Abstract
The mfl-riboswitch regulates expression of ribonucleotide reductase subunit in Mesoplasma florum by binding to 2′-deoxyguanosine and thereby promoting transcription termination. We characterized the structure of the ligand-bound aptamer domain by NMR spectroscopy and compared the mfl-aptamer to the aptamer domain of the closely related purine-sensing riboswitches. We show that the mfl-aptamer accommodates the extra 2′-deoxyribose unit of the ligand by forming a more relaxed binding pocket than these found in the purine-sensing riboswitches. Tertiary structures of the xpt-aptamer bound to guanine and of the mfl-aptamer bound to 2′-deoxyguanosine exhibit very similar features, although the sequence of the mfl-aptamer contains several alterations compared to the purine-aptamer consensus sequence. These alterations include the truncation of a hairpin loop which is crucial for complex formation in all purine-sensing riboswitches characterized to date. We further defined structural features and ligand binding requirements of the free mfl-aptamer and found that the presence of Mg2+ is not essential for complex formation, but facilitates ligand binding by promoting pre-organization of key structural motifs in the free aptamer.
INTRODUCTION
Riboswitch-RNAs are regulating early events in gene expression. It is generally assumed that the binding of a low molecular weight ligand to an aptamer domain and subsequent allosteric conformational transitions of an immediately downstream sequence in the 5′-untranslated region of mRNAs modulate gene expression. Riboswitch-RNAs that act on the level of transcription or on the level of translation have been described. Structural characterization so far has mainly focused on the aptamer domains bound to their ligands and contributed immensely to our understanding of the extraordinary and unexpected binding specificity and affinity found for many riboswitches (1–4). In addition, thorough studies on ligand binding kinetics of a transcriptionally active riboswitch binding to FMN as well as the pbuE-aptamer binding to 2-aminopurine have provided first insight into a general understanding of riboswitch mechanism (5–8).
By NMR spectroscopy, the change in dynamics between the conformational ensembles characterizing the free state of the guanine-sensing aptamer domain (4,9–11) and the ligand-bound complex (12,13) have been mapped and revealed key insight into structural pre-organization and Mg2+ requirements for ligand binding at atomic resolution. Purine-sensing riboswitch aptamers are among the structurally best-characterized riboswitch elements, with various crystal structures available for adenine- or guanine-sensing aptamer domains in complex with a large number of purine analogs (3,14–17). The class of purine riboswitches comprises transcriptionally operating off-switches (e.g. xpt-pbuX from Bacillus subtilis) as well as transcriptional on-switches (pbuE from Bacillus subtilis) and a translational on-switch (addVV from Vibrio cholerae), demonstrating the diversity of riboswitch-mediated gene regulation. In spite of this regulatory diversity employed by purine riboswitches, the respective sensory aptamer domains are highly conserved for all purine riboswitches in terms of primary, secondary and tertiary structure. In addition to small differences in the overall architecture of the ligand-bound state, which is characterized by a distinct organization of three helices P1, P2 and P3 around the three-way-junction serving as the binding pocket (Figure 2a) the adenine-sensing pbuE aptamer and the guanine-sensing xpt-pbuX aptamer show different Mg2+-requirements for ligand-induced structure formation and distinct folding trajectories (9,18–20). These findings show that structural features exhibited in the ligand-bound structures are, although important, insufficient to comprehensively explain the binding requirements for riboswitch function.
Figure 2.
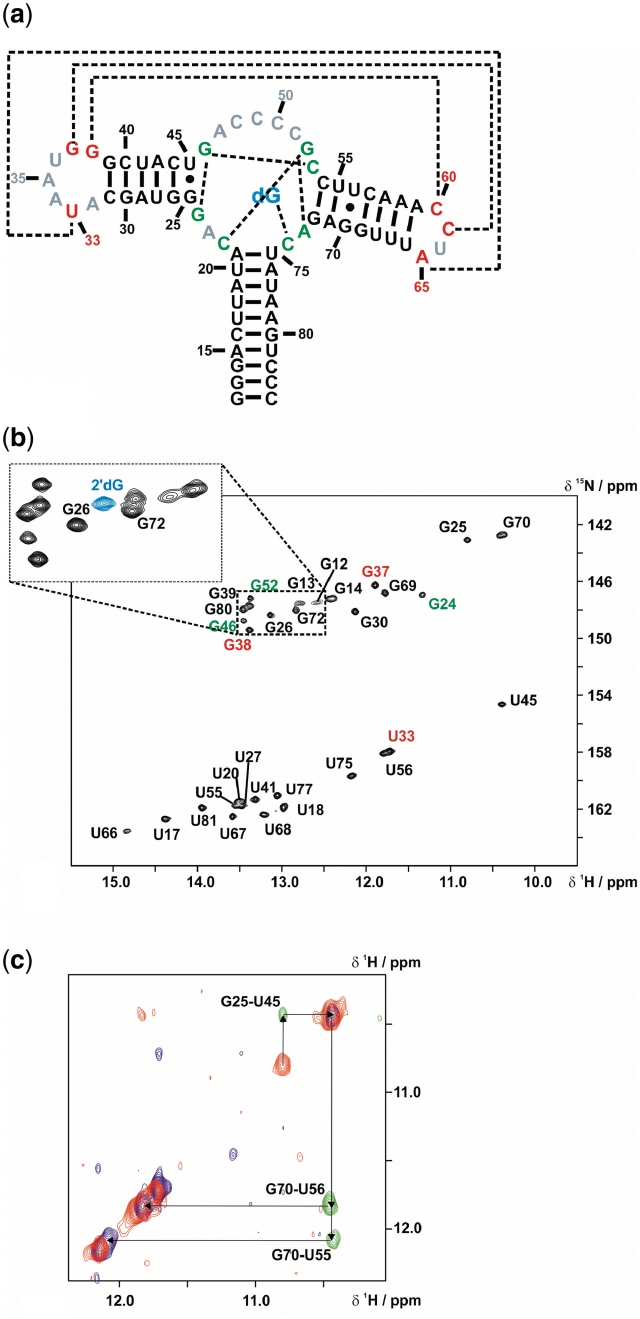
NMR characterization of the mfl-aptamer-2′-deoxyguanosine complex. (a) Secondary structure of the wild-type mfl-aptamer with the native ligand 2′-dG, resonances in non-helical regions detectable by NMR are colour-coded (red: loop regions; green: binding pocket region). Long-range interactions are indicated by dashed lines. (b) 1H,15N-HSQC of the mfl-aptamer in complex with 2′-dG. Signal annotations are colour-coded corresponding to the region of their assigned residues. The small section of the guanosine imino proton region of a 15N- HSQC with 15N-labelled ligand 2′-dG shows the ligand imino proton resonance in blue. (c) Spectral region of the double-half-filtered NOESY showing an overlay of the 14N(ω1, ω2)-edited signals in blue, the 15N(ω1, ω2)-edited signals in red and the 14N(ω1),15N(ω2)-edited signals in green. With a 15N-guanosine-labelled sample, ambiguities arising from spectral overlap were resolved.
