Abstract
In recent years, both homing endonucleases (HEases) and zinc-finger nucleases (ZFNs) have been engineered and selected for the targeting of desired human loci for gene therapy. However, enzyme engineering is lengthy and expensive and the off-target effect of the manufactured endonucleases is difficult to predict. Moreover, enzymes selected to cleave a human DNA locus may not cleave the homologous locus in the genome of animal models because of sequence divergence, thus hampering attempts to assess the in vivo efficacy and safety of any engineered enzyme prior to its application in human trials. Here, we show that naturally occurring HEases can be found, that cleave desirable human targets. Some of these enzymes are also shown to cleave the homologous sequence in the genome of animal models. In addition, the distribution of off-target effects may be more predictable for native HEases. Based on our experimental observations, we present the HomeBase algorithm, database and web server that allow a high-throughput computational search and assignment of HEases for the targeting of specific loci in the human and other genomes. We validate experimentally the predicted target specificity of candidate fungal, bacterial and archaeal HEases using cell free, yeast and archaeal assays.
INTRODUCTION
Gene targeting, the site-specific manipulation of the genome, is the holy grail of gene therapy and genetic engineering. It promises to markedly reduce the risks associated with viral vector-mediated gene insertion, most notably, the risks of induced oncogene overexpression and insertional mutagenesis (1). Gene manipulation at a locus of choice is best facilitated by the introduction of a site-specific double-stranded DNA break (DSB). The default repair of the DSB by non-homologous end joining (NHEJ) can lead to gene disruption. In the presence of an appropriate donor template, the break can be repaired by homologous recombination (HR) leading to gene correction or gene insertion at the desired locus. Indeed, in recent years, much effort has been invested in the engineering of site-specific DNA endonucleases that can cleave desired loci in the human genome and induce gene targeting. Impressive results came from the use of zinc-finger nucleases (ZFNs), chimeric enzymes consisting of an endonuclease domain that is artificially linked to a site-specific array of zinc-finger domains (2). Of special note is the use of ZFNs engineered for the specific ex vivo disruption of the HIV coreceptor CCR5 in the T lymphocytes of AIDS patients, now under clinical trials (3) (see: http://clinicaltrials.gov/ct2/show/NCT00842634). Another promising option is presented by meganucleases, engineered homing endonucleases, selected to cleave a locus of choice [e.g. XPC (4), RAG (5)]. ZFNs and meganucleases have also been used in crop bio-engineering (6), in the production of model cell lines (7,8) animal models (9), induced pluripotent stem cells (10,11) and more.
The obvious advantage of using engineered endonucleases is the ability to target almost any gene of choice. However, the production of site-specific ZFNs and meganucleases is burdensome, lengthy and expensive. Moreover, the rate and distribution of off-target cleavage for these enzymes is difficult to predict (12). Importantly, safety assessments for engineered endonucleases are hindered by the fact that an enzyme selected to cleave a human locus will seldom cleave the homologous gene of an animal model because of sequence divergence. Here we show that native homing endonucleases (HEases), with their predictable target range and conservation of target sites in animal models, may present a promising alternative to engineered endonucleases for gene targeting.
HEases are a large and diverse class of site-specific nucleases found in Archaea, Bacteria and Eukarya and in their respective viruses (13,14). HEase genes (HEGs) are selfish genetic elements that reside as open reading frames (ORF) within self-splicing introns or as an endonuclease domain within inteins (15). An HEase promotes the horizontal propagation of its respective intron/intein into an intron-less or intein-less allele by cleaving the vacant allele to induce HR or reverse transcription, which results in effectively copying the intron/intein together with the HEG into the same position in the vacant allele (Figure 1). Importantly for their use in gene therapy, HEases possess the ability to introduce highly specific breaks in the human genome due to their long target sequences (14–40 bp). Indeed, native HEases are able to induce either site-specific NHEJ or site-specific HR in mammalian genomes engineered so as to contain the HEase’s target site (16–19). However, native HEases have not been used for gene therapy to date, probably because of the common misperception that they have no targets in the human genome. Instead, as mentioned above, selected HEases were subjected to rational engineering and directed evolution, so that they could target disease-associated genes (4,20). Nevertheless, basic research into native HEases has revealed some features with implications for their potential use in gene therapy.
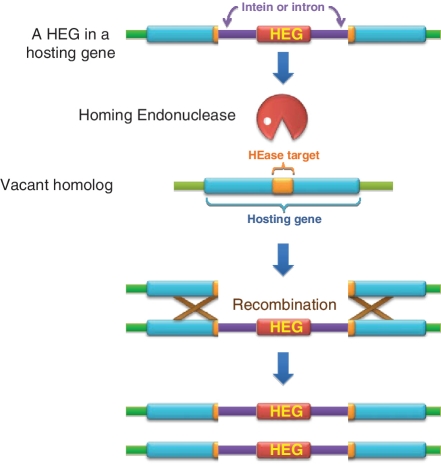
Figure 1.
The Homing process. The homing endonuclease (HEase) is expressed from the HEG (red), residing in an intron or as an in-frame domain of an intein (purple) in a hosting gene (cyan). It cleaves the target site (orange) in a vacant homolog of the hosting gene to induce homologous recombination (gene conversion or double crossover), turning the vacant homolog into a HEG-carrying one.
Importantly, the target sites of HEases are not stringently defined: some nucleotides along the target site can be substituted while cleavage efficiency is retained (21). Without a general way to predict the plasticity in HEase target recognition, it might have been very difficult to apply HEases as tools for gene therapy. However, accumulating evidence suggests that HEase plasticity is, at least to some extent, predictable based on the evolutionary considerations (22–26). It has long been appreciated that inteins and self-splicing introns tend to be found in conserved motifs of essential genes (27). Hence, any partial or inaccurate intron/intein deletion or mutation at a splicing motif is detrimental to the host, which is left with a persistent disruption in a critical gene. The self-serving localization of the intron/intein is also beneficial for the HEase. An HEase promotes the copying of its respective intron/intein into its target site and therefore, HEase targets coincide with intron/intein insertion sites (Figure 1). In particular, a conserved insertion site is also a conserved HEase target site. Therefore, the host cannot easily evade the parasite by altering the target sequence.
Nevertheless, even highly conserved loci usually include some variable sites. HEases have therefore evolved so that they rely on conserved positions within the target sequence for robust target recognition (24–26). In particular, when the hosting gene encodes a protein, selection on this gene acts mainly to conserve its amino acid translation rather than the coding nucleotide sequence. Synonymous substitutions are therefore frequent even in sequences coding conserved protein motifs. Thus, HEases are expected to evolve tolerance to silent target mutations. Indeed, Kurokawa et al. (22) have demonstrated for an array of intron-encoded HEases residing in protein-coding genes, that single silent mutations in the target sites are far better tolerated than mutations that alter the coded polypeptide. Scalley-Kim et al. (23) have examined the specificity profile of the I-AniI HEase and found it to be strongly correlated with wobble versus non-wobble positions and also with the degree of degeneracy inherent in individual codons. At the focus of the above studies were HEases of the LAGLIDADG family, which is the most abundant structural family of HEases. However, similar evolutionary considerations may apply to the binding and cleavage specificity of GIY–YIG HEases (28,29) which have a highly distinct mode of DNA interaction.
