Abstract
Small nuclear and nucleolar RNAs that program pre-mRNA splicing and rRNA processing have a signature 5′-trimethylguanosine (TMG) cap. Whereas the mechanism of TMG synthesis by Tgs1 methyltransferase has been elucidated, we know little about whether or how RNP biogenesis, structure and function are perturbed when TMG caps are missing. Here, we analyzed RNPs isolated by tandem-affinity purification from TGS1 and tgs1Δ yeast strains. The protein and U-RNA contents of total SmB-containing RNPs were similar. Finer analysis revealed stoichiometric association of the nuclear cap-binding protein (CBP) subunits Sto1 and Cbc2 with otherwise intact Mud1- and Nam8-containing U1 snRNPs from tgs1Δ cells. CBP was not comparably enriched in Lea1-containing U2 snRNPs from tgs1Δ cells. Moreover, CBP was not associated with mature Nop58-containing C/D snoRNPs or mature Cbf5- and Gar1-containing H/ACA snoRNPs from tgs1Δ cells. The protein composition and association of C/D snoRNPs with the small subunit (SSU) processosome were not grossly affected by absence of TMG caps, nor was the composition of H/ACA snoRNPs. The cold-sensitive (cs) growth defect of tgs1Δ yeast cells could be suppressed by mutating the cap-binding pocket of Cbc2, suggesting that ectopic CBP binding to the exposed U1 m7G cap in tgs1Δ cells (not lack of TMG caps per se) underlies the cs phenotype.
INTRODUCTION
2,2,7-trimethylguanosine (TMG) cap structures are characteristic of the small nuclear (sn) RNAs that program mRNA splicing (U1, U2, U4 and U5). TMG caps are also found on telomerase RNA and small nucleolar (sno) RNAs. TMG is formed from m7G caps by the enzyme Tgs1 (1), which catalyzes two successive methyl transfer reactions from AdoMet to the N2 atom of 7-methylguanosine (2–7). Unlike m7G caps, the TMG cap is conspicuously not essential for viability of eukaryal cells. A tgs1Δ mutant of fission yeast Schizosaccharomyces pombe grows normally (4). The tgs1Δ mutation of budding yeast Saccharomyces cerevisiae causes a growth defect at cold temperatures, though tgs1Δ cells grow as well as TGS1 cells at 34°C (1,5). The tgs1Δ mutants of budding and fission yeast lack any detectable TMG caps on their U1, U2, U4 and U5 snRNAs and snoRNAs, as gauged by IP-Northerns using anti-TMG antibody (1,4), signifying that there is no Tgs1-independent route to generate TMG caps. tgs1Δ yeast cells have apparently normal steady-state levels of snRNAs, and they display no aberration in the sedimentation profiles of their spliceosomal snRNPs (1). The initially surprising conclusion that fungi grow in the absence of Tgs1 suggested there might be backup mechanisms to ensure the function of the many essential TMG-capped RNAs when the TMG modification is missing. This idea was confirmed by synthetic genetic array analysis in budding yeast, which revealed that the effects of ablating the TMG cap are buffered by spliceosome assembly factors that are themselves inessential for vegetative growth (5,8). Thus, nature has overlaid redundancy on an ancient eukaryal-specific RNA modification (the TMG cap) that participates in a defining step of eukaryal RNA biogenesis (spliceosome-catalyzed intron removal). This raises the important question: what aspects (if any) of the snRNP structure are perturbed when Tgs1 and TMG caps are absent?
We address this issue presently by comparing the protein and RNA compositions of RNPs purified from TGS1 and tgs1Δ strains of S. cerevisiae. There is an extensive literature concerning the protein content of individual yeast snRNPs and yeast spliceosome assembly intermediates (9–15), the protein contents of yeast snoRNPs (16,17) and genome-wide proteomic analyses of the physical associations underpinning the yeast protein ‘interactome’ (18,19). Our analysis of snRNPs and snoRNPs purified from wild-type TGS1 cells verifies many known associations and reveals new ones. We find that compositions of the splicesosomal snRNPs and box C/D and H/ACA snoRNPs are not altered significantly in tgs1Δ cells, with the exception that the yeast nuclear cap-binding protein (CBP) gains a specific association with the U1 snRNP, as a stoichiometric subunit, likely via binding to the residual m7G cap of U1 snRNA in tgs1Δ cells. Affinity-purification of CBP confirms its association with U1 snRNP and reveals a novel connection to snoRNP assembly intermediates. Ectopic CBP binding to U1 appears pertinent to the physiology of tgs1Δ cells, because we show that a hypomorphic mutation in the cap-binding pocket of CBP can suppress the tgs1Δ cold-sensitive growth defect.
MATERIALS AND METHODS
Yeast strains
Yeast TAP-fusion strains were purchased from Open Biosystems. Strains SMB1-TAP (Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 SMB1-TAP::HIS3MX6) or NAM8-TAP, MUD1-TAP, CBC2-TAP, LEA1-TAP, CBF5-TAP and NOP58-TAP were mixed with tgs1Δ cells (Matα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tgs1Δ::natR) and diploids were selected on an agar medium lacking histidine and containing nourseothricin (100 mg/l). Sporulation and dissection yielded His+ nourseothricin-resistant haploid progeny containing the desired TAP-fusion genes and the chromosomal tgs1Δ allele. Western blotting of whole-cell extracts with anti-TAP antibodies (Open Biosystems) confirmed that the respective TAP-tagged proteins were expressed.
Tandem affinity purifications
Yeast cells (e.g. TGS1 SMB1-TAP and tgs1Δ SMB1-TAP) were grown in YPD medium at 30°C. The culture volumes were incrementally increased to keep the cells in logarithmic growth phase to a final volume of 6 l. Cells were harvested by centrifugation at 4°C when A600 reached 2.0–2.5. All subsequent steps were performed at 4°C. Cells were washed twice with water and once with the AGK buffer (10 mM HEPES-KOH pH 7.9, 1.5 mM MgCl2, 200 mM KCl, 10% glycerol and 0.5 mM DTT). The cell pellets were then suspended in 16–20 ml AGK buffer; the cell suspensions were frozen in drops in liquid nitrogen and stored at −80°C. To prepare whole-cell extracts, the frozen pellets were ground with a mortar and pestle in liquid nitrogen to a fine powder, which was transferred to a beaker. The thawed cell pastes were gently stirred for 30 min followed by centrifugation at 18 000 rpm for 30 min in a Sorvall SS34 rotor to remove cell debris. The supernatants were centrifuged at 100 000g for 1 h. The extracts (∼12 ml) were then dialyzed against buffer D (20 mM HEPES–KOH, pH 7.9, 0.2 mM EDTA, 50 mM KCl, 0.5 mM DTT and 20% glycerol). After mixing with an equal volume of 20 mM Tris–HCl, pH 8.0, 150 mM NaCl and 0.2% NP40, the extracts were added to IgG-Sepharose resin (400 µl slurry) that had been equilibrated in IPP100 buffer (10 mM Tris–HCl, pH 8.0, 100 mM NaCl and 0.1% NP40) and mixed gently for 2 h on a nutator. The mixtures were poured into columns (Poly-Prep, 0.8 × 4 cm; BioRad) and the resins were washed thrice with 10 ml IPP100 and once with 10 ml TEV cleavage buffer (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 0.1% NP40, 0.5 mM EDTA and 1 mM DTT). The columns were closed and the IgG-Sepharose resins with bound protein were suspended in 1 ml TEV cleavage buffer and 10 µl of recombinant TEV protease (1.33 mg/ml) was added. The suspensions were gently mixed for 14–16 h. TEV eluates were collected and the resins rinsed with 1 ml TEV cleavage buffer. To the TEV eluates (2 ml), 3 ml of IPP150-CBB buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP40 and 10 mM β-mercaptoethanol) and 6 µl of 1 M CaCl2 were added. The TEV eluates were then added to calmodulin-Sepharose (300 µl slurry) that had been washed with IPP150-CBB buffer. The suspension was gently mixed for 1 h and then the resin was washed thrice with 10 ml aliquots of IPP150-CBB buffer and once with 1 ml of IPP150-CBB (low NP-40) buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.02% NP40 and 10 mM β-mercaptoethanol). Proteins bound to the calmodulin resin were eluted with 2 ml IPP150-CEB buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 0.02% NP40, 10 mM β-mercaptoethanol and 20 mM EGTA). Aliquots of the TAP preparations (1 ml for TGS1 SMB1-TAP, tgs1Δ SMB1-TAP, TGS1 NAM8-TAP, tgs1Δ NAM8-TAP, TGS1 MUD1-TAP, tgs1Δ MUD1-TAP, TGS1 CBC2-TAP and tgs1Δ CBC2-TAP; 1.5 ml for TGS1 LEA1-TAP and tgs1Δ LEA1-TAP; and 1.3 ml for TGS1 CBF5-TAP, tgs1Δ CBF5-TAP, TGS1 NOP58-TAP and tgs1Δ NOP58-TAP) were precipitated with TCA (25%). The proteins were resuspended in 0.1 M Tris–HCl (pH 8.0) and resolved by SDS–PAGE (8–16% acrylamide gradient gels). Polypeptides were visualized by staining the gels with Coomassie blue dye.
