Abstract
Retrotransposons contribute significantly to the evolution of eukaryotic genomes. They replicate by producing DNA copies of their own RNA, which are integrated at new locations in the host cell genome. In the gene-dense genome of the social amoeba Dictyostelium discoideum, retrotransposon TRE5-A avoids insertional mutagenesis by targeting the transcription factor (TF) IIIC/IIIB complex and integrating ∼50 bp upstream of tRNA genes. We generated synthetic TRE5-A retrotransposons (TRE5-Absr) that were tagged with a selection marker that conferred resistance to blasticidin after a complete retrotransposition cycle. We found that the TRE5-Absr elements were efficiently mobilized in trans by proteins expressed from the endogenous TRE5-A population found in D. discoideum cells. ORF1 protein translated from TRE5-Absr elements significantly enhanced retrotransposition. We observed that the 5′ untranslated region of TRE5-A could be replaced by an unrelated promoter, whereas the 3′ untranslated region of TRE5-A was essential for retrotransposition. A predicted secondary structure in the RNA of the 3′ untranslated region of TRE5-A may be involved in the retrotransposition process. The TRE5-Absr elements were capable of identifying authentic integration targets in vivo, including formerly unnoticed, putative binding sites for TFIIIC on the extrachromosomal DNA element that carries the ribosomal RNA genes.
INTRODUCTION
Transposable elements are genetic entities that actively move from one chromosomal location to another. They may be considered molecular parasites or pieces of selfish DNAs with a basic ‘survival’ strategy of spreading among genomes and maintaining active populations within a given host (1,2). Although this view of mobile elements is attractive, it is incomplete because there are many examples of transposable elements providing value to their host genomes. Transposable elements have shaped eukaryotic genomes, for example, by causing illegitimate recombination events, offering alternative splice sites, introducing new promoters or enhancers to existing genes or creating new genes by shuffling exons (3–5).
Retrotransposons amplify their genomes by producing DNA copies of their own RNA by means of RNA-directed DNA polymerases (reverse transcriptases) (6). Integrated retrotransposons are no longer mobile, but produce progeny that can insert into new genomic locations. Because the integration of cDNA copies into the host cell genome is a default mechanism in retrotransposition, it presents a constant threat of insertional mutagenesis. One strategy used by transposable elements to assure their own long-time ‘survival’, particularly in compact genomes, is the active targeting of genomic loci where integration is unlikely to harm the host. This targeting is well-documented for the genomes of yeasts and social amoebae, in which different retrotransposons populate the vicinities of tRNA genes [(7,8); this study].
In the social amoeba Dictyostelium discoideum, the first tRNA gene-associated repetitive element was identified by Dingermann and coworkers (9) in the late 1980s. The DRE element, later renamed TRE5-A, could be classified as a non-long terminal repeat (non-LTR) retrotransposon by its amino acid-sequence similarity with other members of this class of retrotransposon (10). TRE5-A elements integrate ∼50 bp upstream of tRNA genes, always in the same orientation, with the 5′-end of the mobile element facing the 5′-end of the tRNA gene (9,11). With the complete D. discoideum genomic sequence becoming available (12), six more tRNA gene-associated non-LTR retrotransposons were discovered. Considering that their preferred integration sites were either upstream (5′) or downstream (3′) of tRNA genes, these elements are now named TRE5 (A-C) and TRE3 (A-D), respectively (8,13).
A full-length, replication-competent TRE5-A.1 element encodes two open reading frames (ORFs) and has the general structure of AB-ORF1/ORF2-BC, where A-, B-, and C-modules comprise 5′ and 3′ untranslated regions (UTRs) (14). The 200-bp A-module has promoter activity and directs transcription of plus-strand RNA of TRE5-A (15), which is the supposed template of the retrotransposition process. The B-module is a 290 bp sequence that was duplicated from the 5′-UTR to the 3′-UTR in TRE5-A (14). The 5′ B-module defines the translation start of the ORF1 protein, but otherwise its function in TRE5-A retrotransposition is unknown. Duplication of the B-module in TRE5-A seems to be irrelevant for retrotransposition, because the related D. discoideum TRE5-B and TRE5-C retrotransposons lack 3′ B-modules (8). The 134-bp C-module defines the 3′-end of TRE5-A and TRE5-C, but not TRE5-B elements; it is a weak promoter that mediates the transcription of TRE5-A in the antisense direction (15). The function of antisense RNA in the TRE5-A retrotransposition is unknown. TRE5-A.2 is a non-autonomous derivative of TRE5-A.1 that is characterized by a large deletion in ORF2 and the general structure AB-ORF1-BC (16).
We have recently shown using an in vivo retrotransposition assay that an active population of TRE5-A elements resides in the D. discoideum genome (17,18). The ‘TRE trap’ assay allowed us to analyze the properties of pre-defined integration target sites. To initiate tRNA transcription, the RNA polymerase III-specific transcription factor IIIC (TFIIIC) binds in a sequence-specific manner to the B box, an intragenic promoter, and recruits TFIIIB to bind upstream of the tRNA gene (19). We found that a functional B box promoter is essential for integration site selection in vivo (18), suggesting that TFIIIC is involved in target site selection. The TRE5-A-encoded ORF1 protein is able to bind to TFIIIB subunits in vitro (20). Thus, we hypothesized that TFIIIC is essential for the tRNA gene-targeted integration of TRE5-A because it positions TFIIIB at the 5′-end of the tRNA gene, while integration site selection actually occurs via protein–protein interactions between DNA-bound TFIIIB and the TRE5-A ORF1 protein.
The TRE trap assay relies on the retrotransposition activity of natural TRE5-As in the D. discoideum genome, but it cannot be used to characterize structural properties of the mobile element required for retrotransposition. We took advantage of the observation that non-autonomous TRE5-A.2 elements are efficiently mobilized in trans by the ORF2 protein produced by full-length TRE5-A.1 elements (17,18). We generated synthetic TRE5-A retrotransposons (TRE5-Absr elements) that were tagged with a blasticidin-resistance gene that could be functionally expressed only after a complete retrotransposition cycle. We analyzed the retrotransposition frequencies of various structural variants of the TRE5-Absr element and found that the A-module of the TRE5-Absr element could be replaced by a heterologous promoter, but that a putative hairpin structure formed within the C-module was essential for retrotransposition. We observed that TRE5-Absr elements, similarly to endogenous TRE5-A elements, were capable of identifying tRNA genes as integration targets. Interestingly, we observed repeated integration of TRE5-Absr near a formerly unnoticed B box located on the extrachromosomal palindromic DNA that carries the ribosomal RNA genes.
