systems [as in many previous analyses the predicted WT structure was not present (32,33,74) and, in our own experiments, ∼1% RT was achieved in tissue culture without the WT structure], it does have a pronounced stimulatory effect on RT efficiency (3- to 4-fold for SINV, 10- to 14-fold for VEEV). As with the gammaretrovirus 3′ pseudoknot, the precise mechanism by which the stem–loop affects RT remains to be determined. Possibilities include direct interaction with the ribosome (including pausing and/or promotion of conformational changes in the ribosome); provision of a physical block that preferentially occludes release factor from the A-site in favour of tRNAs; or an indirect action via some trans-acting factor. The function of the RNA structure may simply be to achieve a higher RT level that is optimal for the virus. Alternatively, the structure may provide a regulatory mechanism, perhaps allowing different RT levels to be achieved in different hosts or at different stages in the viral cycle.
The long ‘loop’ length of the predicted structures is noteworthy. While long-distance base pairings have been demonstrated to play important regulatory roles in RNA viruses (reviewed in ref. 75), the distances involved in the RT base pairings identified here are very much smaller and a genome-scale regulatory role seems unlikely. Furthermore, our loop-deletion mutant promoted even higher RT than the WT construct, suggesting that the presence of a long loop region, or any sequence motifs therein, play little if any role in stimulating efficient RT. Thus, we hypothesize that evolutionary selection simply acts to place the 3′ component of the stem in a convenient location (e.g. with regards to minimizing interference with the encoded amino acid sequence). Although we refer to the region between the two components of the stem as a ‘loop’, it should not be taken to imply that this region does not fold. In fact the region generally is predicted to fold, and the fact that it can fold may indeed be functionally important—perhaps just to provide stability to the basal stem. However, in most cases, the nature of the fold seems to be relatively unimportant as it is not well-conserved between related sequences.
How can our results be reconciled with previous results which indicated that only the immediately 3′-adjacent 1–3 nt were relevant for RT in these viruses? There are several possibilities. Previous analyses of the RT cassette in SINV alphavirus, and also tobacco rattle tobravirus, were performed in in vitro systems (32,33). However, RT efficiency may vary considerably between in vitro and cell culture systems, depending on the absence or presence and abundance of various relevant near-cognate tRNA species (34), and potentially also on the concentration of various trans-acting factors, salt concentrations, temperature, ribosome loading density and intracellular architecture. Thus a high RT efficiency measured in vitro for a short insert does not mean that the full complement of elements that stimulate efficient RT in cell culture or in vivo has been recapitulated faithfully. Such factors may also explain why our in vitro experiments produced much lower RT efficiencies than previous in vitro experiments, and highlight the importance of our experiments in mammalian cell culture (32,76,77). Although ref. (32) compared, in vitro, the RT efficiency for a short insert (that excluded the predicted structure) with a long insert (comprising the entire nsP3 + nsP4-coding sequences), such comparisons between inserts of very different sizes are not always straightforward, in part because the different protein products may be degraded at different rates, and because chance base pairings with the construct sequence could affect RT efficiency differently for the long and short inserts. In contrast, guided by our computational analysis, we were able to make small but targeted substitutions that allowed for more precise comparisons in the context of a long VEEV insert that included the predicted RNA structure elements. Accurate measurements of the RT efficiency in alphavirus-infected cells are not readily obtainable due to the multiple cleavage products of the non-structural polyprotein and rapid degradation of excess nsP4 (31,47,78). Nonetheless, in ref. (31), 5- to 8-fold less nsP34 was found in WT SINV-infected cells than in cells infected with mutant viruses in which the UGA was replaced by a Ser, Trp or Arg codon, thus suggesting a WT RT efficiency in the range 12.5–20%. Our measurement of ∼7% for WT VEEV and SINV sequences in the dual luciferase construct suggests that there may be additional factors that affect alphavirus RT.
Interestingly, although RT for the VEEV and SINV cassettes was much more efficient in cell culture than in vitro, there was little difference between the two systems for the MuLV RT cassette (Figure 6). While the action of some cellular trans-acting stimulatory factor cannot be ruled out (albeit presumably not interacting with the loop region, given the increased RT observed when the loop was deleted), other possible explanations include: (i) the different stop codons and nucleotide contexts involved (UGA-C in VEEV and SINV; UAG-G in MuLV) and hence the different pools of potential stop codon-decoding tRNAs and (ii) the nature of the 3′ structure (a compact pseudoknot in MuLV but an extended stem–loop in VEEV and SINV) with possible consequences for the ease with which the structure may fold in different environments. Similar differences in RT efficiency between in vitro and cell culture systems were noted for Colorado tick fever coltivirus which, like SINV and VEEV, utilizes a UGA RT codon with a predicted 3′-adjacent stem–loop structure (65).
