Abstract
RET protein functions as a receptor-type tyrosine kinase and has been found to be aberrantly expressed in a wide range of human diseases. A highly GC-rich region upstream of the promoter plays an important role in the transcriptional regulation of RET. Here, we report the NMR solution structure of the major intramolecular G-quadruplex formed on the G-rich strand of this region in K+ solution. The overall G-quadruplex is composed of three stacked G-tetrad and four syn guanines, which shows distinct features for all parallel-stranded folding topology. The core structure contains one G-tetrad with all syn guanines and two other with all anti-guanines. There are three double-chain reversal loops: the first and the third loops are made of 3 nt G-C-G segments, while the second one contains only 1 nt C10. These loops interact with the core G-tetrads in a specific way that defines and stabilizes the overall G-quadruplex structure and their conformations are in accord with the experimental mutations. The distinct RET promoter G-quadruplex structure suggests that it can be specifically involved in gene regulation and can be an attractive target for pathway-specific drug design.
INTRODUCTION
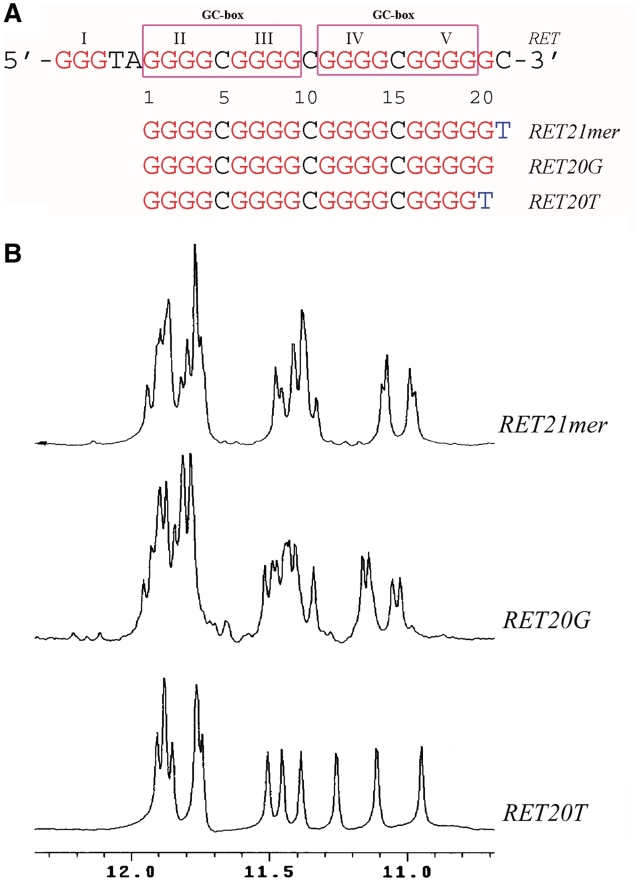
The RET proto-oncogene encodes a receptor-type tyrosine kinase that plays an important role in the initiation and progression of several human cancers, especially thyroid cancer (1–4). For example, the multiple endocrine neoplasia type 2 (MEN 2), an inherited cancer syndrome characterized by medullary thyroid carcinoma (MTC) and pheochromocytoma (PC), was caused by germline mutations in the exon region, encoding one of three specific cysteine residues in the extracellular domain of the RET protein. Moreover, RET protein levels was found to be overexpressed in MTC and PC, a common feature for these cancer cells (5–8). Thus, RET protein has been investigated as a potential therapeutic target in preclinical approaches for the treatment of RET-associated cancers. It was revealed from the studies of the transcriptional regulation of the RET proto-oncogene that the human RET promoter had three GC boxes corresponding to three Sp1 binding sites in the proximal promoter region. Two of these GC boxes, locating between −59 and −25 upstream of the transcription start site, are essential for functional transcription activity of RET basal promoter (9,10). This proximal promoter region of the human RET protooncogene containing five guanine tracts (Figure 1) was reported to form stable parallel-stranded G-quadruplex in the presence of K+ (11), a high-order DNA structure which is related to the activities of promoter sequences.
Figure 1.
(A) The promoter sequence of the RET oncogene and its modifications. RET20G is the wild-type sequence containing the latter four consecutive G-stretches; RET21mer is RET20G sequence with additional T at the 3′-end; RET20T is the mutant of RET20G with the replacement of G at 3′-terminal by T. The five G-stretches are colored in red and numbered using Roman numerals; Two GC-boxes are labeled. (B) The imino proton regions of 1D 1H NMR spectra of RET21mer, RET20G and RET20T with sample concentration at 0.3 mM. NMR buffer conditions: 25°C, 20 mM K-phosphate, 80 mM KCl, pH 6.8.
The G-quadruplexes are generally formed in DNA and RNA sequences containing repeated short guanine-rich tracts by the stacking interaction of successive G-G-G-G tetrads (G-tetrads) and stabilized by bound monovalent Na+ or K+ cation (12). The arrangements of G-quadruplexes can be tetramolecular, bimolecular or intramolecular, which are possible by virtue of the changes in strand polarities and also the sequence and topologies of the loops (13,14). It has been reported that G-quadruplex structures can be formed in vitro in the human telomere ends (15–19) and the promoter regions of different oncogenes, such as c-Myc (20,21), c-kit (22,23), VEGF (24) and Bcl-2 (25,26). The G-quadruplex-interactive agents such as TMPyP4 and telomestatin are used to induce and stabilize G-quadruplex structure formed in G-rich sequence, for example, of the c-Myc and RET promoters, to inhibit their promoter activities and regulate their expression (11,20). This suggests that these G-quadruplexes can serve as therapeutic targets, and it is worthwhile to make considerable efforts to identify evidence in support of an in vivo functional role for G-quadruplexes (27–29).
Here, we report the NMR solution structure for the predominant G-quadruplex structure formed in the RET promoter region. The solution structure provides not only the molecular details of this G-quadruplex but also important insights for its loop conformations and interactions with the core tetrad structures.
