Abstract
Deficiency in Artemis is associated with lack of V(D)J recombination, sensitivity to radiation and radiomimetic drugs, and failure to repair a subset of DNA double-strand breaks (DSBs). Artemis harbors an endonuclease activity that trims both 5′- and 3′-ends of DSBs. To examine whether endonucleolytic trimming of terminally blocked DSBs by Artemis is a biologically relevant function, Artemis-deficient fibroblasts were stably complemented with either wild-type Artemis or an endonuclease-deficient D165N mutant. Wild-type Artemis completely restored resistance to γ-rays, bleomycin and neocarzinostatin, and also restored DSB-repair proficiency in G0/G1 phase as measured by pulsed-field gel electrophoresis and repair focus resolution. In contrast, cells expressing the D165N mutant, even at very high levels, remained as chemo/radiosensitive and repair deficient as the parental cells, as evidenced by persistent γ-H2AX, 53BP1 and Mre11 foci that slowly increased in size and ultimately became juxtaposed with promyelocytic leukemia protein nuclear bodies. In normal fibroblasts, overexpression of wild-type Artemis increased radioresistance, while D165N overexpression conferred partial repair deficiency following high-dose radiation. Restoration of chemo/radioresistance by wild-type, but not D165N Artemis suggests that the lack of endonucleolytic trimming of DNA ends is the principal cause of sensitivity to double-strand cleaving agents in Artemis-deficient cells.
INTRODUCTION
Artemis nuclease is a phosphoprotein that has been shown to play a role in hairpin opening in V(D)J recombination (1,2) and more recently in the regulation of G2/M, and S phase cell cycle checkpoints (3,4). Artemis is also required for the repair of a subset of chemo/radiotherapy-induced DNA double-strand breaks (DSBs) that are rejoined very slowly in normal cells. These DSBs may largely overlap the fraction of DSBs whose repair requires ATM- and 53BP1-dependent phosphorylation of the heterochromatin maintenance protein KAP-1 (5–7). Previous in vitro studies with oligomeric substrates have shown that Artemis nuclease activity is DNA-PK-dependent at DNA ends and that this activity can remove 3′-PG blocking lesions commonly found at DSB termini (8,9). Thus, it is reasonable to propose that lack of such endonucleolytic trimming accounts for both the repair deficiency and the increased cytotoxicity of radiation and radiomimetic agents toward Artemis-deficient cells. Yet, the resulting repair deficiency is subtle, affecting only 10–20% of DSBs, raising the question of whether cell cycle or other regulatory functions of Artemis might be equally or more important determinants of chemo/radiosensitivity.
Artemis belongs to SNM1 family of nucleases and possesses metallo-β-lactamase and β-CASP domains at its amino terminus. Mutation of an aspartic acid residue to asparagine (D165N) selectively abrogates the endonucleolytic function of Artemis without affecting its exonuclease activity or phosphorylation status (8,10). Though D165 is not found in sequences of the available metallo-β-lactamase crystal structures, the abrogation of endonucleolytic activity by this mutation suggests that it may be located in the active site of Artemis (10).
To investigate whether the endonucleolytic activity of Artemis functions in chemo/radioresistance, patient-derived CJ179 cells defective for Artemis were complemented with lentiviral vectors expressing wild-type or D165N Artemis. The D165N mutation eliminates Artemis-mediated endonucleolytic processing of 3′-PG DSB ends in vitro (8). To establish the role of Artemis nuclease activity in DNA repair and cellular survival after DNA damage, clonogenic and DSB-repair assays were carried out with these cells, following treatment with radiation or radiomimetic drugs. Earlier studies (5,6) investigating complementation of Artemis defect by exogenous protein expression were carried out with transiently expressing cell lines due to difficulties in expressing Artemis in cells (11). In contrast to these studies, we have been successful in stably complementing Artemis-deficient fibroblasts with wild-type or D165N mutant Artemis, allowing us to explore the effect of such expression on the critical endpoint of survival following radiation or genotoxic chemical treatment. In the current study, low-level Artemis expression was found to be sufficient for complementing the survival/repair defect of Artemis-deficient cells while even high levels of the D165N Artemis failed to do so. Moreover, the residual DNA DSBs that remained unrejoined due to Artemis deficiency juxtaposed with promyelocytic leukemia (PML) nuclear bodies. Taken together, these results indicate that the endonucleolytic end-processing activity of Artemis is essential for promoting DSB repair and cell survival.
MATERIALS AND METHODS
Cell lines and complementation
Normal 48BR and patient-derived Artemis-deficient CJ179 hTERT-immortalized human fibroblasts originally from Dr Penny Jeggo, were obtained from Dr Lynn Harrison, Louisiana State University, and were cultured in minimum essential medium (MEM-α) supplemented with 10% fetal bovine serum and antibiotics (GIBCO). Cells were constantly maintained at 37°C in 5% CO2 and a humidified atmosphere. Normal Epstein–Barr virus-transformed human lymphoblastoid cells (patient 1646) were from Dr James Lupski, Baylor College of Medicine (12).
Lentiviral constructs were prepared in the 693-2 lentiviral backbone harboring the hygromycin resistance gene (13) carrying either wild-type or D165N mutant Artemis cDNA fused to a c-myc epitope tag on the carboxyl terminus. Artemis wild-type, D165N and empty lentivector DNAs were transfected along with packaging plasmids, pLP1, pLP2 and pLP-VSVG into 293FT cells with Lipofectamine 2000 (Invitrogen). Medium containing packaged lentivirus was collected 48 h post transfection, and centrifuged 5 min at 1200 rpm in a clinical centrifuge at room temperature. The supernatant was filtered through 0.45 µm filters (Novagen), then ultracentrifuged at 20 000 rpm for 2.5 h at 4°C in a SW28 rotor (Beckman). Resulting viral pellets were resuspended with 200 µl of Hanks balanced salt solution and stored in 50 µl aliquots at −80°C. The 48BR and CJ179 cell lines were seeded at ∼75% confluence in 25 cm2 dishes and incubated for 24 h, then infected with 5 µl of concentrated lentivirus with 4 µg/ml polybrene in 1 ml medium without serum for 8 h, then fed with fresh medium and incubated for 48 h. Cells were then selected in medium containing 100 µg/ml hygromycin for 7 days. Cells were expanded under selection to produce cryogenic stocks and 2.5 × 106 cell aliquots harvested and stored at −80°C as cell pellets for western analysis.