Besides the known structure-stimulated RT cases discussed above, RNA structure also plays an integral role in the recoding of UGA codons for selenocysteine insertion. In eukaryotes, this process is dependent on an RNA stem–loop structure containing specific nucleotide motifs, known as the SECIS element, usually located in the 3′-UTR of the corresponding mRNAs (5,6). In certain cases, an additional stem–loop structure close to the recoded stop codon has also been identified (79,80). For example, in the human SEPN1 gene there is a phylogenetically conserved 16 bp stem (with a 1 nt symmetric bulge) and a 5 nt loop, separated from the UGA by a 6 nt spacer. Interestingly, this structure has been shown to stimulate RT in cell culture (but not in vitro) even when the SECIS element is absent. Howard et al. located potential 3′-adjacent structures for at least 5 of 36 human selenocysteine-encoding UGA codons analyzed. However, their initial computational selection involved RNA-folding of just nucleotides +1 to +60 of the human sequence—an analysis which would have missed most of the 3′ RNA structures predicted in this report. Thus 3′-adjacent structures may be a feature of a larger proportion of selenocysteine RT sites than these, though it does not appear to be an essential feature for selenocysteine RT (61).
The various motifs that stimulate RT in eukaryotic cells have been previously classified by Beier and Grimm and by Harrell et al. (34,81). Beier and Grimm define the classes Type I (generally UAG-CAA-UYA; includes tobamovirus replicase, and benyvirus and pomovirus CP extension), Type II (generally UGA-CGG or UGA-CUA; includes alphavirus replicase, tobravirus, pecluvirus, furovirus and pomovirus replicase, and furovirus CP extension), and Type III (generally UAG-G, plus a compact pseudoknot in gammaretroviruses and possible but as yet relatively uncharacterized structures in the luteoviruses and tombusviruses). There are exceptions to the rule (e.g. enamovirus UGA-G, various pomovirus cases with atypical stop codons, and so on). One reason for this may be that the required level of RT may vary between different viruses, and may also be modulated by other sequence elements (e.g. 5′ nucleotide and/or amino acid context) so that, in certain cases, deviations from one of the ‘canonical’ RT motifs may be tolerated. With this proviso, our results suggest that the definition of the Type II motif should, in general though perhaps not ubiquitously, be modified to include a 3′ RNA structure component. Our discovery in alphaviruses and phylogenetically supported predictions for many plant viruses and the Drosophila gene kelch, together with the small number of previously identified cases of structure-stimulated RT, now suggest that 3′ RNA structures as a component of efficient RT cassettes in eukaryotes (especially those that lack a CAR-YYA tobamovirus-like stimulator), rather than being exceptional, may in fact be the norm.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust (grant number 088789 to A.E.F.); Science Foundation Ireland (grant number 08/IN.1/B1889 to J.F.A.); and the National Institutes of Health (grant number R01 GM079523 to J.F.A.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Mike Howard (University of Utah) for his kind gift of plasmid pDluc, and Chris Anderson (University of Utah) for help with tissue culture analyses. The authors also thank Lynn Cooley, their collaborator for experimental analysis of kelch RT (work in progress), for her support.