MATERIALS AND METHODS
Sample preparation
Unlabeled and site-specific low-enrichment (2% 15N-labeled) oligonucleotides were synthesized on an ABI 394 DNA synthesizer and purified by HPLC (30). They were dialyzed successively against 40 mM KCl solution and against water. The strand concentration of the NMR samples was typically 0.2–3 mM; the solutions contained 80 mM KCl and 20 mM potassium phosphate (pH 6.8).
Circular dichroism
Circular dichroism (CD) spectra were recorded at 25°C on a JASCO-815 spectropolarimeter using a 1-cm path length quartz cuvette with a reaction volume of 400 μl. DNA concentration was 7 μM. The DNA oligonucleotides were prepared in a pH 6.8 buffer containing 20 mM potassium phosphate and 5, 10, 20 and 100 mM KCl, respectively. The samples were heated to 95°C for 5 min and cooled down to room temperature overnight. For each sample, an average of three scans was taken, the spectrum of the buffer was subtracted.
NMR experiments
Experiments were performed on 600 MHz Varian and 800 MHz Bruker spectrometers at 25°C. Resonances were assigned unambiguously by using site-specific low-enrichment labeling and through-bond correlations at natural abundance (31,32). The NMR experiments for samples in water solution were performed with Watergate or Jump-and-Return water suppression techniques. The acquisition data points were set to 2048 × (250–512) (complex points). All spectra were processed by using the program NMRPipe (33). The 45° or 60° shifted sine-squared functions were applied to NOESY and TOCSY spectra. The fifth-order polynomial functions were employed for the baseline corrections. The final spectral sizes are 2048 × 1024. The 1H chemical shifts were referenced to 2, 2-dimethylsilapentane-5-sulfonic acid (DSS). Peak assignments and integrations were achieved using the software Sparky (http://www.cgl.ucsf.edu/home/sparky/). The NOE peaks were integrated using the peak fitting function and volume integration of Sparky.
The 31P NMR spectra were collected on a DNA sample at 1.5 mM in D2O (20 mM potassium–phosphate buffer, 80 mM KCl, pH 6.8) at 25°C and were referenced to an external standard of 85% H3PO4, including the one dimensional proton-decoupled phosphorus spectrum, and two dimensional heteronuclear 31P-1H Correlation Spectroscopy (COSY). Assignments of the individual 31P resonance were accomplished by a combination of two dimensional 1H-1H NOESY, COSY, TOCSY and heteronuclear 31P-1H COSY (Supplementary Figure S8).
Distance geometry and simulated annealing calculations
The distances between non-exchangeable protons were estimated based on the NOE cross-peak volumes at 50, 75, 100, 150, 200 and 250 ms mixing times, with the upper and lower boundaries assigned to ±20% of the estimated distances. The cytosine base proton H5–H6 distance (2.45 Å) was used as a reference. Exchangeable proton restraints are based on NOESY data sets at two mixing times (50 and 200 ms) in H2O. Cross-peaks involving exchangeable protons were classified as strong (strong intensity at 50 ms), medium (weak intensity at 50 ms) and weak (observed only at 200 ms), and distances between protons were then restrained to 3.0 ± 0.9 Å, 4.0 ± 1.2 Å and 6.0 ± 1.8 Å, respectively. The distances involving the unresolved protons, e.g. methyl protons, were assigned using pseudo-atom notation to make use of the pseudo-atom correction automatically computed by X-PLOR.
The structure of RET20T was calculated on the basis of NMR restraints by using X-PLOR (NIH version) (34) to embed and optimize 100 initial structures. An arbitrary extended conformation was first generated for the single-stranded RET20T sequence. Substructure embedding was performed to produce a family of 100 distance geometry (DG) structures. Then the 100 structures were subjected to simulated annealing regularization. The experimentally obtained distance restraints and G-tetrad hydrogen-bonding distance restraints were included during the calculation. All distance restraints were specified with the SUM averaging option in X-PLOR.
Distance restrained molecular dynamics calculations
The 10 best structures were selected and subjected to distance restrained molecular dynamics calculations in XPLOR with a distance-dependent dielectric constant. The G-tetrads within the quadruplex were restrained with distances corresponding to ideal hydrogen bond geometry. Each individual hydrogen bond was restrained using two distance restraints (heavy atom–heavy atom and heavy atom–proton). Hydrogen bond distance restraints were also applied to H1 (in G16):O4 (in T20) and NH2 (in C5):N3 (in G3), with larger distance bounds (±0.3 Å). The force constants were scaled at 30 and 100 kcal mol−1 Å−2 for NOE and hydrogen bond distance restraints, respectively. A total of 411 NOE-distance restraints (Table 1), of which 138 are from interresidue NOE interactions, were incorporated into the NOE-restrained structure calculation.
Table 1.