Mass spectrometry
Individual lanes of the stained SDS–polyacrylamide gels were sliced horizontally into 10–12 gel segments (of approximately 0.7 × 0.3 cm each). In situ trypsin digestion of polypeptides in each gel slice was performed as described (20). The tryptic peptides were purified using a 2-µl bed volume of Poros 50 R2 (Applied Biosystems, CA, USA) reversed-phase beads packed in Eppendorf gel-loading tips (21). The purified peptides were diluted to 0.1% formic acid and then subjected to nano-liquid chromatography coupled with tandem mass spectrometry (nanoLC–MS/MS) analysis as follows. Peptide mixtures (in 20 µl) were loaded onto a trapping guard column (0.3 × 5 mm Acclaim PepMap 100 C18 cartridge from LC Packings, Sunnyvale, CA, USA) using an Eksigent nano MDLC system (Eksigent Technologies, Inc. Dublin, CA, USA) at a flow rate of 20 µl/min. After washing, the flow was reversed through the guard column and the peptides eluted with a 5–45% acetonitrile gradient over 85 min at a flow rate of 200 nl/min, onto and over a 75-micron x 15-cm fused silica capillary PepMap 100 C18 column (LC Packings, Sunnyvale, CA, USA). The eluant was directed to a 75-micron (with 10-micron orifice) fused silica nano-electrospray needle (New Objective, Woburn, MA, USA). The electrospray ionization needle was set at 1800 V. A linear ion quadrupole trap-Orbitrap hybrid analyzer (LTQ-Orbitrap, ThermoFisher, San Jose, CA, USA) was operated in automatic, data-dependent MS/MS acquisition mode with one MS full scan (450–2000 m/z) in the Orbitrap analyzer at 60 000 mass resolution and up to five concurrent MS/MS scans in the LTQ for the five most intense peaks selected from each survey scan. Survey scans were acquired in profile mode and MS/MS scans were acquired in centroid mode. The collision energy was automatically adjusted in accordance with the experimental mass (m/z) value of the precursor ions selected for MS/MS. Minimum ion intensity of 2000 counts was required to trigger an MS/MS spectrum; dynamic exclusion duration was set at 60 s.
Initial protein/peptide identifications from the LC–MS/MS data were performed using the Mascot search engine (Matrix Science, version 2.3.02) and the Saccharomyces Genome Database (SGD, 6,717 sequences; Stanford University, CA, USA). The search parameters were as follows: (i) one missed cleavage tryptic site was allowed; (ii) precursor ion mass tolerance = 10 ppm; (iii) fragment ion mass tolerance = 0.8 Da; and (iv) variable protein modifications were allowed for methionine oxidation, cysteine acrylamide derivatization and protein N-terminal acetylation. MudPit scoring was typically applied using significance threshold score P < 0.01. Decoy database search was always activated and, in general, for merged LC–MS/MS analysis of a gel lane with P < 0.01, false discovery rate averaged ∼1%.
Scaffold (Proteome Software Inc., Portland, OR, USA), version 3_00_07 was used to further validate and cross-tabulate the MS/MS-based peptide and protein identifications. Protein and peptide probability was set at 95% with a minimum peptide requirement of 1. We utilized Scaffold to investigate proteins that were identified in individual gel lanes, as well as data obtained by merging information obtained from all the gel slices in a given gel lane. Total numbers of assigned spectra in pairs of TAP samples that were mass analyzed in back-to-back fashion were utilized to comment on the relative amounts of proteins identified in the paired samples.
RNA analysis
Aliquots (200 µl) of the eluates from calmodulin-Sepharose were extracted with phenol–chloroform and ethanol precipitated. RNAs were resuspended in TE (10 mM Tris-HCl, pH 7.4, 1 mM EDTA), formamide dye was added and nucleic acids were separated on 6% polyacrylamide/7 M urea gels in the TBE buffer. RNAs were visualized by staining using a silver staining kit (BioRad).
Cbc2 mutants and tests of their function
CEN LEU2 plasmid pRS415-Cbc2-TAP expressing a Cbc2-TAP fusion protein under the transcriptional control of the native CBC2 promoter was constructed as follows. First, we PCR-amplified a 550-bp segment from yeast genomic DNA of the region upstream of the CBC2 ORF using primers that introduced Not1 and BamH1 sites at the 5′ and 3′ ends, respectively. Second, we PCR amplified the 1.17-kbp Cbc2-TAP ORF using primers that introduced BamHI and XmaI restriction sites upstream of the start codon and downstream of the stop codon, respectively. Third, we PCR-amplified a 390-bp segment of genomic DNA 3′ of the CBC2 ORF with primers that introduced XmaI and XhoI restriction sites at the 5′ and 3′ end, respectively. The upstream, coding and downstream DNA segments were inserted sequentially into the pRS415 plasmid. Mutations Y24A and Y49A were introduced into Cbc2-TAP by two-stage PCR overlap extension with mutagenic primers. The mutated DNA fragments were digested with BamHI and XmaI and inserted into pRS415-Cbc2-TAP to replace the wild-type CBC2 gene. The inserts in each of the pRS415-based plasmids were sequenced completely to confirm that no unwanted coding changes were acquired during amplification and cloning. To assay the function of wild-type and mutated CBC2 alleles, isogenic cbc2Δ and cbc2Δ tgs1Δ cells (7) were transformed with the empty CEN LEU2 vector (pRS415), pRS415-Cbc2-TAP, pRS415-Cbc2(Y24A)-TAP or pRS415-Cbc2(Y49A)-TAP. The strains were also transformed in parallel with pUN100-TGS1 (5). Leu+ transformants were selected at 34°C. Cells were grown in liquid culture in SD–Leu medium. Serial dilutions of equal numbers of cells were spotted on SD–Leu agar; the plates were incubated at 18, 20, 25, 30, 34 and 37°C.
RESULTS AND DISCUSSION
Composition of affinity-purified SmB-containing snRNPs from TGS1 and tgs1Δ yeast cells
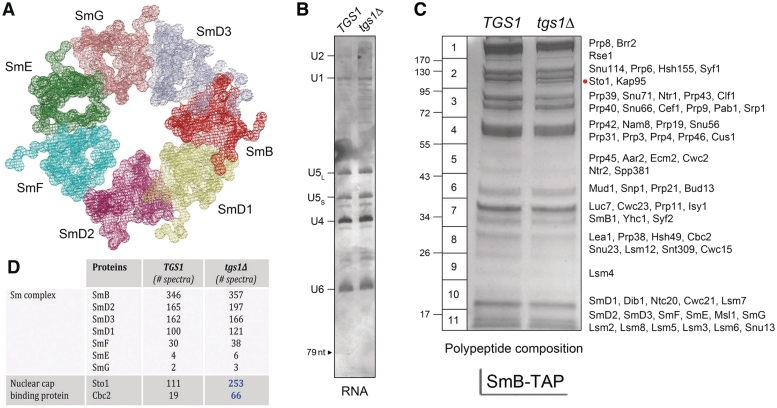
The core protein scaffold of the U1, U2, U4 and U5 snRNPs consists of a toroidal ring composed of seven Sm proteins (Figure 1A) (22–24). To analyze the impact of Tgs1 ablation on spliceosomal snRNPs, we constructed isogenic TGS1 and tgs1Δ yeast strains bearing chromosomal TAP-tagged SMB1 genes and then used the TAP method (12) to isolate SmB-containing complexes from soluble whole-cell extracts (25) of TGS1 and tgs1Δ cells. We analyzed in parallel samples of the purified SmB complexes isolated from equivalent numbers of TGS1 and tgs1Δ cells. The U RNA contents of deproteinized samples were gauged by urea–PAGE and silver staining (Figure 1B). We observed a typical spliceosomal U snRNA profile in the TGS1 SmB-TAP preparation (15), in which the U2 (1175-nt), U1 (568-nt), U5L (214-nt), U5S (179-nt), U4 (160-nt) and U6 (112-nt) species were most prominent and resolved according to their known sizes. (The large U2 snRNA yielded a prominent ‘negatively stained’ species during exposure to the silver reagent, the intensity of which faded with time as the other U RNAs acquired their positive stain; thus, the U2 band is not well defined in the image shown in Figure 1B.) Although the U6 snRNP does not contain SmB, the U6 snRNP is recovered during the TAP procedure by virtue of its association with the U4 and U5 snRNPs. The instructive point of the RNA analysis was that the amounts and electrophoretic mobilities of the spliceosomal U snRNAs from tgs1Δ SmB complexes were indistinguishable from the TGS1 U snRNAs (Figure 1B). We conclude that absence of TMG caps did not grossly affect the synthesis of the U snRNAs or their assimilation into SmB-containing snRNPs.