In this work, we reasoned that the predictability of HEase target recognition may allow the discovery of formerly unidentified HEase targets in the human and other genomes, for the benefit of gene therapy and genetic engineering. We first demonstrate that HEases residing in protein-coding genes are often tolerant of concomitant synonymous substitutions at all wobble positions in their target site. This is a generalization of previous reports (22–26) that allows finding HEase targets in formerly unidentified loci in the human and other genomes. To apply this principle to as many HEases as possible, we searched all public sequence databases for novel HEGs. At this stage, we relied on another property of naturally occurring HEGs, which is that the gene coding the enzyme and the target sequence of the enzyme are found on the same locus. In particular, the target sequence flanks the intron/intein insertion site (Figure 1). This property allowed us to predict a putative target sequence for each newly discovered HEase. The results of this search were compiled in the form of the HomeBase database. Finally, we experimentally validated the predicted specificity range of candidate fungal, bacterial and archaeal HEases using cell-free, yeast and archaeal assays. Thereby, a large arsenal of naturally occurring HEases was compiled together with their predicted target ranges, providing a diverse toolbox of specific cutters for the genetic manipulation of large and complex genomes.
MATERIALS AND METHODS
Experimental methods
Strains, plasmids and oligonucleotides
Supplementary Table S1 lists the strains, plasmids, oligonucleotides and PCR primers used in this study. For extra-cellular cleavage assays we inserted the native, mutated or predicted target-sites of PI-SceI and PI-PspI into pGEM-Teasy (Promega, Figure 2a and b) or into the PfoI site of pDELT (Figure 2d). pDELT was constructed by cleaving pRS304 (30) with SmaI and inserting a 427 bp long PCR segment of the Saccharomyces cerevisiae LYS2 gene, amplified using the OI3 and B82 primers.
Figure 2.
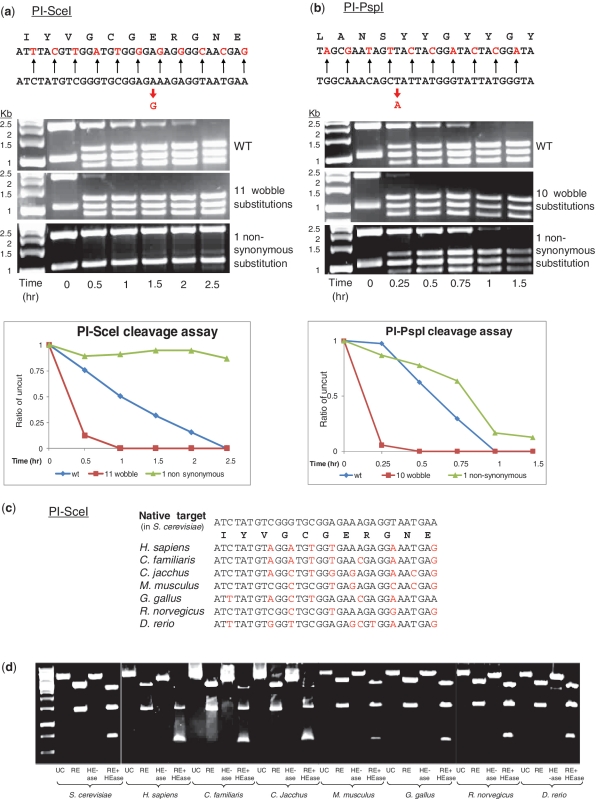
Characterization of HEase plasticity in target recognition and its use for finding HEase targets in the human and other genomes. (a) and (b): HEase cleavage is tolerant of concomitant mutations at all wobble positions along the target site. The cleavage efficiency of the HEases PI-SceI (from S. cerevisiae) (a) and PI-PspI (from Pyrococcus species GB-D) (b) was assayed on different targets cloned on a bacterial plasmid. The cloned vectors were then fragmented by a restriction enzyme for the sake of visual clarity. Both HEases cleave targets mutated at all wobble positions (top DNA sequence) with higher efficiency than they cleave their native targets (bottom DNA sequence). Conversely, a single non-synonymous mutation (red arrow) is sometimes sufficient to abolish cleavage by PI-SceI and to reduce cleavage by PI-PspI. (c) and (d): PI-SceI can cleave its predicted targets from the human ATP6V1A1 gene and its homologs in the genomes of animal models. (c) Alignment of the native target of the PI-SceI HEase from S. cerevisiae with the predicted targets in the human ATP6V1A1 gene and its homologs in the genomes of animal models. (d) Results of an in vitro cleavage assay demonstrating that PI-SceI can cleave its predicted targets from the genomes of diverse organisms. UC, uncut; RE, cut by a restriction enzyme (XbaI); HEase, cut by an HEase, RE + HEase, cut by XbaI AND by PI-SceI. S. cerevisiae, Saccharomyces cerevisiae; H. sapiens, Homo sapiens; C. familiaris, Canis familiaris; C. jacchus, Callithrix jacchus; M. musculus, Mus musculus; G. gallus, Gallus Gallus; R. norvegicus, Rattus norvegicus; D. rerio, Danio rerio.
The archaeal homing assay was conducted using the Haloferax volcanii strain WR532 (H133) ΔpyrE2 (31). The Archaea were transformed with either the pTA131 (32) or the pTA1.1 plasmids, or the pTA1.1hum plasmid. pTA1.1 and pTA1.1hum are derivatives of pTA131. pTA1.1 carries between the EcoRI and SpeI sites a 1.1 kb long PCR segment of H. volcanii POLB gene lacking the POLB intein (33) that was amplified using the Hvol 1.1 F and R primers. The pTA1.1hum carries a similar segment in which a BmgBI fragment including the native target of the POLB HEase was replaced by a fragment carrying the predicted human target.
The S. cerevisiae strains used for the yeast HEase assay are all derivatives of OI50 (Supplementary Table S1). In the following explanation about the construction of the derivatives, X stands for either one of: (i) Botrytis cinerea PRP8 HEase native target; (ii) The B. cinerea PRP8 HEase predicted human target; (iii) The B. cinerea PRP8 HEase predicted mouse target; (iv) Nostoc RNR HEase native target; (v) Nostoc RNR HEase predicted N. punctiforme target; (vi) Nostoc RNR HEase predicted Synechococcus target. Yeast strains with YDEUH prefix (YDEUH + X target) were constructed by transforming OI50 with NcoI-cleaved pDEUH derivatives (pDEUH + X target). pDEUH is pRS303 (30) carrying a 315-bp long PCR segment of the S. cerevisiae URA3 gene (amplified using the OI5 and B45 PCR primers) at its HincII site. The pDEUH derivatives each have a different HEase target at the PfoI site of pDEUH. Yeast strains with YDELT prefix (YDELT + X target) were constructed by transforming OI50 with HpaI-cleaved pDELT derivatives (pDELT + X target). The pDELT derivatives each have a different HEase target in the PfoI site of pDEUH. As explained above, pDELT is pRS304 (30) carrying a 427-bp long PCR segment of the S. cerevisiae LYS2 gene.
Yeast strains with YDEUHLT prefix (YDEUHLT + X targets) were constructed by transforming YDEUH + X target with HpaI-cut pDELT + X target. Final constructs of the form: YDEUHLT + X target + pGML + Y HEase, were constructed by transforming YDEUHLT + X targets with a pGML10 (34) derivative encoding the Y HEase (either B. cinerea PRP8 HEase or Nostoc RNR HEase). The B. cinerea PRP8 HEase was amplified from the B. cinerea strain B05.10, a kind gift from Professor Annika Bokor (35), using the primers: B. cinerea-HEN-F and R. The forward primer includes an ATG start codon and an SV40 nuclear localization signal (NLS). The amplified B. cinerea PRP8 HEase was inserted between the XbaI and XmaI sites of pGML10. The Nostoc RNR HEase was amplified from Nostoc (Anabaena) sp. PCC 7120, a kind gift from Professor Sammy Boussiba (36), using the primers: Nostoc-HEN-F and R. Here again, the forward primer includes an ATG start codon and an SV40 NLS. The amplified Nostoc RNR HEase was inserted between the BamHI and EcoRI sites of pGML10.