Structural statistics for the RET20T G-quadruplex structure
| NMR distance and dihedral constraints | |
| Distance restraints | |
| Total NOEs | 359 |
| Intraresidue | 221 |
| Interresidue | 138 |
| Sequential (|i−j| = 1) | 84 |
| Non-sequential (|i−j| > 1) | 54 |
| Hydrogen bonds | 52 |
| Total dihedral angle restraints | 42 |
| Structural statistics | |
| Violations (mean and SD) | |
| Number (>0.2 Å) | 0 |
| Maximum violations (Å) | 0.158 ± 0.026 |
| Distance constraints (Å) | 0.044 ± 0.0016 |
| Dihedral angle constraints (°) | 0.90 ± 0.09 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.005 ± 0.0001 |
| Bond angle (°) | 1.00 ± 0.01 |
| Impropers (°) | 0.60 ± 0.01 |
| Average pairwise r.m.s.d. of all heavy atoms (Å) | |
| All heavy atoms except C15 | 0.55 ± 0.15 |
| All residues | 0.75 ± 0.26 |
Dihedral angle restraints were used to restrict the glycosidic torsion angle (χ) for the experimentally assigned syn configuration, i.e. G1, G7, G11 and G17 tetrad-guanines [60 (±35)°], and C5 and G16 in the loop [60 (±35)°], as well as for some of the experimentally assigned anti-configuration bases, i.e. G2, G3, G4, G6, G8, G9, G12, G13, G14, G18 and G19 [240(±70)°]. Dihedral angle restraints were also used to restrain the sugar backbone torsion angles β, γ and ε (35). Based on the J-coupling constants of 31P(n)-H5′/H5″(n) and H3′ (n − 1) − 31P(n) obtained from 31P-1H COSY (Supplementary Figure S6), the β angles were restrained to the β-t conformation at 180(±70)° for all the residues, except for G11 and G17 whose β angles were restrained to −60(±20)°. The ε angles were restrained to 90(±20)° for G6, C10 and G14. Based on the relative intensities of H3′–H5′/H5″ and H4′–H5′/H5″, the g angles of the majority of residues with resolved H5′/H5″ are in the regular γ+ conformation (∼60°) or sometimes in the γ- conformation (∼60°), because for each residue, the H3′–H5′ (or H3′–H5″) NOE is clearly stronger than the H3′–H5″ (or H3′–H5′) NOE, except for G14 which shows similar intensities for H3′–H5″ and H3′–H5″, and thus falls in the γ-t region (35). Thus only the γ angle of G14 was restrained to 170(±80)°. The force constants of dihedral angle restraints were 10 kcal mol−1 rad−2 for χ and 5 kcal mol−1 rad−2 for β, γ and ε.
Restrained molecular dynamics calculations were initiated at 300 K, and the temperature was gradually increased to 1000 K in 4 ps. The system was equilibrated at 1000 K for 20 ps, and was then slowly cooled to 300 K in 10 ps. The coordinates saved every 0.2 ps during the last 2.0 ps were averaged. The resulting average structure was subjected to minimization until the energy gradient of 0.1 kcal mol−1 was achieved. A soft planarity restraint (weight of 5 kcal mol−1 Å−2) was imposed on the G-tetrads before the heating process and was removed at the beginning of the equilibration stage. The time steps for all processes of heating, cooling, and equilibration were equal to 0.5 fs. The 10 best molecules were selected based both on their minimal energy terms and number of NOE violations.
Data deposition
The coordinates for the d[(G4C)3G4T] quadruplex have been deposited in the Protein Data Bank (accession code 2L88).
RESULTS AND DISCUSSIONS
Screening sequence RET20T as the predominant G-quadruplex in the RET promoter
A recent study has shown that the four consecutive guanine repeats II–V (Figure 1A) in the G-rich strand of RET promoter form the intramolecular G-quadruplex in the presence of K+ solution (11). The imino proton NMR spectra in Figure 1B indicates the presence of multiple G-quadruplex forms for the wild-type sequence of RET promoter RET20G. To obtain the sequence of RET promoter suitable for NMR G-quadruplex structure studies, we mutated the nucleotide residues flanking the forth G-tracts and got the two sequences RET21mer and RET20T (Figure 1). Fortunately, the sequence RET20T forms the most stable G-quadruplex structure, as demonstrated by NMR spectrum (Figure 1B). RET20T gives a well-defined spectrum that has many overlapping 1H NMR resonance signals with the wild-type sequence of RET20G (Figure 1). Therefore, the RET20T sequence with a mutant G to T at 3′-end was chosen as the sequence for NMR structure determination.
CD signature
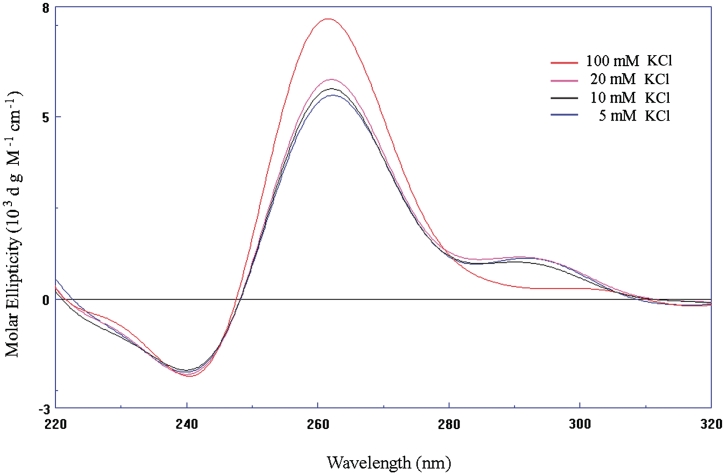
The CD spectra (Figure 2) for RET20T in K+ solution were in agreement with the results from NMR of the formation of G-quadruplex structure. The CD profile for RET20T containing 100 mM KCl almost displayed a single-positive absorption at 260 nm and negative absorption around 240 nm, which is characteristic of all-parallel G-quadruplex structure (36). While RET20T with low concentration of KCl (5, 10 and 20 mM) exhibited a negative minimum around 240 nm, a single positive maximum around 260 nm, and a weak plateau from 280 to 300 nm, indicative of the formation of two or more folds of G-quadruplex in the solution (37). These data revealed that high concentration of K+ favors the all-parallel G-quadruplex structure for the sequence RET20T.
Figure 2.
CD spectra of RET20T in 5, 10, 20, and 100 mM KCl solution recorded at 25°C.
NMR spectral assignments
The spectral line widths of the modified sequence RET20T (2–4 Hz for the sharpest peaks at 25°C) are indicative of a monomeric intramolecular structure, this is supported by the concentration-independent of the well-resolved quality of 1H spectra for RET20T (Supplementary Figure S1), where the line widths of imino proton signals are almost same to each other in both cases at the concentration of 0.2 and 3 mM of RET20T NMR sample. The result is also in agreement with the previous data (11), which showed that the G-quadruplex structure formed by the RET promoter sequence was a parallel-type intramolecular structure by comparative CD and DMS footprinting studies. This is further corroborated by the NMR stoichiometry titration experiment (38) (Supplementary Figure S2).