REFERENCES
- 1.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 2.Atkins JF, Baranov PV. The distinction between recoding and codon reassignment. Genetics. 2010;185:1535–1536. doi: 10.1534/genetics.110.119016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JD, Ryan MD. Ribosome “Skipping”: “Stop-Carry On” or “StopGo” Translation. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. 101–121. [Google Scholar]
- 4.Namy O, Rousset JP. Specification of Standard Amino Acids by Stop Codons. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. 79–100. [Google Scholar]
- 5.Gladyshev VN, Hatfield DL. Selenocysteine Biosynthesis, Selenoproteins, and Selenoproteomes. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. 3–27. [Google Scholar]
- 6.Berry MJ, Howard MT. Reprogramming the Ribosome for Selenoprotein Expression: RNA Elements and Protein Factors. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. 29–52. [Google Scholar]
- 7.Krzycki JA. Translation of UAG as Pyrrolysine. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. 53–77. [Google Scholar]
- 8.Gesteland RF, Weiss RB, Atkins JF. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- 9.Nirenberg M. Foreword. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York: Springer; 2010. pp. v–vi. [Google Scholar]
- 10.Bossi L, Roth JR. The influence of codon context on genetic code translation. Nature. 1980;286:123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- 11.Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl Acad. Sci. USA. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 16.Cridge AG, Major LL, Mahagaonkar AA, Poole ES, Isaksson LA, Tate WP. Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2006;34:1959–1973. doi: 10.1093/nar/gkl074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner AM, Weber K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat. New Biol. 1971;234:206–209. doi: 10.1038/newbio234206a0. [DOI] [PubMed] [Google Scholar]
- 18.Hofstetter H, Monstein HJ, Weissmann C. The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim. Biophys. Acta. 1974;374:238–251. doi: 10.1016/0005-2787(74)90366-9. [DOI] [PubMed] [Google Scholar]
- 19.Gesteland RF, Wolfner M, Grisafi P, Fink G, Botstein D, Roth JR. Yeast suppressors of UAA and UAG nonsense codons work efficiently in vitro via tRNA. Cell. 1976;7:381–390. doi: 10.1016/0092-8674(76)90167-7. [DOI] [PubMed] [Google Scholar]
- 20.Pelham HR. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978;272:469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- 21.Pelham HR. Translation of tobacco rattle virus RNAs in vitro: four proteins from three RNAs. Virology. 1979;97:256–265. doi: 10.1016/0042-6822(79)90337-4. [DOI] [PubMed] [Google Scholar]
- 22.Philipson L, Andersson P, Olshevsky U, Weinberg R, Baltimore D, Gesteland R. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978;13:189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- 23.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl Acad. Sci. USA. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlova M, Yueh A, Leung J, Goff SP. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell. 2003;115:319–331. doi: 10.1016/s0092-8674(03)00805-5. [DOI] [PubMed] [Google Scholar]
- 25.Goff SP. Genetic reprogramming by retroviruses: enhanced suppression of translational termination. Cell Cycle. 2004;3:123–125. [PubMed] [Google Scholar]
- 26.Robinson DN, Cooley L. Examination of the function of two kelch proteins generated by stop codon suppression. Development. 1997;124:1405–1417. doi: 10.1242/dev.124.7.1405. [DOI] [PubMed] [Google Scholar]
- 27.Steneberg P, Samakovlis C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001;2:593–597. doi: 10.1093/embo-reports/kve128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergstrom DE, Merli CA, Cygan JA, Shelby R, Blackman RK. Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics. 1995;139:1331–1346. doi: 10.1093/genetics/139.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss EG, Rice CM, Strauss JH. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl Acad. Sci. USA. 1983;80:5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li GP, Rice CM. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J. Virol. 1989;63:1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, Rice CM. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol. 1993;67:5062–5067. doi: 10.1128/jvi.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urban C, Zerfass K, Fingerhut C, Beier H. UGA suppression by tRNACmCATrp occurs in diverse virus RNAs due to a limited influence of the codon context. Nucleic Acids Res. 1996;24:3424–3430. doi: 10.1093/nar/24.17.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- 36.Zerfass K, Beier H. Pseudouridine in the anticodon GΨA of plant cytoplasmic tRNATyr is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res. 1992;20:5911–5918. doi: 10.1093/nar/20.22.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tork S, Hatin I, Rousset JP, Fabret C. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004;32:415–421. doi: 10.1093/nar/gkh201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills NM, Gesteland RF, Atkins JF. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng YX, Yuan H, Rein A, Levin JG. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J. Mol. Biol. 1999;288:837–852. doi: 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- 43.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 44.Brault V, van den Heuvel JF, Verbeek M, Ziegler-Graff V, Reutenauer A, Herrbach E, Garaud JC, Guilley H, Richards K, Jonard G. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 1995;14:650–659. doi: 10.1002/j.1460-2075.1995.tb07043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown CM, Dinesh-Kumar SP, Miller WA. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J. Virol. 1996;70:5884–5892. doi: 10.1128/jvi.70.9.5884-5892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruyère A, Brault V, Ziegler-Graff V, Simonis MT, Van den Heuvel JF, Richards K, Guilley H, Jonard G, Herrbach E. Effects of mutations in the beet western yellows virus readthrough protein on its expression and packaging and on virus accumulation, symptoms, and aphid transmission. Virology. 1997;230:323–334. doi: 10.1006/viro.1997.8476. [DOI] [PubMed] [Google Scholar]