Here, we investigated the 2′-deoxyguanosine (2′-dG) riboswitch from Mesoplasma florum (mfl-aptamer), classified as a close relative of the purine-sensing riboswitches. It binds specifically to a purine nucleoside rather than to the purine nucleobase (21). The ligand 2′-dG is recognized by Watson–Crick base pairing to C74. The predicted secondary structure of the aptamer domain closely resembles all other purine riboswitch aptamer domains, although a considerable number of conserved nucleotides are altered (Figure 1). The mfl-aptamer recognizes and binds the additional sugar moiety of the ligand 2′-dG with a highly specialized binding pocket, which accommodates binding of a larger ligand by exhibiting a greater structural plasticity than the highly rigid, ligand-enclosing binding pocket of the guanine- and adenine-sensing aptamer domains. The differences between the mfl-aptamer and the xpt-aptamer binding pockets have been previously analysed by a mutational series accompanied by ITC studies and X-ray crystallography (22). The crystal structure of an ‘intermediate’ of this mutational series has been solved for an xpt/mfl-hybrid aptamer (GdG-3 hybrid aptamer) bound to 2′-dG. However, the GdG-3 hybrid aptamer contains the native xpt-loops L2 and L3 and thus does not describe the loop–loop interaction in the native mfl-aptamer.
Figure 1.
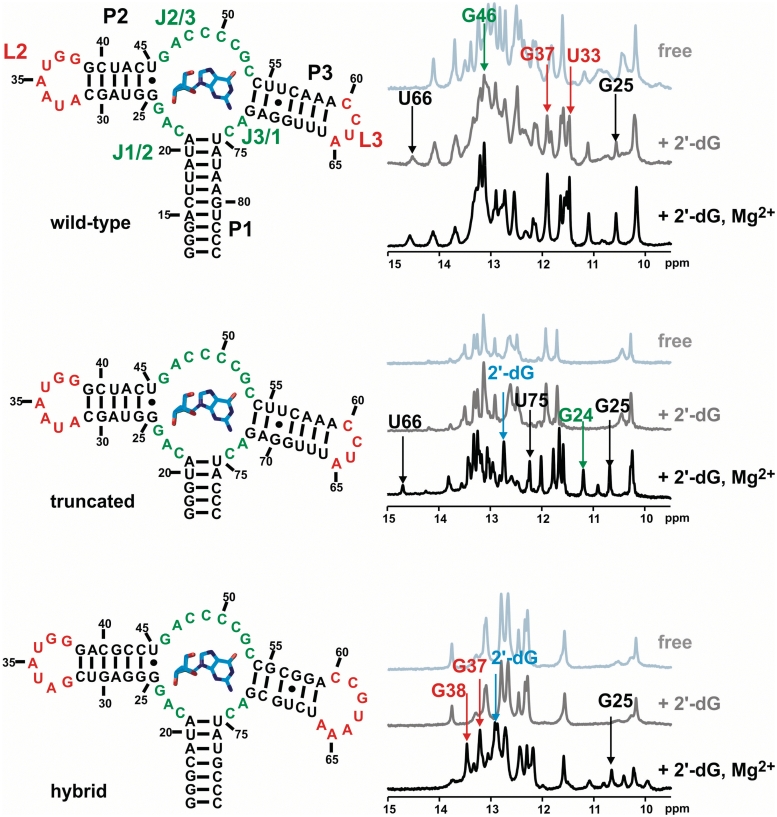
Secondary structures of the three RNA constructs wild-type (upper left), truncated (middle left) and hybrid (lower left) aptamer. On the right side, the corresponding 1D-NMR titration spectra are shown (light grey: free aptamer, dark grey: 2′-dG added, black: 2′-dG and 5 mM Mg2+ added). Arrows indicate key resonances as reporters of ligand-binding induced folding. Nucleotide numbering is according to the purine aptamer nomenclature originally proposed (33).
We characterized the structure of the ligand-bound complex as well as the free mfl-aptamer and the role of Mg2+ in ligand binding and folding processes. In particular, our studies focus on the mode of ligand binding as a prerequisite for understanding the biological function of the wild-type mfl-aptamer and on the structural aspects of the interesting loop–loop interaction, which is unique among purine aptamers up to now. To address the question of RNA structural requirements for ligand binding, we studied three different RNA constructs: (i) the wild-type aptamer consisting of nucleotides 15–81 of the native mRNA sequence (numbering consistent with common purine riboswitch scheme) flanked by three 5′-guanosine residues and three 3′-cytidine residues; (ii) a P1-truncated wild-type aptamer consisting of nucleotides 20–76 within the same flanking sequences as (i) and (iii) an xpt/mfl-hybrid aptamer consisting of the ligand binding pocket of the mfl-aptamer (nucleotides 19–26, 44–54 and 72–77) and the xpt-aptamer scaffold, namely the native xpt-aptamer P2 and P3 helices lengths and L2 and L3 loops (Figure 1). We compared the ligand binding requirements for these RNA constructs, characterized the structural rearrangements induced by ligand binding as well as their Mg2+-dependence by NMR spectroscopy and determined the thermal stabilities by CD spectroscopy. The ligand binding modes for all three investigated constructs are found to be identical to the general purine riboswitch binding mode, although, for the native mfl-aptamer, small but essential unique features exist in the binding pocket architecture. More pronounced differences between the native aptamer and the purine aptamer consensus folds are present in the peripheral elements of the complex, especially in the loop–loop interaction motif.
Our results demonstrate that ligand binding and formation of the key tertiary structure motif are not Mg2+-dependent, the presence of Mg2+, however, facilitates ligand binding by shifting the conformational ensemble of the free aptamer towards a pre-organized, ligand-binding competent population. Furthermore, Mg2+ reduces residual conformational heterogeneity for the ligand-bound complex, mainly by stabilizing the parallel packing of P2 and P3 and the loop–loop interaction.
MATERIALS AND METHODS
RNA
13C,15N- and 15N-labelled nucleotides as well as 13C,15N-labelled 2′-deoxyguanosine were purchased from Eurisotop/Saarbrücken. All RNAs were prepared by in-vitro transcription from linearized plasmids using T7-polymerase as described (23). RNA concentrations were determined by UV spectroscopy with an extinction coefficient of 635000 M−1 cm−1 at 260 nm for the mfl-aptamer, 540800 M−1 cm−1 for the P1-truncated aptamer and 598000 M−1 cm−1 for the xpt/mfl-hybrid aptamer. RNAs were refolded into a homogeneous conformation by thermal denaturation of the RNA at high concentration (0.2–0.5 mM) followed by dilution to 0.05 mM and rapid cooling on ice. Stable and homogeneously folded aptamer–ligand complexes formed at a [ligand]:[RNA]-ratio of 2:1 in the presence of 2 mM Mg2+. Samples were exchanged into NMR-buffer (25 mM potassium phosphate, 50 mM potassium chloride at pH 6.2) using centrifuge concentration devices. D2O samples were prepared by freeze-drying of RNA samples in appropriate amounts of NMR-buffer and resolubilization in 99.98% D2O. Homogeneity of all samples was confirmed by native acrylamid gel electrophoresis.