Extra-cellular cleavage assays
A quantity of 1 µg of a plasmid [pGEM derivative (Figure 2a and b) or pDELT derivative (Figure 2d)] carrying a native, mutated or predicted target of PI-SceI (Figure 2a and d) or PI-PspI (Figure 2b) were subjected to cleavage using 1 U of each enzyme as provided by New Englands Biolabs (at 29 pmol/U for PI-SceI and 80 fmol/U for PI-PspI), in a 50 µl reaction at 37°C (PI-SceI) or 65°C (PI-PspI). Aliquots were extracted every 30 min (Figure 2a), 15 min (Figure 2b) or after a 16 h over-night incubation (Figure 2d). PI-SceI was heat-inactivated (20 min, 65°C) and all samples were fragmented by a restriction endonuclease [BspHI (Figure 2a and b) or XbaI (Figure 2d) for 1 h in a 10 µl reaction] prior to gel elecrophoresis.
Archaeal homing assay
For the transformation of H. volcanii a liquid culture (1.5 ml; OD600nm of 1.5) was centrifuged at 3500 g for 5 min. The supernatant was discarded, the cells were resuspended in 200 µl spheroplasting solution (1 M NaCl, 27 mM KCl, 50 mM Tris–HCl pH 8.2, 15% sucrose) and incubated at room temperature for 5 min. A quantity of 20 µl of 0.5 M EDTA was added and cells were incubated at room temperature for 10 min. Then, 10 µl of purified plasmid DNA was mixed with 15 µl spheroplasting solution and 5 µl of 0.5 M EDTA was added to the cells, followed by incubation of 5 min at room temperature. Subsequently, 240 µl of PEG solution (60% PEG 600 in spheroplasting solution) was added and cells were incubated for an additional 20 min at room temperature. Following the incubation, 1 ml of regeneration solution (3.4 M NaCl, 175 mM MgSO4, 34 mM KCl, 5 mM CaCl2, 50 mM Tris–HCl pH 7.2, 15% sucrose) was added and cells were centrifuged at 3500g for 7 min. The supernatant was discarded and cells were resuspended in HY medium supplemented with 15% sucrose and left to incubate without shaking overnight at 37°C. The cultures were then washed and plated on selective media. The presence of an intein on the exogenic plasmids indicates that homing has taken place and in particular that efficient cleavage has occurred. Intein presence was tested by PCR using the RP2 and M13F primers. The standard errors were calculated based on a binomial distribution with an added pseudo-count of 0.5 successes and 0.5 failures.
Yeast HEase assay
Prior to the recombination assay, dilutions of four independent colonies of each strain were plated on YEPD medium (1% yeast extract, 2% Bacto Peptone, 2% dextrose, 2% Bacto-agar) and on YEP-GAL medium (1% yeast extract, 2% Bacto Peptone, 2% galactose, 2% Bacto-agar) in order to assess the toxicity of each enzyme (Supplementary Figure S3). For the recombination assay, each of the four colonies of each strain was grown overnight at 30°C in a synthetic complete (SC) liquid medium supplemented with 2% galactose (for strains without an HEase expression plasmid), or SC-Leucine liquid medium supplemented with 2% galactose (for strains with an HEase expression plasmid, marked with the LEU2 gene). Cells were then pelleted, diluted and plated on YEPD and on SD-Ura (synthetic complete medium − uracil + 2% dextrose + 2% bacto-agar) and SD-Ura-Lys (synthetic complete medium − uracil − lysine + 2% dextrose + 2% bacto-agar) to assess recombination rate (implying HEase activity rate). HEase activity rate is defined as the average fold increase in colony formation on the selective medium of the strain with target X and HEase Y with respect to the average colony formation on the selective medium of the strains without any HEase. The confidence intervals for the fold increase were calculated by Monte Carlo sampling of pairs of simulated measurements from two normal distributions having the same mean and variance as the actual measurements. The 95% confidence intervals used are the 2.5th and 97.5th quantiles of the emerging distribution of ratios.
Construction of the HomeBase HEase database
Search for HEGs in DNA databases (BLAST-1)
A homology search for novel HEGs was conducted across all available DNA data sets. A set of known HEGs was used as queries for BLAST searches. Protein sequences of known HEases in introns and inteins were retrieved from manually curated databases. A total of 289 sequences of HEGs in Group I introns were downloaded from the Group I Intron Sequence and Structure Database (37) (GISSD; http://www.rna.whu.edu.cn/gissd). Three hundred twenty five sequences of HEGs in inteins were downloaded from INBASE (38) (http://www.neb.com/neb/inteins.html). In both introns and inteins, the manual curation of these databases ensures that these protein sequences do not include exonic or exteinic parts. This is essential for our purpose because otherwise the BLAST searches will retrieve many homologs of the hosting gene instead of novel HEGs. This sequence set, totaling 614 HEGs will be subsequently referred to as the ‘known HEGs set’. Using translated BLAST (tblastn), the known HEase protein sequences were used as queries to search against all possible six frames translations of the non-redundant nucleotide database nt and the non-redundant environmental (metagenomic) nucleotide database env_nt, both downloaded from the NCBI web site (http://www.ncbi.nlm.nih.gov/ftp/). We retained all hits of E-value < 10. This stage will be subsequently referred to as BLAST-1.
For each hit sequence in the BLAST-1 results there are often several high scoring pairs (HSPs), pairwise alignments of a query subsequence to a hit subsequence (which is translated in one of the six possible reading frames). Furthermore, a novel HEG sequence is usually homologous to several of the known HEGs, therefore several queries yield overlapping HSPs in the same hit locus. The number of HSPs per hit sequence is often large for whole chromosome sequences, which may contain several loci of HEase homology. To divide such hit sequences to distinct loci, all HSPs from all queries on the same hit sequence were clustered. Overlapping and neighboring HSPs <2000 bases apart, were clustered and 1000 bases were added on either side of every cluster. These hit subsequences are the putative novel HEGs and their adjacent intron/intein and exon/extein sequences. This procedure often resulted in a cluster of several HEG-containing intron/inteins in the same host gene.
Defining splice sites (HEase target sites) based on vacant homologs (BLAST-2)
For any gene that harbors a HEG inside an intron/intein, a vacant homolog is a homologous sequence of that gene without the intron/intein. A second BLAST search was conducted for each putative HEG-containing sequence in order to identify such a vacant homolog. Each subsequence resulting from the clustering of the BLAST-1 HSPs was used as the query for the second BLAST search, hereby termed BLAST-2. In this translated BLAST (tblastx) each of the three possible translations of the strand that was aligned to the known HEG in BLAST-1 was searched against the same non-redundant DNA databases as above. Each BLAST-2 hit was checked for several criteria to determine if it contained a bona fide vacant homolog (Supplementary List S1). After identifying the first intron/intein in the BLAST-2 query, we repeated the procedure using only BLAST-1 HSPs that do not overlap the intron/intein. Only BLAST-2 HSP pairs that contain these BLAST-1 HSPs were considered as potentially defining introns/inteins. Thereby, several additional non-overlapping introns/inteins were often identified in the same BLAST-2 query until no BLAST-1 HSPs remained.
The final outputs of the algorithm are the target sequence flanking these splice sites in the query (the pHEG-containing sequence) and the ORF of the HEase (in introns) or of the intein, which codes for the HEase. This output constitutes the HomeBase database.