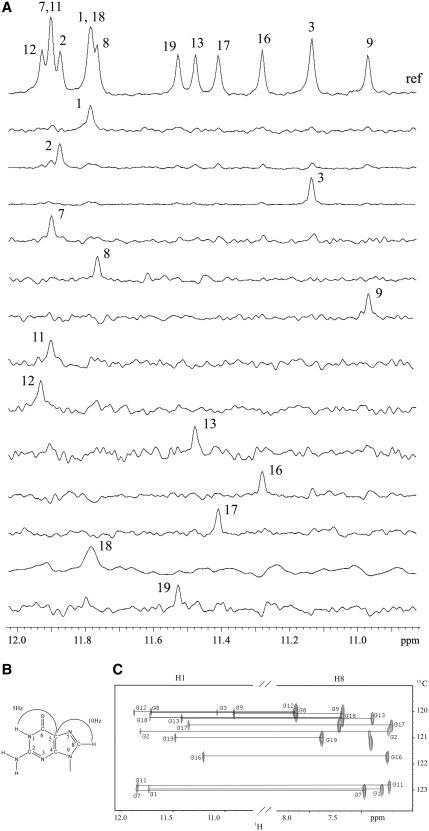
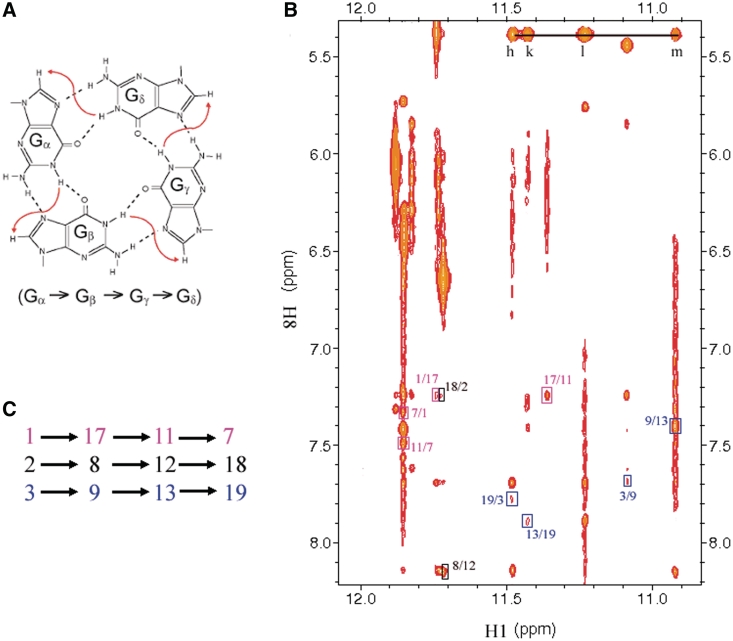
Guanine imino and H8 protons of RET20T were unambiguously assigned by using the site-specific low-enrichment labeling and natural abundance through bond correlation strategies (31,32) (Figure 3A and C). Resonances for cytosine residues were unambiguously assigned by C-to-T substitutions. The non-exchangeable base and sugar protons of the RET20T sequence were assigned using standard protocols by through space (two dimensional 1H-1H NOESY) and through bond (two dimensional 1H-1H COSY, 1H-1H TOCSY and 13C-1H HSQC) experiments in 2H2O K+ solution. In case of spectral overlapping, resonance assignments were facilitated by guanosines (G) to inosine (I) substitutions at positions 4, 14 (double-site RET20T4-14I mutant shown in Supplementary Figures S3, S4 and S5) and position 16 (RET20T-16I mutant shown in Supplementary Figure S6 and S7). The expanded 1H-1H NOESY cross-peaks (mixing time 200 ms) including the classical H8/H6-H1′ sequential connectivities were shown in Figure 3A. The complete assignments of base and sugar proton chemical shifts are listed in Supplementary Table S1. The intensities of intraresidue H8-H1′ NOE cross-peaks (Figure 4B) indicate syn glycosidic conformation for G1, C5, G7, G11, G16 and G17, in contrast to other residues, which adopt anti conformation.
Figure 3.
(A) Imino proton spectra with assignments indicated over the reference spectrum (ref) on the top. Guanine imino protons were assigned in 15N-filtered spectra of samples, 2.0% 15N-labeled at the indicated positions. (B) A schematic indicating long-range J couplings used to correlate imino proton and H8 proton within the guanosine base. (C) H8 proton assignments of RET20T sequence by through-bond correlations between guanosine imino and H8 protons via 13C (at 5-position) at natural abundance, using long-range J couplings shown in (B).
Figure 4.
Non-exchangeable proton assignments of RET20T Sequence. (A) Expanded 1H-1H NOESY spectrum (200 ms mixing time) correlating base and sugar H1′ protons. The line connectivities trace NOEs between a base proton (H8 or H6) and its own and 5′-flanking sugar H1′ protons. Intraresidue base to sugar H1′ NOEs are labeled with residue numbers. The peak assigned to G17 is broadened at 25°C, which reflects a motion at the bottom of structure. (B) Stacked plot of short mixing time (100 ms) NOESY spectrum. The strong intraresidue guanosine H8-H1′ cross-peaks (syn glycosidic bonds) are labeled and can be distinguished from weak cross-peaks (anti-glycosidic bonds). (C) Imino proton NMR spectrum of RET20T sequence after 60 min in D2O solution. Assignments of slowly exchanging imino protons are listed over the spectrum.