NMR spectroscopy
NMR spectra were recorded on Bruker AV600, AV700, AV800, AV900, AV950 spectrometers equipped with cryogenic 5 mm TXI HCN probes and z-axis gradients and on Bruker DRX600 and AV900 MHz spectrometers equipped with RT HCN probes with x,y,z-axis gradients. All spectra were processed and analysed with the software topspin 1.3–3.0 (Bruker Biospin) and Sparky 3.114 (24). All experiments on exchangeable protons were recorded in 90% NMR-buffer/10% D2O at 278 K or 283 K, while all experiments on non-exchangeable protons were recorded in 99.98% D2O at 298 K. 15N-HSQC, HNCO (25–28) HNN-COSY (29), 15N-edited 3D NOESY-HSQC and 1H,1H-2D NOESY experiments in H2O were performed using the soft WaterGATE water suppression technique (30). Amino proton-optimized spectra and NOESY spectra with a X-double half filter (31), were performed using the 3-9-19 water suppression scheme (32).
CD spectroscopy
Melting curves monitored by the CD signal of 5 µM RNA samples in NMR-buffer were recorded on a Jasco J-810 spectropolarimeter equipped with a Peltier temperature control element and analysed with Sigmaplot. Molecular ellipticity of RNA samples was monitored at 263 nm from 4 to 90°C at a temperature gradient of 1°C/min and a sampling rate of 0.2°C. Raw melting data were smoothed by a negative exponential algorithm and the first derivative was calculated numerically from smoothed data. Local minima in the derivative function were defined as melting temperatures.
RESULTS
Mg2+-dependence of RNA folding, ligand binding and thermal stability for wild-type mfl-aptamer and sequence variants
For the three RNA constructs (construct 1: wild-type mfl-aptamer, construct 2: truncated P1-aptamer and construct 3: xpt/mfl hybrid aptamer; Figure 1), binding of 2′-deoxyguanosine (2′-dG) was tested by analysing ligand-induced changes of the chemical shifts of the characteristic imino proton resonances in 1D-NMR spectra (Figure 1). The wild-type aptamer is ligand binding competent in the absence of Mg2+, as shown for example by the reporter signal of the G25 imino proton, which is exclusively observed in the RNA–ligand complex. In contrast, both constructs 2 and 3 require Mg2+ as a cofactor for ligand binding, as the imino proton spectra of both constructs do not change upon ligand titration compared to the free RNAs. However, in the presence of Mg2+, RNA–ligand complex formation for constructs 2 and 3 can be observed by NMR.
Thermal melting of RNA structures monitored by CD spectroscopy was measured under conditions comparable to those for NMR, concerning relative concentrations of RNA, 2′-dG and Mg2+. For constructs 2 and 3, ligand binding is Mg2+-dependent and by NMR titrations, a [Mg2+]:[RNA] ratio of 20:1 was found to result in complete complex formation. To also test the highest absolute Mg2+ concentration used in some NMR experiments, melting was additionally monitored at 5 mM Mg2+. As expected, high molar Mg2+ excess (103 equivalents) results in significant thermal stabilization of all three constructs (T ∼70.9–79.7°C, Table 1). Furthermore, for constructs 2 and 3, melting curves show two transitions at lower Mg2+ conditions, the first of which is either not present or not resolved at 5 mM Mg2+.
Table 1.
Melting temperatures for the three aptamer constructs as determined by CD spectroscopy
| Aptamer construct (5 µM) | Free RNA | Complex (+2 eq. 2′-dG) | Free RNA+ 0.1 mM Mg2+ | Free RNA + 5 mM Mg2+ | Complex (+2 eq. 2′-dG and 0.1 mM Mg2+) |
|---|---|---|---|---|---|
| Construct 1:wild-type | 57.5°C | 57.9°C | 58.1°C | 70.9°C | 58.1°C |
| Construct 2:P1-truncated | 40.2°C/63.4°C | n.d. | 42.3°C/66.3°C | 79.7°C | 67.8°C |
| Construct 3:xpt/mfl hybrid | 9.8°C/63.6°C | n.d. | 16.4°C/65.2°C | 74.2°C | 19.1°C/64.6°C |
Melting curves reveal no significant changes in the transition point for the wild-type aptamer, irrespective of whether ligand or 100 µM Mg2+ or both are present. Apparently, neither 2′-dG nor moderate Mg2+ concentrations influence the thermal stability of the aptamer’s secondary structural elements.
The truncated aptamer shows two melting transitions; we tentatively assign the first transition at ∼40°C to the melting of misfolded RNA species, which are also observable as additional resonances in the 15N HSQC. The first transition is followed by the melting of the natively folded, binding-competent RNA at ∼63°C, which is, surprisingly, slightly larger than the thermal stability of construct 1, although still comparable (ΔT ∼5.9°C). Addition of 100 µM Mg2+ marginally increases the melting point of the high-temperature transition of construct 2 by ∼2.9°C. Ligand binding at these conditions results in a further small melting point elevation by ∼1.5°C. The xpt/mfl hybrid aptamer also shows two melting transitions; in the free form the first transition occurs at ∼9.8°C and the second at ∼63.6°C. The lower-temperature transition appears to be significantly stabilized by 100 µM Mg2+, since the melting temperature increases by ∼6.6°C Here, the lower-temperature transition could be assigned to the melting of the loop–loop interaction by NMR. The respective resonances are detectable in 15N HSQC spectra at 10°C in the presence of 20 equivalents of Mg2+, but not without Mg2+.
Imino proton resonance assignment of the wild-type aptamer–ligand complex
Imino protons are reliable reporters of RNA structure and structural changes on a broad dynamic timescale, since they are only observable by NMR if they are protected from chemical exchange with the solvent. To address the question of how Mg2+ and 2′-dG influence aptamer folding and RNA dynamics, we assigned the imino protons of the wild-type mfl-aptamer (construct 1). By a combination of different NMR methods (2D-NOESY, 15N-filtered NOESY, 15N-edited NOESY, double-half-filter NOESY, 15N-HSQC, HNN-COSY, 3D-NOESY-15N-HSQC) and selective isotope labelling (15N-cytidine-labelled RNA, 15N-uridine-labelled RNA, 15N-cytidine-15N-guanosine-labelled RNA, uniformly 15N-labelled RNA), all imino protons protected from solvent exchange have been assigned and the assignment was further cross-validated in the different experiments. Specifically, analysis of double-half filtered NOESY spectra recorded for 14N-uridine/15N-guanosine-labelled RNA allowed the unambiguous assignment of NOEs in highly crowded spectral regions (Figure 2).