MATERIALS AND METHODS
Plasmids
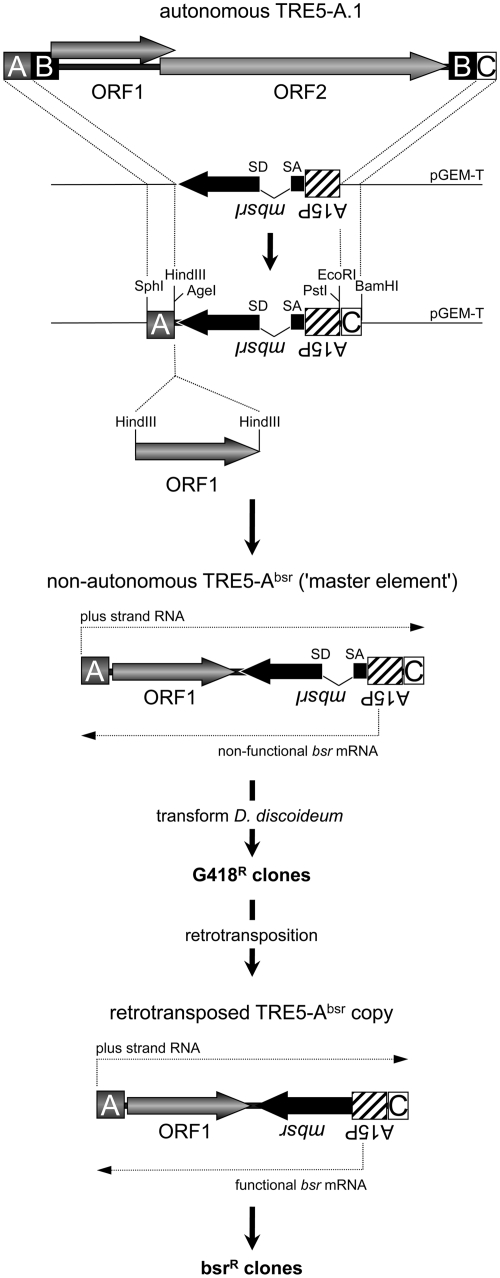
The cloning strategy to generate pTRE5-Absr vectors is briefly illustrated in Figure 1. An expression cassette containing the D. discoideum actin15 promoter (A15P), a gene encoding the blasticidin S resistance gene (bsr) and the actin8 terminator (A8T) was isolated as a PCR fragment from the plasmid pBSR1 (21) and subcloned into pGEM-T (Promega). The bsr gene carries a single BglII restriction site at the nucleotide position 27. This site was used to introduce a 74-bp intron derived from the D. discoideum S17 gene (22), including authentic splice donor and splice acceptor sites, in the opposite transcriptional direction of the bsr gene. The redundant BglII site at the 3′-end of the intron was removed by site-directed mutagenesis to ensure the translation of an authentic bsr gene product after splicing. An A-module of TRE5-A.1 or the actin6 promoter (A6P) was used as the 5′-UTRs for TRE5-Absr elements. We used either the C-module of TRE5-A.1 or the A8T as transcription terminators for the TRE5-Absr elements. Parallel with the published nomenclature for a widely used neomycin resistance marker used to follow retrotransposition in cultured cells (23), we named the D. discoideum selection marker mbsrI, representing a minus-strand bsr gene disrupted by an inverse intron. ORF1 sequences from TRE5-A.1 or TRE5-B were introduced into a single HindIII site located between the TRE5-Absr promoter and the mbsrI gene (Figure 1).
Figure 1.
TRE5-Absr element-based retrotransposition assay. A full-length, retrotransposition-competent TRE5-A.1 element is shown. The 5′-UTR is comprised of the A- module and about half of the B-module. The 3′-UTR contains a second B-module and the C-module. The TRE5-A-encoded proteins are indicated as ORF1 and ORF2. The A- module and the C-module were cloned on a plasmid flanking the mbsrI gene, which is expressed constitutively from the actin15 promoter (A15P). Recognition sites for restriction enzymes important for the assembly of differently structured TRE5-Absr elements are indicated. TRE5-Absr elements were transformed into D. discoideum cells and integrated stably into the genome (these sequences are referred to as ‘master elements’). Transformants were resistant to G418. Master elements do not generate bsrR clones because the mbsr gene is interrupted by the intron. After the retrotransposition of master elements, the TRE5-Absr copies lack the intron in the mbsr gene, which is functionally expressed and confers resistance to blasticidin. The numbers of visible bsrR clones were counted and normalized to 100 master elements per genome and 107 cells.
A splice donor mutant of the mbsrI gene was generated by changing the splice-site sequence GTAAGT to AAAGCT.
By site-directed mutagenesis, a premature translation stop codon was introduced into the ORF1-encoding sequence at the position corresponding to the amino acid isoleucine-10 (designated I10*).
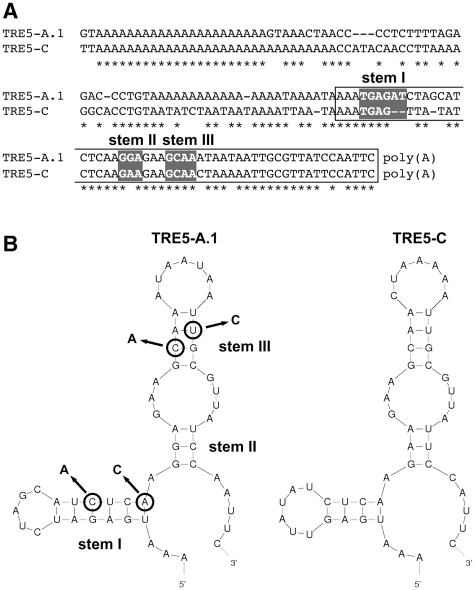
A putative stem–loop structure in the C-module was disrupted by the introduction of point mutations into predicted stem I (C38A and A35C) or stem III (C26A and T15C). Folding of the first 54 RNA nucleotides of the C-module of TRE5-A.1, counted in the antisense direction (corresponding to nucleotides 5647–5594) was performed using the program mfold (version 2.3) set at 22°C (24). Similarly the 3′-terminal 51 nt of the TRE5-C RNA were analyzed using mfold.
Retrotransposition assay
Dictyostelium discoideum cells were grown in HL5 medium (25) and transformed by the calcium phosphate procedure (26) using two plasmids: a TRE5-Absr plasmid and pISAR (27). The latter enables G418 selection (10 µg/ml) on transformed D. discoideum cells and isolation of transformants that stably integrated both plasmids. After transformation, either single G418R colonies were picked and cultured separately, or resistant clones from one Petri dish were pooled and cultured together. From these cell lines, genomic DNA and total RNA were prepared as described (28). The average copy number of integrated TRE5-Absr master elements per cell was determined by quantitative PCR using a primer pair specific for the mbsrI gene from the TRE5-Absr elements and a primer pair specific for the single-copy gene gpdA encoding glyceraldehyde-3-phosphate dehydrogenase (28).
For retrotransposition experiments, polyclonal cell lines with similar numbers of master TRE5-Absr elements were used. All cells were cultured after transformation and prior to blasticidin selection for ∼25 days. In retrotransposition experiments, individual polyclonal cell lines were split and cultured in five 10-cm Petri dishes containing ∼5 × 106 cells in HL5 medium supplemented with 5 µg/ml blasticidin. After ∼10 days, blastR colonies containing retrotransposition-competent TRE5-Absr master elements became visible on Petri dishes. In some experiments, several blastR clones were isolated and cultured separately in HL5 medium containing 5 µg/ml blasticidin. Retrotransposition activity was determined as the number of blastR clones per 107 cells, corrected for the copy number of 100 TRE5-Absr plasmids integrated into the genome (master elements) and presented as means ± SD. All experiments were repeated three times with independent cell lines. TRE5-Absr elements were analyzed by PCR using the primers bsr1 (5′-CTGCCGAAATGATTTCTCC-3′), bsr2 (5′-AAAGATCTGGATCAATTTAACATTTCTC-3′), bsr5 (5′-GGAATTCGGCTGCAGGTCGACGGATCCTC-3′) bsr6 (5′-AAAGCTTAAAGCTTTTTATTAATTTCGGGTATATTTGAGTG-3′) and bsr3′seq (5′-GTGTAGGGAGTTGATTTCAGACTATGCACC-3′), as indicated in the figures.