Determination of G-quadruplex folding topology
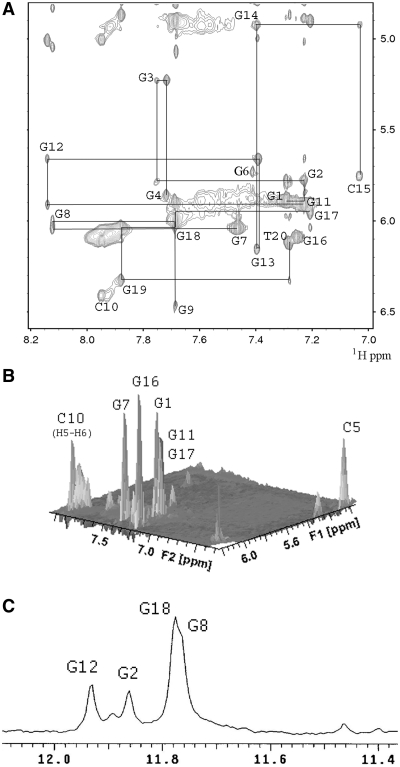
Analysis of characteristic NOEs between imino and H8 protons (Figure 5B) revealed the formation of an intramolecular G-quadruplex involving three G-tetrads, G1•G7•G11•G17, G2•G18•G12•G8, and G3•G19•G13•G9 (Figures 5A and C), which is further confirmed by the NOEs in the H1 region (Supplementary Figure S9). The hydrogen-bond directionalities of the three G-tetrads are clockwise, anti-clockwise and anti-clockwise, respectively. The glycosidic conformations of guanines around the first G-tetrad are syn-syn-syn-syn, while those of the other two G-tetrads are anti-anti-anti-anti. These glycosidic conformations are consistent with the all parallel-stranded G-quadruplex core containing four G-tracts (G1-G2-G3, G7-G8-G9, G11-G12-G13 and G17-G18-G19) oriented in one direction. All of the three loops are of the double-chain reversal type. The first and the third loops are made of three nucleotides (G4–C5–G6 and G14–C15–G16), while the second loop is composed of a single nucleotide (C10). In this G-quadruplex fold G2, G8, G12 and G18 are in the central G-tetrad, consistent with imino protons of these residues being the most protected from exchange with water (Figure 4C).
Figure 5.
Determination of G-quadruplex topology for the promoter sequence RET20T in K+ solution. (A) Characteristic guanosine imino-H8 NOE connectivity patterns around a Gα•Gβ•Gγ•Gδ tetrad as indicated by arrows. (B) Guanosine imino-H8 connectivities observed for G1•G17•G11•G7 (Magenta), G2•G8•G12•G18 (black) and G3•G9•G13•G19 (blue) G-tetrads. (C) Interstrand NOEs between imino H1 and aromatic H8 of unimolecular guanine bases within the same layer of G-tetrads (boxed in magenta, black and blue corresponding to the three tetrad mentioned in (B), respectively).
Solution structure of RET20T G-quadruplex in the presence of K+
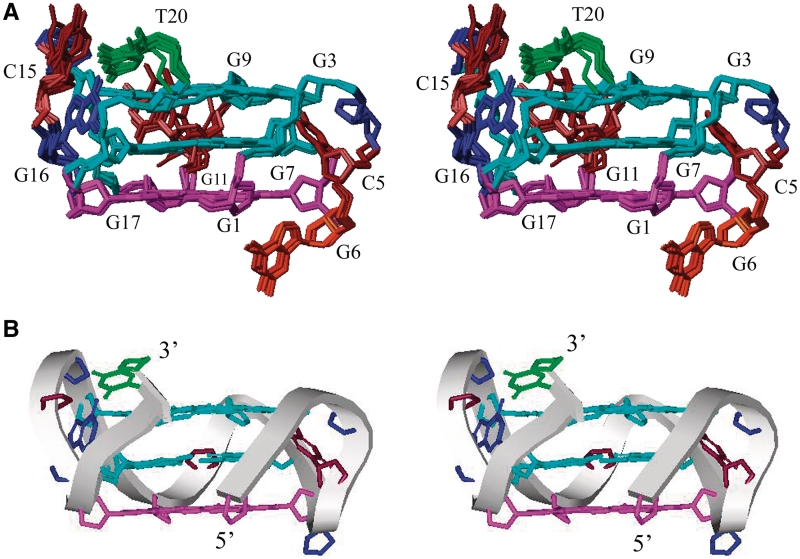
The structure of the RET20T quadruplex was calculated on the basis of NMR restraints (Table 1) by using the X-PLOR program (34) (see ‘Materials and Methods’ section). Ten superimposed lowest energy refined structures of the RET20T quadruplex are shown in Figure 6A. Ribbon view of a representative refined structure of the RET20T quadruplex are shown in Figure 6B.
Figure 6.
Stereoview of the RET20T G-quadruplex structure in K+ solution. (A) 10 superimposed refined structures. Guanosine bases in G-tetrad core are colored cyan (anti) and magenta (syn). Bases in loops are in brown for cytosines (C5, C10 and C15), orange–red for G6, blue for G16, and green for T20. For clarity, the bases G4, G6 and G14 are only shown the backbone in the figure and (B) Ribbon view of a representative structure.