Figure 1.
Composition of SmB-containing RNPs from TGS1 and tgs1Δ cells. (A) Structure of the Sm ring of human U1 snRNP (23). (B) The RNA content of SmB-TAP preparations from TGS1 and tgs1Δ cells was analyzed by denaturing PAGE. The silver-stained gel is shown. The U snRNAs are indicated at left. The position of a 79-nt marker analyzed in parallel is denoted by the arrowhead at left. (C) The polypeptide composition of SmB-TAP preparations from TGS1 and tgs1Δ cells was analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown. The individual gel slices subjected to in situ proteolysis and LC–MS/MS analysis are demarcated on the left, along with the positions and sizes (kDa) of marker polypeptides analyzed in parallel. The identities of the relevant polypeptides in the gel slices are indicated on the right. The Sto1 polypeptide enriched in the tgs1Δ sample is denoted by the red dot. (D) The numbers of peptide spectra assigned with >95% confidence to the Sm ring subunits and the nuclear CBP subunits are tabulated. (See Supplementary Table S1 for a fuller account of the MS analysis.).
The protein contents of the purified SmB complexes were resolved by SDS–PAGE and stained with Coomassie blue dye (Figure 1C). Here, we see that the amounts of protein in each sample were quite similar in TGS1 versus tgs1Δ, as were the overall polypeptide profiles. Closer inspection highlighted a ∼100 kDa polypeptide (denoted by the red dot in Figure 1C) that was apparently more abundant in the tgs1Δ sample. The contents of the indicated horizontal slices of the TGS1 and tgs1Δ lanes were digested in situ with trypsin and the resulting peptides were analyzed by LC–MS/MS and correlated to specific peptide fragments identified in the S. cerevisiae proteome. The principal polypeptide components of the slices identified by mass spectrometry are specified to the right of the gel in Figure 1C. The total numbers of peptide spectra assigned to individual yeast proteins with >95% confidence were compiled and are presented in Figure 1D and Supplementary Table S1 according to the known functions and physical interactions of the proteins. (We omitted from this compilation any intrinsic ribosomal proteins, heat-shock proteins and protein chaperones.)
The first key finding was that the SmB contents of the TGS1 and tgs1Δ preparations were nearly identical (346 and 357 SmB spectra, respectively), signifying that pairwise comparison of the spectral counts for other SmB-associated proteins provides a simple index of whether any are depleted or enriched in the tgs1Δ. (We operationally defined a 2-fold difference in the paired samples as the criterion of enrichment/depletion of SmB-associated proteins for which at least 10 peptide spectra were detected in one of the samples.) A second important finding was that the peptide counts for the six other yeast Sm ring subunits were closely comparable in TGS1 and tgs1Δ (Figure 1D). Thus, the absence of TMG caps on the U snRNAs apparently did not affect the assembly of the seven-subunit yeast Sm complex.
The defining protein subunits of the individual yeast snRNPs were easily identified in the SmB-TAP preparations (most with spectral counts >100) and, in nearly all cases, their abundance did not vary more than 2-fold in TGS1 versus tgs1Δ cells (Supplementary Table S1). This applied to: (i) all 10 specific subunits of the yeast U1 snRNP: Prp39, Prp40, Snu71, Snu56, Snp1, Mud1, Luc7, Prp42, Nam8 and Yhc1; (ii) the U2-specific proteins Lea1, Rse1, Hsh155, Hsh49, Cus1 and Msl1; (iii) the major U5-specific proteins Snu114 and Prp8; (iv) most constituents of the U4–U5–U6 tri-snRNP and (iv) most of the many other yeast splicing factors that were associated with SmB-containing complexes (Supplementary Table S1). Also, the Lsm2-8 subunits of the U6 snRNP were equally abundant in the TGS1 and tgs1Δ SmB preparations (Supplementary Table S1), signifying that the absence of TMG caps did not adversely affect the physical interactions of the U4 and U5 snRNPs with the U6 snRNP.
Two minor components of the U5 snRNP—Aar2 and Lin1—met the criterion for under-representation in the tgs1Δ SmB preparation (Supplementary Table S1). For example, whereas 19 Aar2 spectra were present in the TGS1 sample, only 5 were detected in tgs1Δ (Supplementary Table S1). Aar2 is an essential protein that was characterized as a distinctive component of a 16 S U5 snRNP that included Prp8 and Snu114 and copurifed with the U1 snRNP1; in contrast, Aar2 was not found in the U4–U5–U6 tri-snRNP (26). Lin1 is inessential. Two proteins associated with the U4–U5–U6 tri-snRNP were under-represented in the tgs1Δ SmB preparation: Spp381 and Prp38 (Supplementary Table S1). Spp381 and Prp38 interact with each other genetically and physically (11,27). Four non-snRNP splicing factors were also under-represented by ≥2-fold in the tgs1Δ SmB fraction: Prp21, Cwc2, Bud13 and Ecm2 (Supplementary Table S1). Other proteins involved in RNA transactions that were partially depleted from the tgs1Δ SmB fractions were the essential RNA helicase Ded1 (32 peptide spectra in TGS1 versus two spectra in tgs1Δ) and Pbp1 (23 versus nine peptide spectra). Ded1 is implicated in both translation initiation and pre-mRNA splicing (28) and was identified previously as a constituent of the yeast penta-snRNP (14). Pbp1 is an inessential protein that interacts physically and genetically with poly(A)-binding protein Pab1 (29). Pab1 itself was associated with purified SmB, and its levels were comparable in the TGS1 and tgs1Δ preparations (61 and 72 spectra, respectively) (Supplementary Table S1).
Three yeast proteins were enriched in the tgs1Δ SmB preparation compared to TGS1: Brr1, Sto1 and Cbc2 (Supplementary Table S1 and Figure 1D). Brr1 is an inessential protein that facilitates U snRNP biogenesis when yeast cells are grown at 16–17°C (30). Only 1 Brr1 peptide spectrum was identified in the TGS1 SmB sample, but this value increased to 12 spectra in the tgs1Δ preparation (Supplementary Table S1). The increased Brr1 association with Sm-containing snRNPs in tgs1Δ cells is noteworthy in light of the strong synthetic growth defect of a tgs1Δ brr1Δ double-mutant over a range of temperatures (25–37°C) at which the respective single mutants grow normally (5). It is possible that Brr1 association with snRNPs lacking TMG caps helps to buffer the effects of their loss.
Sto1 and Cbc2 are the large and small subunits of the heterodimeric yeast nuclear CBP. Although the cap-binding pocket is located within the Cbc2 subunit (called CBP20 in mammals), heterodimerization with the large Sto1 subunit (CBP80 in mammals) is required for CBP binding to m7G caps (31,32). Both CBP subunits were associated with SmB-containing snRNPs in TGS1 cells (111 and 19 peptide spectra for Sto1 and Cbc2, respectively) and their abundance increased 2.5- and 3-fold (to 253 and 66 spectra, respectively) in the tgs1Δ SmB preparation (Figure 1D). Indeed, the MS analysis indicated that the ∼100 kDa polypeptide seen as increased in abundance in tgs1Δ SmB by SDS–PAGE (Figure 1C) corresponded to Sto1.
Our results suggest that the residual 5′ m7G caps of some of the spliceosomal U snRNAs in tgs1Δ cells are accessible to binding by nuclear CBP in the context of their native snRNPs. We hypothesize that the CBP enrichment seen in tgs1Δ snRNPs is a direct consequence of a change in the snRNA cap structure from TMG to m7G, rather than CBP being recruited indirectly to tgs1Δ Sm-containing snRNPs by other proteins. The CBP subunits are known to make many protein–protein interactions, chief among them being to the karyopherin (importin) complex that participates in RNP trafficking (33–35). We readily recovered the Kap95 (β subunit) and Srp1 (α subunit) components of the yeast karyopherin heterodimer in the SmB preparation from TGS1 cells (110 and 25 peptide spectra, respectively). Because Kap95/Srp1 levels did not change significantly in the tgs1Δ SmB fraction (91 and 31 peptide spectra) (Supplementary Table S1), it seemed unlikely that karyopherin was responsible for the observed CBP enrichment. We entertained the thought that enrichment for CBP in tgs1Δ SmB complexes could have occurred during the preparation of the yeast whole cells extracts, via post-facto binding of free soluble CBP to the accessible m7G caps of the U snRNAs. However, if this was true, we might also expect to detect enrichment of the yeast cap-binding translation factor Cdc33 (eIF4E) in the tgs1Δ SmB fraction. In fact, we detected no Cdc33-derived peptides in the SmB samples purified from either TGS1 or tgs1Δ extracts.
Given the initial findings that CBP enrichment is the most prominent, and arguably the most interpretable, difference in spliceosomal snRNPs in cells that lack TMG caps, we proceeded to investigate whether individual snRNPs were equally accessible to CBP and to query the relevance of CBP enrichment to the cold-sensitive growth defect characteristic of S. cerevisiae tgs1Δ mutants. These experiments are presented and discussed below.
