Abstract
microRNAs play an important roles in cell growth, differentiation, proliferation and apoptosis. They can function either as tumor suppressors or oncogenes. We found that the overexpression of miR-192 inhibited cell proliferation in A549, H460 and 95D cells, and inhibited tumorigenesis in a nude mouse model. Both caspase-7 and the PARP protein were activated by the overexpression of miR-192, thus suggesting that miR-192 induces cell apoptosis through the caspase pathway. Further studies showed that retinoblastoma 1 (RB1) is a direct target of miR-192. Over-expression of miR-192 decreased RB1 mRNA and protein levels and repressed RB1-3′-UTR reporter activity. Knockdown of RB1 using siRNA resulted in a similar cell morphology as that observed for overexpression of miR-192. Additionally, RB1-siRNA treatment inhibited cell proliferation and induced cell apoptosis in lung cancer cells. Analysis of miRNA expression in clinical samples showed that miR-192 is significantly downregulated in lung cancer tissues compared to adjacent non-cancerous lung tissues. In conclusion, our results demonstrate that miR-192 is a tumor suppressor that can target the RB1 gene to inhibit cell proliferation and induce cell apoptosis in lung cancer cells. Furthermore, miR-192 was expressed at low levels in lung cancer samples, indicating that it might be a promising therapeutic target for lung cancer treatment.
INTRODUCTION
microRNAs (miRNAs) are single-stranded non-coding small RNAs of ∼22 nt that can regulate gene expression in animals, plants and viruses (1). miRNAs are first transcribed by RNA polymerase II as primary miRNAs (pri-miRNAs) that are several thousand nucleotides long (2,3). Pri-miRNAs are processed by the microprocessor complex, which is composed of the RNase III type enzyme, Drosha and the double-stranded RNA binding protein, DiGeorge syndrome critical region gene 8 (DGCR8), to generate ∼70 nt precursor miRNAs (pre-miRNAs) with hairpin-shaped structures (4,5). These pre-miRNAs are exported to the cytoplasm by exportin-5 (Exp-5) and the cofactor Ran-GTP (6). In the cytoplasm, pre-miRNAs are processed into 22 nt mature miRNA duplexes by the RNase III Dicer enzyme (7). Mature miRNAs are incorporated into ‘miRNA-containing RNA-induced silencing complex (miRISC), which induce either cleavage or translational repression of targeted mRNAs (1,8). The miRNA database (miRBase16.0) contains 1048 records, and the number of known miRNAs is still growing (http://microrna.sanger.ac.uk) (9).
miRNAs play an important roles in cell growth, differentiation, proliferation, apoptosis and cell death. miRNAs associated with tumorigenesis act as either tumor suppressors or oncogenes. For example, let-7 in lung cancer (10) and the miR15a/16 cluster in CLL act as tumor suppressors (11). In contrast, the miR-17-92 cluster in malignant lymphoma (12) and miR-155/BIC in Burkitt lymphoma (13) act as oncogenes.
miR-192 was first cloned by Lagos-Quintana et al. (14) and later confirmed by Lim et al. (15). The miR-192 gene is located on human chromosome 11 and is transcribed as a cluster with miR-194 (16). miR-192 sequence mutations have been identified in some hepatocellular carcinoma (HCC) tissues, but may not represent the primary mechanism of hepatocarcinogenesis (17). The expression of miR-192 can be regulated by hepatocyte nuclear factor-1a (HNF-1a) (16), transforming growth factor β (TGF-β) and p53 (18,19). In the kidney, miR-192 controls TGF-β-induced Col1a2 expression by downregulating the E-box repressor survival of motor neuron protein interacting protein 1 (SIP1) (20), and miR-192 targets WNK1 in regulation of sodium and potassium balance (21). In breast cancer, both miR-192 and bone morphogenetic protein-6 can inhibit delaEF1 expression to prevent breast cancer cell migration (22). In colon cancer, miR-192 targets transcriptional thymidylate synthase (TYMS) to influence 5-fluorouracil resistance (23) and targets DHFR to regulate cellular proliferation through the p53-microRNA circuit (24). Additional studies have shown that p53 induces miR-192 expression and down-regulates the genes that regulate G1 and G2 checkpoints, resulting in cell cycle arrest in G1 or G2 (18,25).
RB1 was the first described tumor suppressor. It can stabilize the constitutive heterochromatin to maintain the overall chromatin structure. It can bind the transcription factor E2F1 and regulate the expression of many genes. One of the functions of RB1 is to inhibit apoptosis (26,27). Knockdown of RB-induced apoptosis can be canceled by overexpression of miR-17–92 in lung cancer cells (28). Lung cancer is the leading cause of death throughout the world. miRNA might play the important roles in lung cancers (29,30).