Identification of de novo integration events
Approximately 1000 blastR colonies derived from the TRE5-Absr element 8 (configuration A6P-ORF1-mbsrI-C) were pooled, and genomic DNA was prepared from the pooled colonies. First, different PCRs were performed using tRNA gene-specific primers (listed below) and the mbsrI-specific primer bsr5. Next, nested PCR was performed using the same tRNA gene-specific primer and the mbsrI-specific primer bsr3′seq. Leu(UAA)rev, 5′-GCAAGAGGCGAGATTCGAACTC-3′; Gly(GCC)rev, 5′-GTATGCTGGGAATCGAACCC-3′; Asp(GUC)rev, 5′-CCCTGTCCGGGAATTGAACCC-3′; Lys(UUU)rev, 5′-CCAAAGGGGGGCTCGAACCC-3′; Glu(UUC)rev, 5′-CCATTCGGGAATCGAACCCGAGG-3′; Val(AAC)rev, 5′-CAGAGCGGGAATCGAACCCACGAC-3′.
RESULTS
TRE5-Absr elements amplify by retrotransposition
The TRE trap assay (17,18) was based on the analysis of the de novo integration of natural TRE5-A elements upstream of a ‘bait’ tRNA gene at a pre-determined chromosomal locus. We were interested in developing an alternative assay that would allow us to analyze the unbiased de novo retrotransposition of TRE5-A elements into their natural integration sites. Unfortunately, it has been impossible to maintain a full-length, genetically tagged TRE5-A.1 on plasmids. However, recent observations of the frequent retrotransposition of endogenous TRE5-A.2s encouraged us to design genetically tagged, non-autonomous TRE5-As that lack the ORF2 region, but are mobilized in trans by ORF2 protein produced from retrotransposition-competent TRE5-A.1 elements (Figure 1).
Engineered selectable markers such as the neomycin resistance gene, which become functional only after a complete retrotransposition event, have a long and successful tradition in the in vivo characterization of retrotransposons (23,29–32). In contrast to mammalian cells, a single chromosomal copy of a neomycin resistance gene is unable to confer sufficient resistance to G418 in D. discoideum cells. Thus, this marker seemed unsuitable to establish a retrotransposition assay for our purpose. Instead, we designed a reporter cassette based on the blasticidin resistance gene bsr. The bsr gene was expressed from the minus strand of a TRE5-A element (mbsr). It was disrupted by an intron that was placed in the inverse transcriptional direction of mbsr (mbsrI), but in the plus-strand orientation of the TRE5-A retrotransposon. These elements were named TRE5-Absr (Figure 1).
We generated several D. discoideum strains harboring chromosomal TRE5-Absr copies through co-transformation of TRE5-Absr-containing plasmids with a vector that confers G418 resistance. We refer to the transformed TRE5-Absr elements as ‘master elements’, which constitutively produce mbsrI RNA from the strong D. discoideum actin15 promoter; however, the intron cannot be spliced out of the mbsrI RNA, preventing functional mbsr from being expressed (Figure 1). On the other hand, the mbsrI intron could be spliced out of plus-strand RNA derived from the TRE5-Absr promoter, and the resulting mRNA transcript could be reverse transcribed and, finally, integrated (Figure 1). From the retrotransposed TRE5-Absr copy, functional mbsr mRNA could be expressed. As a consequence, each cell exhibiting at least one productive retrotransposition event by a TRE5-Absr element could gain blasticidin resistance (referred to as blastR clones).
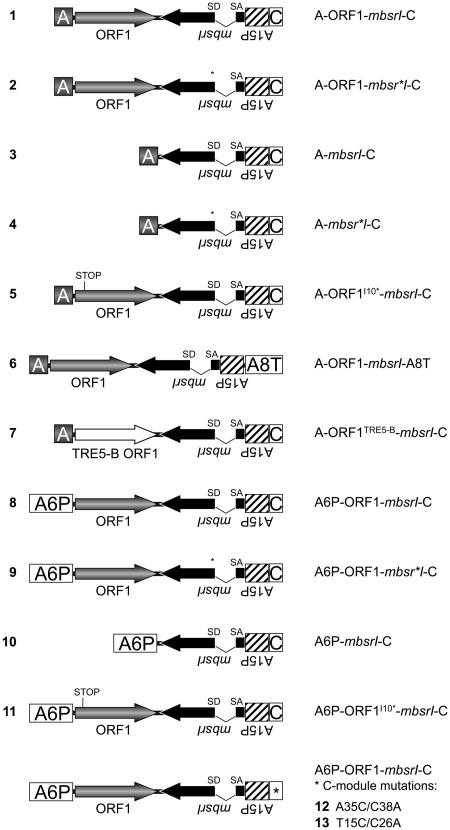
We designed the set of TRE5-Absr elements listed in Figure 2. For convenience, we designated numbers for each TRE5-Absr element to address the different structural variants. TRE5-Absr plasmids were co-transformed into D. discoideum cells with a plasmid that confers G418 resistance. Transformations yielded 50–100 G418R colonies per Petri dish, which were pooled, and the average copy number of integrated TRE5-Absr master elements per cell within a pool was determined by quantitative PCR (data not shown). Only master cell lines with similar numbers of stably integrated TRE5-Absr master elements were used for retrotransposition assays. These cells lines were stored frozen to avoid uncontrolled accumulation of retrotransposition events during prolonged culture periods. For retrotransposition assays polyclonal cell lines were reactivated from the stocks and subjected to selection in blasticidin-containing medium. BlastR clones were counted and numbers were normalized for 107 cells and 100 mbsrI master copies in each cell line.
Figure 2.
Overview of the TRE5-Absr elements generated in this study. Refer to the Figure 1 legend for details. A6P: actin6 promoter; A8T: actin8 terminator.
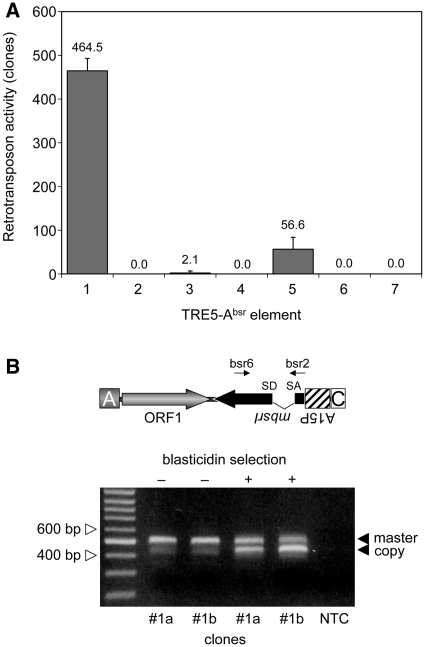
The TRE5-Absr element with the structure A-ORF1-mbsrI-C (element 1; Figure 2) produced ∼460 blastR colonies per 107 cells (Figure 3A). Hence, the non-autonomous TRE5-Absr element was readily mobilized in trans by the ORF2 protein delivered by endogenous TRE5-A.1 elements. We consistently observed that the blastR clone numbers increased with prolonged culture periods, suggesting that retrotransposition of the master element was a continuing process in the cell population of the established cell lines (data not shown).
Figure 3.