To assess the accuracy of prediction, we sampled 30 putative HEGs and manually inspected all stages of the automatic pipeline, including: confidence in the BLAST-1 homology and conservation of known HEase motifs; confidence and accuracy of the BLAST-2 alignment to the vacant homology, especially of the exon/extein boundaries after correction of gaps/overlaps (where possible we compared the prediction to annotation of intron position and intron/intein splice sites consensus). We found 92 ± 4% (mean ± SE) of the results to have reliable homology to a known HEG and 75 ± 7% to be true HEGs with correct identification of the target site. For the 222 results of the second iteration, we estimate an accuracy level of 25 ± 7%. The overall number of known HEGs was estimated by querying all protein records available in the Entrez engine. The search term used was ‘homing endonuclease’ [All Fields] NOT txid33208 [Organism:exp]. (33 208 is the taxonomy identifier for Metazoa). The number of predicted HEGs that were previously annotated as HEGs was determined by checking the GenBank records containing them. All CDS features overlapping the containing intron/intein were examined. In addition, if a CDS contained a ‘protein_id’ tag, the corresponding GenPept record was obtained through Entrez and all features found were included in the search. For all the features selected, the text of the following tags was examined: ‘note’, ‘product’, ‘gene’, ‘gene_synonym’ for CDS features and ‘note’, ‘region_name’ and ‘product’ for features found in the protein. A HEG was considered to be previously annotated if any of the tags contained the term ‘homing endonuclease’.
The HomeBase database
The HomeBase database is the unified set of HEGs including our novel predictions and the known HEGs from INBASE and GISSD. To infer the target sequences from these databases, we download the exon or extein sequences from INBASE or GISSD respectively and extracted the first seven amino acids from each exon/extein.
We classified HomeBase records into HEG families by homology (blastp) of the translated ORF of the putative HEG against a set of HEG sequences with family annotation collected from the INBASE, GISSD and Entrez Protein databases. This set includes representatives from the LAGLIDADG, HNH, GIY–YIG and His–Cys families. Each HomeBase HEG was assigned the family annotation of the top BLAST hit, except in ambiguous cases where the top hit from a different family had an E-value that was close to the top hit E-value by a factor <10.
Identifying candidate HEases for specific targets
To reveal the potential utility of the HomeBase collection for genetic intervention in humans, we used translated BLAST (tblastn) to search for a match between the translations of HomeBase target sequences and all possible six-frame translations of the human genome. Resulting hits were sorted by their similarity. We also required that five out of the six central residues be identical and that the sixth residue be at least similar. We classified the hits as genic or intergenic depending on whether or not they reside within an annotated gene or the 5000 bases flanking it (ftp://ftp.ncbi.nih.gov/gene/DATA/gene2accession.gz). Furthermore, we classified genic hits into coding exons versus non-coding introns and flanking sequence (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/mapview/seq_gene.md.gz) to identify hits that may be the result of true protein homology (Supplementary Table S2). To estimate the number of off-target hits for each HEase in the human genome, we counted the number of BLAST hits whose E-value is three times worse (larger) than the E-value of the top hit.
RESULTS
Native HEases can efficiently cleave targets with concomitant silent mutations at all wobble positions
It has previously been shown that HEases encoded by genes residing in introns/inteins of protein-coding genes can cleave variants of their target sites bearing single or a few synonymous substitutions (22,23). Here, we demonstrate that at least some HEases can efficiently cleave targets with concomitant silent mutations in all codons of the target sequence. This is a result of the evolutionary history of the HEases and their hosts: mutations in the target site that prevent insertion of the intron/intein would be advantageous for the host, however, as most molecular parasites occupy critical positions within essential genes, most mutations of this sort are lethal and only silent mutations are tolerated. HEases thus adapted by being able to cut variants of the target site carrying synonymous substitutions. The far-reaching implication of this finding is that one can use translated BLAST and find many formerly unidentified HEase targets in the human and other genomes (see below). To demonstrate this general principle, we examined the target specificity of HEases stemming from two different domains of life: PI-SceI, an HEase from the yeast S. cerevisiae and PI-PspI from the archaeon Pyrococcus species GB-D. We assessed the cleavage efficiency of these HEases on their native targets as well as on targets where all wobble positions underwent transitions. Both enzymes were found to cleave a target bearing 10 (PI-PspI) or 11 (PI-SceI) silent mutations even better than they do their original target, while a single non-silent substitution completely eliminated cleavage by PI-SceI and reduced cleavage by PI-PspI (Figure 2a and b). Tolerance of wobble substitutions does not imply promiscuity. Cleavage by PI-SceI is almost completely prevented by non-synonymous substitutions in any one of nine different positions along its target (21). Thus, HEase target recognition combines tolerance for mutations in synonymous positions with increased specificity for the non-synonymous positions. [albeit restricted; e.g. tolerance to some non-synonymous substitutions (21,23), Figure 2b].
Our results imply that HEases residing in protein-coding genes can cleave many DNA sequences having the same translation as their native target. This increases the odds of finding an HEase that can cleave any disease-associated gene or pre-specified genomic safe harbor by several orders of magnitude. For example, a 24-bp long DNA sequence is found at random every 424 ≈ 3*1014 bp while its eight amino acid long translation is found at random every 208 ≈ 2.5*1010 codons. It is therefore expected that within a large enough array of HEases at least some will be found able to cleave the human genome at desired loci. Furthermore, we recall that HEGs reside within introns or inteins found in conserved motifs. Therefore, in many instances, finding an HEase target in the human genome is not a random event but rather a result of evolutionary motif conservation from the HEase hosting microbe to humans. For example, the PI-SceI HEase originates from an intein lying in the vacuolar membrane ATPase (VMA) gene of the budding yeast S. cerevisiae. The S. cerevisiae VMA is a homolog of the human vacuolar ATPase ATP6V1A1 and has a high degree of sequence similarity in the regions flanking the PI-SceI-intein insertion site (Figure 2c; note that the intein is present only in the yeast—there are no inteins in humans). Indeed, PI-SceI can specifically cleave a PCR-amplified segment of the human ATP6V1A1 locus including the predicted target (Figure 2d). We note that vacuolar ATPases are recognized as a potential therapeutic target for the treatment of osteoporosis (39). In particular, the vacuolar ATPase inhibitor, FR167356, has been shown to prevent bone resorption in ovariectomized rats (40). It is, therefore, implied that PI-SceI may be used for ATP6V1A1 disruption in desired tissues. Moreover, because the target recognition of native HEases is based on evolutionary conservation, we predicted that PI-SceI would be able to cleave not only the human ATP6V1A1 but also the ATPase gene homologs in animal models (Figure 2c), thus facilitating pre-clinical efficacy and safety assessments. We cloned the predicted target from different animals in a bacterial plasmid and subjected the vector to PI-SceI cleavage. We observed efficient cleavage of all ATP6V1A1 homologs by PI-SceI (Figure 2d).
Construction of HomeBase, the HEase database
Acknowledging the potential applicability of native HEases, we set out to develop a high-throughput approach for the detection of HEGs in sequence databases and the prediction of HEase targets in the human and other genomes. Our algorithm for the construction of the ‘HomeBase’ HEase database is depicted in Figure 3a. As a crude first step, we used manually curated collections of known HEGs (the above-mentioned INBASE and GISSD databases) as queries in a translated BLAST to find putative HEGs in the genomic and metagenomic DNA databanks (metagenomic DNA databanks hold DNA sequences of uncultured organisms, mostly from environmental samples). This BLAST search is conducted with a very permissive similarity threshold (E-value < 10).
Figure 3.