In the current study, we found that miR-192 is downregulated in lung cancer tissue compared with respective non-cancerous lung tissue. Overexpression of miR-192 inhibits cell proliferation and promotes cell apoptosis in lung cancer cells, and miR-192 inhibits tumorigenesis in a nude mouse model in vivo. Further analysis showed that RB1 is an important target of miR-192 in lung cancer cells.
MATERIALS AND METHODS
Cell culture and tissue sample collection
A549, NCI-H460 (H460 cell) and 95D cells (all of them were human lung cancer cell lines) were maintained in Ham's F12 (F12) or RPMI-1640 (1640) medium; HeLa (cervical cancer cell line) and HEK293 (Human Embryonic Kidney 293 cell line) cells were maintained in Dulbecco's Modified Eagle's medium (DMEM). All media were supplemented with 10% fetal bovine serum (FBS, Hyclone, USA) and 1% antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin sulfate). Cells were grown in an atmosphere of 5% CO2 at 37°C. All cell lines were a kind gift from Dr Duanqing Pei's Lab at the Guangzhou Institute of Biomedicine and Health, Guangzhu, China 510530.
The A549-luc-2 cell line was constructed in our lab by transfecting the pGL4 vector (Promega, USA) into A549 cells and selecting cell lines with stable expression of the luciferase protein. The cell line was maintained in 1640 medium supplemented with 10% FBS and 800 U/ml of G418 antibiotic.
Seventeen lung cancer tissues and respective non-cancer lung tissues were obtained from the Guangzhou Institute of Respiratory Disease, the First Affiliated Hospital of Guangzhou Medical College (Guangzhou, China) in 2008 with informed consent and Institutional Review Board (IRB) permission. All clinical and biological data were available for the samples. Samples were separated from the surgical patient samples and immediately snap-frozen in liquid nitrogen and stored in liquid nitrogen until use.
Synthesis and transfection of siRNAs, miRNA mimics and miRNA inhibitors
siRNAs, miRNA mimics and miRNA inhibitors were designed and synthesized by Guangzhou RiboBio (RiboBio, China). miRNA inhibitors were all nucleotides with 2′-O-methyl modification. Three siRNAs targeting on RB1 gene were designed and synthesised, the most effective siRNA (siRB1) identified by qPCR was applied for the further experiments. The sequence of siRB1 is:
sense: 5′-CACCCAGGCGAGGUCAGAAdTdT-3′;
anti-sense: 3′-dTdTGUGGGUCCGCUCCAGUCUU-5′.
Twenty-four hours prior to transfection, cells were plated onto a 96-well plate (Greiner, Germany) at ∼40–60% confluence. Transfection was performed with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's protocol. The medium was replaced 4–6 h after transfection with new culture medium, except in the case of HEK293 cells, for which the medium was not replaced after transfection.
Apoptosis assay
A549, HeLa, and HEK293 cells were transfected with 50 nM of the miRNA mimics. Forty-eight hours after transfection, 20 µl of MTT (5 mg/ml) was added to each well of a 96-well plate and the cells were cultured for 4 h. The MTT medium mixture was then discarded and 150 µl of dimethyl sulfoxide (DMSO) was added to each well. Absorbance was measured at 570 nm using a multi-well spectrophotometer (Bio-Tek, USA).
Cell viability was measured using the Celltiter-Glo luminescent cell viability assay kit (Promega, USA) according to the manufacturer's protocol. Briefly, A549, HEK293, HeLa, H460 and 95D cells were plated onto 96-well plates. Forty-eight hours after transfection, the Celltiter-Glo reagent was added and luminescent intensity was read by a Microplate Luminometer (Turner Biosystems, USA) using the respective Promega protocols.
Cell apoptosis was also detected using an Annexin V-PE apoptosis detection kit (BD, USA) according to the manufacturer's protocol and FACS analysis (BD, USA).
5-ethynyl-2′-deoxyuridine assay
A549 and H460 cells were transfected with miRNA mimics in 96-well plates. Forty-eight hours after transfection, 5-ethynyl-2′-deoxyuridine (EdU) (100 µM) (Cell Light EdU DNA imaging Kit, Guangzhou RiboBio, China) was added and the cells were cultured for an additional 2 h. The cells were then stained according to the following protocol: discard the EdU medium mixture, add 4% paraformaldehyde to fix cells at room temperature for 30 min, wash with glycine (2 mg/ml) for 5 min in a shaker, add 0.2% Trion X-100 for 10 min, wash with PBS for two times, add click reaction buffer (Tris–HCl, pH 8.5, 100 mM; CuSO4, 1 mM; Apollo 550 fluorescent azide, 100 µM; ascorbic acid, 100 mM) for 10–30 min while protecting from light, wash with 0.5% Triton X-100 for three times, stain with Hoechst (5 µg/ml) for 30 min at room temperature, wash with 0.5% Triton X-100 for five times, and, finally, add 150 µl PBS. Images were taken and analyzed using High Content Imaging Pathway 855 (BD, USA). EdU positive cell was caculated with (EdU add-in cells/Hoechst stained cells) × 100%.
