Abstract
Organisms growing at elevated temperatures face the challenge of maintaining the integrity of their genetic materials. Archaea possess unique chromatin proteins for gene organization and information processing. We present the solution structure of Sso7c4 from Sulfolobus solfataricus, which has a homodimeric DNA-binding fold forming a swapped β-loop-β ‘Tai-Chi’ topology. The fold is reminiscent of the N-terminal DNA-binding domain of AbrB and MazE. In addition, several amide resonances in the heteronuclear single quantum coherence spectra of Sso7c4 are shifted and broadened with the addition of small amounts of duplex DNA oligomers. The locations of the corresponding amides in the Sso7c4 structure define its DNA-interacting surface. NMR spectra of DNA titrated with the protein further indicated that Sso7c4 interacts with DNA in the major groove. Taken together, a plausible model for the Sso7c4–DNA complex is presented, in which the DNA double helix is curved around the protein dimer.
INTRODUCTION
Sulfolobus solfataricus is a thermoacidophilic crenarchaeon that grows optimally at 80–85°C and pH 2–4 (1). It contains a number of relatively abundant small basic DNA-binding proteins of 7-, 8- and 10-kDa families. One of the 7-kDa family members, named Sso7d, has been thoroughly studied (2,3) and has provided insights into non-specific DNA binding by the only known Sulfolobus chromatin protein–DNA complex structure (4–6). Little is known about the 8-kDa family members, except that they are apparently composed of two species (Sso8a and Sso8b). The 10-kDa group comprises three unrelated species (Sso10a, Sso10b and Sso10b2) (7). NMR and X-ray studies revealed that Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled-coil rod (8,9). Sso10b (renamed Alba) is a conserved archaeal chromatin protein interacting with sir2 and tightly binds to double-stranded DNA (dsDNA) modulated by acetylation but without apparent sequence specificity (10,11), while Sso10b2 is the smaller isoform of Sso10b (12).
From sequence alignment, another small DNA-binding protein, Sso7c4, exhibited similarities with two transcriptional repressors (Ecrep 6.8 and Ecrep 7.3) of Escherichia coli (13), the repressor protein ACCR in Agrobacterium tumefaciens (14) and the initiation factor aIF-1A from the archaeon Methanococcus jannaschii (15). Hence, Sso7c4 might have a regulatory role in gene transcription in archaea (16).
Here, we used multidimensional NMR spectroscopy to determine the solution structure of Sso7c4. The structure revealed a homodimeric DNA-recognition fold, which belongs to the SpoVT-AbrB superfamily. To gain insights into the molecular basis of DNA binding by Sso7c4, we studied its complex with DNA using NMR spectroscopy. An analysis of the backbone and side-chain chemical-shift differences between the free and bound forms of the protein shows the ‘footprint’ of the protein surface in contact with the DNA, which provides a model of Sso7c4 complexed with dsDNA.
MATERIALS AND METHODS
Expression and purification
The S. solfataricus P2 (EMBL accession number CAC23286) Sso7c4 gene was amplified from S. solfataricus chromosomal DNA. The gene was cloned into the pET-30 vector (Novagen) with NdeI and XhoI sites to allow for expression of the recombinant protein with an N-terminal sequence and then transformed into E. coli BL21(DE3). Protein expression was induced by the addition of 0.4 mM isopropyl-beta-D-thiogalactoside (IPTG) at 310 K for 3 h. The cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl) and broken by microfluidization. The lysate was then centrifuged at 40 000g. The supernatant was heated to 338 K for 10 min in a water bath, and denatured proteins were precipitated by centrifugation at 40 000g. The supernatant was applied to an SP-Sepharose high-performance 26/10 column (Hi-Load, Amersham-Pharmacia). A gradient of 0–1 M NaCl in buffer A was used to elute cationic proteins. Fractions corresponding to a distinct absorbance peak were analyzed by SDS–PAGE and shown to contain essentially homogenous Sso7c4 protein. Finally, the pure fraction was dialyzed against deionized water and lyophilized for longer storage. Eletrospray (ES)/mass spectrometry (MS) identified that the first-methionine residue was removed.