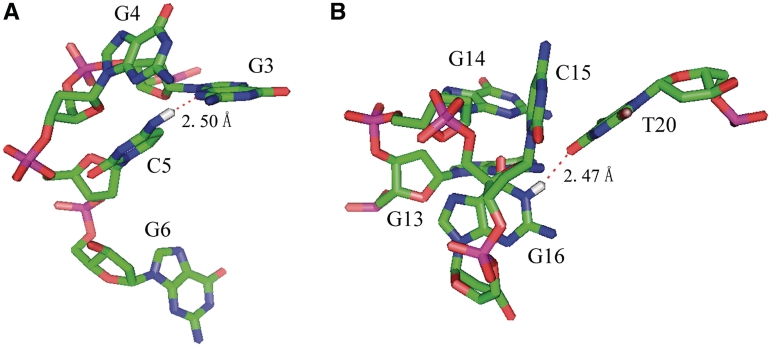
As shown in Figure 8A, the G-quadruplex consists of three G-tetrads linked with four parallel right-handed G-strands (G1-G2-G3, G7-G8-G9, G11-G12-G13 and G17-G18-G19) that are connected by three double-chain reversal side loops (G4–C5–G6, C10, G14–C15–G16). The first loop is of the double-chain-reversal type, and it is composed of three nucleotide segment G4-C5-G6 with unique topology features. C5 flanked by the groove of the three G-tetrad core structure adopts syn conformation for the glycosidic bonds from the NOE intensities of H1′-H6. This conformation is stabilized by the hydrogen bond between atoms N3 (in G3) and NH42 (in C5), which is covered by the G4 located at the top of the core structure. The observation of NOEs between H41, H42 (in C5) and the sugar protons of G4, H5, H6 (in C5) and H1 (in G2 and G3) confirms the alignment (in Figure 7A). While G6 in the loop is very flexible as shown in Figure 6A, for there are almost no NOEs between G6 and its neighboring nucleotides. The second loop is also of double-chain reversal type and is bridged by only 1 nt C10. The hird linker G14–C15–G16 still forms the double-chain reversal loop, which connects columns G11–G13 and G17–G19. This loop is stabilized by the hydrogen bond between H1 (in G16) and O4 (in T20), well explaining the up-field shifted 11.2 ppm imino proton of G16, typical of a guanosine imino proton hydrogen bonded to a thymine oxygen accepter. G14 stacks over the top G-tetrad G3•G9•G13•G19, consistent with the NOE between the average amino proton of G14 and imino protons of G3, G9, G13 and G19 (Figure 5B). C15 stacks over the G16:T20 base pair, which further stabilize the hydrogen bond between G16 and T20. The alignment of G14–G16 and T20 is further confirmed by the observation of NOEs between methyl group of T20 and H1 protons (in G16 and G19), NH2 (in G14), H5 (in C15) (in Figure 7B).
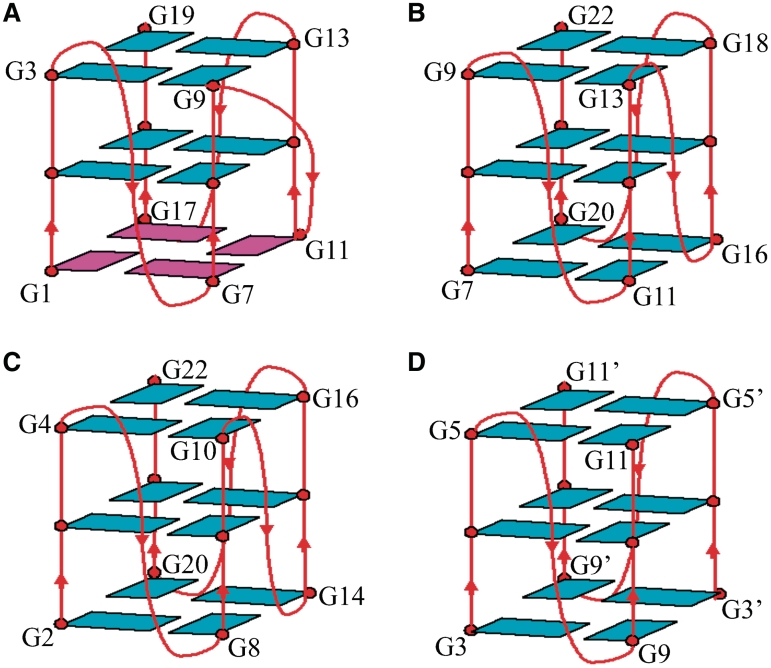
Figure 8.
Schematic structures of all parallel G-quadruplexes formed by (A) the promoter sequence of oncogene RET in K+ solution (this work), (B) the promoter sequence of c-Myc in K+ solution, (C) the four-repeat human telomeric d[AGGG(TTAGGG)3] sequence in K+ solution and (D) the two-repeat human telmoreic d[TAGGGTTAGGGT] sequence in K+. Loops are colored red; anti and syn guanines are colored cyan and magenta, respectively.
Figure 7.
Detailed loop structures of the RET20T G-quadruplex in K+ solution. (A) The conformation of the first loop G4–G6. The amino group of C5 forms hydrogen bond with N3 in G3, which is further protected by G4. (B) Loop conformation for the segment G14–G16 and T20. H1 (in G16) in the third loop forms hydrogen bond with O4 (in T20), which stabilizes the conformation of the double-chain reversal loop in RET20T quadruplex structure.
Mutational analysis of RET20T for loop segments
We have carried out systematic analysis of RET20T promoter sequences with mutated residues in the loop regions to determine their functional role in RET20T G-quadruplex formation and stability. We incorporated two mutations in the first and third loops, G4 and G14 with inosine, resulting in much higher quality of the imino signals (Supplementary Figure S3B). Further analysis of the spectra of the sequence RET20T4-14I indicated no changes on the overall quadruplex topology from the NOE pattern (Supplementary Figure S4). As shown in Supplementary Figure S6, replacement of unpaired residue C15 by T15 (i.e. RET20T-15T) also has no impact on the imino signals in the spectrum. Replacement of the central residue C5, which is involved in the special conformation of G3–G4–C5, by T5 (i.e. RET20T-5T) impaired the quality of imino proton signals, indicative of a role for the hydrogen bond formed between N3 (in G3) and NH2 (in C5) in stabilizing the double-chain reversal loop G4–C5–G6 of the G-quadruplex. Substitution of G16I resulted in a single conformation with the same general fold from the NOE pattern (Supplementary Figure S7). The 3′-terminal residue T20, which is involved in the formation of hydrogen bond with G16, could be substituted with A and C (Supplementary Figure S6) without impact on the general folding of the structure from the NOE pattern (data for RET20T-20A shown in Supplementary Figure S10, data for RET20T-20C are not shown). This could be explained by the flexibility of the terminal residue T20. A20 and C20 instead of T20 still could form hydrogen bond with G16 by N1 in A20 (NOE between H2 in A20 and H1 in G16) and N3 in C20 with H1 in G16.