Retrotransposition activity of the TRE5-Absr elements expressed from the A-module promoter. (A) Retrotransposition activity of the indicated TRE5-Absr elements. Polyclonal G418R cell lines expressing the indicated TRE5-Absr elements were cultured in five parallel 10-cm Petri dishes, each containing ∼5 × 106 cells, in HL5 medium supplemented with 5 µg/ml blasticidin. Retrotransposition activity was determined as the number of bsrR clones per 107 cells, normalized to a copy number of 100 integrated TRE5-Absr plasmids. The retrotransposition frequencies are presented as means ± SD. This experiment was repeated three times with similar results. (B) Analysis of genomic DNA to determine the structure of the mbsrI gene before and after the selection of G418-resistant transformants carrying TRE5-Absr master elements with blasticidin. Clones #1a and #1b were grown in medium containing G418, but not blasticidin (–). Blasticidin was added to parallel cultures (+). Single bsrR clones were isolated, and PCR was performed on genomic DNA using the mbsrI-specific primers bsr6 and bsr2 as indicated. The upper band (521 bp) was derived from stably integrated master elements, and the lower band (447 bp) was derived from newly retrotransposed copies. As a control, the same PCR was performed without template DNA (NTC).
We introduced a point mutation into element 1 to disrupt the splice donor site in the mbsrI intron, resulting in element 2. We anticipated that this element would not be able to generate blastR clones because the intron would not be removed from the plus-strand RNA produced from the A-module promoter and, consequently, prevent the expression of a functional mbsr gene product from the mbsrI gene in the retrotransposed TRE5-Absr copy. As shown in Figure 3A, element 2 did not produce blastR clones in any of our experiments, indicating that the removal of the intron may be involved in the retrotransposition of TRE5-Absr elements.
To provide direct evidence that the intron was removed by splicing from retrotransposed TRE5-Absr elements, we allowed element 1 to amplify and then isolated blastR clones. The structure of the mbsrI gene in genomic DNA extracted from individual clones was analyzed by PCR using mbsrI exon-specific primers (Figure 3B). The PCR product obtained from the master element was expected to be 521 bp in length. This PCR fragment was readily obtained from G418R clones prior to blasticidin selection (as examples, see clones #1a and #1b in Figure 3B). The 521-bp band was gel-purified and sequenced. The DNA sequence at the intron boundaries of the 521-bp band demonstrated the expected topology of the master element (data not shown). After selection of G418R clones #1a and #1b in blasticidin medium, PCR analysis of blastR clones revealed an enriched 447-bp fragment, which was reminiscent of the retrotransposed TRE5-Absr element after removal of the intron. Direct sequencing of the 447-bp fragment verified the splicing of the mbsrI gene in the retrotransposed TRE5-Absr element (data not shown). We should note that the weak 447-bp band was also visible in PCR on genomic DNA prepared from pools of G418R clones of retrotransposition-competent elements, prior to blasticidin selection (see the first two lanes in Figure 3B); this presumably resulted from ongoing retrotransposition activity by the TRE5-Absr population. The 447-bp band was not observed in experiments with elements 2 or 3, which displayed a strongly reduced or absent retrotransposition activity (Figure 3A and data not shown).
The TRE5-A ORF1 protein displays a cis preference for its encoding RNA
For the human L1 retrotransposon, it has been shown that the retrotransposon-encoded proteins bind to their own encoding transcripts directly after translation, thereby drastically enhancing the retrotransposition frequency of the retrotransposon (33). This observation has been termed cis preference. We tested the importance of the ORF1 protein-encoding sequences to the retrotransposition activity of TRE5-Absr elements. Element 3, which lacked the ORF1 sequence, exhibited a retrotransposition frequency that was 0.5% of element 1’s frequency. The retrotransposition activity of element 3 was significantly above the background, as the element containing the splice-donor mutant (4) did not produce any blastR clones (Figure 3A). Thus, the ORF1 protein provided in trans by endogenous TRE5-A elements supported the retrotransposition of TRE5-Absr elements in D. discoideum cells rather poorly, meaning that the ORF1 protein displayed a strong cis preference when translated from an retrotransposition-competent element. Element 5 containing an ORF1 sequence disrupted by a premature translation stop codon exhibited 12% of the retrotransposition frequency of the control element 1, yet it was 27-fold more active than element 3 that lacked any ORF1-encoding sequence (Figure 3A). This suggested that binding of ORF1 protein to its own element’s ORF1-encoding RNA enhanced the retrotransposition activity, predicting an RNA-sequence-specific binding property of the ORF1 protein.
Heterologous ORF1 protein does not support TRE5-A retrotransposition
In vitro two-hybrid assays suggested that integration site selection by TRE5-A may involve direct protein–protein interactions between the TRE5-A ORF1 protein and TFIIIB subunits (20). The TRE5-A-related retrotransposon TRE5-B exhibits an integration preference very similar to that of TRE5-A (8). The TRE5-B ORF1 protein shows only 25% sequence identity to the TRE5-A ORF1, but is nevertheless capable of binding to TFIIIB subunits in vitro (20). This finding encouraged us to test whether TRE5-A and TRE5-B retrotransposons can generate mixed pre-integration complexes. We cloned the TRE5-B-derived ORF1 DNA sequence into TRE5-Absr to generate element 7. This element was completely inactive in our assay (Figure 3A), indicating either that ORF1 proteins display RNA binding activities that are specific to their own retrotransposon or that heterologous ORF1 proteins may not be able to mediate interactions with TRE5-A ORF2 proteins required for retrotransposition.
The A-module is dispensable for retrotransposition of TRE5-Absr elements
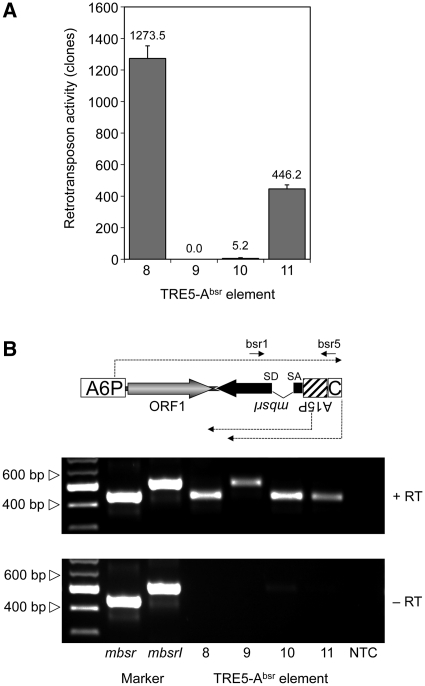
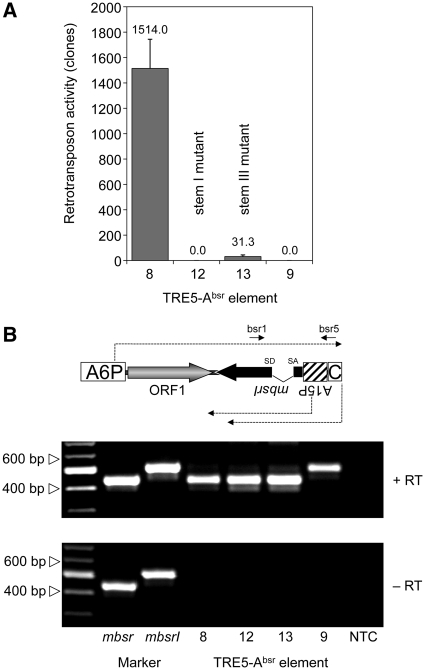
Non-LTR retrotransposons are believed to retrotranspose through a process called target-primed reverse transcription (TPRT) (34–37). The TPRT model states that the retrotransposon-encoded ORF2 protein binds to the 3′-end of the retrotransposon RNA, nicks the integration target DNA, and uses the emerging 3′ hydroxyl group as a primer for cDNA synthesis. If we assume that TRE5-A amplifies by a TPRT mechanism, then the 3′-end of the TRE5-Absr element should be essential for retrotransposition, while the 5′-UTR may be required to supply a promoter activity that directs the transcription of the retrotransposon and produces substrates for TPRT-mediated retrotransposition. We show here that the A-module was required as an active promoter to support the expression of TRE5-Absr elements (a prerequisite for retrotransposition), but it was not per se essential for retrotransposition (Figure 4A). The D. discoideum actin6 gene promoter (A6P) could readily replace the A-module and serve as the 5′- UTR to support the retrotransposition of TRE5-Absr. This element (8) was highly active in our retrotransposition assays and yielded roughly 3-fold excess of blastR clones compared with element 1 (Figure 4A). This result probably reflected the stronger promoter activity of the actin6 promoter compared with that of the A-module (O.S. and T.W., unpublished results). As expected, elimination of the splice donor site from element 8 completely abolished its ability to generate blasticidin-resistant colonies (element 9; Figure 4A), although both elements were comparably well expressed (Figure 4B).