For the RNA–ligand complex of the native mfl-aptamer bound to 2′-dG, 15 uridine imino protons out of 17 uridine residues are exchange protected and can be detected and all of the 16 guanosine residues are detected. In addition, the imino proton resonance of the ligand 2′-dG is observed at a 1H chemical shift of 12.9 ppm and a 15N chemical shift of 142.7 ppm. The only imino protons subject to rapid solvent exchange belong to residues U36 in loop L2 and U64 in loop L3. This observation is in accordance with the purine aptamer consensus tertiary structure model: residue U36 is solvent-exposed in all purine aptamer crystal structures, whereas U64 is part of loop L3 that is truncated in the mfl-aptamer domain compared to other purine-sensing aptamer domains which contain a heptaloop at this position. Solvent exposure and additional dynamics are also indicated by very strong H5/H6 aromatic resonances with narrow linewidths for U36 and U64 (data not shown), a feature commonly assigned to fast internal dynamics largely decoupled from the slow global dynamics expected for an RNA of such size (34). All other imino protons are observed as a single set of well-dispersed resonances (Figure 2b) with similar linewidths, showing that the aptamer–ligand complex is homogeneously folded. For most nucleotides, analysis of chemical shifts reveals that they are involved in Watson–Crick base pairing (35). Canonical Watson–Crick base pairing is further confirmed by HNN-COSY spectra for all imino proton resonances assigned to the helical regions P1, P2 and P3 (see Supplementary Figure S1). Resonances with unusual upfield chemical shifts, both in the 1H and the 15N dimensions, can be assigned to those nucleotides for which the secondary structure model predicts non-canonical interactions: U56 and G70 form a wobble base pair in the center of P3, and G25 and U45 form a wobble base pair directly adjacent to the binding pocket. As previously reported, the presence of this wobble pair substantially enhances 2′-dG binding affinity of the aptamer (22) and is now confirmed to be present also in solution in the wild-type mfl-aptamer. Furthermore, residues G24 and U33 show imino proton signals indicative of non-canonical base pairing. For U33, the hydrogen bond acceptor nitrogen can be identified as A65 N7 forming a reverse Hoogsteen base pair with U33 (see Supplementary Data, Supplementary Figure S2). By contrast, the G24 imino proton signal can be detected but lacks a cross peak in the HNN-COSY. Thus, either the hydrogen bond acceptor is a carbonyl or hydroxyl group or else the G24 imino proton is solvent protected by being buried inside a hydrophobic environment but not involved in hydrogen bonding.
Binding pocket architecture of the wild-type mfl-aptamer
2′-dG is recognized and bound by the mfl-aptamer by means of the universal purine riboswitch molecular recognition mode, namely the formation of a Watson–Crick base pair between the ligand 2′-dG and the pyrimidine Y74, here C74, as observed in the HNN-COSY experiment performed for 15N-cytidine-labelled RNA in complex with 15N-labelled 2′-dG. (Supplementary Data, Supplementary Figure 2b). In the purine aptamer consensus tertiary structure, the ligand position is furthermore fixed by U51 through formation of a Watson–Crick-sugar-edge base pair. These additional hydrogen bonding interactions are confirmed both by the crystal structures and by NMR data of the xpt-aptamer complex bound to hypoxanthine (3,4) or guanine (14) and for the pbuE-aptamer complex bound to adenine (14) or 2,6-diaminopurine (4). The crystal structure of the GdG-3 hybrid aptamer (22) displays the respective residue, C51, occupying a similar position as its counterpart U51 in the xpt-aptamer crystal structure, but at a slightly larger distance to the 2′-dG sugar edge (3.2 versus 2.8 Å). Hence, the hydrogen bonds suggested by the X-ray structure of the GdG-3 hybrid aptamer to be formed (C51 amino proton to the 2′-dG N3 and the 2′-dG amino protons to the C51 N3) cannot be detected in an HNN-COSY and therefore not be confirmed by NMR. Our NMR data rather suggest the respective nitrogen-nitrogen distances to be larger than 3 Å, leading to a 2hJNN coupling too small to be detected by the HNN-COSY experiment (36,37). In addition, no NOE cross signal from the 2′-dG amino protons to the C51 amino protons is observed, although this could be attributed to line broadening of the 2′-dG amino protons in general due to chemical exchange. The 2′-dG amino protons resonate at 1H chemical shifts of 8.9 and 6.8 ppm, respectively, and have linewidths of 53 and 60 Hz. Cytidine amino protons in Watson-Crick base pairs, in contrast, have smaller linewidths of ∼40 to 50 Hz. In summary, our NMR data show that the C51-2′-dG interaction is considerably weaker than the corresponding U51-ligand interaction in the purine-sensing aptamer domains.
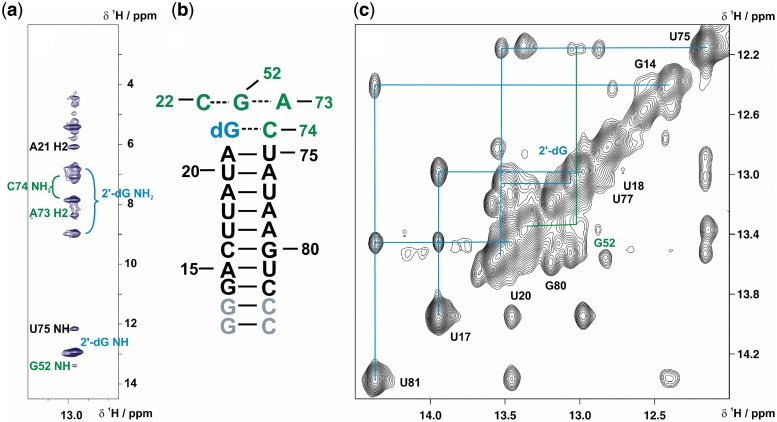
The nucleobase moiety of 2′-dG stacks between the P1 closing base-pair U75-A21 and a Watson–Crick base pair formed by the residues C22 and G52 of the junction regions, as shown by the continuous sequential walk in the imino proton region of NOESY spectra extending from G13 up to G52 (Figure 3b). The stacking interaction with the upper P1 base-pair, A21-U75, is further indicated by a strong upfield chemical shift of the A21 H2 resonance, which results from ring current shielding by the 2′-dG base moiety. In striking contrast to all other purine-binding aptamers, the binding pocket of the mfl-aptamer has to accommodate the extra 2′-deoxyribose moiety. According to the crystal structure of the GdG-3 hybrid aptamer, this is accomplished by a structural rearrangement mainly of J2/3, which usually forms the lid of the binding pocket, here leading to a less complete encapsulation of the ligand (22). The 2′-deoxyribose moiety is oriented towards P1, occupying the minor groove of the base-pairs U20-A76 and A21-U75. In the NMR solution spectra of the wild-type mfl-aptamer, we observe strong indications for a similar structural arrangement. NOESY spectra of non-exchangeable protons of the complex reveal contacts between the ligand H5′/H5′′ protons and the H2 protons of A21 and even A76 (data not shown). Further NOE contacts are observed from the ligand to residues of the binding pocket, namely from the H2′ proton to the G52 imino proton and to the C22 aromatic H6 proton. Another NOE contact observed between the ligand imino proton and A73 H2 (Figure 3a), additionally confirms the base moiety position within the binding pocket. However, differences are observed in the upper part of the binding pocket: an NOE cross signal from the G24 imino proton is observed to the imino protons of G46, strongly suggesting that G24 forms a base triple with G46 and C53 (see Supplementary Data, Supplementary Figure S3), forming the apical plane of the binding pocket. Moreover, G24 displays NOE contacts to U45 of the basal base pair of P2 instead of P3. Finally, C2 of G24 has a downfield 13C chemical shift indicating a hydrogen bond involvement of the exocyclic amino group (see Supplementary Data, Supplementary Figure S4). The formation of the base triple G24-G46-C53 seems to be an exclusive feature of the mfl-aptamer-2′dG complex, since in all other purine aptamer–ligand complexes described to date, including the GdG-3 aptamer bound to 2′-dG, the apical base pair of the binding pocket is formed by nucleotides A23-G46-C53 (3,14,22).