Comparison of tandem affinity purified U1 snRNPs from TGS1 and tgs1Δ yeast cells
The yeast U1 snRNP is composed of the Sm ring plus ten additional U1-specific protein subunits (9,10). In order to examine the effects of the loss of TMG caps on U1 snRNP structure, we separately TAP-tagged two of the U1 subunits: Mud1 and Nam8. Nam8 is inessential for yeast vegetative growth, but is essential for yeast sporulation as it promotes splicing of a small set of specific yeast mRNAs (with non-consensus 5′ splice sites or non-consensus branch point sequences) that encode proteins required for meiotic recombination and cell division (36). Mud1, the yeast homolog of mammalian U1A, is also inessential for vegetative growth of S. cerevisiae (37). Ablation of Mud1 has no apparent effect on splicing in vitro or in vivo when wild-type U1 snRNA is present. However, Mud1 becomes important when yeast rely on a mutated U1 snRNA, leading to the suggestion that Mud1 helps maintain the U1 snRNP in an active conformation (37). Note that nam8Δ and mud1Δ both display strong synthetic growth defects with tgs1Δ (5).
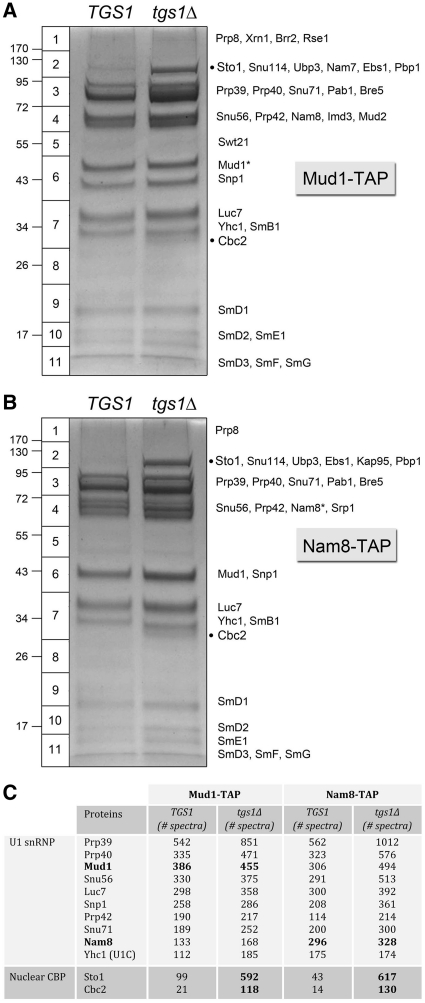
Here, we constructed isogenic TGS1 and tgs1Δ yeast strains bearing chromosomal TAP-tagged NAM8 or MUD1 genes. The fact that the NAM8-TAP tgs1Δ and MUD1-TAP tgs1Δ strains grew as well as their NAM8-TAP TGS1 and MUD1-TAP TGS1 counterparts (data not shown) verified that the TAP-tag did not compromise the function of either of these two U1 subunits. We then used the TAP method to isolate Mud1- or Nam8-containing complexes from soluble whole-cell extracts of TGS1 and tgs1Δ cells. The protein contents of the purified complexes were resolved by SDS–PAGE and stained with Coomassie blue dye (Figure 2A and B). The principal polypeptides of the indicated horizontal slices of the TGS1 and tgs1Δ lanes were determined by in situ proteolysis and LC–MS/MS and are listed next to the Mud1-TAP and Nam8-TAP gels in Figure 2A and B. Two themes emerged from the results. First, the polypeptide profiles were similar in TGS1 versus tgs1Δ, except for the ∼100 kDa and ∼30 kDa polypeptides (denoted by dots) that were uniquely enriched in the tgs1Δ preparations. These two species correspond to Sto1 and Cbc2. Second, the profiles of the Mud1-TAP and Nam8-TAP preparations were nearly identical to each other, except for the presence of a major polypeptide in slice 6 in Mud1-TAP that was absent in Nam8-TAP and a novel polypeptide in slice 4 in Nam-TAP that was absent from Mud1-TAP. The novel species corresponded to the tagged polypeptide themselves (Mud1* and Nam8* in Figure 2A and B) that displayed slower electrophoretic mobility because of the residual 5-kDa calmodulin-binding domains appended to their C-termini.
Figure 2.
Composition of affinity-purified U1 snRNPs from TGS1 and tgs1Δ cells. The polypeptide compositions of Mud1-TAP (A) and Nam8-TAP (B) preparations from TGS1 and tgs1Δ cells were analyzed by SDS–PAGE. The Coomassie blue-stained gels are shown. The individual gel slices subjected to proteomics analysis are demarcated on the left, along with the positions and sizes (kDa) of marker polypeptides analyzed in parallel. The identities of the relevant polypeptides in the gel slices are indicated on the right. The CBP subunits Sto1 and Cbc2 that were enriched in the tgs1Δ sample are denoted by black dots. The numbers of peptide spectra assigned to the U1-specific snRNP subunits and the nuclear CBP subunits are tabulated in (C). (See Supplementary Table S2 for a fuller account of the MS analysis.).
The total numbers of peptide spectra assigned to individual yeast proteins in the Mud1-TAP and Nam8-TAP preparations are shown in Figure 2C and Supplementary Table S2. The major components in all samples (yielding between 100 and 1000 peptide spectra each) were the 10 known U1-specific snRNP subunits: Prp39, Prp40, Snu71, Snu56, Snp1, Mud1, Luc7, Prp42, Nam8 and Yhc1 (Figure 2C). The other major constituents were the core Sm proteins (Supplementary Table S2). In the Mud1-TAP samples, the TGS1 and tgs1Δ preparations yielded 385 and 455 Mud1 spectra, respectively (tgs1Δ/TGS1 ratio of 1.2). In the Nam8-TAP samples, the TGS1 and tgs1Δ preparations yielded 296 and 328 Nam8 spectra, respectively (tgs1Δ/TGS1 ratio of 1.1). Ratio-normalized comparisons of the spectral counts for non-tagged U1 snRNP proteins were used to gauge whether any were depleted or enriched in the tgs1Δ preparations (applying the same 2-fold difference in the paired samples as the criterion of enrichment/depletion). By this test, the U1 snRNPs from tgs1Δ cells were not depleted for any of the normal constituents of U1 snRNPs isolated from TGS1 cells. Thus, the absence of TMG caps on the U1 snRNA did not affect the assembly of the U1 snRNP.
Nuclear CBP is a stoichiometric component of affinity-purified U1 snRNPs from tgs1Δ yeast
The striking finding was that yeast CBP, which is a minor constituent of U1 snRNPs in TGS1 cells, became a stoichiometric subunit of U1 snRNPs in tgs1Δ cells (Figure 3A and B). In the tgs1Δ Mud1-TAP sample, we detected 592 Sto1 spectra and 118 Cbc2 spectra, compared with 99 Sto1 and 21 Cbc2 spectra in the TGS1 preparation, respectively. The tgs1Δ/TGS1 ratios of 6.0 for Sto1 and 5.6 for Cbc2, when normalized to the Mud1 counts, represent 5-fold enrichments for nuclear CBP in the Mud1 affinity-purified tgs1Δ U1 snRNPs. In the tgs1Δ Nam8-TAP sample, we detected 617 Sto1 spectra and 130 Cbc2 spectra, compared with 43 Sto1 and 14 Cbc2 peptides in the TGS1 sample, respectively. Here, the tgs1Δ/TGS1 ratios of 14.3 for Sto1 and 9.3 for Cbc2, respectively, when normalized to the Nam8 counts, represent an order-of-magnitude enrichment for CBP in the Nam8 affinity-purified tgs1Δ U1 snRNPs. We detected little or no association of Cdc33 with U1 snRNPs purified from the extracts of either TGS1 or tgs1Δ cells, arguing against post-facto exposure of the U1 cap to all available cellular cap binding proteins. [Low levels of translation initiation factor eIF4G were detected in the U1 snRNP preparations (Supplementary Table S2), consistent with a report that yeast eIF4G is present in the nucleus and interacts with the U1 snRNP via the Snu71 subunit (38).] Karyopherin subunits Kap95 and Srp1 were detected in the U1 snRNPs, though their levels did not rise in the tgs1Δ samples that were strongly enriched for nuclear CBP. We surmise that the 5′ cap of the U1 snRNP in tgs1Δ cells is fully accessible to, and occupied by, the nuclear CBP.
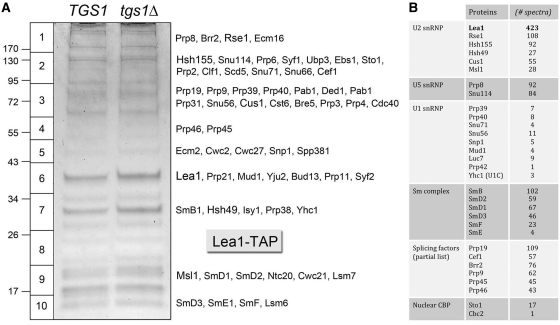
Figure 3.