Multiple conformations in RET promoter sequence
The RET promoter sequence located between −59 and −25 upstream of the transcription start site contains five G-stretches, which contain three/four/five guanosines per stretch (Figure 1A). One could imagine multiple possible ways of G-quadruplex formation using different G-stretches. Indeed, this G-rich strand has the ability to adopt two intramolecular G-quadruplex structures in the presence of K+ with two sets of G-tracts I–IV and II–V. While the DNA polymerase assay indicated that the G-quadruplex formed by the latter sequence was the major form (11). However, even for a given set of four G-tracks, there may be several possible intramolecular G-quaduplex topologies, which differ by G-selection in one G-tract, strand orientations, syn/anti distributions, and/or loop connections (13,14). From the 1H NMR spectra (Figure 1B), the sequence RET21mer and RET20G could form different conformations in the presence K+. While the topology formed by the sequence RET20T, almost a natural fragment derived from the RET promoter, might represent the most stable conformation involved in the biologically relevant G-quadruplex structure.
The RET20T fold represents a new conformation for all parallel-stranded G-quadruplex
We have identified the derivative guanine-rich strand sequence from the RET promoter, RET20T, which is suitable for structural characterization by NMR. The sequence contains four G-tracts with four guanines per tract that have been shown previously to participate in formation of G-quadruplexes (11). We have shown here that RET20T forms intramolecular propeller-type parallel-stranded G-quadruplexes in K+ solution: the core of three G-tetrads is formed by four G-tracts oriented in the same direction, all three loops are of double-chain reversal loop type. The parallel-stranded G-quadruplex topologies with three or two double-chain reversal loops have been reported for the promoter sequence of c-Myc (Figure 8B) (39,40) and for the human telomeric G-rich strands containing the four-repeat d[AG3(T2AG3)3] (Figure 8C) (16) and the two-repeat d[TAG2T2AG3T] (Figure 8D) (16). The core structure and conformations of loops represented here are different and represent some new features. The conformations adopted by the guanines involved in the formation of the core structure: when their G-tetrad cores are aligned, the glycosidic conformations of the first G-tetrad are syn•syn•syn•syn and those of the other two are anti•anti•anti•anti (Figure 8A), while the conformations of the three G-tetrads are all anti•anti•anti•anti in all the other three structures (Figure 8B–D). Moreover, although these structures are made of double-chain-reversal loops, the first and the third loops in structure RET20T still exhibit their distinct alignments which are involved in the specific interactions between the residues in the loops and the core structure (Figure 7).
A common feature of the stable loop conformation in RET20T G-quadruplex and in the promoter G-quadruplexes c-Myc (39,40), Bcl2 (25,26), and in human chl1 intronic G-quadruplexe (41), is the single-nucleotide G3NG3 double-chain reversal loop. Such loop spanning three G-tetrad was first reported by Phan AT et (39) in the G-quadruplex structure formed by the promoter sequence c-Myc and seems essential for the stability for the promoter sequence G-quadruplex structure, such as for likely VEGF (42,43), HIF-1α (44) promoter sequences.
A recent study of glycosidic conformation of guanines in anti-parallel G-quadruplex structure revealed the rules of relationship between the glycosidic conformation and strand orientation: same strand direction, same glycosidic conformation and to form as many 5′-syn-anti steps for glycosidic conformational pattern as possible (45). These rules also seem to be applied in the all-parallel G-quadruplex structure. In the all-parallel G-quadruplex structures, such as shown in Figure 8B–D, the guanines within one G-tetrad display identical glycosidic conformations (all anti guanines) corresponding to their strand orientation. While the all-parallel structure formed by the sequence RET20T (Figure 8A) has the maximum number of the 5′-syn-anti steps (4 syn guanines). The free-energy calculation combining solute entropy estimates indicates that syn-anti and anti-anti steps are close in stability for guanines stacking (45). Moreover, the hydrogen bond between O5′-H5T (in G1) and N3 (in G1) (∼2.0 Å) in the RET20T further contribute the stability of the overall folding (Tm ∼58°C, Supplementary Figure S2). Therefore, the RET20T topology with four syn guanines within one G-tetrad represents a new overall folding for all-parallel G-quadruplex structures.
Biological implications for the oncogene RET G-quadruplex
In the presence of K+, the promoter region of oncogene RET could form G-quadruplex structure. Our NMR data revealed the G-rich strand within the oncogene RET promoter region could form multiple G-quadruplex structures. A major form of the G-quadruplex structure formed by the four G-stretches (II–IV) is determined in the present work. The G-quadruplex is composed of three stacked G-tetrad and 4 syn guanines within one G-tetrad, three double-chain reversal loops which interact with the core structure specifically. The specific alignments make the overall G-quadruplex structure has a distinct pattern of grooves in comparison with the all parallel-stranded G-quadruplex structure formed by the oncogene promoter region of c-Myc (39,40). The specific conformations of the two double-chain reversal loops formed by G4–C5–G6 and G14–C15–G16 impose strictly restrictions of accessibility to the grooves between the columns G1-G2-G3 and G7-G8-G9 and between G11-G12-G13 and G17-G18-G19, respectively. While the neighboring groove formed by the G-tracts G7-G8-G9 and G11-G12-G13, bridged by only 1 nt C10, is accessible to G-quadruplex-interactive agents through hydrogen bond with the edges of guanosines within the G-tetrad. The last groove between columns G1-G2-G3 and G17-G18-G19 (Figure 6) is completely accessible to hydrogen bond recognition by G-quadruplex ligands. Moreover, all syn guanines within the G-tetrad in the quadruplex structure modify the conformations of these grooves when compared to those of c-Myc quadruplex structure and provide the chance for selective recognition by G-quadruplex ligands. Of course, no any restrictions at the 5′-end of RET20T is also completely accessible to those ligands which bind to G-quadruplex structure by stacking over the G-tetrad.