Cell cycle assay
A549 cells were transfected with miR-192 or no-target control, 24 h later, Nocodazole (40ng/ml) was added. Cells were collected after 48 h and stained with propidium iodide (PI) for cell cycle analysis with FACS (BD, USA). Data were collected and analyzed with the CELLQuest and ModFit LT software.
RNA extraction and qRT–PCR
Total RNA was extracted from cell lines and tissue samples using Trizol reagent (Invitrogen, USA) according to the manufacturer's protocol. cDNA was synthesized from 2 µg of total RNA with M-MLV Reverse Transcriptase (Promega, USA) in a 25 µl volume {2 µg total RNA, 400 mM reverse transcription primer [oligo(dT)18 for mRNA genes, random primers for U6 rRNA and miRNA specific primers (Bulge-Loop™ miRNA qPCR Primers from RiboBio, China) for miRNA], 4 U/µl M-MLV, 1 U/µl inhibitor, 0.4 mM dNTP mix}. Real time PCR was carried out with the reagents of a Sybr green I mix (Takara, China) in a 20 µl reaction volume (10 µl Sybr green I mix, 200 mM forward and reverse primer, 2 µl cDNA template) on a MJ Opticon Monitor chromo4 instrument (Bio-Rad, USA) using the following protocol: 95°C for 20 s; 40 cycles of 95°C for 10 s, 60°C for 20 s, 70°C for 1 s. Data analysis were performed using the 2−ΔΔCt method (31).
Western blot
Cells were washed with PBS chilled to 4°C. Whole-cell proteins were extracted with cell lysis buffer (Beyotime, China) that included the protease inhibitor phenylmethanesulfonyl fluoride (PMSF, Beyotime, China) at 4°C for half an hour and separated by 15% sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The proteins and the pre-stained marker (Fermantas, USA) were then transferred onto an Immobilon-P transfer polyvinylidene fluoride membrane (PDVF, pore size: 0.45 μm; Millipore, USA) with a Semi-dry Transfer Cell (Model 755, Bio-Rad, USA) for 90 min, allowing the pre-stained marker to be completely transferred from the gel to the membrane. The gel was discarded and the membrane was incubated at room temperature in 5% non-fat milk prepared with TBS-T buffer. Two hours later, the membrane was cut as required (as β-Actin is ∼37 kDa; RB1, 130 kDa; cleaved caspase7, 20 kDa; PARP, 116 and 89 kDa), placed in a suitable hybridization bag with 1 ml of primary antibody [all primary antibodies were diluted 1:1000 with 1% non-fat milk; the anti-β-Actin antibody was obtained from Sigma, USA; the anti-RB1(H4) antibody, the PARP antibody, and the cleaved caspase7 antibody were all obtained from Cell Signal Technology, USA] and incubated at 4°C overnight. The membrane was washed three times with TBS-T and the secondary antibody was added [goat anti-rabbit antibody for Actin, cleaved caspase7 and PARP; goat anti-mouse antibody for RB1; both of the secondary antibodies were conjugated to horseradish peroxidase (HRP) and were obtained from Santa Cruz, USA] for 2 h at room temperature. The membrane was washed three times with TBS-T buffer and incubated with HRP substrate (BeyoECL plus A/B, Beyotime, China) for 1 min. Blots were developed on X-film (Kodak, Japan).
Luciferase reporter assay
The 3′-UTRs of the predicted target genes were PCR amplified from genomic DNA of A549 cells, which was extracted with a Genomic DNA Extraction kit (Dingguo, China). The PCR products were gel purified, digested and inserted into the digested psicheck-2 plasmid (Promega, USA) between the XhoI and NotI sites. RB1-3′-UTR target site mutations were introduced using the KOD-plus mutagenesis kit (Toyobo, Japan) according to the manufacturer's protocol. All inserted or mutated sequences were confirmed by sequencing. All primers were synthesized by Guangzhou RiboBio (RiboBio, China), and the sequences of the primers are available upon request.
Two hundred nanogram per microliter of plasmid and 50 nM miRNA mimics were co-transfected into A549 cells. Forty-eight hours after transfection, luciferase activity was detected using the Dual-Glo luciferase assay kit (Promega, USA) according to the manufacturer's protocol. Luminescence intensity was read with a Microplate Luminometer using the respective Promega protocol. Transfections were performed in duplicate and repeated three times.