Affinity-purification of Lea1-containing U2 snRNPs from TGS1 and tgs1Δ cells. The polypeptide composition of Lea1-TAP preparations from TGS1 and tgs1Δ cells was analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown in (A). The individual gel slices of the tgs1Δ lane that were subjected to proteomics analysis are demarcated on the left, along with the positions and sizes (kDa) of marker polypeptides analyzed in parallel. The identities of the relevant polypeptides in the tgs1Δ slices are indicated on the right. The numbers of peptide spectra assigned to specific subunits of the U2, U5 and U1 snRNPs, the core Sm complex and the nuclear CBP subunits are tabulated in (B), along with a partial list of spectral counts derived from splicing factors. (See Supplementary Table S3 for a fuller account of the MS analysis.).
These findings and others (38) raise interesting questions concerning the sub-stoichiometric association of CBP with U1 snRNPs from TGS1 cells. The CBP-U1 snRNP association was a consistent finding in our U1 snRNPs purified by TAP-tagging two different intrinsic U1 subunits. Thus, it is unlikely to be either adventitious (e.g. a non-specific contaminant of TAP purification; see below) or reflective of indirect associations via other snRNPs or non-snRNP proteins in the U1 preparations. In that respect, we did find that low levels of Prp8- and Snu114-containing U5 snRNPs co-purified with our TAP-tagged U1 snRNPs (Supplementary Table S2), which is consistent with prior reports (9,10,26). To get a sense of the level of U5 snRNP association with purified U1 snRNP versus their abundance in the total spliceosomal U snRNP pool, we calculated ratios of the U5-specific Snu114 peptide spectral counts to the U1-specific Prp39 spectra in the TGS1 Mud1-TAP (27/542 = 0.05), Nam8-TAP (15/562 = 0.027) and SmB-TAP (507/219 = 2.3) preparations. By this test, we estimated that the U1 snRNP was enriched 45- to 85-fold versus U5 snRNP during the Mud1 and Nam8 affinity purifications. It is therefore doubtful that the basal TGS1 and increased tgs1Δ levels of CBP in the U1 snRNP preparations could be caused by the residual U5 snRNP. There were only trace levels of U2-specific peptides detectable in the U1 snRNP preparations and few or no peptides derived from tri-snRNP components or other splicing factors (Supplementary Table S2). The LC–MS/MS analysis was notable for the presence of substantial amounts of poly(A)-binding protein Pab1 in each of the U1 snRNP preparations, which did not vary significantly between paired TGS1 and tgs1Δ samples (Supplementary Table S2).
There are three distinct scenarios to explain the association of CBP with U1 snRNP in TGS1 cells, whereby: (i) CBP interacts physically with one or more of the U1 proteins, but not with the U1 snRNA TMG cap; (ii) CBP binds to the U1 snRNA TMG cap, albeit with lower affinity than it would to an m7G cap; or (iii) a subpopulation of U1 snRNPs in wild-type cells that has m7G caps instead of TMG caps is bound with high affinity by CBP. Although prevailing models of pre-mRNA splicing invoke a role for CBP in recruiting the U1 snRNP to the 5′ splice site, this is presumed to entail protein–protein interactions between U1 snRNP and a CBP heterodimer bound to the m7G cap of the pre-mRNA substrate (39–43). To our knowledge, there has been little consideration given to the possibility of CBP interaction with the U1 snRNA cap, or the prospect that not all U1 snRNPs have TMG caps in wild-type cells. The favored method to identify and study the spliceosomal U RNAs entails the use of anti-TMG antibodies as affinity purification reagents. Accordingly, snRNAs or snRNPs without hypermethylated caps will be missed or underrepresented and there will be a strong bias toward a population of snRNAs and snRNP that have antibody-accessible 5′ caps. Indeed, the exclusion principle teaches us that anti-TMG cap-affinity purification will systematically ignore any yeast snRNPs or snRNAs that have a cellular protein bound to their cap structures, be it CBP or an as yet unknown yeast TMG-specific cap receptor. In this light, it is worth underscoring that previous proteomics analyses of yeast U1 snRNPs that were affinity purified by a two-step procedure—using anti-TMG antibodies as the first affinity step and Ni-affinity chromatography of His-tagged Snp1 as the second step—did not result in reported detection of the yeast CBP subunits as constituents of the U1 snRNP preparation (9,10).
Affinity purification of Lea1-containing U2 snRNPs from TGS1 and tgs1Δ yeast cells
The yeast U2 snRNP subunit Lea1 (a homolog of metazoan U2A′) has been TAP-tagged and exploited for purification of the yeast U2 snRNP and the identification of additional U2-specific subunits (13,44). Here, we constructed isogenic TGS1 and tgs1Δ yeast strains bearing chromosomal TAP-tagged LEA1 genes. We then used the TAP method to isolate Lea1-containing complexes from soluble whole-cell extracts of TGS1 and tgs1Δ cells. The protein contents of the purified complexes were resolved by SDS–PAGE and stained with Coomassie blue dye, which revealed no gross differences between the two samples (Figure 3B). We performed in situ proteolysis and LC–MS/MS analysis on the Lea1-TAP sample from tgs1Δ cells, with special attention to the presence and abundance of CBP. The polypeptide contents and spectral counts are compiled in Supplementary Table S3; selected components are highlighted in Figure 3A. As expected, Lea1 was the most abundant component (Figure 3B, gel slice 6) with 423 peptide spectra. The other intrinsic U2 snRNP proteins (Rse1, Hsh155, Hsh49, Cus1 and Msl1) were also prominent, signifying that U2 snRNP biogenesis was not grossly perturbed in the absence of TMG caps (Figure 3A). The Lea1-TAP preparation resulted in co-purification of: (i) intrinsic U5 and U6 proteins, presumably as the U4/U5/U6 tri-snRNP; (ii) the multi-subunit ‘Nineteen complex’ of the spliceosome (including the eponymous Prp19 with 102 peptide spectra); (iii) a slew of additional splicing factors and (iv) relatively low levels of all 10 protein subunits of the U1 snRNP (Supplementary Table S1 and Figure 3A). The CBP subunits Sto1 (17 spectra) and Cbc2 (one spectrum) were not enriched in the affinity-purified U2 snRNP from tgs1Δ cells. This is in stark contrast to affinity purified U1 snRNPs from tgs1Δ cells, of which CBP is an apparently stoichiometric component. [There were no Cdc33(eIF4E) spectra detected in the Lea1-TAP sample.] We conclude that nuclear CBP specifically accesses the 5′ m7G cap of U1 snRNA in tgs1Δ cells, whereas the 5′ m7G cap of U2 snRNA (and also of U5) is not bound by the nuclear CBP, conceivably because the U2 and U5 caps are not freely exposed in the context of the snRNP.
Affinity purification of box C/D snoRNPs from TGS1 and tgs1Δ yeast cells
The box C/D family of snoRNAs in budding yeast includes: (i) the TMG-capped U3 snoRNA essential for endonucleolytic processing of pre-18S ribosomal RNA; (ii) the non-essential TMG-capped snR4 snRNA, of as yet unknown function; and (iii) numerous other snoRNAs that direct 2′-O-methylation of ribosomal RNA. The yeast C/D snoRNAs are assembled into a ribonucleoprotein (RNP) composed of four core polypeptides: Nop1 (the 2′-O-methyltransferase subunit, also known as fibrillarin), Nop56, Nop58 and Snu13. The nuclear rRNA-processing steps occur in the context of a pre-ribosomal particle composed of intrinsic ribosomal proteins and a multitude of extrinsic ribosome maturation factors. The nuclear pre-rRNA cleavages flanking the 18S rRNA are guided by base pairing of the 5′ external spacer to the U3 snoRNA (45). U3 snoRNP has been isolated from budding yeast as an even larger ribonucleoprotein complex—with dozens of associated proteins (many designated Utp, for ‘U-Three-Protein’). This complex, called the small subunit processosome, is a ribosome assembly intermediate (17,46).