- 47.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attoui H, Sailleau C, Mohd Jaafar F, Belhouchet M, Biagini P, Cantaloube JF, de Micco P, Mertens P, Zientara S. Complete nucleotide sequence of Middelburg virus, isolated from the spleen of a horse with severe clinical disease in Zimbabwe. J. Gen. Virol. 2007;88:3078–3088. doi: 10.1099/vir.0.83076-0. [DOI] [PubMed] [Google Scholar]
- 49.Firth AE, Chung BY-W, Fleeton MN, Atkins JF. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauss EG, Levinson R, Rice CM, Dalrymple J, Strauss JH. Nonstructural proteins nsP3 and nsP4 of Ross River and O'Nyong-nyong viruses: sequence and comparison with those of other alphaviruses. Virology. 1988;164:265–274. doi: 10.1016/0042-6822(88)90644-7. [DOI] [PubMed] [Google Scholar]
- 51.Kim KH, Rümenapf T, Strauss EG, Strauss JH. Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology. 2004;323:153–163. doi: 10.1016/j.virol.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 55.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reeder J, Steffen P, Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–W324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Firth AE, Atkins JF. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1’ may derive from ribosomal frameshifting. Virol. J. 2009;6:14. doi: 10.1186/1743-422X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simmonds P, Karakasiliotis I, Bailey D, Chaudhry Y, Evans DJ, Goodfellow IG. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008;36:2530–2546. doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 60.Firth AE, Brown CM. Detecting overlapping coding sequences in virus genomes. BMC Bioinformatics. 2006;7:75. doi: 10.1186/1471-2105-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fixsen SM, Howard MT. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J. Mol. Biol. 2010;399:385–396. doi: 10.1016/j.jmb.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 63.Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams MJ, Antoniw JF, Kreuze J. Virgaviridae: a new family of rod-shaped plant viruses. Arch. Virol. 2009;154:1967–1972. doi: 10.1007/s00705-009-0506-6. [DOI] [PubMed] [Google Scholar]
- 65.Jaafar FM, Attoui H, de Micco P, de Lamballerie X. Termination and read-through proteins encoded by genome segment 9 of Colorado tick fever virus. J. Gen. Virol. 2004;85:2237–2244. doi: 10.1099/vir.0.80019-0. [DOI] [PubMed] [Google Scholar]
- 66.Tamada T, Schmitt C, Saito M, Guilley H, Richards K, Jonard G. High resolution analysis of the readthrough domain of beet necrotic yellow vein virus readthrough protein: a KTER motif is important for efficient transmission of the virus by Polymyxa betae. J. Gen. Virol. 1996;77:1359–1367. doi: 10.1099/0022-1317-77-7-1359. [DOI] [PubMed] [Google Scholar]
- 67.Wu B, Pogany J, Na H, Nicholson BL, Nagy PD, White KA. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 2009;5:e1000323. doi: 10.1371/journal.ppat.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei N, Hacker DL, Morris TJ. Characterization of an internal element in turnip crinkle virus RNA involved in both coat protein binding and replication. Virology. 1992;190:346–355. doi: 10.1016/0042-6822(92)91221-f. [DOI] [PubMed] [Google Scholar]
- 69.White KA, Skuzeski JM, Li W, Wei N, Morris TJ. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology. 1995;211:525–534. doi: 10.1006/viro.1995.1434. [DOI] [PubMed] [Google Scholar]
- 70.Demler SA, de Zoeten GA. The nucleotide sequence and luteovirus-like nature of RNA 1 of an aphid non-transmissible strain of pea enation mosaic virus. J. Gen. Virol. 1991;72:1819–1834. doi: 10.1099/0022-1317-72-8-1819. [DOI] [PubMed] [Google Scholar]
- 71.Watson MT, Tian T, Estabrook E, Falk BW. A small RNA resembling the beet western yellows luteovirus ST9-associated RNA is a component of the California carrot motley dwarf complex. Phytopathology. 1998;88:164–170. doi: 10.1094/PHYTO.1998.88.2.164. [DOI] [PubMed] [Google Scholar]
- 72.Walter CT, Pringle FM, Nakayinga R, de Felipe P, Ryan MD, Ball LA, Dorrington RA. Genome organization and translation products of Providence virus: insight into a unique tetravirus. J. Gen. Virol. 2010;91:2826–2835. doi: 10.1099/vir.0.023796-0. [DOI] [PubMed] [Google Scholar]
- 73.Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 74.Zerfass K, Beier H. The leaky UGA termination codon of tobacco rattle virus RNA is suppressed by tobacco chloroplast and cytoplasmic tRNAsTrp with CmCA anticodon. EMBO J. 1992;11:4167–4173. doi: 10.1002/j.1460-2075.1992.tb05510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller WA, White KA. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Groot RJ, Hardy WR, Shirako Y, Strauss JH. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hardy WR, Strauss JH. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J. Virol. 1988;62:998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Howard MT, Moyle MW, Aggarwal G, Carlson BA, Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA. 2007;13:912–920. doi: 10.1261/rna.473907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harrell L, Melcher U, Atkins JF. Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res. 2002;30:2011–2017. doi: 10.1093/nar/30.9.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.