Figure 3.
RNA-ligand complex architecture determined by NMR. (a) Strip plot of a 15N-edited-NOESY spectrum of the selectively 13C,15N-cytidine-labelled aptamer in complex with 13C,15N-labelled 2′-dG showing the signals from protons within NOE distances to the 2′-dG imino proton. Resonance assignments are colour-coded according to Figure 2. (b) Secondary structure of P1 stem extended by the 2 W-C base pairs formed by 2′-dG-C74 and C22-G52. The latter base pair is predicted to form a base triple with A73, as the ligand-imino proton NOE to the A73 H2 indicates stacking of these residues. (c) Imino proton region of NOESY-spectrum (800 MHz, mixing time = 250 ms). Signal connectivities due to NOE cross peaks of the P1 imino protons is sketched. The imino proton assignments are colour-coded corresponding to the secondary structure of the aptamer (Figures 2 and 3 b).
Architecture of helices P2 and P3 and the loop–loop interaction
In the wild-type mfl-aptamer, key residues involved in tertiary interactions are mutated or deleted compared to the purine aptamer consensus sequence. Most obviously, L3 is truncated and therefore the structural basis for the loop–loop interaction is substantially different. Another important aspect is the swap in lengths between P2 and P3 in the wild-type mfl-aptamer with respect to all other purine riboswitches. Compensation for the helix length discrepancy may be accomplished by a larger degree of P3 twisting as well as by the truncation of L3 and the adjustment of the binding pocket architecture described above. Due to this altered architecture of the binding pocket, the lower part of P3 can virtually slip one plane down, which corresponds to a spatial adjustment of ∼3.2 Å. The morphology of the loop–loop interaction in the native mfl-aptamer-2′-dG complex is unknown, since no high-resolution structural data are available and homology-based predictions are likely to fail due to truncation of L3 with respect to all other purine aptamers.
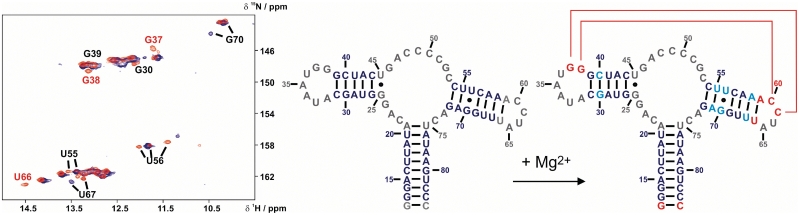
Our NMR data unambiguously reveal the presence of two inter-loop G-C Watson–Crick base pairs, G37-C61 and G38-C60, both by sequential NOESY and inter-residual HNN-COSY cross signals (see Supplementary Data, Supplementary Figures S1 and S2). The imino proton of U66, which forms the closing base pair of helix P3, shows a NOESY-cross peak to the G38 imino proton, which is stacked upon helix P3 and shows a further NOE cross signal to the G37 imino proton. The Watson–Crick-like nature of the G38-C60 and G37-C61 base-pairs is confirmed in the HNN-COSY experiment and by the aromatic–anomeric connectivities between C60 and C61 in the NOESY spectra of non-exchangeable protons. Additionally, an A:U reversed Hoogsteen base pair is unambiguously assigned, formed between U33 in L2 and A65 in L3. The G37 imino proton also shows a cross signal to an adenosine H2 resonance in the NOESY. This H2 proton likely belongs to A35. The A35 H2 resonates in the same region as A79, A58 and A71, indicating that the A35 residue stacks between purine residues (38). As expected, the A:A base pair formed by A33 and A66 in the xpt-aptamer is missing in the mfl-aptamer, which lacks both nucleotides in its sequence. However, the G38 imino proton chemical shift is similar for the xpt-aptamer and the mfl-aptamer, whereas U34 and G37 show distinct chemical shifts in the mfl-aptamer compared to the xpt-aptamer (Figure 1). Thus, it is more likely that of the 2 quartets present in the loop–loop interaction motif of the xpt-aptamer, only the one involving G38 is present in the mfl-aptamer. Therefore it is conceivable that the A66:A33-Hoogsteen base pair is replaced by a U33:A65 Hoogsteen base pair in the mfl-aptamer (see Supplementary Data, Supplementary Figure S5 for a preliminary model). Further structural information derived by NMR is the unusual carbon chemical shift of the U66 C2 carbonyl group (see Supplementary Data, Supplementary Figure S4). The downfield shift of 152 ppm indicates this carbonyl group to be a hydrogen bond acceptor, while C2 carbonyl groups of uridine residues involved in canonical Watson–Crick base pairs resonate around 150 ppm.
Secondary and tertiary structure stabilization of the free aptamer and the ligand-bound complex by Mg2+
In the free form, the mfl-aptamer is already well-structured, but shows considerable more conformational heterogeneity on the NMR timescale compared to the ligand-bound form as indicated by larger imino proton signal line widths (for example, 35.4 Hz for the U81 imino proton signal in the free form versus 24.5 Hz in the ligand-bound complex) and additional signals in the 15N-HSQC (Figure 4). Such signals arise from additional, low-populated conformations (∼10%) with lifetimes in the higher millisecond regime or longer, as they can be detected as separate resonances by NMR. The secondary structures of the three helices appear largely preformed, especially in the P1 helix which constitutes the longest canonical A-helix in the construct. Imino proton resonances in helical regions that are not detectable in the free aptamer belong to G25, U45 and U75, which form the closing base pairs of P1 and P2 next to the completely unstructured binding pocket. All imino proton signals from residues constituting the binding pocket as well as the loops L2 and L3 are not detectable due to fast solvent exchange. For some residues, however, slower dynamic processes are observable, as two sets of signals are observed. Interestingly, U18 of helix P1 seems to adopt at least three distinct conformations, of which the one corresponding to the single, ligand-bound conformation present in the complex is favoured by Mg2+ addition to the free RNA. Another intrinsically dynamic region in the free aptamer is located around the U56-G70 wobble base pair of P3. This wobble base pair exists in at least two distinct conformations, propagating to the adjacent base pairs and thus rendering the entire P3 stem flexible. Titration of Mg2+ to the free aptamer results in stabilization of the loop-closing base pairs of P2 (C31-G39) and P3 (A59-U66) and those adjacent to them, including residues G37 and G38, which form the inter-loop base pairs. In contrast, G25, U45 and U75 remain absent even at high Mg2+ concentrations, indicating that the binding pocket is indeed not pre-organized. Imino proton resonance intensities of U20, G30, G37, G38, G39, U66 and U67, increase up to ratios of 10 equivalents of Mg2+ to RNA, while resonance intensities of imino protons in the already well-structured regions decrease slightly and resonances arising from alternative and minor conformations decrease rapidly. In particular, this effect is observed for the G-U wobble base pair formed by U56 and G70 in the center of P3. Along with the increasing intensities of the loop imino proton signals, G37 and G38, alternative resonances for U56 and G70 as well as U55 disappear and the resonances belonging to the complex-like conformation dominate. In agreement with previous reports for Mg2+-induced folding of the xpt-aptamer by FRET studies (39), the concentration range where a population shift in the RNA conformational ensemble occurs lies between 5 and 10 equivalents. At very high Mg2+ excess, new additional conformations become populated and linewidths increase again, consistent with a general increase in the rate of imino proton solvent-exchange facilitated by Mg2+.