Formation of G-quadruplex structures in the c-Myc (20) and c-kit (46) promoter regions has been shown to deregulate the gene expression and that in the Bcl2 (47) has been suggested to activate the gene expression. The same role may be involved in the RET oncogene. The unique all-parallel structure formed by RET promoter regions with specific ligand binding sites analyzed as above suggests that it could be an attractive target for pathway-specific drug design.
ACCESSION NUMBER
2188.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Basic Research Program of China under (No. 2009CB918600 and 2011CB966300); National Science Foundation of China under (No. 30800176, 20872169 and 20921091); Science and Technology Commission of Shanghai Municipality, China under, Pujiang talents awards (No. 08PJ1411700); State Key laboratory of Magnetic Resonance and Atomic and Molecular Physics; Wuhan Center for Magnetic Resonance, Wuhan Institute of Physics and Mathematics, CAS, under (No. T152902).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Yihua Yu (in East China Normal University) and Prof. Guoqiang Song (in Shanghai Institute of Materia Medica) for their help in collecting NMR data.
REFERENCES
- 1.Takahashi M, Buma Y, Iwamoto T, Inaguma Y, Ikeda H, Hiai H. Cloning and expression of the RET proto-oncogene encoding a tyrosine kinase with two potential transmembrane domains. Oncogene. 1988;3:571–578. [PubMed] [Google Scholar]
- 2.Kodama Y, Asai N, Kawai K, Jijiwa M, Murakumo Y, Ichihara M, Takahashi M. The RET proto-oncogene: A molecular therapeutic target in thyroid cancer. Cancer Sci. 2005;96:143–148. doi: 10.1111/j.1349-7006.2005.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouvaraki MA, Shapiro SE, Perrier ND, Cote GJ, Gagel RF, Hoff AO, Sherman SI, Lee JE, Evans DB. RET proto-oncogene: a review and update of genotype–phenotype correlations in hereditary medullary thyroid cancer and associated endocrine tumors. Thyroid. 2005;15:531–544. doi: 10.1089/thy.2005.15.531. [DOI] [PubMed] [Google Scholar]
- 4.Arighi E, Borrello MG, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 2005;16:441–467. doi: 10.1016/j.cytogfr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Santoro M, Rosati R, Grieco M, Berlingieri MT, D’Amato GL, de Franciscis V, Fusco A. The RET proto-oncogene is consistently expressed in human pheochromocytomas and thyroid medullary carcinomas. Oncogene. 1990;5:1595–1598. [PubMed] [Google Scholar]
- 6.Sawai H, Okada Y, Kazanjian K, Kim J, Hasan S, Hines OJ, Reber HA, Hoon DS, Eibl G. The G691S RET polymorphism increases glial cell line–derived neurotrophic factor–induced pancreatic cancer cell invasion by amplifying mitogen-activated protein kinase signaling. Cancer Res. 2005;65:11536–11544. doi: 10.1158/0008-5472.CAN-05-2843. [DOI] [PubMed] [Google Scholar]
- 7.Veit C, Genze F, Menke A, Hoeffert S, Gress TM, Gierschik P, Giehl K. Activation of phosphatidylinositol 3-kinase and extracellular signal-regulated kinase is required for glial cell line-derived neurotrophic factor-induced migration and invasion of pancreatic carcinoma cells. Cancer Res. 2004;64:5291–5300. doi: 10.1158/0008-5472.CAN-04-1112. [DOI] [PubMed] [Google Scholar]
- 8.Putzer BM, Drosten M. The RET proto-oncogene: a potential target for molecular cancer therapy. Trends Mol. Med. 2004;10:351–357. doi: 10.1016/j.molmed.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Munnes M, Patrone G, Schmitz B, Romeo G, Doerfler W. A 5'-CG-3'-rich region in the promoter of the transcriptionally frequently silenced RET protooncogene lacks methylated cytidine residues. Oncogene. 1998;17:2573–2583. doi: 10.1038/sj.onc.1202165. [DOI] [PubMed] [Google Scholar]
- 10.Andrew SD, Delhanty PJ, Mulligan LM, Robinson BG. Sp1 and sp3 transactivate the RET proto-oncogene promoter. Gene. 2000;256:283–291. doi: 10.1016/s0378-1119(00)00302-4. [DOI] [PubMed] [Google Scholar]
- 11.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel DJ, Phan AT, Kuryavyi V. Survey and summary human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burge S, Parkinson GN, Hazel P, Todd AT, Neidle S. Survey and summary Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 17.Zhang N, Phan AT, Patel DJ. (3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K + solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-Myc transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-Myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuryavyi V, Phan AT, Patel DJ. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010;38:6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human Bcl2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the Bcl-2 P1 promoter. J. Am. Chem. Soc. 2005;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 29.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends in Cell Biol. 2009;19:412–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan AT, Patel DJ. A site-specific low-enrichment 15N, 13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 32.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Book X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. New Haven, CT: Yale University Press; 1993. [Google Scholar]
- 35.Wijmenga SS, Van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- 36.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 37.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 38.Phan AT, Gueron M, Leroy JL. Nuclear Magnetic Resonance of Biologica Macromolecules, Pt A. Vol. 338. San Diego, CA: Academic Press Inc.; 2001. pp. 341–371. [Google Scholar]
- 39.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-Myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-Myc promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 41.Kuryavyi V, Patel DJ. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010;18:73–82. doi: 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo K, Gokhale V, Hurley HL, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36:4598–4607. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Armond R, Wood S, Sun DY, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1 alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 45.Cang XH, Sponer J, Cheatham TE., 3rd Explaining the varied glycosidic conformational, G-tract length and sequence preferences for anti-parallel G-quadruplexes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEB J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.