Figure 4.
Retrotransposition activity of TRE5-Absr elements expressed from a heterologous promoter. (A) Retrotransposition activity of the indicated TRE5-Absr elements. Polyclonal G418R cell lines containing comparable numbers of TRE5-Absr master elements were cultured in five parallel 10-cm Petri dishes (5 × 106 cells per dish) in HL5 medium and 5 µg/ml blasticidin. Retrotransposition activity was determined by counting bsrR clones. Values were normalized to 107 cells and 100 integrated TRE5-Absr master elements. The retrotransposition frequencies are presented as means ± SD. This experiment was repeated three times with similar results. (B) Expression of TRE5-Absr elements. TRE5-Absr elements produce plus-strand RNA derived from the actin6 promoter (A6P) and two types of minus-strand RNAs derived from either the actin15 promoter (A15P) or the C-module. Total RNA was isolated from pools of G418R clones transformed with TRE5-Absr elements 8–11. The synthesis of cDNA was initiated using primer bsr5, which binds downstream of the actin15 promoter. PCR on cDNA preparations was performed using the primers bsr1 and bsr5 as indicated. PCR fragments produced from plasmid-borne mbsr and mbsrI were used as size markers. Note that, in element 9, the splice donor site is disrupted, resulting in a larger PCR product. The C-module is such a weak promoter that its derived RNA transcripts are not detectable in this RT–PCR.
The experiments using TRE5-Absr elements driven by the actin6 promoter confirmed our previous results indicating both the trans-complementation and cis preference of the ORF1 protein. First, omitting the ORF1 sequence from element 8, resulting in element 10, led to a 99.5% drop in retrotransposition activity (Figure 4A). Second, element 11, containing an ORF1 sequence with a premature translation stop codon, was roughly 90-fold more active than the element lacking ORF1. Furthermore, element 11 exhibited only 35% of the activity of element 8, which contained an intact ORF1 sequence.
The C-module is essential for TRE5-Absr retrotransposition
When we replaced the C-module of element 1 with the unrelated transcription terminator of the D. discoideum actin8 gene (A8T) to yield element 6, no blasticidin-resistant clones were obtained in any of our experiments (Figure 3A). The C-module of TRE5-A.1 is a 137-bp sequence. The TRE5-A.2 element has, in addition to the large ORF2 deletion, a 72-bp deletion in the BC-module sequence, leaving only the 3′-terminal 87 bp of the C-module in the TRE5-A.2 elements intact. Because the TRE5-A.2 elements are nevertheless retrotransposition-competent (17,18), the structural information within the 3′-terminal 87 bp of the C-module RNA is essential for retrotransposition. Although this part of the TRE5-A.1 C-module shares only 78% identical nucleotides with the C-module of the related retrotransposon TRE5-C (Figure 5A), both C-module-derived RNAs may form strikingly similar secondary structures. As shown in Figure 5B, the program mfold predicted the formation of a secondary structure within the C-module RNAs from TRE5-A (dG = −20.08 kcal/mol at 22°C) and TRE5-C (dG = −12.52 kcal/mol at 22°C). To evaluate the significance of the predicted secondary structure in the TRE5-A.1 C-module to the retrotransposition of the element in vivo, we introduced point mutations into stem I or stem III, creating TRE5-Absr elements 12 and 13. As shown in Figure 6A, mutations in stem III (element 13) reduced the retrotransposition activity by 98% compared with the control element 8, whereas element 12, bearing the mutation in stem I, was completely inactive. The loss of the retrotransposition capacity of elements 12 and 13 was not due to a general destabilization of the TRE5-Absr-derived transcripts, as demonstrated by RT–PCR (Figure 6B). Taken together, these data indicate that a critical secondary structure in the 3′-UTR may be important for the retrotransposition of the TRE5-A element.
Figure 5.
Prediction of a secondary structure in the C-module. (A) Alignment of the C-modules from retrotransposons TRE5-A.1 and TRE5-C. The sequences used for secondary structure prediction are indicated in white boxes. Nucleotides predicted to participate in secondary structure formation (stems I–III) are shown in gray boxes. (B) The program mfold predicted similar secondary structures for the C-modules from retrotransposons TRE5-A and TRE5-C. The predicted folding of the first 54 and 51 nt, respectively, of the C-modules is shown, counted from the 3′-ends of the elements. Double mutations predicted to disrupt the secondary structure of the TRE5-A C-module in TRE5-A.1 are indicated by arrows (stem I: C38A and A35C; stem III: C26A and T15C). The resulting TRE5-Absr elements are numbered 12 and 13.
Figure 6.
Point mutations in the C-module abolish retrotransposition. (A) Retrotransposition activities of TRE5-Absr elements carrying C-module mutations. In this experiment polyclonal G418R cells expressing the indicated TRE5-Absr master elements were cultured in blasticidin-containing medium. Retrotransposition activity was determined by counting the numbers of bsrR clones per 107 cells, normalized to a copy number of 100 integrated TRE5-Absr master elements. The retrotransposition frequencies are presented as means ± SD. (B) Expression of TRE5-Absr elements carrying C-module mutations. Total RNA was isolated from pools of G418R clones transformed with TRE5-Absr elements 8, 9, 12 and 13. The synthesis of cDNA was performed using primer bsr5 and PCR on cDNA preparations was carried out using the primers bsr1 and bsr5 as indicated. PCR fragments produced from plasmid-borne mbsr and mbsrI were used as size markers.