Figure 4.
Mg2+-induced structural changes for the mfl-aptamer. Left overlay of 15N-HSQCs: free mfl-aptamer recorded in the absence (blue) of Mg2+ and at 10 equivalents [Mg2+]:[RNA] (red). Signals only observable upon addition of Mg2+ and signals with significant chemical shift perturbations are annotated. Middle: observable base-pairs in the mfl-aptamer in the absence (blue) of Mg2+ and, right, at 10 equivalents [Mg2+]:[RNA]. Red nucleotides represent the secondary and tertiary structural elements detectable only in the presence of Mg2+ and light blue nucleotides represent residues for which resonances significantly shift in the presence of Mg2+, respectively.
As observed earlier for the xpt-aptamer, the binding pocket is largely unstructured in the absence of ligand, while the loop–loop interaction is preformed also in the free mfl-aptamer. In the case of the mfl-aptamer, this loop–loop interaction is, however, only detected in the presence of Mg2+ or at low temperatures (below ∼0°C). Formation of the loop–loop interaction is monitored by the G37 and G38 imino proton resonances. The A:U Hoogsteen base pair, however, is not detectable in the absence of ligand even at higher Mg2+ concentrations up to 10 mM. Comparison of the G37 and G38 chemical shifts in the free aptamer versus the complex reveals that the conformations are different in these folds, indicating that further structural rearrangements in the loop–loop interaction take place upon ligand binding. However, the existence of loop–loop interactions in the free aptamer, also if only transiently formed, greatly reduces the entropic cost of P2 and P3 alignment upon ligand binding-induced folding. This is also supported by the behaviour of the alternative U56/G70 resonances, which shift to the complex-like form if Mg2+ is added to the free aptamer (Figure 4).
The Mg2+ dependence of imino proton signal intensity changes is much more uniform for the ligand-bound complex, which already in the absence of Mg2+ contains all the structural elements of the tertiary fold. With the exception of the lower part of P1, all imino proton resonance intensities increase uniformly upon Mg2+ addition. This concerted behaviour indicates that the presence of Mg2+ leads to a less dynamic structure. The P3 region reveals the most pronounced structural heterogeneity in the ligand-bound complex in the absence of Mg2+, as similarly observed for the free RNA. Signals representing alternative conformations disappear at an 8-fold molar excess of Mg2+ over RNA, the ratio at which the Mg2+-induced effect on all resonances is saturated. Also for the loop–loop interactions, represented by G37, G38 and U33, resonances increase equally and with the same Mg2+ concentration dependence. Resonance intensities of the loop closing base pairs C31-G39, G30-C40, A59-U66 and A58b-U67 increase alongside. The general structure stabilization effect of Mg2+ is also transmitted to P1, as observed by the intensity increase of the U20 imino proton resonance.
DISCUSSION
Mg2+ dependence of ligand binding and folding
As observed previously for the xpt-aptamer and the pbuE-aptamer (4,9), the mfl-aptamer does not require Mg2+ for ligand binding and folding into the final structure of the complex, as all tertiary interactions are formed and the ligand is bound without additional Mg2+. Mg2+ titrations of free RNA and ligand-bound complex, however, reveal a supporting role of Mg2+ in terms of structural motif stabilities and a restraining role in terms of dynamics. The conformational ensemble of the free aptamer shifts towards a higher populated state of pre-organized species at least at moderate Mg2+ concentrations. For the RNA–ligand-complex, residual minor conformations are drastically reduced. In the absence of ligand, Mg2+ stabilizes the global fold as reported by the presence of the loop–loop imino signals and the increase in intensity for the upper P2 and P3 base-pair imino signals. Mg2+-dependent formation of the (at least temporarily) stable loop–loop interaction by the free mfl-aptamer directly reduces the degrees of freedom for P2 and P3 and thereby the dynamics of these peripheral structural elements. On the other hand, Mg2+ does not affect the dynamics of the binding pocket on the NMR time scale, as the imino proton resonances of U75, U45, G25, G24, G46 and G52 (representing the binding pocket) all remain absent even at non-physiologically high Mg2+ concentrations in the absence of 2′-dG. The highly flexible nature of the ligand binding pocket reflects the capability of the aptamer to allow fast ‘screening’ of potential ligands by the predominantly accessible C74.
Although for the wild-type mfl-aptamer, the RNA–ligand complex displays significantly reduced conformational dynamics in the presence of Mg2+, divalent cations are apparently not essential for ligand-binding competence. However, if P1 is truncated such that only the two upper base pairs (U20-A76 and A21-U75) remain together with the three G-C closing base pairs obligatory for in-vitro transcription, ligand binding by construct 2 is rendered Mg2+-dependent. This change of Mg2+-dependence, which is induced by a reduced P1 helix length, strongly emphasizes the direct mutual influence of ligand binding and P1 stability, which ultimately is the key to riboswitch regulatory function (19,40,41). For the truncated aptamer, the role of Mg2+ could either be a non-specific stabilization of a compact ensemble of structures which are at least partly capable of ligand binding, thereby compensating for the loss of the native enthalpic contributions of P1 stacking interactions, or the specific stabilization of the parallel alignment of P2 and P3 via the loop–loop interaction, thereby partly pre-forming the binding pocket and facilitate ligand binding. The latter role of Mg2+ has been described in the context of an xpt-aptamer loop mutant, where the native loop–loop interactions are weakened (10). Still, even at 5 mM Mg2+, the conformational ensemble of the ligand-bound complex is heterogeneous in case of the truncated aptamer (construct 2). A loss of ligand-binding competence has also been observed for the purine aptamer domains if the identity of the upper closing base pairs is altered, and stacking interactions between P1 and the ligand are reduced (42). In summary, the sequence as well as the thermodynamic stability of P1 appears to be crucial for ligand binding in the wild-type mfl-aptamer. The hybrid aptamer represents another example where Mg2+ compensates for native RNA interactions. This construct is still capable of 2′-dG binding and folds into the tertiary structure stabilized by the loop–loop interaction and stacking of P2 and P3 on the binding pocket. However, in contrast to the wild-type mfl-aptamer, ligand binding is strictly Mg2+-dependent for the hybrid aptamer. For the wild-type aptamer bound to 2′-dG, we observe a loss of fast conformational exchange if Mg2+ is added. Taken together, these observations imply a role for Mg2+ in aptamer folding which is certainly not completely non-specific in the sense that any compact structure is equally stabilized, but rather an assisting role in driving conformational ensembles to a thermodynamic minimum by restricting conformational dynamics. The folding trajectory seems to be directly encoded in the primary sequence of the aptamer, since only the native sequence is able to fold into the ligand-bound complex upon ligand addition alone. Conservation of secondary (hybrid aptamer) or tertiary (truncated aptamer) structure alone is not sufficient to retain the native folding trajectory, which in turn can be rescued by Mg2+, thus presumably providing alternative folding pathways and reaching at least partly the same final state.