Selection of clones
From each derivative cell line, approximately 20 clones were isolated. Total RNA was extracted from each clone using Qiagen RNeasy minikit and converted to cDNA using high-capacity RNA to cDNA kit (Applied Biosystems) following the protocols provided by the manufacturer. Amplification of cDNA was performed on an ABI 7900HT Real time q-PCR instrument using SYBR green detection (Applied biosystems). Relative Artemis levels were determined after normalizing to β-actin levels using SDS 2.2.2 software. The primers used were 5′-ACAGAGCCTCGCCTTTGCCG-3′, 5′-CACCATCACGCCCTGGTGCC-3′ for β-actin and 5′-AGTACGGAGCCAAAGTATAAACCACT-3′, 5′-TCCGGGTATGGAACTTTGTGC-3′ for Artemis cDNA amplification (Synthesized by IDT). Highest and lowest Artemis expressing clones among those screened were identified and the protein levels were confirmed by western as described below. The selected clones were further used in survival/repair assays.
Western blots
Direct western blotting of cell lysates was attempted with a variety of antibodies to Artemis but was found to be insufficiently sensitive and specific for Artemis detection; therefore, an immunoprecipitation (IP) step was added as previously described (14). Briefly, ∼2.5 million cells were harvested and sonicated on ice with three 30-s pulses in 750 µl of TBST [25 mM Tris–HCL pH 7.4, 130 mM NaCl, 3 mM KCl, 0.1% (v/v) Tween-20] supplemented with 1 μM leupeptin, 1 μM pepstatin A and 1 μM aprotinin protease inhibitors.
Lysates were cleared by centrifugation at 14 000 rpm for 15 min at 4°C and incubated with equilibrated protein A/G beads (Protein A/G UltraLink Resin, Thermo Scientific) for 1 h, then the extract was transferred to a new tube. Extracts were incubated with 1 µl of SCIDA1024 rabbit antiserum (14) for 1 h at 4°C, then 20 µl of equilibrated A/G beads were added and tumbled overnight at 4°C. The A/G beads were pelleted in a microfuge at 5000 rpm for 30 sec at 4°C and washed three times with 500 µl extract buffer. Fifty micro-liter Laemmli buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.004% bromphenol blue, 0.125 M Tris–HCl pH 6.8) was added to the washed beads and heated for 10 min at 98°C and centrifuged at 14 000 rpm for 15 min. Approximately one million cell equivalents (20 µl) was loaded onto 8% SDS–PAGE gels and resolved at 150 V and transferred to a nitrocellulose membrane with 100 V for 90 min in methanol transfer buffer, then incubated in TBST, 10% (w/v) powdered milk (blocking buffer), 1 h at 22°C, with rocking. Membranes were probed with chicken IgY anti-human Artemis antibody (Abcam, ab14289) at a 1:2000 dilution in 8 ml blocking buffer overnight at 4°C with tumbling, then washed in TBST 10 min × 3, and incubated in peroxidase-conjugated donkey anti-chicken IgY (H+L) (Gallus Immunotech Inc.,) at 1:5000 dilution in 8-ml blocking buffer, 1 h, RT, tumbling then washed in TBST 10 min × 3 and developed with ECL plus (GE Healthcare).
Cell survival assays
Confluent cells were further maintained in 0.5% serum for 5 days. The cells were then irradiated [MDS Nordion Gammacell 40 research irradiator (ON, Canada), with a 137Cs source delivering a dose rate of 1.05 Gy/min] or treated with either bleomycin or neocarzinostatin (NCS) for 4 h. Following treatment, cells were washed with PBS and 500–20 000 cells were seeded into 10-cm dishes in fresh medium. Cells were then incubated for 12 days before they were fixed with methanol (100%) and stained with 1% crystal violet, rinsed with water and air-dried. Visible colonies were counted manually.
Immunofluorescence
Cells were grown to confluence on tissue culture-treated 4-well chamber slides (Nunc lab tek) and further maintained at 0.5% serum for 24 h. Cells were then either irradiated (2 or 4 Gy) or treated with NCS (6 nM) for 1 h. After treatment, cells were fixed with 3% paraformaldehyde and permeabilized with 0.5% Triton X-100/PBS. Blocking was done using Casein blocker (Bio-Rad). Primary antibodies used for immunostaining were anti-γ-H2AX (Upstate), anti-53BP1 (gift of Dr David Gewirtz, Virginia Commonwealth University, originally from Dr Thanos Halazonetis, University of Geneva) at 1:500 dilution, anti-PML (PG-M3, Santacruz biotech) at 1:200 dilution, anti-γ-H2AX rabbit polyclonal (Novus biologicals) at 1:400 dilution and anti-hMre11 rabbit polyclonal (Novus biologicals) 1:200 dilution. The secondary antibodies were Alexa Fluor 488/568 goat anti-mouse/anti-rabbit (Invitrogen Molecular Probes) at 1:500 dilutions. Slides were mounted with Vectashield mounting medium containing DAPI (4′,6 diamidino-2-phenylindole). Images were captured using Olympus fluoview 500 confocal microscope, using a 430nm diode laser with a 605nm band pass filter, a 510nm laser with a 530nm band pass filter and a 660-nm laser with 605-nm band pass filter. Foci from approximately 100 cells were scored for each time point in 2–3 independent experiments for each cell line. Focus diameter for the five largest foci in each cell was measured in one direction parallel to the equatorial plane of the image field.