Mouaikel et al. (1) showed that tgs1Δ cells lack TMG caps on their U3 snoRNAs. A pertinent question is whether lack of TMG caps affects the composition of the U3 snRNP or its association with the SSU processosome, and whether the residual m7G caps of U3 snoRNAs (or other capped C/D snoRNAs) in tgs1Δ cells are bound by nuclear CBP. To address this issue, we made isogenic TGS1 and tgs1Δ yeast strains bearing chromosomal TAP-tagged NOP58 genes and then tandem affinity-purified Nop58-containing complexes from soluble whole-cell extracts of TGS1 and tgs1Δ cells. The SDS–PAGE analysis revealed similar polypeptide compositions (Figure 4B). Proteolysis and LC–MS/MS analysis were performed on the Nop58-TAP sample from tgs1Δ cells. Nop58 was the most abundant component (Figure 4B, gel slice 4) with 678 peptide spectra (Figure 4A). The box C/D snoRNP core proteins Nop56 (507 spectra) and Nop1 (265 spectra) were copurifed with Nop58-TAP, as was the small snoRNP subunit Snu13. (The relatively low yield of eight Snu13 peptides was not caused by tgs1Δ; analysis of the corresponding gel segment from the TGS1 sample yielded only six Snu13 spectra.) The full complement of proteins comprising the yeast SSU processosome was copurified with Nop58-TAP (Figure 4). So too were many other nucleolar proteins involved in rRNA biogenesis, including constituents of the larger 90S pre-ribosome complex (Supplementary Table S4). We conclude that neither the composition of the U3 snRNP nor its association with the SSU processosome is grossly affected in the absence of TMG caps. Moreover, we detected no peptide spectra derived from CBP subunits Sto1 or Cbc2 (or from Cdc33) in the Nop58-TAP sample from tgs1Δ cells, signifying that the residual U3 m7G cap was not accessible to nuclear CBP.
Figure 4.
Affinity-purification of Nop58-containing C/D snoRNPs from TGS1 and tgs1Δ cells. The polypeptide composition of Nop58-TAP preparations from TGS1 and tgs1Δ cells was analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown in (A). The individual gel slices of the tgs1Δ lane that were subjected to proteomics analysis are demarcated on the left, along with the positions and sizes (kDa) of marker polypeptides analyzed in parallel. The identities of the relevant polypeptides in the tgs1Δ slices are indicated on the right. The numbers of peptide spectra assigned to the box C/D snoRNP core subunits and the constituents of the SSU processosome are tabulated in (B). (See Supplementary Table S4 for a fuller account of the MS analysis.).
Affinity purification of H/ACA snoRNPs from TGS1 and tgs1Δ yeast cells
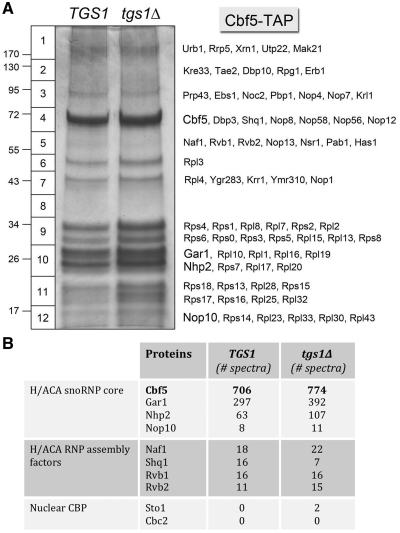
The yeast H/ACA snoRNAs guide site-specific pseudouridylation of rRNA and snRNA and the nucleolytic processing of 18S rRNA (47). H/ACA snoRNAs assemble into a snoRNP composed of four protein subunits: Cbf5 (pseudouridine synthase), Gar1, Nhp2 and Nop10. The yeast snR8, snR11, snR31, snR33, snR35 and snR42 H/ACA snoRNAs are modified by TMG caps in TGS1 cells, but not in tgs1Δ cells (1). To evaluate whether lack of TMG caps affects the composition of H/ACA snoRNPs, and if the residual m7G caps of H/ACA snoRNAs in tgs1Δ cells are bound by nuclear CBP, we tandem affinity-purified Cbf5-containing complexes from soluble whole-cell extracts of TGS1 and tgs1Δ cells. SDS–PAGE analysis revealed similar polypeptide compositions (Figure 5A). Proteolysis and LC–MS/MS analysis were performed on the Cbf5-TAP sample from TGS1 and tgs1Δ cells. Cbf5 was the most abundant component in the TGS1 and tgs1Δ preparations (Figure 5A, gel slice 4) with 706 and 774 peptide spectra, respectively (Figure 5B). The H/ACA snoRNP core proteins Gar1 (297 and 392 spectra in TGS1 and tgs1Δ, respectively), Nhp2 (63 and 107 spectra) and Nop10 (8 and 11 spectra) copurifed with Cbf5-TAP (Figure 5A and B). Also copurifying with TAP-Cbf5 were: (i) the essential H/ACA snoRNP assembly factors Shq1 and Naf1, which bind directly to Cbf5; and (ii) the Rvb1–Rvb2 complex that functions with Hsp90 in H/ACA RNP assembly (Figure 5B) (47). Thus, we conclude that the assembly and composition of core H/ACA snoRNPs were not grossly affected by the absence of TMG caps. The Cbf5-TAP purifications also captured a large number of nucleolar proteins implicated in rRNA biogenesis (Supplementary Table S5), suggesting that we had copurified ribosome assembly and processing intermediates with which H/ACA snoRNPs were associated. Nuclear CBP was not a significant constituent of the Cbf5-TAP preparation from tgs1Δ cells, which yielded only two peptide spectra derived from Sto1 and no peptides from Cbc2 (or from Cdc33). In summary, our proteomics analysis of bulk and individual snRNPs and snoRNPs underscores the specific gain of nuclear CBP as a component of the U1 snRNP.
Figure 5.
Affinity-purification of Cbf5-containing H/ACA snoRNPs from TGS1 and tgs1Δ cells. (A) The polypeptide composition of Cbf5-TAP preparations from TGS1 and tgs1Δ cells was analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown. The individual gel slices subjected to proteomics analysis are demarcated on the left. The identities of the relevant polypeptides are indicated on the right. (B) The numbers of peptide spectra assigned to the H/ACA snoRNP core subunits, H/ACA RNP assembly factors and the nuclear CBP subunits are tabulated. (See Supplementary Table S5 for a fuller account of the MS analysis).
Affinity purification of yeast nuclear cap binding protein
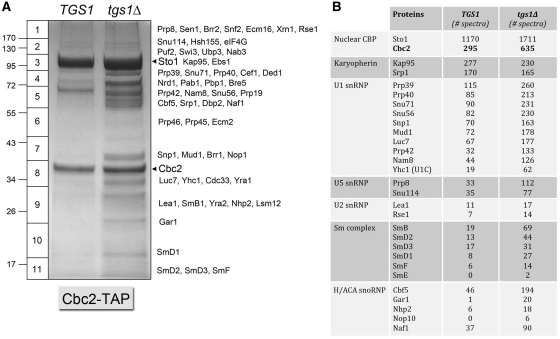
Nuclear CBP is implicated in multiple aspects of cellular RNA metabolism, including splicing, polyadenylation, termination, export and translation. CBP associates with the RNA polymerase II transcription elongation complex, presumably via binding to the m7G cap of nascent mRNAs (41). There are many reported physical and genetic interactions of the yeast CBP subunits Sto1 and Cbc2 with other yeast proteins involved in RNA metabolism (34,42,48,49). Here we applied the TAP method to affinity purify the cap-binding Cbc2 subunit of yeast CBP from extracts of TGS1 and tgs1Δ cells (Figure 6B). SDS–PAGE analysis revealed two predominant polypeptides, in gel slices 3 and 8, corresponding to CBP subunits Sto1 and Cbc2, respectively. Consistent with the more intense dye staining in the tgs1Δ lane, LC–MS/MS analysis recovered about 2-fold higher levels of Sto1 and Cbc2 peptide spectra in the tgs1Δ sample (1711 for Sto1 and 635 for Cbc2) than in the TGS1 sample (1170 for Sto1 and 295 for Cbc2). The Kap95/Srp1 karyopherin heterodimer and the U1 snRNP subunits were the most abundant proteins copurifying with nuclear CBP (Figure 6A and Supplementary Table S6). RNA analysis by denaturing PAGE and silver staining showed that U1 snRNA was the predominant species in the Cbc2-TAP preparations (Supplementary Figure S1). The U2, U4 and U6 snRNAs were hardly detectable by silver staining and were clearly under-represented in Cbc2-TAP, compared to the SmB-TAP preparation analyzed in parallel that contained a comparable level of U1 snRNA (Supplementary Figure S1). Diagnostic subunits of the U5 and U2 snRNPs were detectable in the Cbc2-TAP preparations, albeit in lower abundance than the U1 snRNP subunits (Figure 6A). It was noteworthy that each of the U1 subunits was 2- to 4-fold more abundant in the tgs1Δ sample versus TGS1 (Figure 6A), as was the U1 snRNA (Supplementary Figure S1), consistent with the higher level of input CBP. The same trend was seen for the core subunits of the Sm ring (Figure 6A). In contrast, the karyopherin signal was the same in the paired samples (Figure 6A), suggesting that a higher fraction of the CBP pool in tgs1Δ cells interacts with partners other than karyopherin.
Figure 6.