CD-melting curve analysis of the wild-type mfl-aptamer shows that neither addition of ligand nor of Mg2+ to the free aptamer increases the melting temperature. This finding implies that the complex of mfl-aptamer bound to 2′-dG, even in the presence of Mg2+, displays no greater thermal stability than the free mfl-aptamer itself. This is rather surprising, as for the complex, numerous additional hydrogen bonds are identified by NMR. One may speculate that the additional hydrogen bonds forming mainly tertiary interactions do not melt in a cooperative manner and thus are not detected by CD melting analysis. The fact that secondary structure melting occurs at lower temperature for the wild-type than for the P1-truncated construct 2 further supports the notion of the functional importance of the native P1 structure, which might be thermodynamically destabilized by non-optimal stacking and thus allowing for a finely tunable switch, the crucial event which must be preceded by P1 melting before any alternative structure can be formed.
Tertiary structure of the wild-type mfl-aptamer—2′-deoxyguanosine complex
For the mfl-aptamer, no high-resolution structure is available to date. Due to the size of the RNA (70 nucleotides), several limitations inherent to solution NMR spectroscopy are encountered: Intrinsically low chemical shift dispersion and proton density hamper the unambiguous identification of spatial distance restraints, and flexible parts of the RNA lead to line broadening of a considerable number of resonances, thereby preventing a complete assignment. Although thus lacking a three-dimensional model for the system under investigation, we could confirm many of the secondary and tertiary interactions suggested by in-line and chemical probing results as well as homology models of purine-sensing aptamer structures by extensively analyzing a large set of applicable NMR experiments. The key recognition mode shared by all purine riboswitches characterized so far is also employed by the mfl-aptamer. C74 forms a Watson–Crick base pair with 2′-dG, thereby rejecting all adenine-type purine compounds. This interaction is directly demonstrated by solution NMR by the 2′-dG imino-C74 N3 cross peak in an HNN-COSY spectrum. The helical folds of the three stems P1, P2 and P3 are readily detected in NOESY spectra by the three sequential NOE-connectivities of the respective imino proton signals. Watson–Crick base pairing is confirmed by the hydrogen bond-mediated scalar coupling between the two nitrogens involved in the imino-type hydrogen bonds in the HNN-COSY experiment (see Supplementary Data, Supplementary Figure S2).
Long-range base pairs are formed within the binding pocket by G46-C53 and G52-C22, the latter base pair stacks upon the purine moiety of the ligand as shown by the NOE cross signal between the 2′-dG imino proton and the G52 imino proton (Figure 3b). This motif is also in accordance with the purine aptamer consensus tertiary fold. In all crystal structures available to date, several base triples between residues of the three junction-regions form the highly structured binding pocket. A base quartet is formed by each purine ligand, C74, U51 and U47, where U51 forms hydrogen bonds to the sugar-edge site of the purine ligand. This structural feature of all other purine aptamers relies on the free N9 imino group of the purine ligand which is absent in 2′-dG. Thus, the C51-ligand interaction has to be different in the mfl-aptamer-2′-dG complex, and in fact, its presence cannot be confirmed by NMR.
How exactly the mfl-aptamer achieves the discrimination of 2′-dG versus the smaller guanine is challenging to explain, especially as the relative metabolite concentrations of 1.9 × 10−4 M for guanine and 5.2 × 10−7 M for 2′-dG (as measured in E. coli) would greatly favour guanine over 2′-dG binding (43). The fact that the hybrid aptamer is still able to bind to 2′-dG supports the hypothesis that ligand specificity is largely accomplished by C51 as the key residue inside the binding pocket, whereas ligand binding affinity is considerably affected by the adjacent P2 basal G-U wobble base pair (22), and P2 and P3 mainly serve to stabilize the structural scaffold. For large parts of the binding pocket, our NMR data are consistent with the GdG-3 hybrid aptamer crystal structure, where the hydrogen bond to the ligand-containing base triple formed by U47 is disrupted due to a U47A mutation in the mfl-aptamer versus the purine aptamer consensus sequence. We do not observe any interactions of this adenosine residue to other nucleotides. In contrast, the resonances of the A47 H2- and H8-protons are unusually strong, suggesting additional dynamics similar to the observation for residues U36 and U64. Moreover, both the H2 and the H8 protons of A47 resonate at the downfield edge of all adenosine resonances, which is a result of the absence of shielding ring-current effects in the local environment (38). From our NMR data, we therefore propose that A47 is not fixed by tertiary contacts, but remains flexible in solution. Interestingly, the crystal structure of the GdG-3 hybrid aptamer reveals two different possible conformations of residues A47, U48 and U49 in the binding pocket, supporting the notion of an at least partly disordered binding pocket. According to the GdG-3 hybrid aptamer crystal structure, part of the disrupted hydrogen bonding network in the ligand binding pocket is restored by the 2′-dG ribose moiety (22). Specifically, in the xpt-aptamer-guanine complex, C50 interacts with the P1 closing base-pair A21-U75 and U49 interacts with the next base-pair, U20-A76, thereby ‘fixing’ the lid of the binding pocket tightly. While NMR data for the xpt-aptamer-guanine complex confirm at least the U49-interaction with the U20-A76 base pair both by the presence of an exchange-protected U49 imino proton and the corresponding U20 NH-A76 N3 cross signal in the HNN-COSY spectra, neither the presence of a C49-U20-A76 interaction nor a C50-A21-U75 interaction is supported by any NMR data of the wild-type mfl-aptamer-2′-dG complex. In fact, we observe interactions of the H5′ and H5′′ protons of the 2′-dG to the two upper base-pairs of P1 (U20-A76 and A21-U75). A further characteristic feature of the purine aptamer–ligand complexes is the parallel alignment of P2 and P3, stabilized by the loop–loop interaction between L2 and L3. In the xpt-aptamer, the loop–loop interaction is mediated by two inter-loop base quartets, consisting of the W-C-base pair G37-C61 hydrogen bonded to a reversed Hoogsteen base pair U34-A65 and another W-C base pair, G38-C60, which interacts with the A33-A66 reversed Hoogsteen base pair (3,14). For the mfl-aptamer, the two Watson–Crick type base pairs G37-C61 and G38-C60 are observed, with G38 showing an NOE cross signal to U66, the closing base pair of P3. Thus, we directly confirm the compact parallel alignment of P2 and P3 for the wild-type mfl-aptamer-2′-dG complex. However, no indication for base quartet formation is found by NMR, although we observe the additional inter-loop U33-A65 Hoogsteen base pair. Interestingly, U33 is the only loop residue displaying reduced scission in the in-line probing assay performed for the wild-type mfl-aptamer (21). Instead, the closing base pair of L3, A59-U66, likely adopts a conformation different from canonical A-helix form, as indicated by the significant C2 downfield chemical shift. Thereby, it might provide a compensation for the deleted L3 nucleotides in terms of inter-helical structure formation. Compared to the xpt-aptamer, a stabilization of the loop–loop interaction by further inter-loop base pair formation is impossible in case of the mfl-aptamer due to the numerous nucleotide deletions within L3. This truncated loop still has to accomplish a 180°-turn, thus leaving little conformational space in addition to the three inter-loop base pairs already formed. Moreover, the absence of the signals of two solvent-exposed uridine imino protons and evidence for fast dynamics in the aromatic resonances of U36 and U64 as well as at least 2 more adenine residues from L2 imply that the loop–loop architecture contains at least those four extremely flexible residues and is considerably less compactly structured than in the xpt-aptamer. However, since parallel stacking of P2 and P3 as well as the base pairs G37-C61 and G38-C60 are already formed in the absence of ligand if Mg2+ is available, the loop–loop interaction fulfills a crucial structural role in the mfl-aptamer and thus is worth further investigations.