Fraction of activity released DSB-repair assay
Pulsed-field gel electrophoresis (PFGE) was used to quantify DSB repair as described previously (15) with minor variations. Briefly, subconfluent cells were cultured on a 150cm dish and labeled for 24 h with 0.2 µCi/ml [methyl-3H]thymidine (20 Ci/mmol, Perkin-Elmer). Confluence-arrested cells were serum starved (0.5%) for 24 h prior to irradiation. After irradiation samples were incubated for noted repair times and then trypsinized and resuspended in L buffer (0.1 M EDTA, 0.01 M Tris–Cl, 0.02 M NaCl) at a concentration of 2 × 107 cells/ml. Of this suspension, 250 µl was mixed with 250 µl of 2% low-melting agarose (Nusieve GTG) in a 15ml conical tube maintained at 45°C. About 60 µl of cell agarose mixture was transferred to a sample CHEF disposable plug mold (Bio-Rad) and allowed to solidify at 4°C. The plugs were removed and incubated in the digestion mixture (0.1 M EDTA, 0.01 M Tris–Cl, 0.02 M NaCl, 1% w/v Sarkosyl, 0.1 mg/ml proteinase K). The plugs were incubated for 24 h at 50°C with a change of digestion mixture after 3 h. They were then washed with TE (Tris–EDTA) over a period of 3 h. The plugs were further incubated with 40 µg/ml PMSF for 30 min at 50°C followed by three washes over a period of 3 h. The plugs were inserted into the wells of a 1% agarose gel (Bioline, DNA type grade) and run in 0.5 × TBE for 65 h at 1.5 V/cm with switch times varying between 60 and 3600 s. The temperature was maintained at 14°C throughout the run. Each lane of the plug was cut into five equal slices and the fraction of radioactivity released fraction of activity released (FAR) from the plugs and remaining in the plugs was measured by liquid scintillation.
RESULTS
D165N mutant Artemis fails to rescue chemo/radiosensitivity
Previous in vitro studies have shown that in the presence of DNA-PK, Artemis has endonucleolytic activity toward both the 3′ and 5′ termini of DNA ends (2,8), which may participate in the slow processing of 3′-PG-terminated DSBs (9). To investigate the biological relevance of Artemis endonucleolytic activity, wild-type and D165N mutant Artemis were expressed in patient-derived CJ179 cells, as well as in normal 48BR cells, using lentiviral vectors. In the absence of recombinant DNA vectors, the CJ179 cells are Artemis-null and fail to express any Artemis transcript (5).
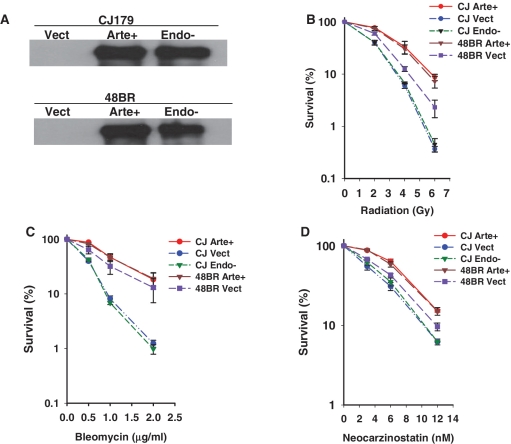
Artemis expression levels were first evaluated by western blots of Artemis immunoprecipitated from extracts of the complemented and mock-complemented lines (Figure 1A). In cells harboring Artemis-encoding viral constructs, levels of wild-type and D165N Artemis were comparable, and were much greater than the level of native Artemis in normal fibroblasts, which was below the level of detection. However, the presence of an Artemis transcript in normal 48BR cells was verified by gel electrophoresis following real-time q-PCR, which showed a product of the expected size that was not evident in the Artemis-deficient CJ179 cells (Supplementary Figure 1).
Figure 1.
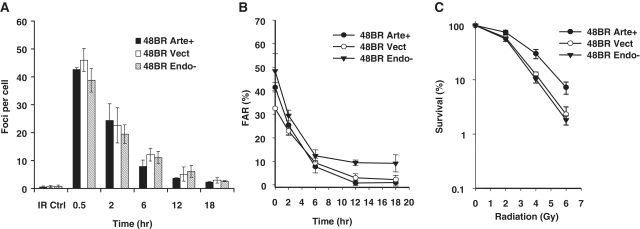
Chemo/radiosensitivity of Artemis deficient and D165N Artemis mutant fibroblasts. (A) Expression levels of wild-type and D165N mutant Artemis in complemented Artemis-deficient CJ179 cells (CJ Arte+, CJ Endo−), and in 48BR normal fibroblasts (48BR Arte+, 48BR Endo−). CJ Vect and 48BR Vect are the respective empty-vector-infected cell lines. Artemis was immunoprecipitated using rabbit polyclonal anti-hArtemis antibody and immunoblotted using chicken polyclonal IgY anti-hArtemis (Abcam). Endogenous Artemis in 48BR cells is below the level of detection. (B–D) Clonogenic survival assays were performed on confluence-arrested serum-starved cells treated with radiation (B), bleomycin (C) or NCS (D). Error bars show SEM from at least three independent experiments.
Radiation, bleomycin and NCS are known to induce free radical-mediated DSBs of diverse structure, many of which bear 3′-PG termini (16–18). The toxicity of these agents was therefore evaluated in Artemis-deficient CJ179 cells carrying integrated viral constructs expressing wild-type Artemis, D165N mutant Artemis or no protein. The Artemis-deficient CJ179 fibroblasts showed significant hypersensitivity to all three agents (Figure 1B–D), as reported previously for other, unrelated Artemis-deficient cell lines (9). Complementation with wild-type Artemis but not D165N mutant Artemis rescued this sensitivity. Furthermore, this overexpression of wild-type Artemis increased the chemo/radioresistance of normal 48BR cells, as compared to that of 48BR cells carrying the empty vector. Taken together, these results suggest a direct role of Artemis endonucleolytic activity in survival following radiation or radiomimetic drug treatment.