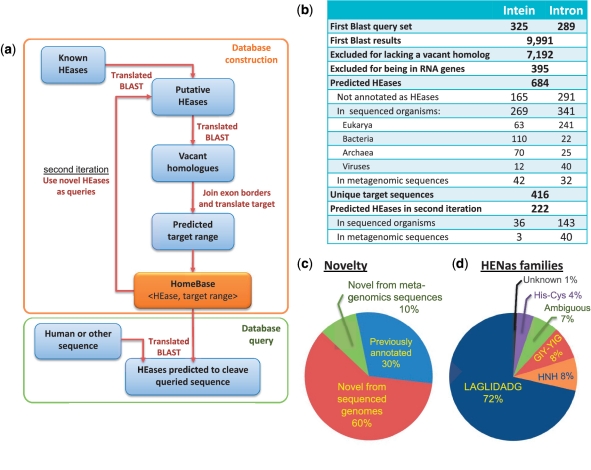
The HomeBase algorithm and database. (a) Database construction involves: (i) the identification of putative HEGs in both genomic and meta-genomic databanks using translated BLAST searches; (ii) The identification of vacant homologs using a second translated BLAST; (iii) The identification of exon boundaries and the prediction of the target based on the vacant homologs and the translation of the target to define the target range. The HomeBase database consists of a set of HEG sequences and the predicted target range of each HEase. The user may then query HomeBase with (e.g.) a human gene of interest and receive a list of HEases, which are predicted to cleave the query sequence based on a translated BLAST search against the targets in the database. (b) Statistics of the HomeBase database. (c) A pie diagram depicting the percentage of previously annotated versus newly discovered HEGs within the results of the HomeBase algorithm. (d) Classification of HomeBase HEGs into structural families.
We next used several filters to separate true HEGs from false hits. First, we screened for a feature that is unique to HEGs—the presence of a vacant homolog—defined as a homolog of the HEG-hosting gene that lacks the intron or intein in the respective locus (Figure 1). HEG-hosting genes are expected to have vacant homologs because the phylogenetic distribution of mobile introns and inteins is typically non-monophyletic (41,42); if a certain gene in a certain species codes a mobile intron, it is highly probable that there exists a related species in which the homologous gene is intronless. Conversely, any false hit of the initial BLAST search that is not encoded in an intron or an intein will be excluded for lack of a vacant homolog. HEG-less introns and inteins may have vacant homologs, but these were excluded for being shorter than a pre-determined length threshold (see ‘Materials and Methods’ section). These filters allowed us to use a very low homology threshold in search for novel genes, while keeping the frequency of false hits relatively low (Figure 3b and c). Thereby many highly divergent novel HEGs can be discovered. HEGs residing in non-coding RNA genes were excluded from HomeBase because their target plasticity is not based on synonymous/non-synonymous positions.
Using this method we have been able to find 684 HEGs (Figure 3b and c), 70% of which have not been annotated before as HEGs (60% in sequenced genomes and 10% in metagenomic sequences). In a manual inspection of a random sample of 30 predictions we found 75 ± 7% (mean ± SE) to be true HEGs with correct identification of the target site (see ‘Materials and Methods’ section). We then complemented our original data with those of others (37,38,43) to conclude with a final database of approximately a thousand distinct HEGs. To facilitate the applicability of our database, we developed a web-server, which allows one to search a DNA sequence of choice (e.g. a human gene) for putative targets of HEases. The web-server can be freely accessed at: http://homebase-search.tau.ac.il/.
The significant addition of novel HEGs to the set of known HEGs can be used to expand the query set of the HomeBase algorithm. We conducted a second iteration of the entire pipeline using the union of the original query set with the 684 HEase-coding sequences that were identified in the first iteration of HomeBase. This procedure extended the predictions by 222 additional HEGs (Figure 3b) demonstrating that the potential of novel HEGs in the extant sequence databases has not yet been exhausted. We nevertheless hold the results of the second iteration as a distinct set and not united with our initial 684 results, because, as can be expected, we observed significantly lower prediction accuracy after the iterative process (See ‘Materials and Methods’ section).
We note that the NCBI database holds 1386 sequences annotated as HEGs, only 213 of which (15%) were retrieved by our algorithm. This is expected because of our stringent demand for the presence of a vacant homolog in the databases. Importantly, these vacant homologs facilitate the identification of the target site of each HEase. As the intron/intein insertion site marks the target sequence of the resident HEase, we assigned each HEase with a predicted target site composed of the DNA sequences flanking the inferred splice sites from the exon/extein side (Figure 1). The exact boundaries of each target could not be automatically predicted. HEase targets vary in length and many are asymmetrically positioned with respect to the intron/intein insertion site. As a partial remedy, each HomeBase record holds seven codons 5′ and seven codons 3′ to the splice site, which is enough to encompass the target of any characterized HEase. Any predicted HEase target (e.g. in the human genome) would align to a significant and central subsequence of these 14 codons, hopefully encompassing the true target of the HEase (see ‘Materials and Methods’ section). Importantly, HomeBase assigns each HEase not with a target sequence but rather with a target range. As discussed above, an HEase encoded within a protein-coding gene is expected to cleave many DNA sequences that have the same translation as its native target (Figure 2). We, therefore, use the amino acid translation of each target to define the HEase target range, increasing the odds of finding HEase targets in the human and other genomes by several orders of magnitude. Our algorithm has retrieved 416 unique target ranges for the 684 HEases identified (Figure 3b).
After the first translated BLAST used to identify putative HEGs and the second translated BLAST used to find vacant homologs, we applied a third translated BLAST to find HEase targets of therapeutic or biotechnological uses. At this stage, the amino acid target ranges are aligned to the targeted genome, human or other. First, we searched for hits in and around human genes associated with hereditary and other diseases, as listed in NCBI’s OMIM database (http://www.ncbi.nlm.nih.gov/omim). Importantly, the match in the human gene needs not be in the translated reading frame of the target gene or even on the sense strand. Moreover, a match in an intron or in the 5′-UTR can be equally useful (for inserting a cDNA under the endogenous promoter but upstream to the deleterious mutations).
Table 1 presents selected results of potential medical use. The selection exemplifies the special emphasis that we have given to those targets that were found in the human genome as a result of genuine homology and conservation of the targeted protein motif from the native microbe to humans: these targets are also found in the homologous loci of animal models and are thus useful for pre-clinical experiments.
Table 1.
Selected results of medical significance
| Source of HEasea | HEase family | Human gene | Therapeutic relevance | Target site identityb | Off-target hitsc |
|---|---|---|---|---|---|
| Botrytis cinerea | LAGLIDADG | PRPF8 | Retinitis pigmentosa | 11/11 | 0 |
| Haloferax volcanii | LAGLIDADG | POLD1 | Colon and colorectal cancer | 9/10 | 0 |
| Metagenomic | LAGLIDADG | FANCA | Fanconi anemia | 8/9 | 1 |
| Metagenomic | GIY–YIG | LMNA | Dilated cardiomyopathy | 8/9 | 8 |
| Trichodesmium erythraeum IMS101 | LAGLIDADG | SMAR-CAL1 | Schimke immunoosseous dysplasia | 11+1/12 | 14 |
| Metagenomic | LAGLIDADG | VCP | Inclusion body myopathy | 10+1/12 | 1 |
| Metagenomic | LAGLIDADG | CYP11B2 | Low renin hypertension | 8/8 | 3 |
| Hypoaldosteronism | |||||
| Metagenomic | LAGLIDADG | SPG7 | Spastic paraplegia 7 | 9+1/11 | 3 |
aOften, a family of related HEases can each cleave the same given target. Here, we list representatives of such families. Our online database and server http://homebase-search.tau.ac.il/, hold all members of such an HEase family, when relevant.
bX+Y/Z: X identities and Y similarities out of a Z amino acid long alignment between the translation of the indigenous target and the translation of a putative target sequence in the human gene. An eight amino acids long sequence is predicted to be unique within a random sample of the size of the six-frame translation of the human genome. The targets of many HEases are not longer than 8 × 3 = 24 bp.
cOff-target hits: number of BLAST hits in the human genome whose E-value was less than 3 times worse (larger) than the E-value of the desired hit.