TRE5-Absr elements integrate upstream of tRNA genes
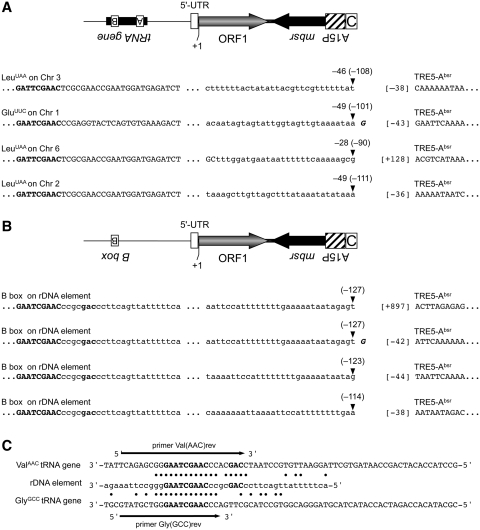
In order to isolate and characterize the de novo integration of TRE5-Absr elements, we took advantage of the strong preference of TRE5-A to integrate upstream of tRNA genes. TRE5-Absr element 8, driven by the actin6 promoter, was allowed to amplify, and blastR clones were selected. Because TRE5-A integration is orientation-specific, PCR on genomic DNA isolated from pooled blastR clones could be designed using a forward primer specific to a certain tRNA gene family and a reverse primer that binds in the mbsr gene region of integrated TRE5-Absr elements. Several obtained PCR fragments were cloned and sequenced, and four examples of de novo integration of TRE5-Absr upstream of tRNA genes are shown in Figure 7A. The 5′-ends of three integrated TRE5-Absr elements were located within the actin6 promoter, 38–43 bp upstream of the ORF1 start codon. This result indicated that the mobile TRE5-Absr elements left the actin6 promoter behind, as expected from a canonical RNA polymerase II promoter, except for a 5′-UTR of roughly 40 bp. This observation indicated that the three analyzed integration sites contained complete TRE5-Absr elements. These elements integrated within a range of 46–49 bp upstream of tRNA genes, representing the authentic integration preference of TRE5-A (17,18). A fourth analyzed integration site exhibited the 5′ deletion of the TRE5-Absr element, an observation typical of TRE5-A elements (17,18). This integration included a shorter distance to its targeted tRNA gene (28 bp) than expected, which seemed to be caused by a target-site deletion. Such deletions have frequently been observed after the de novo integration of 5′ truncated TRE5-A elements in the TRE trap assay (18).
Figure 7.
De novo integration sites for TRE5-Absr elements. Examples of integration sites upstream of tRNA genes on four different D. discoideum chromosomes (A) and four independent integration events into the same position on the extrachromosomal rDNA element (B). Element 8 was allowed to amplify, bsrR clones were pooled and integrants were isolated by PCR on genomic DNA using tRNA gene family-specific forward primers and an mbsrI-specific reverse primer. PCR fragments were cloned and sequenced. Indicated in bold letters is the B box sequence of the isolated tRNA gene. The distances from the integrated TRE5-Absr element to the first nucleotide of the tRNA gene are indicated by negative numbers (e.g. –46). The corresponding distances to the B boxes of the targeted tRNA genes are provided in round brackets (e.g. –108). The first nucleotides of the integrated TRE5-Absr elements are indicated by numbers in square brackets left to the TRE5-Absr sequence. The +1 nt of the TRE5-Absr element was defined as the start of translation for ORF1, meaning that ‘[–38]’ refers to the 5′-UTR derived from the actin6 promoter. (C) Similarity of tRNA gene-specific primers with sequences on the rDNA element. Shown are the complete sequences of the D. discoideum ValAAC and GlyGCC tRNA genes. Note that the DNA sequences are written in the same orientation as in panel B. The B boxes are written in bold. A section of the rDNA element is shown with black dots indicating sequence identities to the two tRNA genes. The arrows indicate the tRNA-specific primers used to search for TRE5-Absr integrations in the D. discoideum genome.
TRE5-Absr elements target the extrachromosomal rDNA element
In D. discoideum, the ribosomal RNA genes are carried on a multicopy extrachromosomal DNA segment that is estimated to contribute ∼20% of total nuclear DNA (38). This mirror-symmetric, palindromic element holds the rRNA genes, but contains neither tRNA genes nor natural TRE integrations (12,38). We detected TRE5-Absr integrants on the rDNA element during PCR screening for tRNA-associated integration sites (Figure 7B), due to sequence homologies between the tRNA gene-specific primers Val(AAC)rev and Gly(GCC)rev, which partially overlap with the B boxes of their target tRNA genes, and a short sequence on the rDNA element (Figure 7C). The four TRE5-Absr copies were found to be inserted, in the correct orientation, 114–127 bp upstream of a sequence that matches the consensus sequence of tRNA gene-associated B boxes, which is GTCnnnnGTTCRAnYC (n = any nucleotide, R = purine, Y = pyrimidine). The distances of the integrated TRE5-Absr elements to the putative B box on the rDNA palindrome agreed with previously determined distances of de novo integrations of endogenous TRE5-As into the TRE trap containing either a complete tRNA gene or an isolated B box (18), suggesting that the TRE5-Absr element had targeted a functional B box on the rDNA element.
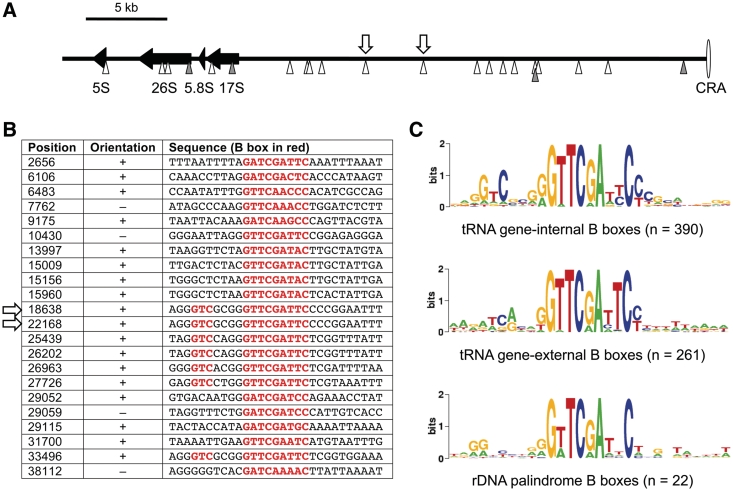
This observation was unexpected because the rDNA elements were not known to contain functional TFIIIC binding sites or integrations of TRE retrotransposons. Searching for additional B box-like sequences on the rDNA element, we noticed a total of 22 such sequences distributed along one ∼40 kb arm of the palindromic rDNA element (Figure 8A). The identified B boxes are listed in Figure 8B. The sequences did not reveal any homology to D. discoideum tRNA genes, except for the B boxes, meaning that the origin of the isolated B boxes on the rDNA palindrome remains elusive. The consensus sequence of all B boxes located on the rDNA palindrome seemed to be more related to tRNA gene-external B boxes, which are found ∼41 bp downstream of 66% of all D. discoideum tRNA genes (18), than to tRNA gene-internal B boxes. The latter are characterized by a 5′-GTC-3′ signature located 5–7 bp upstream of the most conserved GTTCRAnYC motif (18) (Figure 8C). Inspection of the TRE5-Absr integration site on the rDNA element revealed the target sequence 5′-AGGGTCGCGGGTTCGATTCCCC-3′ (B box sequence underlined), suggesting that TRE5-Absr integrated at a B box with similarity to tRNA gene-internal B boxes. Of the 22 B boxes determined on the rDNA palindrome, seven have the characteristic 5′-GTC-3′ signature of intragenic B boxes. The exact position of the TRE5-Absr integration sites could not be determined because it is part of a duplication of at least 4 kb listed in the published rDNA palindromic sequence [GenBank AY171066; (38)], resulting in two indistinguishable integration sites at positions 18638 and 22168 (Figure 8A).
Figure 8.
Isolated B boxes on the extrachromosomal rDNA palindromic sequence. (A) The left arm of the palindromic sequence (∼40 kb in length) is shown, with the ‘central region of asymmetry’ (CRA) displayed to the right. Black arrows represent the rRNA genes. Arrow heads below the black line indicate B boxes located on the upper (white) and lower (gray) DNA strand. The white arrows above the black line indicate the two indistinguishable TRE5-Absr integration sites shown in detail in Figure 7B. (B) Sequences of the B boxes located on the left arm of the rDNA palindromic sequence. Position numbers refer to GenBank entry AY171066. The consensus B box motif GTCnnnnGTTCRAnYC is highlighted in red. The arrows indicate the two indistinguishable TRE5-Absr integration sites. (C) WebLogo (53) alignment of intragenic B boxes (upper panel), extra B boxes found downstream of most D. discoideum tRNA genes (middle panel) and isolated B boxes found in the rDNA element (lower panel).