Endonucleolytic activity of Artemis is essential for DNA DSB rejoining in cells
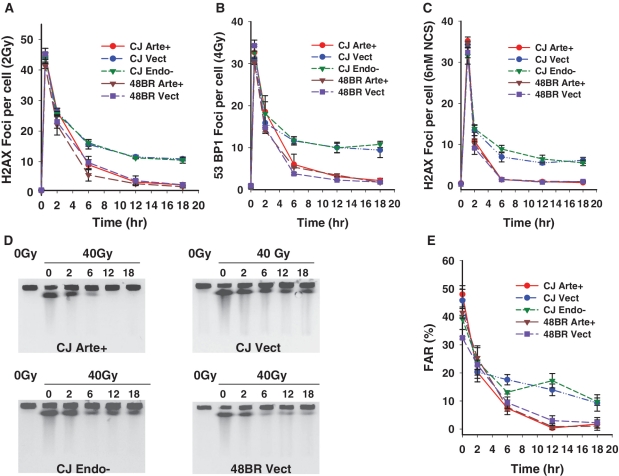
To more directly assess the role of Artemis’ endonuclease activity in DSB repair, γ-H2AX and 53BP1 foci were quantified as surrogate markers for residual DNA DSBs in Artemis-deficient/proficient cell lines following irradiation or NCS treatment. These assays were performed using non-replicating G0/G1 cells (Supplementary Figure S2) to avoid spontaneous focus formation at stalled replication forks. The formation and loss of γ-H2AX and 53BP1 foci was similar in all cell lines at 30 min and 2 h post-irradiation. However, at 6–18 h, a significant fraction of foci were seen to persist in Artemis-deficient CJ179 fibroblasts, while nearly all of the foci resolved in normal 48BR cells (Figure 2A and B and Supplementary Figure S3). Stable complementation of CJ179 with wild-type Artemis corrected this defect, resulting in wild-type focus levels, while the D165N mutant Artemis completely failed to rescue the DSB repair defect, indicating that resolution of γ-H2AX and 53BP1 foci following radiation requires Artemis endonucleolytic activity. Essentially identical complementation results were obtained in cells treated with NCS (Figure 2C). Although the kinetics of γ-H2AX and 53BP1 foci were nearly identical, a higher radiation dose was required to induce a comparable number of 53BP1 foci. Staining with a mouse monoclonal antibody to γ-H2AX (data not shown) confirmed that some γ-H2AX foci did not contain detectable 53BP1, as has been previously reported (19).
Figure 2.
Dependence of cellular DSB rejoining on Artemis endonucleolytic activity. (A) γ-H2AX foci were scored in confluence-arrested serum-starved cells following 2 Gy γ irradiation, and the results plotted as the number of foci/cell. (B) 53BP1 foci were scored following 4 Gy γ irradiation. (C) γ-H2AX foci were scored as in (A) following treatment with 6 nM NCS for 1 h. Error bars represent the SEM from three independent experiments except for (B), which shows data from two experiments. (D) PFGE was performed on DNA from confluence-arrested serum-starved normal (48BR) or Artemis-deficient (CJ179) cells that were complemented with wild-type or D165N mutant Artemis. Cells were irradiated and then incubated for the indicated times (in hour) to allow repair before analysis by PFGE. (E) FAR values were plotted after subtraction of the FAR value for unirradiated cells. Fluoromicrographs of a typical repair focus experiment are shown in Supplementary Figure S1. PFGE experiments with NCS were precluded by the large amount of drug required.
The above results were confirmed by a PFGE assay that measures DSBs directly but requires use of higher doses of radiation (40 Gy, Figure 2D and E). Consistent with the repair focus studies above, 10–20% of the DSBs remained unrejoined in Artemis deficient and D165N mutant-complemented CJ179 cells. Taken together, these data show that repair of this subset of DNA DSBs requires Artemis endonucleolytic activity at both low and high doses.
Low-level Artemis expression is sufficient to restore radiosurvival
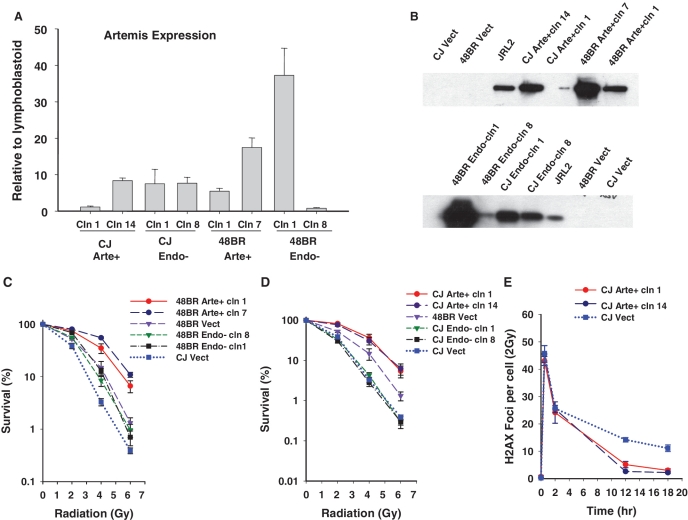
A possible confounding factor in interpreting these studies, as well as other studies using transient expression (6), is the elevated level of Artemis transgene expression, which is much higher than endogenous levels in normal fibroblasts. To address this concern, clonal isolates having various levels of Artemis expression were derived from cultures of 48BR and CJ179 cells transduced with vectors encoding wild-type or D165N Artemis.