For each human target the table indicates the therapeutic relevance, as well as the natural host species of the HEase, the level of identity between the predicted (translated) HEase target and the (translated) human sequence, the classification into HEase families, and an estimate of the number of off-target cleavage sites in the human genome (see ‘Materials and Methods’ section). A list of the top 66 HomeBase hits in human genes is given in Supplementary Table S2. These hits are mostly from LAGLIDADG HEases (79%) and some GIY–YIG (15%), HNH (3%) and His–Cys (3%). The number of off-target cleavage sites for these HEases ranges from 0 to 112, with a median of 9.
We note that some HEases have targets only in inter-genic human loci (Supplementary Figure S1). Some of these loci may prove to be genomic safe harbors, allowing for a stable, safe and efficient transgene expression (e.g. for the treatment of recessive diseases). Finally, the targets of many HEases in HomeBase diverge slightly from a desired sequence (e.g. a disease-associated human locus). These enzymes may be chosen as scaffolds for enzyme engineering to achieve the necessary specificity and efficacy.
Validation of HomeBase predictions
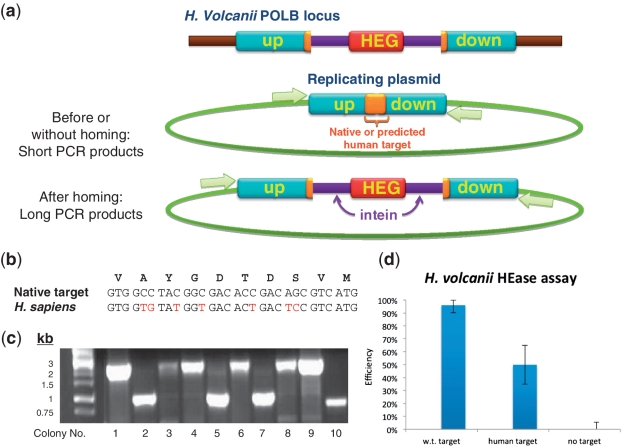
When a putative HEase is found in an organism with highly developed genetic tools, the activity of the enzyme can be assayed in the native organism (44). For example, HomeBase predicts that the PolB intein of the halophilic archaeon H. volcanii encodes an active HEase. Our prediction of the intein borders coincides with the INBASE predictions (38). We have previously identified its endonuclease domain and experimentally validated its endonuclease activity. Using an archaeal plasmid assay, we have also demonstrated that the H. volcanii HEase can cleave its predicted target with high efficiency. The assay involves transforming the archaeon with a plasmid bearing the HEase target as an integral part of a PolB vacant allele. When the plasmid enters the archaeal cell, the endogenously expressed HEase can recognize and cleave its target and induce intein homing into the plasmid via homologous recombination (Figure 4a). Intein homing can be detected by PCR amplification using target flanking, plasmid-specific primers. In the context of the present HomeBase gene targeting application, we now substituted the native target of the enzyme with its predicted target in the human homolog of PolB, the DNA polymerase delta gene (PolD1, Figure 4b). We find that high cleavage efficiency is retained (Figure 4c and d). We note that PolD1 in eukaryotes is involved in DNA repair and that mutations in this gene in humans are associated with colon cancer and with sporadic colorectal carcinomas (45).
Figure 4.
The PolB HEase of H. volcanii can cleave a target sequence from the human gene PolD1. (a) A PCR is preformed on H. volcanii individual colonies transformed with a plasmid bearing a vacant PolB allele coding either the native target of the PolB HEase or a homologous sequence from the human PolD1 gene. The PCR reaction (white arrows denote primers) can amplify either a short product in the absence of homing or a long product if homing has taken place. (b) Nucleotide and amino acid alignments of the target sequence from the H. volcanii PolB gene and the homologous human sequence from the PolD1 gene. (c) Representative results of the PCR assay in cells carrying a plasmid with the human target sequence. A long PCR product indicates that homing has occurred. (d) The relative homing efficiency of the PolB HEase to the plasmid-borne vacant homolog carrying either native (archaeal) or human targets, or no target (n = 30). Error bars represent 95% confidence intervals based on Monte Carlo simulations.
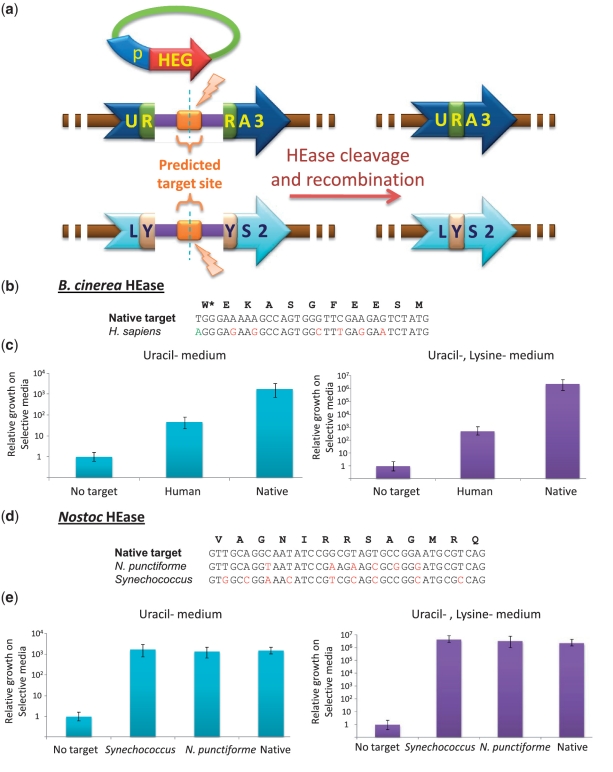
The H. volcanii HEase is a special case in that its activity on the predicted human target could easily be assayed in the natural host, because methods of genetic manipulations are highly developed for this archaeal species. It should also be noted that H. volcanii is halophilic and, therefore, the enzyme is expected to have reduced activity in human cells [although extremophilic HEases can be readily adapted to mesophilic conditions by genetic engineering (46)]. Therefore, we wanted to develop a more robust validation method that would allow us to verify the activity of HEases on their predicted targets in a eukaryotic setting. Following Chames et al. (47), we designed an assay in the budding yeast S. cerevisiae, wherein the predicted HEase target is inserted between the truncated repeats of the genes encoding the metabolic enzymes Ura3 and Lys2. The HEG is plasmid borne and is expressed from a galactose-induced promoter. Upon HEase cleavage, the truncated repeats recombine and reconstitute the metabolic genes, allowing the yeast to grow on medium lacking uracil and lysine (Figure 5a).
Figure 5.