Tumor formation in BALB/c nude mice
All experimental procedures involving animals were performed according to the institutional ethical guidelines for animal experiments at the Guangzhou Institute of Medicine and Health, Chinese Academy of Sciences. A549-luc-2 cells were pre-treated with 50 nM of miR-192 mimics or a non-targeting control, mock control were also included. After 6 h transfection, new medium was added to the cells. Twelve hours after transfection, cells were collected and suspended in 100 µl PBS at a concentration of 1 × 106 cells/ml and injected s.c. into either side of the posterior flank of the same BALB/c athymic nude mouse (5–6 weeks of age, each group with at least four mouse, n = 4) (32). Thirty minutes after cell injection, luciferase substrate was added at a dose of 150 mg/kg and live images of the mouse were obtained using an IVIS200 (Xenogene, USA). These data were classified as Day 0. Luciferase activity was measured every 4 days using the same protocol. Tumor growth was examined every 2 days beginning at Day 4. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers and calculated with the formula V = (L × W2) × 0.5.
Statistical analysis
Data are presented as means ± (SEM) from at least three separate experiments. Multiple group comparisons were performed with ANOVA (P < 0.05).
RESULTS
miR-192 inhibits cell proliferation in A549 cells
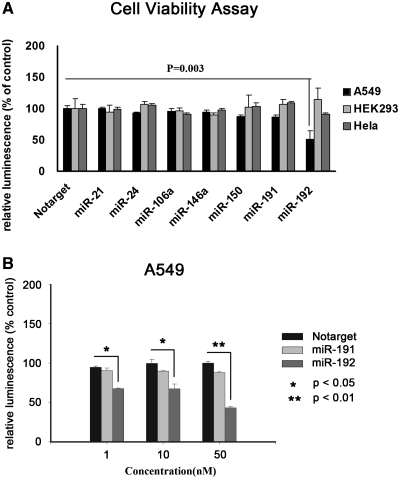
Several miRNAs, including miR-21, 24, 106a, 150 and 192, have been found to be differentially expressed in lung cancers, but the role of these miRNAs in lung carcinogenesis and tumor growth requires clarification (6). We performed a cell viability assay demonstrating that the relative cell viability was dramatically decreased by 48.4% (P < 0.05) when cells were treated with miR-192 (Figure 1A). However, no obvious changes in the growth of HEK293 or Hela cells were observed after treatment with miR-192 or the miRNA mimics (Figure 1A). In addition, we examined the cell viability of A549 cells treated with different doses of miR-192 and miR-191 (a randomly selected control). As shown in Figure 1B, miR-192 suppressed A549 cell viability in a dose-dependent manner. At doses of 1, 10 and 50 nM, treatment with miR-192 significantly (P < 0.05) lowered cell viability by 23.4, 32.5 and 57.2%, respectively, compared with the same concentrations of non-targeting control (Notarget) miRNA. A549 cells are deficient of Mre11A gene, which encodes a nuclear protein involved in homologous recombination, telomere length maintenance and DNA double-strand break repair. But siRNA knockdown of Mre11A did not make Hela cells or HEK293 cells sensitive to miR-192 reduced cell viability (Supplementary Figure S1), so deletion of Mre11a may not be the result of miR-192 reduced cell viability in A549 cells.
Figure 1.
miR-192 inhibits cell viability. (A) Cell viability of A549, HEK293 and HeLa by Celltiter assay. Cells were transfected with 50 nM of miRNA mimics or non-targeting control (Notarget) in 96-well plate. Cell viability assays were performed 48 h after transfection, Celltiter-Glo reagent was added and luminescent intensity was measured by a Microplate Luminometer. (B) Cell viability with different concentrations of miRNA mimics. A549 cells were transfected with 50, 10 and 1 nM miRNA mimics and Notarget control in 96-well plate. Cell viability assays were performed 48 h after transfection. Relative luminescence was calculated with luminescencemiRNA/luminescenceNotarget. Data were the average of three independent experiment runs; bars, SEM. *P < 0.05; **P < 0.01, compared with Notarget control.
miR-192 induces cell apoptosis in A549 cells
Previous reports have shown that expression of miR-192 can arrest cells in G1 phase and reduce S phase entry in colon cancer cells (18,25). We conducted an EdU cell proliferation assay to determine whether miR-192 suppresses the entry of A549 cells into S phase. A comparison of treatments with miR-192 and with the notarget control miRNA (Figure 2A) indicated that the proportion of cells incorporating EdU was decreased by 42.9 ± 5.3% (P < 0.05). Moreover, we examined cell apoptosis in A549 cells using an AnnexinV-PE apoptosis detection kit and found that levels of cell apoptosis were elevated by ∼5-fold in cells exposed to miR-192 versus the Notarget control (Figure 2B). Further experiments revealed that miR-192 induced the expression of poly (ADP-ribose) polymerase (PARP) and of cleaved Caspase-7 (Figure 2C), members of a well-known apoptosis pathway. Cell cycle assay results showed that miR-192 arrested cell in G1 phase, at 18.63 ± 1.96% compared with 7.33 ± 0.52 of Notarget control (Figure 2D). Taken together, our data indicates that miR-192 suppresses A549 cell growth via inhibition of proliferation and enhancement of apoptosis.