Relative Artemis expression levels were first evaluated by real-time q-PCR and then confirmed by IP/western. All transgene-expressing clones produced detectable levels of Artemis protein, which in all cases correlated with Artemis mRNA levels. While the Artemis levels in all clones were still higher than the undetectable level in 48BR cells, they were comparable to and in some cases lower than the endogenous level in normal lymphoblastoid cells (Figure 3A and B). Moreover, the CJ179 clone expressing the lowest level of wild-type Artemis was as radioresistant (Figure 3D) and as repair-proficient (Figure 3E) as the highest expressing clone (showing 7-fold higher expression by q-PCR), and both were more radioresistant than normal 48BR Vect cells. In contrast, even high levels of Artemis D165N expression (∼8-fold higher than the level in CJ Arte+ clone 1 by q-PCR; Figure 3A) conferred no detectable radioresistance (Figure 3D).
Figure 3.
Effect of wild-type/D165N Artemis expression levels on radiosurvival and repair. (A) Artemis cDNA was amplified using real time q-PCR and the relative quantities of Artemis expression for indicated clones (cln) using normal lymphoblastoid cells as calibrator sample and β-actin as endogenous control are plotted. Error bars indicate SEM for 3 independent experiments. All 20 isolated CJ Endo− clones had similar levels of expression. (B) Artemis protein level for selected clones was verified by IP/Western blot. JRL2 = normal lymphoblastoid cells. (C and D) Clonogenic survival assays were performed on confluence-arrested serum-starved clones following exposure to radiation. The data points for CJ Vect and 48BR Vect are same for (C) and (D). Error bars represent SEM for three independent experiments. (E) γ-H2AX foci were scored in confluence-arrested serum-starved high/low wild-type Artemis expressing CJ179 cells. Error bars represent standard deviation for two independent experiments.
Overall, these results show that a relatively modest amount of wild-type Artemis is sufficient to correct the repair defect in Artemis-deficient cells and increase radioresistance, and suggest that the restoration of radioresistance is dependent on endonuclease activity and is not an artifact of overexpression.
Persistent radiation-induced foci in Artemis-deficient cells grow larger with time and juxtapose with PML-nuclear bodies
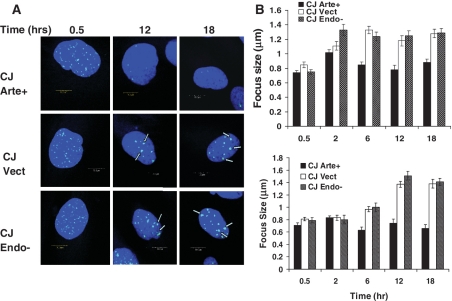
In addition to a larger number of persistent foci, cells lacking functional Artemis showed a significant time-dependent increase in the average diameter of the residual foci, from ∼0.8 µm at 30 min to 1.4 µm at 12–18 h (Figure 4). This increase was seen for both 53BP1 and γ-H2AX foci, and for both Artemis-deficient and Artemis D165N-complemented cells. These results are consistent with hypersensitivity in these cells being due to defective repair processes. Moreover, these data suggest that the unrepaired DSBs promote persistent ATM activation and continued accumulation of repair factors such as Mre11 in the vicinity of the break site (Supplementary Figure S4).
Figure 4.
Growth of persistent radiation-induced foci in Artemis-deficient cell lines. (A) Fluorescence microscopy of Artemis-deficient and D165N mutant-complemented CJ179 fibroblasts, showing increase in the size of γ-H2AX foci that persist 12–18 h post-irradiation. (B) Average size of 53BP1 (upper panel, dose of 4 Gy) or γ-H2AX (lower panel, dose of 2 Gy) foci. Focus diameter on fluoromicrographs (A) was measured in one direction parallel to the equatorial plane of the image field. The error bars represent SEM for 35 foci. The arrows are pointing toward large foci.
PML protein is a tumor suppressor that along with Daxx, SP100 and CBP has been identified as the main constituent protein within sub-nuclear compartments also referred to as nuclear bodies (20,21). PML is phosphorylated by ATM (22) and PML-NBs have also been shown to co-localize with Mre11 and p53 at the sites of radiation-induced foci specifically at later time points (21).
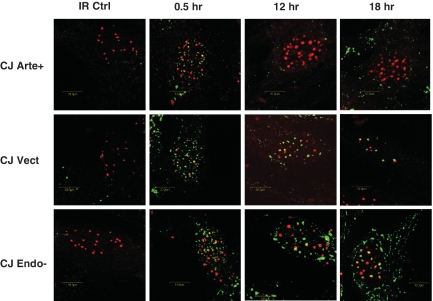
To investigate whether PML NBs and associated proteins may be recruited to the residual DNA DSBs, PML and γ-H2AX interaction was analyzed in cells by double immunostaining and confocal microscopy. As shown in Figure 5 and Supplemental Figure S5, a subset PML NBs were found to be juxtaposed to the persisting DNA DSBs in Artemis-deficient cell lines 12–18 h after irradiation. These results indicate that such positioning occurs even at relatively low levels of initial DNA damage, and suggest that the determining factor for co-localization is the persistence of a significant number of unresolved DSBs.
Figure 5.
PML and γ-H2AX partial co-localization and juxtapositioning after 2 Gy γ-radiation exposure. Double PML/γ-H2AX staining was performed after paraformaldehyde fixation. Confocal analysis of representative cells is shown. Green fluorescence: γ-H2AX (polyclonal anti γ-H2AX, Novus biologicals); red fluorescence: PML (monoclonal anti-PML, Santa Cruz, PGM-3). IR Ctrl = no irradiation.