DISCUSSION
TRE5-Absr elements mimic authentic TRE5-A retrotransposition events
In this study, we established genetically tagged TRE5-A retrotransposons that copy the natural retrotransposition of the TRE5-A elements present in D. discoideum cells. The TRE5-Absr elements were able to integrate, in an orientation-specific manner, ∼50 bp upstream of tRNA genes. TRE5-Absr elements could only amplify and generate blasticidin-resistant colonies if the artificial intron was removed from the plus-strand RNA. This provided experimental evidence for the hypothesis that TRE5-A is, in fact, a retrotransposon. In line with the typical structure of a non-LTR retrotransposon, which suggests that TRE5-A amplifies by canonical TPRT, we found that the 5′-UTR of TRE5-A could be replaced by an unrelated promoter. This result supports the idea that the primary function of the 5′-UTRs of non-LTR retrotransposons is to ensure the production of plus-strand RNA as the substrate for retrotransposition. Similar results have been obtained for other non-LTR retrotransposons, such as the Drosophila I element (39) and the human L1 element (23).
Considering the relatively high retrotransposition activity of TRE5-A in D. discoideum cells (17,18), it is surprising that the D. discoideum genome does not contain processed pseudogenes resulting from the reverse transcription and integration of cellular mRNAs by TRE5-A-derived proteins. We showed here that the 3′-end of the TRE5-A element is essential for productive retrotransposition, and we demonstrated that the putative stem–loop structure of the C-module is presumably recognized by a TRE5-A-encoded protein as an integration substrate. Interestingly, conserved stem–loop structures have also been reported to occur in the 3′-UTRs of the eel UnaL2 and insect R2 elements (40,41) and may therefore reflect a general property of non-LTR retrotransposons. The recognition of the stem–loop structure of the C-module may contribute to the high specificity of the TRE5-A-encoded proteins for their own RNA substrates and, thus, readily explain the absence of processed pseudogenes in the D. discoideum genome. This high substrate specificity of TRE5-A proteins may be considered an adaptation of the retrotransposon to its host cell, which may not tolerate genome expansion by the acquisition of processed pseudogenes.
Amplification of non-LTR retrotransposons by TPRT frequently results in 5′ deletions of the integrated retrotransposon copies. This is explained by the limited processivity of the retrotransposon-encoded reverse transcriptases and/or the premature termination of reverse transcription, forced by microhomologies between retrotransposon-derived RNA and the target DNA (42,43). We found here that integrated TRE5-Absr elements may become 5′ deleted similar to endogenous TRE5-A elements after de novo integration into the TRE trap (17,18). Completely integrated non-LTR retrotransposons often insert extra nucleotides at the 5′ integration site, often a single guanosine that may be derived from reverse transcription of the 7-methyl guanosine cap presumed to be present on RNA derived from non-LTR retrotransposons (44,45). This phenomenon is also a property of both endogenous TRE5-As (18) and TRE5-Absr elements (Figure 7).
The previously described TRE trap assay (17,18) and the TRE5-Absr element-based retrotransposition assay complement each other. The TRE trap assay has the advantage of analyzing the retrotransposition of natural TRE5-As, yet it is restricted to the observation of de novo integration at particular ‘bait’ sequences. The TRE5-Absr element-based retrotransposition assay, on the other hand, provides an opportunity to observe the unbiased integration of TRE5-A and is, therefore, able to detect any accessible target sites (discussed below). In addition, the TRE5-Absr element-based retrotransposition assay is a valuable tool for the analysis of host factors involved in retrotransposition of TRE5-A. It may be used to test the in vivo relevance of the protein–protein interactions assumed to mediate target-site selection (20) or to analyze mutants defective in host functions that are assumed to be involved in productive retrotransposition, such as DNA repair.
The TRE5-A ORF1 protein exhibits cis preference
Experiments using the human L1 element as a model have demonstrated that both the ORF1 and the ORF2 protein form complexes with L1 RNA, resulting in large preintegration complexes (46). Both L1 proteins display strong cis preferences, and there is evidence that very little, if any, free L1-derived protein accumulates in the cytoplasm. The cis preference of L1 proteins may have evolved to favor the retrotransposition of elements with intact ORFs, while efficient complementation in trans would present the risk of retrotransposing defective copies (33).
In our experiments using the D. discoideum TRE5-A element, we found evidence for both trans-complementation and cis preference of the ORF1 protein. TRE5-Absr elements lacking ORF1 exhibited drastically reduced retrotransposition activities compared with elements containing translatable ORF1 sequence (compare elements 1 and 3 or 8 and 10 in Figures 3 and 4). This result raised the question whether elements 3 and 10, lacking ORF1, amplified at lower rates because cellular ORF1 protein was either rate-limiting or insufficiently capable of trans complementation. We conclude that cellular ORF1 protein was not rate-limiting because element 11, which was driven by the strong actin6 promoter but could not increase the amount of cellular ORF1 protein due to a premature translation stop codon, produced similar amounts of retrotransposed copies as element 1. The latter was expressed at much lower rate from the A-module promoter (data not shown). Thus, it seemed more likely that cellular ORF1 protein complemented TRE5-Absr retrotransposition in trans, however, leading to much lower retrotransposition rates compared to TRE5-Absr elements providing ORF1 protein in cis.
TRE5-Absr elements 5 and 11, containing untranslatable ORF1 sequences, were 27- and 85-fold, respectively, more active than their ORF1-less counterparts 3 and 10 (compare elements 1 and 5 or 8 and 11 in Figures 3 and 4). This result can only be explained assuming that the ORF1 protein displays sequence-specific binding to the ORF1-encoding part of the TRE5-A retrotransposon in trans. This interesting property of the TRE5-A ORF1 protein may offer an attractive explanation for the observed cis preference of the ORF1 protein, but this has to be addressed in future experiments.
We cannot draw conclusions on the properties of the TRE5-A ORF2 protein, since the ORF2-encoding sequence could not yet be stably incorporated into TRE5-Absr-containing plasmids. However, D. discoideum cells seem to contain sufficient amounts of (free) TRE5-A-derived ORF2 protein to bind to TRE5-Absr-derived RNA and support its retrotransposition in trans. We speculate that ORF2 protein is able to specifically bind to TRE5-A-derived RNAs by recognizing the unique secondary structure of the C-module. Whether the ORF2 protein of TRE5-A also displays a cis preference remains an open question.
TRE5-A targets the extrachromosomal rDNA element
Previous data (18,20) predict that TRE5-A integration targets must be occupied by the TFIIIC/TFIIIB complex. The TRE5-Absr elements described in this study can be used as tools to map putative RNA polymerase III transcription complexes to certain genomic loci. Thus, the observed de novo integration of TRE5-Absr into the extrachromosomal rDNA palindrome predicts that functional TFIIIC/TFIIIB complexes are formed on previously unsuspected B boxes. We have previously shown that isolated B boxes are principal targets for TRE5-A integration in the context of the TRE trap (18). The TRE5-Absr integration events on the rDNA element concerned two identical target sites, which exactly matched the B box consensus GTCnnnnGTTCRAnYC, indicating that at least one of these two binding sites is occupied by TFIIIC and TFIIIB. At present, we do not know whether the other 20 B boxes also represent bona fide TFIIIC binding sites. Studies on the genomic dispersal of components constituting the RNA polymerase III complex have revealed unexpected ‘extra TFIIIC’ (ETC) sites in budding yeast and ‘chromosome-organizing clamps’ in fission yeast. Both elements possess intrinsic chromatin-boundary activity (47–51). This activity can be exhibited by partially assembled transcription complexes containing only DNA-bound TFIIIC that may function both as heterochromatin barriers and insulators against gene activation. In light of these observations, it is attractive to assume that TFIIIC binding sites contribute to the structural organization of the chromatin of D. discoideum rDNA elements. Noma et al. (50) noted that TFIIIC bound to ETC sites may recruit certain genomic loci to the nuclear periphery, and Thompson et al. (52) found that functional RNA polymerase III complexes induce nucleolar clustering of tRNA genes. The latter authors suggested that tRNA gene localization in the nucleus is, to a certain extent, dependent on the transcription of rRNA genes. Thus, the TFIIIC binding sites on the D. discoideum rDNA element may contribute to the sub-nuclear co-localization of rRNA and tRNA genes as hot-spots for transcription by RNA polymerases I and III.