A yeast assay demonstrating the activity of different HEases on their native targets as well as on targets of therapeutic or biotechnological uses. (a) The yeast assay for HEase activity [Following Chames et al. (48)]. An HEase target site is inserted between truncated Ura3 repeats and between truncated Lys2 repeats. Upon HEase cleavage a recombination event reconstitutes the respective metabolic markers. (b and c) The B. cinerea PRP8 HEase can cleave the human PRPF8 gene. (b) Nucleotide alignment of the B. cinerea PRP8 HEase-target and the homologous sequence from the human PRPF8 gene. The asterisk indicates that the adenine (green A) in the human sequence is the last nucleotide of an intron. The cDNA of human PRPF8 has a thymidine at this position (and is part of a tryptophan codon—W). The target used in our assay has adenine to show cleavage of the genomic sequence. (c) Relative activity of the B. cinerea PRP8 HEase on its native target and on its human target from the PRPF8 gene (log scale). Relative activity is the ratio between the growth rates of strains with or without the HEase-expressing plasmid. (d and e) The Nostoc RNR HEase can cleave its predicted target as well as targets in related cyanobacteria of biotechnological use. (d) Nucleotide alignment of the Nostoc species PCC7120 RNR HEase-target and the homologous sequence from the RNR genes of N. punctiforme and Synechococcus. (e) Relative activity of the Nostoc species PCC7120 RNR HEase on its native target and on sequences from the RNR genes of N. punctiforme and Synechococcus (log scale). Error bars represent 95% confidence intervals based on Monte Carlo simulations. N. punctiforome, Nostoc punctiforome.
The intein encoded in the gene for the splicing factor PRP8 of the fungus B. cinerea has recently been shown to be an active HEase (35). According to the HomeBase paradigm, this enzyme should be able to cleave the PRP8 human homolog PRPF8, mutations in which cause the progressive blindness disorder retinitis pigmentosa (48). The amino acid sequences of the fungal and human genes share a high degree of similarity, while the nucleotide sequences have diverged (Figure 5b). We designed two yeast constructs, one carrying the native B. cinerea target between the truncated repeats of the metabolic markers and the other carrying the predicted human target. Enzyme activity is highest for the natural target but is also very high for the human target [respectively: ∼1800- and ∼50-fold increase in efficacy; Figure 5c and Supplementary Figure S2 showing similar results using the homologous target sequence from the mouse PRPF8 gene].
Botrytis cinerea is a fungus, as is the budding yeast. However, the yeast assay can demonstrate the activity of HEases from diverse phyla. We applied this assay to cyanobacteria to demonstrate this ability. The ribonucleotide reductase (RNR) of Nostoc species PCC7120 encodes an intein with characteristic HEase motifs. We used the yeast assay to evaluate the Nostoc HEase activity on its predicted target. In spite of the large phylogenetic distance between S. cerevisiae and Nostoc, the cyanobacterial enzyme cleaved its predicted target in yeast with extremely high efficiency (Figure 5d and e). We note that cyanobacteria biotechnology is an exponentially growing field with implications in biofuel production (49) and bioremediation (50,51). Many Nostoc strains and species of related cyanobacteria encode vacant RNR genes, lacking the intein and therefore possess an intact HEase target. While the nucleotide sequences diverge between different species, the translation is highly conserved (Figure 5d). We found that the cleavage efficiency of the HEase from Nostoc species PCC7120 of targets from related species of proven biotechnological value, such as N. punctiforme (52) and Synechoccus (49) is as high or higher than its activity on the endogenous target (Figure 5e). The Nostoc RNR HEase exemplifies how the HomeBase paradigm could and should be extended far beyond the scope of gene therapy alone. Finally, we emphasize that both the B. cinerea PRP8 HEase and the Nostoc RNR HEase showed no significant toxicity to the budding yeast (Supplementary Figure S3) while being nearly as potent as the I-SceI golden standard (Supplementary Figure S4). This result demonstrates the exquisite specificity of these enzymes, because even a single un-repaired DSB is strictly lethal in S. cerevisiae (53).
DISCUSSION
Attempts to engineer HEases for gene targeting have so far focused on a small group of enzymes (54), while the plethora and diversity of native HEases have been overlooked. Our HomeBase platform lists for the first time approximately a thousand different HEases alongside their predicted targets. The computational pipeline developed here is a set of tailor-made methods for HEase discovery and characterization that rely on their unique biological and evolutionary properties: the presence of vacant homologs, their setting within introns/inteins, and their predictable tolerance for silent mutations in their target sequences.
Enzyme engineering can now begin by choosing a scaffold HEase whose target is most similar to the sequence at the target locus of choice. We have also developed methods for preliminary validation of HEase activity in a eukaryotic setting. Candidate screening in yeast can help in the detection of degenerated HEases that are naturally common (13,55,56). Notably, evolutionary considerations may sometime only help to approximate the intricacies of enzyme specificity, as can be revealed, for example, by yeast surface display (57) or by cleavage assays with large randomized target libraries (23). When suboptimal specificity is revealed, the yeast selection system can be used for directed evolution of selected HEases (58). However, we have shown here that native HEases could sometimes themselves be used for the gene targeting of disease-associated genes and genes of biotechnological relevance. HEases possess exquisite specificity that has evolved through billions of years. The long target sequences of HEases allow the recognition of unique sites while considerations of sequence conservation allow an approximation of the plasticity in target recognition. In particular, for those HEases that reside in protein-coding genes, we have established that a translated BLAST could be used to find cleavable targets in the human and other genomes. Even when an HEase has more than a single target in a genome of choice, the off-target effect may be confined and predictable based on evolutionary considerations. This predictability is however limited as some tolerance of non-synonymous substitutions can be expected (21,23).
We believe that the safety concerns regarding the use of site-specific endonucleases in gene therapy are well addressed by native HEases. This is all the more true for a subgroup of HEases whose predicted human targets are found in homologs of the microbial HEase-hosting gene. These enzymes are shown to cleave the same locus in humans and in any animal model of choice (Figure 2c and d), thus facilitating pre-clinical efficacy and safety assessments. The Nostoc RNR exemplifies how such enzymes could also be used in the biotechnology industry. In this study we focused on members of the LAGLIADG HEase family, comprising ∼80% of all HEases. Future studies should test the applicability of our conclusions to all HEases as may be implied by previous reports (28,29). Finally, the genes for native HEases are readily available and can be incorporated in therapeutic and other vectors with relative ease and low costs compared to the engineered alternative. We therefore believe that the under-recognized diversity and plasticity of native HEases should become a valuable tool in the fields of gene therapy and genetic engineering.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Israel Science Foundation (to M.K.); the German-Israeli Science Fund (to M.K.); the Israel Cancer Research Fund (to M.K.); the Israel Science Foundation (to T.P.); the National Evolutionary Synthesis Center (NESCent) National Science Foundation #EF-0905606t (to T.P.); the Binational Science Foundation (to U.G.); the McDonnell Research Foundation (to U.G.); fellowship of the Edmond J. Safra Bioinformatics Program (to E.P.); fellowship of the Converging Technologies Program of the Israeli Council for Higher Education (to D.B.). Funding for open access charge: the Israel Cancer Research Fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Annika Bokor for providing us the B. cinerea strain B05.10 and Prof. Sammy Boussiba for providing us with Nostoc (Anabaena) sp. PCC 7120 and members of the Kupiec, Gophna and Pupko lab for discussions and encouragement.