Overexpression of D165N mutant Artemis renders normal cells repair deficient
If, as proposed, the primary role of Artemis in chemo/radioresistance is endonucleolytic trimming of DSB ends, then overexpression of mutant Artemis might be expected to displace the normal enzyme and prevent such trimming. At 2 Gy, or at equally toxic concentrations of bleomycin or NCS, no such dominant-negative effect was seen, as D165N-overexpressing 48BR cells were as chemo/radioresistant as empty-vector controls (Figure 6C and Supplementary Figure S6), and were fully competent in resolution of repair foci (Figure 6A). However, in FAR assays, overexpression of D165N mutant Artemis conferred upon normal fibroblasts a slight repair defect at high doses of γ-radiation (40 Gy, Figure 6B). Conversely, overexpression of wild-type Artemis rendered normal cells more radioresistant (Figure 6C), but did not produce a detectable change in repair, with all measurable DSBs being rejoined within 12 h according to both focus formation (Figure 6A) and FAR assays (Figure 6B).
Figure 6.
Overexpression of D165N mutant Artemis confers high-dose DSB-repair deficiency. (A) γ-H2AX focus assay was performed after 2 Gy γ irradiation. (B) PFGE analysis of normal fibroblasts overexpressing wild-type or D165N mutant Artemis after exposure to 40 Gy γ-rays. The panel displays FAR versus repair time (for 48BR Vect versus 48BR Arte+ P > 0.3 at all times; for Vect versus Endo− P = 0.04 at 12 h and P = 0.16 at 18 h, by t-test). (C) Clonogenic survival was determined following treatment with radiation (for Vect versus Arte+ P = 0.04 at 4 Gy and P = 0.07 at 6 Gy; for Vect versus Endo− P > 0.3 at all doses). The data for 48BR Vect and 48BR Arte+ are same as shown in Figures 1 and 2, and are shown here separately for the sake of clarity and comparison with 48BR Endo−. Error bars represent the SEM from three independent experiments except for (A), which shows data from two experiments.
DISCUSSION
Artemis deficiency has pleiotropic effects in human cells following exposure to DNA damaging agents, including defects in regulation of cell cycle checkpoints (3,4) and in apoptotic DNA fragmentation (23). However, other studies suggest that Artemis is epistatic with ATM in promoting radiosurvival and DSB repair, even in growth-arrested cells that should not be subject to cell cycle effects (5). Specifically, in ATM- or Artemis-deficient cells a small fraction of DSBs (10–20%) remains unrejoined, even after several days (5,14).
Experiments using defined substrates and purified enzymes have shown that in the presence of DNA-PK, Artemis gains an endonucleolytic activity toward DNA ends that is inherently capable of resolving DSBs bearing terminal blocking groups such as 3′-PGs (8–10,24,25). PFGE as well as focus-formation assays in noncycling contact-arrested fibroblasts (Figure 2) show that wild-type but not endonuclease-deficient Artemis completely restores DSB-repair proficiency, in agreement with recent observations in G2 cells where Artemis was expressed transiently (6). Taken together, these results strongly suggest that the primary function of Artemis in DSB repair is the endonucleolytic processing of DNA ends.
More importantly, survival assays with stably complemented Artemis cell lines (Figure 1) show that only endonuclease-proficient Artemis is able to restore chemo/radioresistance. These data suggest that lack of endonucleolytic activity, and by inference DSB end processing, is the principal cause of chemo/radiosensitivity in Artemis-deficient fibroblasts. Although an effect of the D165N mutation on recruitment to damaged DNA (26), or on other unknown functions of Artemis, cannot be strictly excluded, the mutant enzyme retains its 5′→3′ exonuclease activity (8), and is phosphorylated by DNA-PK as efficiently as the wild-type protein (10). These data suggest that D165N Artemis has intact secondary structure and retains proficiency in assembling with DNA-PK at DNA ends. Recently, it was shown that most (but not all) of the exonuclease activity in His6-affinity-purified Artemis could be eliminated by ion-exchange chromatography, suggesting that the exonuclease might be a contaminant (27). However, other studies showed that extensively purified Artemis protein retained exonuclease activity (28), and that antibodies raised against Artemis protein gel-purified from Escherichia coli inactivate the exonuclease, suggesting that this exonuclease activity is intrinsic (8).
The finding that endonuclease-deficient Artemis, even when overexpressed, has no effect at all on chemo/radiosurvival (Figure 1), combined with evidence that Artemis-deficient fibroblasts retain functional G1 and G2 checkpoints (14,29), would also appear to exclude a checkpoint defect as a major contributor to the chemo/radiosensitivity of Artemis-deficient fibroblasts. Although checkpoint deficiencies have been reported for tumor cells in which Artemis expression has been knocked down with siRNA (3,4), there is no evidence that these deficiencies account for radiosensitivity.
Previous studies have shown that a small number of DSBs remain unrejoined in irradiated cells lacking Artemis (5,14), but the exact nature of these repair-resistant breaks remains incompletely defined. The three DNA-damaging agents used here, (radiation, NCS and bleomycin) all generate 3′-PG-terminated DNA DSBs, but in different contexts. Bleomycin treatment gives rise to DSBs having either blunt ends or single-base 5′-overhangs, with 5′-phosphate and 3′-PG termini at both ends of the break. NCS-induced DSBs have at one end a 5′-phosphate and a 3′-phosphate on a 2-base 3′ overhang. The opposite end has a 5′-aldehyde and either a 3′-PG (∼20%) or a 3′-phosphate (∼80%) on a one-base 3′ overhang (17,18). Radiation induces 3′-PG-terminated DNA DSBs in various contexts, presumably with random stagger between breaks in opposite strands and with the fraction of 3′-PG ends estimated at anywhere from 10% to 50% (16,30). Because the endonucleolytic activities of Artemis/DNA-PK do not require a specific structure at either the 5′ or the 3′ ends and can bypass terminal moieties (8), these activities could serve to remove chemically modified termini such as 3′-PGs and 5′-aldehydes. It is highly unlikely that Artemis’ exonucleolytic activity can resolve such blocked ends, because it acts only at 5′ termini and requires a 5′-phosphate, and also because it is suppressed by DNA-PK (8,28), which is expected to be bound to all DNA ends during DSB repair in G1. Thus, deficiency in endonuclease activity of the D165N mutant Artemis likely causes a failure to process a subset of DSB termini induced by NCS, bleomycin and radiation, thus conferring the same chemo/radiosensitivity as complete lack of Artemis.