Affinity-purification of nuclear cap binding protein from TGS1 and tgs1Δ cells. The polypeptide compositions of Cbc2-TAP preparations from TGS1 and tgs1Δ cells were analyzed by SDS–PAGE. The Coomassie blue-stained gel is shown in (A). The individual gel slices subjected to proteomics analysis are demarcated on the left. The identities of the relevant polypeptides in the gel slices are indicated on the right. The CBP subunits Sto1 and Cbc2 are denoted by arrowheads. The numbers of peptide spectra assigned to CBP, karyopherin, U1, U5 and U2 snRNP components, the core Sm complex and H/ACA snoRNP components are tabulated in (B). (See Supplementary Table S6 for a fuller account of the MS analysis).
The more complete peptide analysis of the Cbc2-TAP samples (exclusive of ribosomal proteins, chaperones, etc.) is compiled in Supplementary Table S6. In addition to the individual U1, U5 and U2 snRNP components, we recovered peptides derived from 22 known pre-mRNA splicing factors. These results presumably reflect the presence of CBP bound to the mRNA cap and/or the U1 snRNP in the context of pre-mRNA-bound spliceosomes. It is in intriguing that splicing factor Swt21 was detected in the tgs1Δ CBP sample (eight peptides), but not in the TGS1 sample, in light of the reports that Swt21, which is inessential per se, has synthetic lethal interactions with both Tgs1 and Sto1 (5,49).
Cbc2-TAP also recovered many yeast proteins implicated in translation, RNA binding, transport and turnover (Supplementary Table S6). In general, these proteins were of similar abundance in the TGS1 and tgs1Δ samples. The subunits of the Swi/Snf chromatin remodeling complex were recovered with CBP in both samples; this interaction has not been reported previously and may be pertinent to recent findings that yeast CBP stimulates RNA polymerase II pre-initiation complex formation at a subset of yeast promoters (50).
A possible connection between CBP and H/ACA snoRNP biogenesis
Notwithstanding the 2-fold difference in applied CBP in the TGS1 and tgs1Δ lanes, it was apparent from the Coomassie blue-stained polypeptide profile (Figure 6B) and the MS analysis (Supplementary Table S6) that the tgs1Δ sample was enriched significantly for multiple polypeptides, many of which are associated with snoRNPs and rRNA biogenesis. For example, all four components of the H/ACA snoRNA core were present in the Cbc2-TAP preparation from tgs1Δ cells (Cbf5, Gar1, Nhp2 and Nop10, yielding 194, 20, 18, and 6 peptide spectra, respectively) at 3- to 20-fold higher levels than in the TGS1 preparation (Figure 6A). Closer inspection revealed that the relative amounts of H/ACA snoRNA core subunits recovered with CBP were highly skewed toward Cbf5 per se, and against Gar1. For example, compare the Cbf5/Gar1 ratio in Cbf5-TAP from tgs1Δ cells (774/392 = 2) to the Cbf5/Gar1 ratio in Cbc2-TAP from tgs1Δ cells (194/20 = 9.7). Cbf5 was also uniquely enriched versus Gar1 among the core H/ACA subunits in Cbc2-TAP from TGS1 cells (where Cbf5, Gar1, Nhp2 and Nop10, yielded 46, 1, 6 and 0 peptide spectra, respectively). In contrast, the Cbf5/Nhp2 ratios in the Cbc2-TAP samples (46/6 = 8 in TGS1 and 194/18 = 11 in tgs1Δ) were similar to that seen in the Cbf5-TAP preparations (706/63 = 11 in TGS1 and 774/107 = 7.2 in tgs1Δ).
Another instructive finding was that the Cbf5-binding H/ACA snoRNP assembly factor Naf1 was also enriched in the Cbc2-TAP preparations [37 and 90 peptide spectra in the TGS1 and tgs1Δ samples, with Cbf5/Naf1 ratios of 1.2 and 2.2, respectively (Figure 6A)] compared to Naf1′s lower abundance in relation to Cbf5 in the TGS1 and tgs1Δ Cbf5-TAP preparations [Cbf5/Naf1 ratios of 39 and 35, respectively (Figure 5B)]. No peptides derived from the H/ACA assembly factor Shq1 were detected in the Cbc2-TAP samples. Collectively, our proteomics results indicate that yeast CBP is associated with a H/ACA snoRNP assembly intermediate that includes Cbf5, Naf1 and Nhp2, but not Gar1 (47). That we detect the association of CBP with a putative Cbf5 assembly intermediate in TGS1 and tgs1Δ cells (and to a greater degree when TMG caps are lacking)—combined with the fact that CBP is virtually absent from the affinity-purified Cbf5 preparation (Supplementary Table S5) that we presume consists predominantly of mature H/ACA snoRNPs—argues that the CBP-associated Cbf5-Naf1-Nhp2 complex captures the snoRNP assembly pathway at a step prior to TMG-capping of the yeast H/ACA snoRNAs. Our results resonate with those of Fortes et al. (42), who identified mutants alleles of CBF5 as synthetic lethal in an otherwise viable sto1Δ cbc2Δ strain.
CBP and box C/D snoRNPs
The Nop1, Nop56 and Nop58 subunits of yeast C/D snoRNAs were present in the tgs1Δ Cbc2-TAP preparation (yielding 17, 20 and 34 peptide spectra, respectively), but either missing or less abundant in the TGS1 Cbc2-TAP sample (where we detected 0, 0 and 5 spectra, respectively) (Supplementary Table S6). It was notable than not a single ‘Utp’ component of the U3 snoRNP-containing SSU processosome was identified in the Cbc2-TAP samples; this being in stark contrast to the Nop58-TAP preparations, where the SSU processosome is abundant (Figure 4). These results signify that mature, functional U3 snoRNPs are not associated with CBP. Given that CBP was not detectable in affinity purified ‘mature’ Nop58-containing C/D snoRNPs, we speculate that CBP interacts transiently with a C/D snoRNP assembly intermediate. Here again, our results underscore the findings of Fortes et al. (42), who identified mutant alleles of NOP58 as synthetic lethal in a sto1Δ cbc2Δ strain and showed that sto1Δ and cbc2Δ mutants accumulated rRNA processing intermediates indicative of a defect in the endonucleolytic steps of 18S rRNA biogenesis.
Is CBP relevant to the tgs1Δ phenotype?
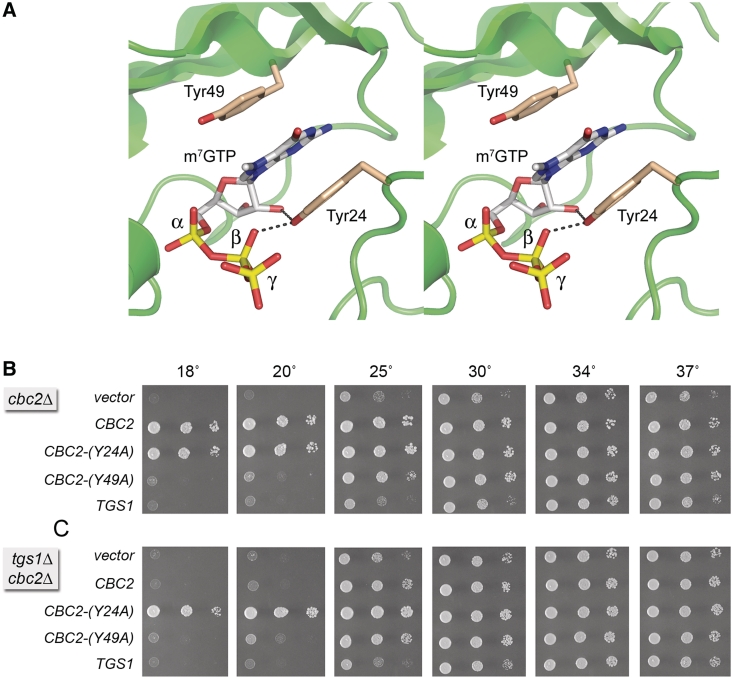
Saccharomyces cerevisiae tgs1Δ cells fail to grow at low temperatures (18–20°C). In light of the proteomics study discussed above, we considered the idea that gain of binding of yeast nuclear CBP to the 5′ m7G cap of U1 snRNA in tgs1Δ cells might account for, or contribute to, the cold sensitive phenotype of tgs1Δ cells. If this model is correct, then we reasoned that a hypomorphic mutation in the Cbc2 subunit that weakens cap binding, without grossly compromising the CBP function, might rescue the tgs1Δ cold sensitivity. Structural and functional studies of human CBP clearly demarcated the cap-binding site within the CBP20(Cbc2) subunit, while affirming that heterodimerization with CBP80(Sto1) is essential for the cap binding activity (31,32). The m7G nucleoside of the cap is sandwiched in a π stack between two conserved tyrosines: Tyr20 and Tyr43 in human CBP20, corresponding to Tyr24 and Tyr49 in yeast Cbc2 (Figure 7A), respectively. Mutations Y20A and Y43A in human CBP20 diminished affinity of the CBC complex for capped RNA and m7GTP ligands, with Y43A having a greater impact than Y20A (31,51).
Figure 7.