Figure 2.
Cell apoptosis induced by miR-192. (A) EdU assay of relative Hoechst stained cells and EdU add-in cells. A549 cells were transfected with 50 nM miRNA mimics, miRNA inhibitor (ant-miR192, fully 2′ O-methyl modified RNA oligo) or Notarget control. Forty-eight hours after transfection, EdU (100 µM) was added and the cells were cultured for 2 h. EdU and Hoechst staining were performed as described in ‘Materials and Methods’ section. At least 200 cells were counted per well. (B) miR-192 induces A549 cell apoptosis. A549 cells were transfected with 50 nM miR-192 mimics, ani-miR192 or Notarget control, 48 h after transfection, apoptosis assay was performed. (C) Western blot analysis of β-Actin, cleaved PARP and Caspase7 after 48 h of treatment with miR-192 or Notarget control. (D) Cell cycle of miR-192 and Notarget-treated A549 cells. Nocodazole (40 ng/ml) were added after 24 h transfection, cells were collected after 48 h and stained with PI and cell cycle was analyzed with FACS. Data were the averages of at least three independent runs; bars, SEM. *P < 0.05; compared with Notarget control.
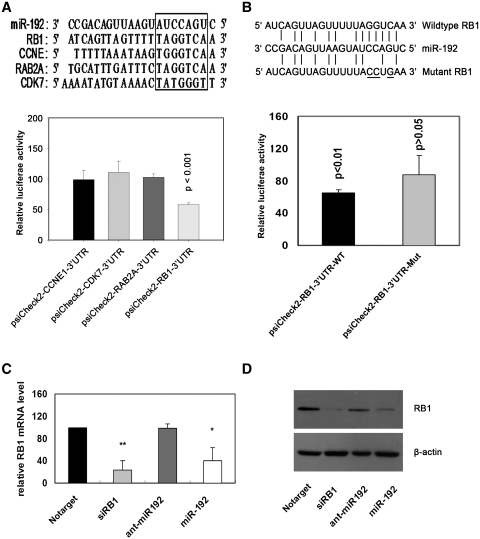
RB1 is a target of miR-192 in A549 cell
We used three open access programs—picTar, TargetScan and miRnaDa—to predict targets of miR-192. Four cell cycle-related genes (CDK7, RB1, CCNE1, RAB2A) that were potential miR-192 targets were selected for further analysis. The 3′-UTRs of these four genes were cloned into the psiCheck-2 plasmid (the predicted binding sites for miR-192 are shown in Figure 3A). The luciferase reporter assay indicated that the activity of the reporter containing the 3′-UTR of the RB1 gene was decreased following treatment with miR-192 mimics, whereas the reporters containing the 3′-UTRs of the other genes were not obviously altered (Figure 3A). We performed site-directed mutagenesis of the reporter containing the 3′-UTR of the RB1 gene as described in Figure 3B and the luciferase reporter assay was performed with the mutated construct. The results demonstrate that the inhibitory effect of miR-192 mimics was abolished in the mutant reporter (Figure 3B). In addition, real time PCR and western blot analysis revealed that the expression of RB1 mRNA and protein was impaired by the treatment with miR-192 mimics in A549 cells (Figure 3C and D). We have also examined whether RB1 reverse the inhibition of cell viability and cell proliferation caused by miR-192 overexpression in the lung cancer cells. The vector of pRB that contains only the RB1-coding sequence was constructed for RB1 expression without miR-192 targeting. The cells were co-transfected with miR-192 mimics and either pRB or pNEG empty vector. The assays clearly showed that ectopic expression of RB1 significantly rescued the suppression of cell viability and cell proliferation caused by miR-192 overexpression (Supplementary Data, Supplementary Figures S2 and S3). Taken together, our results demonstrated that RB1 is a major target of miR-192 in the lung cancer cells.
Figure 3.
RB1 is one of the direct targets of miR-192. (A) The binding site of miR-192 in the predicted target sequences. A549 cells were co-transfected with 50 nM of miR-192 mimics or Notarget control and 200 ng/ml of 3′-UTR reporter plasmid. Luciferase activity was detected at 48 h after transfection. (B) miR-192 binding site in the 3′-UTR of the RB1 mRNA. A mutation was generated in the RB1-3′-UTR sequence in the complementary site for the seed region of miR-192 as indicated. A549 cells were co-transfected with 50 nM miR-192 mimics or Notarget controls and 200 ng/ml of RB1 3′-UTR reporter plasmid or its mutant form. Luciferase activity was detected 48 h after transfection. (C) Total RNA was extracted and real time PCR was performed 48 h after transfection. (D) miR-192 inhibits RB1 protein expression in A549 cells. Total protein was extracted and western blot analysis was performed 48 h after transfection. Relative luciferase activity was calculated with (RlucmiRNA/LucmiRNA)/(RlucNotarget/LucNotarget). Data represented the average of three independent runs; bars, SEM. *P < 0.05; **P < 0.01, compared with Notarget control.