However, even in Artemis-deficient and Artemis-mutant cells, at least 80% of radiation- and NCS-induced DSBs are quickly repaired (Figure 2), raising the question of why a small fraction of breaks seems to require endonucleolytic trimming by Artemis while the majority does not. At least for NCS-induced DSBs, the repair-resistant, Artemis-requiring fraction cannot be a subset of complex DSBs with accompanying base damage, as neither NCS nor bleomycin produces such lesions (17,18). It is also unlikely that there is a one-to-one correspondence between 3′-PG DSBs and DSBs requiring Artemis for repair, as in that case Artemis-deficient cells should be much more sensitive to bleomycin, which forms DSBs that have almost exclusively 3′-PG termini at both ends, than to any of the other agents. If the unrepaired DSBs are those in heterochromatin, as has been proposed for DSBs that require ATM for repair (31), then there must be alternative enzyme(s) [e.g. TDP1; (12,32)] that are biochemically competent to resolve the damaged ends, but are specifically excluded from heterochromatic DSBs. Alternatively, the repair focus-associated protein complexes that are recruited to unrepaired DSBs may effectively shield the breaks from most enzymatic processing, so that any breaks which fail to repair within 1–2 h (including those in heterochromatin) can only be processed by Artemis, due to its close association with DNA-PK (2,24). A recent study of repair joints from a site-specific HO endonuclease-induced DSB in mouse mammary cells has raised yet another alternative: that in heterochromatin Artemis may excise a whole DSB-containing nucleosome, exposing internucleosomal DNA at both ends of the break that can then be more easily joined (33). A final possibility is that a fraction of the initial DSBs may be processed into derivative structures that only Artemis can resolve, for example, hairpins resulting from single-strand resection at inverted repeat sequences, as suggested previously (9).
In both Artemis-deficient and Artemis endonuclease-mutant cell lines, a time-dependent increase in the size of residual γ-H2AX and 53BP1 foci was observed 6–18 h after irradiation (Figure 4). A similar slow increase in focus size has been previously reported for cells exposed to high linear energy transfer radiation (34), which induces complex DSBs that are relatively resistant to repair. One study that followed foci along α-particle tracks in 3D, showed that these foci often coalesce into a few larger foci (35). Exposure to high salt immediately after γirradiation can also promote increased focus size, an effect attributed to inhibition of repair, due to chromatin compaction (36). Thus, in several contexts, persistent unrejoined DSBs appear to promote further H2AX phosphorylation and further aggregation of repair factors, suggesting continued ATM/DNA-PK activation (37) and a gradual spreading of decondensation over larger areas of chromatin (38). Moreover, these persistent breaks were seen to juxtapose with PML-NBs (Figure 5). PML is a known tumor suppressor (22,39) and is a key constituent of PML-NBs along with Daxx, SP100 and CBP. PML−/− cells and mice have been shown to be resistant to lethal effects of γ-radiation. Cds1/Chk2-mediated phosphorylation of PML triggers apoptosis (39) probably through recruitment of Daxx to nuclear bodies (40). PML is phosphorylated by several DNA damage activated kinases such as ATM and ATR (22) and associates with hMre11 and p53 at 12–24 h following irradiation (21). Our results that PML NBs partially juxtapose with late repair foci in Artemis-deficient cells suggest that these foci represent highly persistent unrepaired DSBs that continue to recruit additional DNA-repair machinery for their resolution. Repair-focus expansion may also reflect a search for additional DNA end(s) that can be resolved into a ligatable substrate. However, if repair fails, then the constituent proapoptotic proteins of PML bodies may drive cells toward death.
Our data also show that an overexpression of wild-type Artemis in normal cells adds to the survival advantage of these cells (Figure 6). Although no effects on repair (PFGE or foci) were detected, it is possible that in the presence of excess Artemis, some breaks which can be repaired by multiple redundant pathways may be channeled through an Artemis-dependent pathway. If that pathway were less error-prone than the alternative(s), then the higher fidelity might lead to increased survival without necessarily affecting measurements of DSB rejoining. However, this increase in survival is limited such that once a critical level of Artemis expression is reached in cells, additional Artemis confers no added survival advantage (Figure 3). This result suggests that in the Artemis transgene-expressing cells, the concentration of Artemis at DSBs is no longer a limiting factor in repair.
Conversely, overexpression of D165N mutant Artemis in normal cells made them slightly repair deficient as judged by PFGE (Figure 6), suggesting that the mutant Artemis can effectively compete with endogenous Artemis for DNA binding. The defect in repair appears to be expressed only when the repair system is saturated by very high levels of DNA damage. Although there was a slight trend toward greater radiosensitivity when very high levels of mutant Artemis were expressed in normal cells, it was not statistically significant (Figure 3C). Nevertheless, the dominant-negative effect on repair as well as the increase in radioresistance with overexpression of Artemis is most consistent with Artemis functioning directly in the repair process.
Taken together, the results suggest that a fraction of DSBs with chemically modified termini strictly require Artemis endonuclease activity for repair in G1. The complete failure of endonuclease-deficient Artemis to improve survival suggests that lack of end processing by this endonuclease can fully account for the chemo/radiosensitivity conferred by Artemis deficiency.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute (grants CA40615 to L.F.P. and CA104660 to S.M.Y.); US Department of Energy Office of Science (under contract number DE-AC02-05CH11231). Funding for open access charge: National Institutes of Health Grant (CA40615).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Thanos Halazonetis and David Gewirtz for 53BP1 antibody and Penny Jeggo and Lynn Harrison for CJ179 and 48BR cells.