DNA–RNA gel retardation assay
Plasmid PET-16b was purified by use of a QIAprep Spin Miniprep Kit (Qiagen, GMbH). Yeast RNA was resuspended in RNase-free water. The plasmid DNA (200 ng) or yeast RNA (10 µg) was mixed with increasing amounts of Sso7c4 in 15 µl of 10 mM Tris, 1 mM EDTA buffer, pH 8.0 and the mixtures were incubated at room temperature for 2 min, then subjected to electrophoresis on a 0.5% or 1% agarose gel in the Tris–borate–EDTA buffer provided with the kit.
Surface plasmon resonance
The following two DNA duplexes that were PAGE-purified and 5′-biotin-labeled (MDBio Inc, Taiwan) were used: d(biotin-CGCTATAGCG) (8-mer) and d(biotin-CGCGTACGCGTACGCG) (16-mer). Surface plasmon resonance was measured by use of a BIAcore X instrument (BIAcore AB, Uppsala, Sweden) with the BIAlogue kinetics evaluation program (BIAEVALUATION v3.1, Pharmacia Biosensor). Samples of DNA oligomer in HBS-EP buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20) were applied to flow cells in streptavidin-derivatized sensor chips (BIAcore SA chips) in a BIAcore X optical biosensor system at a flow rate of 2 µl min−1 to achieve long contact times with the surface and to control the amount of DNA bound to the surface. The sensor chips were conditioned with 3 consecutive 1-min injections of 1 M NaCl in 50 mM NaOH, then extensive washing with HBS–EP buffer. Similar amounts of each oligomer were immobilized on the surface by non-covalent capture. The DNA ligand-modified chip surface was regenerated by injecting 20 ml of 1 M NaCl in 50 mM NaOH. The sensor chip surface without a DNA ligand coating was used as the control and was injected with Sso7c4 simultaneously for each binding experiment. Steady-state binding analysis was performed at 25°C by sequential injection of different concentrations of indolicidin over the immobilized DNA surfaces, each for a 10-min period at a flow rate of 20 µl min−1. Solutions of known concentrations of Sso7c4 were prepared in filtered and degassed HBS–EP buffer by serial dilutions from a stock solution in HBS–EP buffer and were injected from 7-mm plastic vials with pierceable plastic crimp caps (BIAcore Inc.). The results were plotted on a sensorgram and expressed as resonance units against time.
NMR spectroscopy
Samples used for NMR data collection contained 1.5–2.5 mM protein, were ∼99% pure, and were stored at pH 5.0 in 20 mM potassium phosphate, 50 mM sodium chloride and 95% H2O/5% D2O or 99.9% D2O. Typical homonuclear and heteronuclear NMR spectra (17,18) were recorded at 310 K on a Bruker Avance-800, Avance-600 or AMX-500 spectrometer with use of cryogenic triple-resonance probes with z-axis pulse-field gradient capability, essentially as described (19).
Determination of the 3D structure
Distance restraints of Sso7c4 were derived primarily from the 200-ms NOESY and 15N-NOESY-HSQC spectra recorded at 310 K in 20 mM phosphate buffer, pH 5.0 and compared to the 100-ms NOESY spectra to assess possible contributions from spin diffusion. Peak intensities were classified as large, medium, small and very small, corresponding to upper-bound interproton distance restraints of 2.5, 3.5, 4.5, and 6.0 Å, respectively. An additional correction of 1.0 Å was added for methylene and methyl groups. Backbone dihedral restraints were inferred from 3JNHα coupling constants from HNHA, with restraint to −120 ± 40° for 3JNHα > 8 Hz and −60 ± 30° for 3JNHα < 6 Hz. 3D structures were generated by use of a simulated annealing and energy minimization protocol in the program Xplor-NIH (20). Hydrogen-bond restraints were included only in the final stage of refinement. Average structures were calculated with the final set of refined structures and were further energy minimized to ensure correct local geometry. The INSIGHTII (Molecular Simulation Inc., San Diego, CA), MOLMOL (21) and PyMol (22) software programs were used to visualize sets of structures and to calculate and draw the electrostatic surface potential of the final 3D models. The convergence of the calculated structures was evaluated in terms of the structural parameters [i.e. the root mean square deviation (rmsd) from the experimental distance and dihedral constraints, the values of the energy statistics (Fnoe, Ftor, Frepel) and the rmsd from the idealized geometry].