RNAi knockdown of RB1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells
The efficiency of siRB1 in the knockdown of RB1 expression was examined by western blot and qPCR (Figure 4B and C). The siRB1 was then used in cell viability, apoptosis and EdU detection assays. The results shown in Figure 4A demonstrated that treatment with RB1 siRNA resulted in a statistically significant inhibition of viability of A549, H460 and 95D cells. Caspase-7 activation was also observed following treatment with siRB1 in these three cell lines (Figure 4B). EdU assay in A549 and H460 cells show that the cell number is significantly reduced in RB1 siRNA-treated cells compared with notarget control. These experiments provided further evidence that RB1 is one of the direct targets of miR-192 in lung cancer cells.
Figure 4.
siRB1 inhibits cell proliferation and induces cell apoptosis (A) Cell viability of A549, H460, 95D cells. Cell viability assays were performed with 50 nM siRB1, miR-192 mimics or Notarget control, 48 h after transfection, Celltiter-Glo reagent was added and luminescent intensity was read by a Microplate Luminometer. (B) Western blot analysis of RB1, cleavge Caspase 7 after treatment with miR-192 mimics, siRB1 and notarget control (NC). (C) RB1 mRNA expression in A549, H460 and 95D cells after transfected with 50 nM miR-192 mimics, siRB1 and Notarget control for 48 h, β-Actin was used as an internal control. (D) EdU assay of Hoechst stained cells and EdU add-in cells. Cells were transfected with miR-192 mimics, siRB1 and Notarget control at 50 nM. EdU assay were performed 48 h after transfection. Data were the averages of three independent runs; bars, SEM. *P < 0.05, compared with Notarget control.
Expressions of miR-192 is downregulated in human lung cancers
Expression of miR-192 has been reported to be suppressed in colon cancers (18). We compared the endogenous expression of miR-192 in human lung cancer and adjacent non-cancerous lung tissues by qRT–PCR. As shown in Figure 5, expression of miR-192 was downregulated by up to 50% (P < 0.001) in 71% (12 out of 17) of lung cancer tissues compared with the corresponding adjacent non-cancerous lung tissues. One lung cancer sample displayed a 2-fold upregulation of miR-192 expression. These results indicate that miR-192 may act as a tumor suppressor gene.
Figure 5.
miR-192 expression in 17 lung cancer tissues. Stem loop RT–PCR analysis of miR-192 in lung cancer tissues and adjacent non-cancerous lung tissues. U6 rRNA was used as an internal control. 2−ΔΔCt method was used to analyze the qPCR data. The positions of 2-fold overexpression or 2-fold downexpression were drawn a dashed line, respectively.
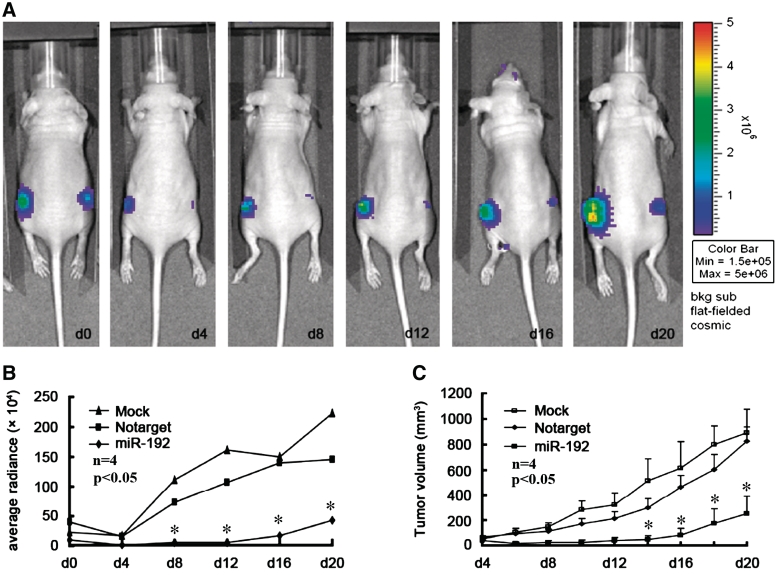
miR-192 inhibits tumorigenicity in vivo
To confirm the tumor suppressor role of miR-192, we established a BALB/c nude mouse xenograft model using a stable cell line derived from human lung cancer A549 cells (A549-luc). The A549-luc cells were pre-treated with miR-192 or a Notarget control and injected s.c. into either posterior flank of the same nude mouse. The luciferase activity and tumor volume were measured every 4 days until Day 20. The fluorescence intensity of A549-luc cells treated with Notarget control miRNA decreased at Day 4 and dramatically increased thereafter until Day 20, whereas the intensity of A549 cells treated with miR-192 mimics increased slowly after Day 4 (Figure 6A and B). The tumor volume in the flanks injected with miR-192 mimic pre-treated cells was also significantly (P < 0.05) smaller than the volume in the flanks injected with Notarget control pre-treated cells at every indicated time point from Day 4 on (Figure 6C). These results indicate that miR-192 significantly inhibits tumorigenicity of A549 cells in vivo in a nude mouse xenograft model.