REFERENCES
- 1.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 2.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA- dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 3.Geng L, Zhang X, Zheng S, Legerski RJ. Artemis links ATM to G2/M checkpoint recovery via regulation of Cdk1-cyclin B. Mol. Cell. Biol. 2007;27:2625–2635. doi: 10.1128/MCB.02072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Zhang X, Geng L, Teng L, Legerski RJ. Artemis regulates cell cycle recovery from the S phase checkpoint by promoting degradation of cyclin E. J. Biol. Chem. 2009;284:18236–18243. doi: 10.1074/jbc.M109.002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, artemis, and proteins locating to γ-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 8.Yannone SM, Khan IS, Zhou R, Zhou T, Valerie K, Povirk LF. Coordinate 5′ and 3′ endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:3354–3365. doi: 10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by artemis nuclease. J. Biol. Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 10.Pannicke U, Ma Y, Hopfner KP, Niewolik D, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 2004;23:1987–1997. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Multhaup M, Karlen AD, Swanson DL, Wilber A, Somia NV, Cowan MJ, McIvor RS. Cytotoxicity associated with artemis overexpression after lentiviral vector-mediated gene transfer. Hum. Gene Ther. 2010;21:865–875. doi: 10.1089/hum.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair. 2005;4:556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Fouladi B, Waldren CA, Rydberg B, Cooper PK. Comparison of repair of DNA double-strand breaks in identical sequences in primary human fibroblast and immortal hamster-human hybrid cells harboring a single copy of human chromosome 11. Radiat. Res. 2000;153:795–804. doi: 10.1667/0033-7587(2000)153[0795:corodd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. Gamma-ray induced deoxyribonucleic acid strand breaks. 3′ glycolate termini. J. Biol. Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 17.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat. Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 18.Dedon PC, Goldberg IH. Sequence-specific double-strand breakage of DNA by neocarzinostatin involves different chemical mechanisms with a staggered cleavage site. J. Biol. Chem. 1990;265:14713–14716. [PubMed] [Google Scholar]
- 19.van Vugt MA, Marcel ATM, Gardino AK, Linding R, Ostheimer GJ, Reinhardt HC, Shao-En Ong, Tan CS, Miao H, Keezer SM, Li J, et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G2/M DNA damage checkpoint. PLoS Biology. 2010;8:1–20. doi: 10.1371/journal.pbio.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 21.Carbone R, Pearson M, Minucci S, Pelicci PG. PML NBs associate with the HMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002;21:1633. doi: 10.1038/sj.onc.1205227. [DOI] [PubMed] [Google Scholar]
- 22.Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harbor Perspectives in Biology. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britton S, Frit P, Biard D, Salles B, Calsou P. ARTEMIS nuclease facilitates apoptotic chromatin cleavage. Cancer Res. 2009;69:8120–8126. doi: 10.1158/0008-5472.CAN-08-4400. [DOI] [PubMed] [Google Scholar]
- 24.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, et al. DNA-PK autophosphorylation facilitates artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weterings E, Verkaik NS, Keijzers G, Florea BI, Wang SY, Ortega LG, Uematsu N, Chen DJ, van Gent DC. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol. Cell. Biol. 2009;29:1134–1142. doi: 10.1128/MCB.00971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drouet J, Frit P, Delteil C, de Villartay JP, Salles B, Calsou P. Interplay between ku, artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J. Biol. Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 27.Pawelczak KS, Turchi JJ. Purification and characterization of exonuclease-free artemis: Implications for DNA-PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair. 2010;9:670–677. doi: 10.1016/j.dnarep.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu J, Li S, Zhang X, Wang L, Niewolik D, Schwarz K, Legerski RJ, Zandi E, Lieber MR. DNA-PKcs regulates a single-stranded DNA endonuclease activity of artemis. DNA Repair. 2010;9:429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krempler A, Deckbar D, Jeggo PA, Löbrich M. An imperfect G2M checkpoint contributes to chromosome instability following irradiation of S and G2 phase cells. Cell Cycle. 2007;6:1682–1686. doi: 10.4161/cc.6.14.4480. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, Zhou X, Taghizadeh K, Chen J, Stubbe J, Dedon PC. GC/MS methods to quantify the 2-deoxypentos-4-ulose and 3′-phosphoglycolate pathways of 4′ oxidation of 2-deoxyribose in DNA: Application to DNA damage produced by γ-radiation and bleomycin. Chem. Res. Toxicol. 2007;20:1701–1708. doi: 10.1021/tx700164y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 32.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double-strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002;276:24323–24330. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 33.Kanikarla-Marie P, Ronald S, De Benedetti A. Nucleosome resection at a double-strand break during non-homologous ends joining in mammalian cells - implications from repressive chromatin organization and the role of ARTEMIS. BMC Res. Notes. 2011;4:13. doi: 10.1186/1756-0500-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibañez IL, Bracalente C, Molinari BL, Palmieri MA, Policastro L, Kreiner AJ, Burlon AA, Valda A, Navalesi D, Davidson J, et al. Induction and rejoining of DNA double strand breaks assessed by H2AX phosphorylation in melanoma cells irradiated with proton and lithium beams. Int. J. Radiat. Oncol. Biol. Phys. 2009;74:1226–1235. doi: 10.1016/j.ijrobp.2009.02.070. [DOI] [PubMed] [Google Scholar]
- 35.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 36.Reitsema TJ, Banáth JP, MacPhail SH, Olive PL. Hypertonic saline enhances expression of phosphorylated histone H2AX after irradiation. Radiat. Res. 2004;161:402–408. doi: 10.1667/rr3153. [DOI] [PubMed] [Google Scholar]
- 37.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 38.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, Kuo C, Bisi JE, Kim MK. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat. Cell Biol. 2002;4:865. doi: 10.1038/ncb869. [DOI] [PubMed] [Google Scholar]
- 40.Zhong S, Salomoni P, Ronchetti S, Guo A, Ruggero D, Pandolfi PP. Promyelocytic leukemia protein (pml) and daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 2000;191:631–640. doi: 10.1084/jem.191.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.