It is shown that 2′-dG binding by the mfl-aptamer results in transcription termination (21). We speculate that the mfl-aptamer is a transcriptionally operating riboswitch, as the presence of a stable hairpin followed by a poly-U stretch directly downstream of the aptamer suggests. However, by secondary structure prediction, no alternative structure forming an anti-terminator in the absence of ligand can be identified. P2 and P3 seem to be stable also without P1 formation, as indicated by the CD melting data for the truncated aptamer. Although not yet proven, these findings strongly suggest that this riboswitch also functions as a transcriptional regulation element where helix P1 is either formed in the presence of ligand, or P1 being disrupted by formation of an anti-terminator in the absence of ligand. The great discrepancy between in vitro measured KD values for 2′-dG binding (80 nM) and concentrations necessary for transcription termination (2 µM) (21) hints at a kinetically controlled riboswitch (5,6).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG (SPP 1258: ‘Sensory and regulatory RNAs in Prokaryotes’ and Wo 901/1-1) and the state Hesse [Center for Biomolecular Magnetic Resonance (BMRZ)]. H.S. and J.W. are members of the DFG-funded cluster of excellence: macromolecular complexes. Funding for open access charge: DFG.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 2.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 4.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 6.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 7.Wickiser JK. Kinetics of riboswitch regulation studied by in vitro transcription. Methods Mol. Biol. 2009;540:53–63. doi: 10.1007/978-1-59745-558-9_5. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck J, Noeske J, Wohnert J, Schwalbe H. Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2010;38:4143–4153. doi: 10.1093/nar/gkq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck J, Furtig B, Noeske J, Wohnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl Acad. Sci. USA. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain N, Zhao L, Liu JD, Xia T. Heterogeneity and dynamics of the ligand recognition mode in purine-sensing riboswitches. Biochemistry. 2010;49:3703–3714. doi: 10.1021/bi1000036. [DOI] [PubMed] [Google Scholar]
- 14.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J. Am. Chem. Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem. Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Priyakumar UD, MacKerell AD., Jr Role of the adenine ligand on the stabilization of the secondary and tertiary interactions in the adenine riboswitch. J. Mol. Biol. 2010;396:1422–1438. doi: 10.1016/j.jmb.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2'-deoxyguanosine. Proc. Natl Acad. Sci. USA. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards AL, Batey RT. A structural basis for the recognition of 2'-deoxyguanosine by the purine riboswitch. J. Mol. Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoldt M, Wohnert J, Gorlach M, Brown LR. The NMR structure of Escherichia coli ribosomal protein L25 shows homology to general stress proteins and glutaminyl-tRNA synthetases. EMBO J. 1998;17:6377–6384. doi: 10.1093/emboj/17.21.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; 2007. [Google Scholar]
- 25.Schleucher J, Sattler M, Griesinger C. Coherence selection by gradients without signal attenuation: application to three-dimensional HNCO experiments. Angew. Chem. Int. Ed. Engl. 1993;32:1489–1492. [Google Scholar]
- 26.Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. 1994;A109:129–133. [Google Scholar]
- 27.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reson. 1992;99:201–207. [Google Scholar]
- 28.Ohlenschlager O, Wohnert J, Bucci E, Seitz S, Hafner S, Ramachandran R, Zell R, Gorlach M. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3 C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Dingley AJ, Nisius L, Cordier F, Grzesiek S. Direct detection of N-H[…]N hydrogen bonds in biomolecules by NMR spectroscopy. Nat. Protoc. 2008;3:242–248. doi: 10.1038/nprot.2007.497. [DOI] [PubMed] [Google Scholar]
- 30.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 31.Wuethrich K, Otting G. Extended heteronuclear editing of 2D 1H NMR spectra of isotope-labeled proteins, using the X (ω1, ω2) double half filter. J. Magn. Reson. 1998;85:586–594. [Google Scholar]
- 32.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-tailored water suppression for H-1 -N-15 Hsqc experiments optimized to retain full sensitivity. J. Magn. Reson. Ser. A. 1993;102:241–245. [Google Scholar]
- 33.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 34.Ferner J, Villa A, Duchardt E, Widjajakusuma E, Wohnert J, Stock G, Schwalbe H. NMR and MD studies of the temperature-dependent dynamics of RNA YNMG-tetraloops. Nucleic Acids Res. 2008;36:1928–1940. doi: 10.1093/nar/gkm1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 36.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 37.Vokacova Z, Sponer J, Sponer JE, Sychrovsky V. Theoretical study of the scalar coupling constants across the noncovalent contacts in RNA base pairs: the cis- and trans-watson-crick/sugar edge base pair family. J. Phys. Chem. B. 2007;111:10813–10824. doi: 10.1021/jp072822p. [DOI] [PubMed] [Google Scholar]
- 38.Cromsigt JA, Hilbers CW, Wijmenga SS. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR. 2001;21:11–29. doi: 10.1023/a:1011914132531. [DOI] [PubMed] [Google Scholar]
- 39.Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49:1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 42.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.