Figure 6.
miR-192 inhibits tumorigenicity in vivo in BALB/c nude mice. (A) Images of one miR-192-treated mice after 20 days implantation. Fluorescence intensity (B) and tumor volume (C) of miR-192 mimics, Notarget control, and mock control-treated mice were measured at the indicated days after treatment (each group with four mouse, n = 4; data are reperest means ± SEM; *P < 0.05).
DISCUSSION
Our results clearly show that miR-192 inhibits cell proliferation of A549, H460 and 95D lung cancer cells and induces cell apoptosis. In colon cancer cells, miR-192 inhibits the cell proliferation depending on the status of p53; the inhibition efficiency is higher in cells with wild-type p53 than in cells with null or mutant p53 (24,25). A549, H460, 95D, HEK293 and HeLa cells all express wild-type p53, but miR-192 expression had no effect on HeLa or HEK293 cell proliferation. These results indicate that the p53 protein may play a partial role in determining which cell lines may be affected by expression of miR-192, but that there are likely other factors involved in the mechanism of miR-192 action (25). Our EdU assay showed that both the total number of cells and the number of cells with newly synthesized DNA dramatically decreased after treatment with miR-192 mimics. EdU was added just 2 h before detection, indicating that at least during this 2-h period the number of DNA synthesizing cells (reflecting cells in S-phase of the cell cycle) was lower in the miR-192-treated samples than in the Notarget-treated controls. This result is similar to that observed in colon cancer cells, in which miR-192 prevents cells from completing the G1-S transition and delays cell cycle progression (18).
miR-192 inhibits cell proliferation and induces cell apoptosis in A549, H460 and 95D cells. The rate of apoptosis of miR-192 mimic-treated A549 cells was more than 4-fold greater than the rate in Notarget or mock control-treated cells. Both PARP and/or Caspase 7 were cleaved in A549, H460 or 95D cells after treatment with miR-192 mimics. These findings indicate that miR-192 induces lung cancer cell apoptosis through the caspase pathway.
RB1 was the first cancer suppressor gene to be identified. It not only plays important roles in childhood retinoblastoma (RB), bladder cancer and osteogenic sarcoma, but also plays a vital role in regulating cell apoptosis. Knockdown of the Rb gene has been shown to induce γ-H2AX foci formation, a marker of DNA damage, and to promote cellular apoptosis in A549 cells (28). Overexpression of the p16 gene induces cleavage of RB and ultimately induces apoptosis in A549 cells (33). In RB-deleted CC42 (Rb−/−), myocytes (CC42 cells were isolated from Rb null mutation mouse stem cells) undergo higher frequencies of apoptosis during mitogen deprivation-induced myogenesis, while overexpression of RB inhibited apoptosis in Rb deleted or wide-type CC42 cells (27). RB can also protect Saos-2 cells from E2F1 overexpression iduced cell apoptosis, but cannot protect E2F1 mutant (1–376 or 406–415 site mutant) overexpression induced cell apoptosis (26). We found that miR-192 regulates RB1 expression at both the mRNA and the protein levels (Figure 4B and C); miR-192 expression and siRB1 knockdown of the RB1 gene both inhibited A549, H460 and 95D cell proliferation and induced cell apoptosis. Additionally, expression of miR-192 repressed RB1 3′-UTR reporter activity. Overexpression of RB1 without 3′-UTR can significantly reverse the inhibition of miR-192. These results confirm that RB1 is one of the direct targets of miR-192 in lung cancer cells.
Some miR-192 targets have been identified in different cells and organs. miR-192 plays a role in the kidney and diabetic nephropathy by regulating SIP1 (20), and it also targets dihydrofolate reductase (DHFR) in colon cancer to control the cell cycle (24). Microarray experiments have shown that 62 genes are downregulated following miR-192 expression, 8 of which have been validated as targets of miR-192 involved in cell cycle control (18). miR-192 may be a critical downstream mediator of TGF-β/Smad3 signaling in the development of renal fibrosis, but the exact targets involved in this process are unknown (19). The miR-192/194 cluster plays a role in the circadian clock at the post-transcriptional level, but again the targets involved are unknown (34). We found RB1 to be a novel target of miR-192 involved in cell apoptosis. Additional targets remain to be identified.