REFERENCES
- 1.Cavazzana-Calvo M, Carlier F, Le Deist F, Morillon E, Taupin P, Gautier D, Radford-Weiss I, Caillat-Zucman S, Neven B, Blanche S, et al. Long-term T-cell reconstitution after hematopoietic stem-cell transplantation in primary T-cell-immunodeficient patients is associated with myeloid chimerism and possibly the primary disease phenotype. Blood. 2007;109:4575–4581. doi: 10.1182/blood-2006-07-029090. [DOI] [PubMed] [Google Scholar]
- 2.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 5.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 7.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl Acad. Sci. USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabaniols JP, Paques F. Robust cell line development using meganucleases. Methods Mol. Biol. 2008;435:31–45. doi: 10.1007/978-1-59745-232-8_3. [DOI] [PubMed] [Google Scholar]
- 9.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One. 2010;5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cathomen T, Schambach A. Zinc-finger nucleases meet iPS cells: zinc positive: tailored genome engineering meets reprogramming. Gene Ther. 2009;17:1–3. doi: 10.1038/gt.2009.151. [DOI] [PubMed] [Google Scholar]
- 12.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 13.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 15.Mills KV, Perler FB. The mechanism of intein-mediated protein splicing: variations on a theme. Protein Pept. Lett. 2005;12:751–755. doi: 10.2174/0929866054864337. [DOI] [PubMed] [Google Scholar]
- 16.Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ZY, He CY, Kay MA. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 18.Gouble A, Smith J, Bruneau S, Perez C, Guyot V, Cabaniols JP, Leduc S, Fiette L, Ave P, Micheau B, et al. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J. Gene Med. 2006;8:616–622. doi: 10.1002/jgm.879. [DOI] [PubMed] [Google Scholar]
- 19.Cornu TI, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 20.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimble FS, Wang J. Substrate recognition and induced DNA distortion by the PI-SceI endonuclease, an enzyme generated by protein splicing. J. Mol. Biol. 1996;263:163–180. doi: 10.1006/jmbi.1996.0567. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa S, Bessho Y, Higashijima K, Shirouzu M, Yokoyama S, Watanabe KI, Ohama T. Adaptation of intronic homing endonuclease for successful horizontal transmission. FEBS J. 2005;272:2487–2496. doi: 10.1111/j.1742-4658.2005.04669.x. [DOI] [PubMed] [Google Scholar]
- 23.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 25.Lucas P, Otis C, Mercier JP, Turmel M, Lemieux C. Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res. 2001;29:960–969. doi: 10.1093/nar/29.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc. Natl Acad. Sci. USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swithers KS, Senejani AG, Fournier GP, Gogarten JP. Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol. Biol. 2009;9:303. doi: 10.1186/1471-2148-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J. Mol. Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Brok-Volchanskaya VS, Kadyrov FA, Sivogrivov DE, Kolosov PM, Sokolov AS, Shlyapnikov MG, Kryukov VM, Granovsky IE. Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res. 2008;36:2094–2105. doi: 10.1093/nar/gkn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allers T, Mevarech M. Archaeal genetics - the third way. Nat. Rev. Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 32.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naor A, Lazary R, Barzel A, Papke RT, Gophna U. In vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon haloferax volcanii. PLoS One. 2011;6:e15833. doi: 10.1371/journal.pone.0015833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iha H, Tsurugi K. Shuttle-vector system for Saccharomyces cerevisiae designed to produce C-terminal-Myc-tagged fusion proteins. Biotechniques. 1998;25:936–938. doi: 10.2144/98256bm01. [DOI] [PubMed] [Google Scholar]
- 35.Bokor AA, van Kan JA, Poulter RT. Sexual mating of Botrytis cinerea illustrates PRP8 intein HEG activity. Fungal Genet. Biol. 2010;47:392–398. doi: 10.1016/j.fgb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Lluisma AO, Karmacharya N, Zarka A, Ben-Dov E, Zaritsky A, Boussiba S. Suitability of Anabaena PCC7120 expressing mosquitocidal toxin genes from Bacillus thuringiensis subsp. israelensis for biotechnological application. Appl. Microbiol. Biotechnol. 2001;57:161–166. doi: 10.1007/s002530100776. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Lu C, Wu QJ, Wang Y, Sun ZT, Deng JC, Zhang Y. GISSD: Group I intron sequence and structure database. Nucleic Acids Res. 2008;36:D31–D37. doi: 10.1093/nar/gkm766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perler FB. InBase: the intein database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan FL, Li X, Lu WG, Li CW, Li JP, Wang Y. The vacuolar ATPase in bone cells: a potential therapeutic target in osteoporosis. Mol. Biol. Rep. 2010;37:3561–3566. doi: 10.1007/s11033-010-0004-7. [DOI] [PubMed] [Google Scholar]
- 40.Niikura K, Nakajima S, Takano M, Yamazaki H. FR177995, a novel vacuolar ATPase inhibitor, exerts not only an inhibitory effect on bone destruction but also anti-immunoinflammatory effects in adjuvant-induced arthritic rats. Bone. 2007;40:888–894. doi: 10.1016/j.bone.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Poulter RT, Goodwin TJ, Butler MI. The nuclear-encoded inteins of fungi. Fungal Genet. Biol. 2007;44:153–179. doi: 10.1016/j.fgb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc. Natl Acad. Sci. USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barzel A, Naor A, Privman E, Kupiec M, Gophna U. Homing endonucleases residing within inteins: evolutionary puzzles awaiting genetic solutions. Biochem. Soc. Trans. 2011;39:169–173. doi: 10.1042/BST0390169. [DOI] [PubMed] [Google Scholar]
- 45.Flohr T, Dai JC, Buttner J, Popanda O, Hagmuller E, Thielmann HW. Detection of mutations in the DNA polymerase delta gene of human sporadic colorectal cancers and colon cancer cell lines. Int. J. Cancer. 1999;80:919–929. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 46.Prieto J, Epinat JC, Redondo P, Ramos E, Padro D, Cedrone F, Montoya G, Paques F, Blanco FJ. Generation and analysis of mesophilic variants of the thermostable archaeal I-DmoI homing endonuclease. J. Biol. Chem. 2008;283:4364–4374. doi: 10.1074/jbc.M706323200. [DOI] [PubMed] [Google Scholar]
- 47.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Gimeno M, Gamundi MJ, Hernan I, Maseras M, Milla E, Ayuso C, Garcia-Sandoval B, Beneyto M, Vilela C, Baiget M, et al. Mutations in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2003;44:2171–2177. doi: 10.1167/iovs.02-0871. [DOI] [PubMed] [Google Scholar]
- 49.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 50.Pandi M, Shashirekha V, Swamy M. Bioabsorption of chromium from retan chrome liquor by cyanobacteria. Microbiol. Res. 2009;164:420–428. doi: 10.1016/j.micres.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Demirel S, Ustun B, Aslim B, Suludere Z. Toxicity and uptake of iron ions by Synechocystis sp. E35 isolated from Kucukcekmece Lagoon, Istanbul. J. Hazard Mater. 2009;171:710–716. doi: 10.1016/j.jhazmat.2009.06.058. [DOI] [PubMed] [Google Scholar]
- 52.Lindberg P, Lindblad P, Cournac L. Gas exchange in the filamentous cyanobacterium Nostoc punctiforme strain ATCC 29133 and Its hydrogenase-deficient mutant strain NHM5. Appl. Environ. Microbiol. 2004;70:2137–2145. doi: 10.1128/AEM.70.4.2137-2145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 55.Koufopanou V, Burt A. Degeneration and domestication of a selfish gene in yeast: molecular evolution versus site-directed mutagenesis. Mol. Biol. Evol. 2005;22:1535–1538. doi: 10.1093/molbev/msi149. [DOI] [PubMed] [Google Scholar]
- 56.Okuda Y, Sasaki D, Nogami S, Kaneko Y, Ohya Y, Anraku Y. Occurrence, horizontal transfer and degeneration of VDE intein family in Saccharomycete yeasts. Yeast. 2003;20:563–573. doi: 10.1002/yea.984. [DOI] [PubMed] [Google Scholar]
- 57.Jarjour J, West-Foyle H, Certo MT, Hubert CG, Doyle L, Getz MM, Stoddard BL, Scharenberg AM. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Z, Zhao H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005;33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.