Mutation of the cap-binding pocket of Cbc2 suppresses the cold-sensitive phenotype of tgs1Δ. (A) Stereo view of the CBP20 subunit of the human nuclear CBP heterodimer (from pdb 1H2T) highlighting the π-stacked sandwich of the m7G cap nucleoside between two conserved tyrosines. The tyrosines are labeled in the figure according to their residue numbers in the homologous Cbc2 subunit of yeast CBP. (B) The growth phenotypes of cbc2Δ cells harboring CEN LEU2 plasmids with wild-type CBC2, the mutant alleles CBC2-Y24A, CBC2-Y49A or wild-type TGS1 were compared by spotting 3-µl aliquots of 10-fold serial dilutions of cells (from liquid cultures grown to mid-log phase at 34°C and adjusted to A600 of 0.1) to SD–Leu agar and incubating the plates at the indicated temperatures. (C) The growth phenotypes of cbc2Δ tgs1Δ cells containing LEU2 plasmids with the indicated genes were assessed as described in (B).
Here, we introduced the corresponding Y24A and Y49A mutations into a plasmid-borne yeast CBC2 gene under the control of its native promoter and then tested their activity in vivo in a cbc2Δ yeast strain in parallel with a wild-type CBC2 plasmid and an empty CEN vector. The cbc2Δ cells grew well on YPD agar at 34°, but were slow growing at 25, 30 and 37°, as gauged by the colony size. cbc2Δ cells failed to grow at 18 and 20° (Figure 7B). Normal growth was restored at all temperatures after transformation of cbc2Δ with the wild-type CBC2 plasmid, but not with a plasmid bearing an extra copy of TGS1 (Figure 7B). The instructive findings were that: (i) Y24A supported growth at all temperatures, as well as wild-type CBC2; (ii) Y49A was hypomorphic in vivo, i.e. the cbc2-Y49A allele rescued the slow growth of cbc2Δ at 30 and 37°, complemented partially at 25°, but did not support growth at 18 or 20° (Figure 7B). Thus, Y24A emerged as a candidate worth testing for its genetic interaction with tgs1Δ at cold temperatures.
CBP2-Y24A suppresses the tgs1Δ cold-sensitive phenotype
Genetic interactions between Tgs1 and Cbc2 were tested by plasmid complementation in a tgs1Δ cbc2Δ double-mutant. The tgs1Δ cbc2Δ cells (vector in Figure 7C) displayed the same cs lethality at 18–20° seen with the cbc2Δ single mutant (TGS1 in Figure 7C) (7) and the tgs1Δ single mutant (CBC2 in Figure 7C). The cbc2-Y49A allele conferred no growth improvement on tgs1Δ cbc2Δ cells at 18 and 20°, as expected. The striking finding was that the active CBC2-Y24A allele completely suppressed the cold-sensitive growth defect caused by tgs1Δ (compare wild-type CBC2 and CBC2-Y24A at 18 and 20°C in Figure 7C).
We infer that the cs phenotype of tgs1Δ is not caused by the lack of TMG caps per se, but rather by the tight ectopic association, at low temperatures, of nuclear CBP with the m7G cap of a normally TMG-capped RNA (with U1 snRNA being the likely culprit). It remains to be determined why this association is deleterious, but we can safely surmise from the data here that an association of CBP with U1 does not significantly affect the composition of the U1 snRNP. An intriguing prospect is that CBP binding to the U1 cap in tgs1Δ cells mislocalizes the U1 snRNP within the nucleus (1). We queried whether an increased dosage of the U1 snRNA gene (SNR19) could alleviate the tgs1Δ cs phenotype, but found that this was not the case, either when the U1 gene alone was introduced into tgs1Δ cells on a 2 µ plasmid, or when then U1 and U2 snRNA genes (SNR19 and SNR20) were introduced together on a 2 µ plasmid (Supplementary Figure S2).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (grants GM52470 to S.S. and GM50288 to B.S.); NCI Cancer Center (grant P30 CA08748 to H.E.-B.). Funding for open access charge: U.S. National Institutes of Health (grant GM52470).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Elizabeth Chang for help with mass spectrometry. S.S. is an American Cancer Society Research Professor.
REFERENCES
- 1.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonné R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 2.Hausmann S, Shuman S. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1) J. Biol. Chem. 2005;280:4021–4024. doi: 10.1074/jbc.C400554200. [DOI] [PubMed] [Google Scholar]
- 3.Hausmann S, Shuman S. Giardia lamblia RNA cap guanine-N2 methyltransferase (Tgs2) J. Biol. Chem. 2005;280:32101–32106. doi: 10.1074/jbc.M506438200. [DOI] [PubMed] [Google Scholar]
- 4.Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res. 2007;35:1411–1420. doi: 10.1093/nar/gkl1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hausmann S, Zheng S, Costanzo M, Brost RL, Garcin D, Boone C, Shuman S, Schwer B. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of TMG caps, snRNP components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 2008;283:31706–31718. doi: 10.1074/jbc.M806127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benarroch D, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Shuman S. Cap analog substrates reveal three clades of cap guanine-N2 methyltransferases with distinct methyl acceptor specificities. RNA. 2010;16:211–220. doi: 10.1261/rna.1872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang J, Schwer B, Shuman S. Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: proteins implicated in pre-mRNA splicing. RNA. 2010;16:1018–1031. doi: 10.1261/rna.2082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Lührmann R, Mann M. Identification of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl Acad. Sci. USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Séraphin B, Rosbash M, Lührmann R, et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschalk A, Neubauer G, Banroques J, Mann M, Lührmann R, Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 13.Caspary F, Shevchenko A, Wilm M, Séraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 15.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 16.Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA binding protein, are present together with Gar1p in all H box/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwacher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18 S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 19.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen L, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 20.Sebastiaan Winkler G, Lacomis L, Philip J, Erdjument-Bromage H, Svejstrup JQ, Tempst P. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 21.Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, MacNulty DE, Carr SA, Tempst P. Micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- 22.Walke S, Bragado-Nilsson E, Séraphin B, Nagai K. Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol. 2001;308:49–58. doi: 10.1006/jmbi.2001.4549. [DOI] [PubMed] [Google Scholar]
- 23.Pomeranz Krummel DD, Oubridge C, Leung AKW, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber G, Trowitzcsh S, Kastner B, Lührmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29:4172–4184. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansari A, Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;15:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottschalk A, Kastner B, Lührmann R, Fabrizio P. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA. 2001;7:1554–1566. [PMC free article] [PubMed] [Google Scholar]
- 27.Lybarger S, Beickman K, Brown V, Dembla-Rajpal N, Morey K, Seipelt R, Rymond BC. Elevated levels of a U4/U6.U5 snRNP-associated protein, Spp381p, rescue a mutant defective in spliceosome maturation. Mol. Cell Biol. 1999;19:577–584. doi: 10.1128/mcb.19.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarn WY, Chang TH. the current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 29.Mangus DA, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noble SM, Guthrie C. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- 31.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 33.Görlich D, Kraft R, Kostka S, Vogel F, Hartman E, Laskey RA, Mattaj IW, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 34.Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 35.Dias SMG, Wilson KF, Rojas KS, Ambrosio ALB, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 2009;16:930–937. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu ZR, Schwer B, Shuman S. Determinants of Nam8-dependent splicing of meiotic pre-mRNAs. Nucleic Acids Res. 2011;39:3427–3445. doi: 10.1093/nar/gkq1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao XC, Tang J, Rosbash M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- 38.Kafasla P, Barrass JD, Thompson E, Fromont-Racine M, Jacquier A, Beggs JD, Lewis J. Interaction of yeast eIF4G with spliceosome components. RNA Biol. 2009;6:563–574. doi: 10.4161/rna.6.5.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis JD, Izaurralde E, Jarmolowski A, McGuinan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 40.Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 41.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interaction involving the yeast nuclear cap-binding complex. Mol. Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj IW. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999;12:2425–2438. doi: 10.1101/gad.13.18.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tardiff DF, Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein KA, Gallagher JEG, Mitchell BM, Granneman S, Baserga SJ. The small subunit processosome is a ribosome assembly intermediate. Eukaryot. Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 48.Shen EC, Stage-Zimmermann T, Chui P, Silver PA. The yeast mRNA-binding protein Npl3 interacts with the cap-binding complex. J. Biol. Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 49.Hage R, Tung L, Du H, Stands L, Rosbash M, Chang TH. A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/pre-mRNA interaction. Mol. Cell Biol. 2009;29:3941–3952. doi: 10.1128/MCB.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res. 2011;39:2188–2209. doi: 10.1093/nar/gkq1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worch R, Jankowska-Anyszka M, Niedzwiecka A, Stepinshi J, Mazza C, Darzynkiewicz E, Cusack S, Stolarski R. Diverse role of three tyrosines in binding of the RNA 5′ cap to the human nuclear cap binding complex. J. Mol. Biol. 2009;385:618–627. doi: 10.1016/j.jmb.2008.10.092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.