Our results demonstrate that miR-192 is downregulated in lung cancer tissues compared to adjacent non-tumor lung tissue. miR-192 is also downregulated in the lungs of rats exposed to cigarette smoke (35), and repressed in colon cancer (36), ulcerative colitis (37), multiple myeloma (38), diabetic nephropathy (39), while miR-192 upregulated in lgA nephropathy (40,41) and in hypertensive nephrosclerosis (42). Furthermore, miR-192 can be downregulated by genotoxic stress in colon cancer cells (18,25). Ectopic overexpression of miR-192 inhibits cell proliferation in vitro and tumorigenicity in vivo in a nude mouse model. These results suggest that miR-192 is a tumor suppressor and indicate that it may be useful as a future drug target for controlling cancer progression.
In summary, our results demonstrate the following: miR-192 is downregulated in human lung cancer tissues compared to adjacent non-cancerous lung tissues; miR-192 mimics inhibit cell proliferation and induce cell apoptosis in lung cancer cells and inhibit tumorigenicity in a nude mouse model in vivo; and RB1 is likely a bona fide target of miR-192. Our data suggest that miR-192 is an important tumor suppressor that could be exploited in lung cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-R-221 and KSCX2-YW-R-244 to B.Z.); Science and Technology Program of Bureau of Science and Technology of Guangzhou Municipality, Guangzhou, China (2007Z1-E4041 to B.Z. and 2007Z1-E0111 to J.H.). Funding for open access charge: Guangzhou Institutes of Biomedicine and Health, CAS.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 3.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 8.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam W, Dahlberg JE. miR-155/BIC as an oncogenic microRNA. Gene Chromosome Can. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 16.Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, Watanabe M. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–1442. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Zhou F, Xu T, Deng H, Ge YY, Zhang C, Li J, Zhuang SM. Analysis of sequence variations in 59 microRNAs in hepatocellular carcinomas. Mutat. Res. 2008;638:205–209. doi: 10.1016/j.mrfmmm.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 19.Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 2010;21:1317–1325. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl Acad. Sci. USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elvira-Matelot E, Zhou XO, Farman N, Beaurain G, Henrion-Caude A, Hadchouel J, Jeunemaitre X. Regulation of WNK1 Expression by miR-192 and Aldosterone. J. Am. Soc. Nephrol. 2010;21:1724–1731. doi: 10.1681/ASN.2009111186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S, Du J, Wang Z, Yan J, Yuan W, Zhang J, Zhu T. Dual mechanism of deltaEF1 expression regulated by bone morphogenetic protein-6 in breast cancer. Int. J. Biochem. Cell Biol. 2009;41:853–861. doi: 10.1016/j.biocel.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 23.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol. Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 24.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate rEdUctase and cellular proliferation through the p53-microRNA circuit. Clin. Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh JK, Fredersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–1852. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Guo K, Wills KN, Walsh K. Rb functions to inhibit apoptosis during myocyte differentiation. Cancer Res. 1997;57:351–354. [PubMed] [Google Scholar]
- 28.Ebi H, Sato T, Sugito N, Hosono Y, Yatabe Y, Matsuyaa Y, Yamaguchi T, Osada H, Suzuki M, Takahashi T. Conterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancer. Oncogene. 2009;28:3371–3379. doi: 10.1038/onc.2009.201. [DOI] [PubMed] [Google Scholar]
- 29.Nana-Sinkam SP, Karsies T, Riscili B, Ezzie M, Piper M. Lung microRNA: from development to disease. Expert Rev. Respir. Med. 2009;3:373–385. doi: 10.1586/ers.09.30. [DOI] [PubMed] [Google Scholar]
- 30.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 2010;10:543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;26:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 33.Katsuda K, Kataoka M, Uno F, Murakami T, Kondo T, Roth JA, Tanaka N, Fujiwara T. Activation of caspase-3 and cleavage of RB are associated with p16-mediated apoptosis in human non-small cell lung cancer cells. Oncogene. 2002;21:2108–2113. doi: 10.1038/sj.onc.1205272. [DOI] [PubMed] [Google Scholar]
- 34.Nagel R, Clijsters L, Agami R. the miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 35.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earle JS, Luthra R, Romans A, Abraham R, Ensor J, Yao H, Hamilton SR. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J. Mol. Diagn. 2010;12:433–440. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 38.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr, Hofmainster C, Alder H, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18:367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest. 2010;90:98–103. doi: 10.1038/labinvest.2009.118. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Kwan BC, Lai FM, Chow KM, Kam-Tao,Li P, Szeto CC. Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis. Markers. 2010;28:79–86. doi: 10.3233/DMA-2010-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am. J. Hypertens. 2010;23:78–84. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.