FUNDING
This work was supported by a grant (WI 1142/5-5) from the Deutsche Forschungsgemeinschaft (DFG). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 2.Orgel LE, Crick FHC. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 3.Han SF, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? BioEssays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 4.Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 5.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke JD, Chapman KB. Retrotransposition mechanisms. Curr. Opin. Cell Biol. 1991;3:502–507. doi: 10.1016/0955-0674(91)90079-e. [DOI] [PubMed] [Google Scholar]
- 7.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 8.Glöckner G, Szafranski K, Winckler T, Dingermann T, Quail M, Cox E, Eichinger L, Noegel AA, Rosenthal A. The complex repeats of Dictyostelium discoideum. Genome Res. 2001;11:585–594. doi: 10.1101/gr.162201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marschalek R, Brechner T, Amon-Böhm E, Dingermann T. Transfer RNA genes: landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science. 1989;244:1493–1496. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- 10.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann J, Schumann G, Borschet G, Gosseringer R, Bach M, Bertling WM, Marschalek R, Dingermann T. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5′-flanking and 3′-flanking regions. J. Mol. Biol. 1991;222:537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- 12.Eichinger L, Pachebat JA, Glöckner G, Rajandream M-A, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szafranski K, Glöckner G, Dingermann T, Dannat K, Noegel AA, Eichinger L, Rosenthal A, Winckler T. Non-LTR retrotransposons with unique integration preferences downstream of Dictyostelium discoideum transfer RNA genes. Mol. Gen. Genet. 1999;262:772–780. doi: 10.1007/s004380051140. [DOI] [PubMed] [Google Scholar]
- 14.Marschalek R, Hofmann J, Schumann G, Gosseringer R, Dingermann T. Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol. Cell. Biol. 1992;12:229–239. doi: 10.1128/mcb.12.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann G, Zündorf I, Hofmann J, Marschalek R, Dingermann T. Internally located and oppositely oriented polymerase II promoters direct convergent transcription of a LINE-like retroelement, the Dictyostelium Repetitive Element, from Dictyostelium discoideum. Mol. Cell. Biol. 1994;14:3074–3084. doi: 10.1128/mcb.14.5.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marschalek R, Hofmann J, Schumann G, Dingermann T. Two distinct subforms of the retrotransposable DRE element in NC4 strains of Dictyostelium discoideum. Nucleic Acids Res. 1992;20:6247–6252. doi: 10.1093/nar/20.23.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck P, Dingermann T, Winckler T. Transfer RNA gene-targeted retrotransposition of Dictyostelium TRE5-A into a chromosomal UMP synthase gene trap. J. Mol. Biol. 2002;318:273–285. doi: 10.1016/S0022-2836(02)00097-9. [DOI] [PubMed] [Google Scholar]
- 18.Siol O, Boutliliss M, Chung T, Glockner G, Dingermann T, Winckler T. Role of RNA polymerase III transcription factors in the selection of integration sites by the Dictyostelium non-long terminal repeat retrotransposon TRE5-A. Mol. Cell. Biol. 2006;26:8242–8251. doi: 10.1128/MCB.01348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 20.Chung T, Siol O, Dingermann T, Winckler T. Protein interactions involved in tRNA gene-specific integration of Dictyostelium discoideum non-long terminal repeat retrotransposon TRE5-A. Mol. Cell. Biol. 2007;27:8492–8501. doi: 10.1128/MCB.01173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaulsky G, Escalante R, Loomis WF. Developmental signal transduction pathways uncovered by genetic suppressors. Proc. Natl Acad. Sci. USA. 1996;93:15260–15265. doi: 10.1073/pnas.93.26.15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bäuerle A, Mutzel R. Nucleotide sequence of the gene for ribosomal protein S17 from Dictyostelium discoideum. Biochim. Biophys. Acta. 1995;1260:223–226. doi: 10.1016/0167-4781(94)00218-r. [DOI] [PubMed] [Google Scholar]
- 23.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hydridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sussman M. In: Methods in cell biology. Spudich JA, editor. Vol. 28. Orlando, FL: Academic Press; 1987. pp. 9–29. [Google Scholar]
- 26.Gaudet P, Pilcher KE, Fey P, Chisholm RL. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007;2:1317–1324. doi: 10.1038/nprot.2007.179. [DOI] [PubMed] [Google Scholar]
- 27.Maniak M, Nellen W. pISAR, a tool for cloning genomic sequences adjacent to the site of vector integration. Nucleic Acids Res. 1989;17:4894. doi: 10.1093/nar/17.12.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas J, Bilzer A, Moll L, Zundorf I, Dingermann T, Eichinger L, Siol O, Winckler T. The carboxy-terminal domain of Dictyostelium C-module-binding factor is an independent gene regulatory entity. PLoS One. 2009;4:e5012. doi: 10.1371/journal.pone.0005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 30.Heidmann T, Heidmann O, Nicolas JF. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc. Natl Acad. Sci. USA. 1988;85:2219–2223. doi: 10.1073/pnas.85.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 32.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 33.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 35.Feng QH, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 36.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell. Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sucgang R, Chen GK, Liu W, Lindsay R, Lu J, Muzny D, Shaulsky G, Loomis WF, Gibbs R, Kuspa A. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res. 2003;31:2361–2368. doi: 10.1093/nar/gkg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen S, Cavarec L, Dhellin O, Heidmann T. Retrotransposition of a marked Drosophila line-like I element in cells in culture. Nucleic Acids Res. 1994;22:1484–1488. doi: 10.1093/nar/22.8.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura Y, Kajikawa M, Baba S, Nakazato S, Imai T, Sakamoto T, Okada N, Kawai G. Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2. Nucleic Acids Res. 2006;34:5184–5193. doi: 10.1093/nar/gkl664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruschak AM, Mathews DH, Bibillo A, Spinelli SL, Childs JL, Eickbush TH, Turner DH. Secondary structure models of the 3' untranslated regions of diverse R2 RNAs. RNA. 2004;10:978–987. doi: 10.1261/rna.5216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin SL, Li W-LP, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogen. Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 43.Zingler N, Willhoeft U, Brose H-P, Schoder V, Jahns T, Hanschmann KMO, Morrish TA, Löwer J, Schumann G. Analysis of 5' junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5' end attachment requiring microhomology-meditaed end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bibillo A, Eickbush TH. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 2004;279:14945–14953. doi: 10.1074/jbc.M310450200. [DOI] [PubMed] [Google Scholar]
- 45.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl Acad. Sci. USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 51.Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, Edwards DJ, Donze D. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Euk. Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]