Ensemble analysis of Sso7c4 backbone stereochemical quality involved use of PROCHECK_NMR (23) and showed that >91.5% of the residues were in favorable or allowed locations on a Ramachandran plot, and no residues were in the disallowed regions. A total of 966 restraints were obtained from analysis of the NMR data and include 890 NOEs, 40 dihedral angle restraints and 36 hydrogen bond distance restraints (two restraints per hydrogen bond, added during the final stages of refinement). Distance geometry and simulated annealing calculations were used to generate an ensemble of 10 conformers consistent with the NMR data (Figure 1C). The structures exhibit good covalent geometry and have no NOE, scalar coupling or dihedral angle violations >0.5 Å, 2 Hz or 5°, respectively. Complete restraint and structural statistics are in Table 1.
Figure 1.
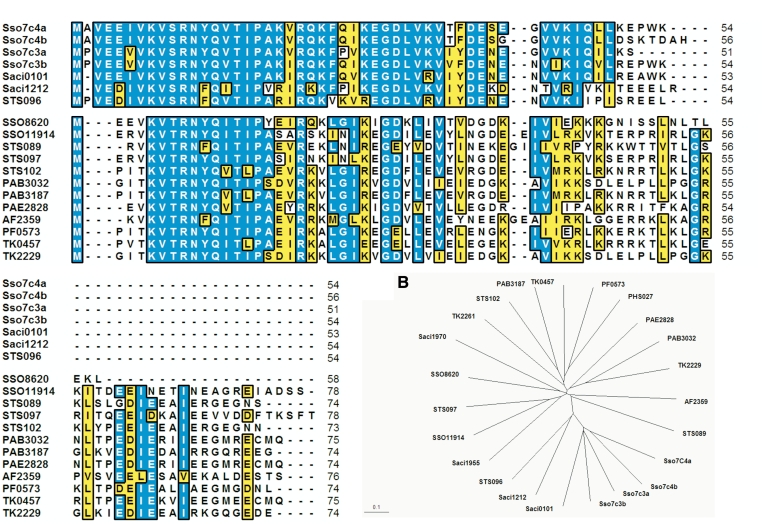
Comparison of Primary Sequences of Sso7c4 protein family in Archaeal kingdom. (A) Sequence alignment of the Sso7c4 family from several archaeas with the corresponding secondary structural elements noted. The alignment was performed with the T-coffee server (30). Sequences were obtained form the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov). Entry codes are P81552 (Sso7c4a and Sso7c4b), P81551 (Sso7c3a and Sso7c3b), NP_376734 (STS096), YP_254819 (Saci_0101), YP_255851 (Saci_1212), NP_343080 (SSO8620), NP_344389 (SSO11914), NP_376639 (STS089), NP_376742 (STS097), NP_376811 (STS102), NP_125810 (PAB3032), NP_126453 (PAB3187), NP_560284 (PAE2828), NP_071181 (AF2359), NP_578302 (PF0573), YP_184642 (TK2229), YP_182870 (TK0457), YP_184674 (TK2261), YP_256551 (Saci1955) and NP_142833 (PHS027). For the group I (Sso7c3, Sso7c4, Saci0101, Saci1212 and STS096) and the group 2, each possessing shorter sequence length near 55 and 75 amino acids, respectively, are aligned. Residues are numbered according to Sso7c4a from S. solfataricus. The residues R11 and R22 are highly conserved across the species and other hydrophobic residues are also important in hydrophobic interactions. (B) The phylogenetic tree for the above alignment. The protein names are colored according to the classification as in (A). The scale bar indicates the distance corresponding to 0.1 amino acid substitutions per site. Sso, S. solfataricus; STS, S. tokodaii; PAB, Pyrococcus abyssi; PAE, P. aerophilum; PF, P. furiosus; TK, Thermococcus kodakaraensis; PHS, P. horikoshii.
Table 1.
Structural statistics on the final set of simulated annealing structures of Sso7c4
| Constraints used | |
| Distance restraints | |
| Intra-residue | 278 |
| Sequential | 244 |
| Medium-range | 70 |
| Long-range | 174 |
| Intermolecular | 124 |
| Hydrogen bonds | |
| Intramolecular | 16 |
| Intermolecular | 20 |
| Total distance restraints | 926 |
| Dihedral angles | |
| Backbone ϕ,φ | 40 |
| Statistics for the final X-PLOR structures | |
| Number of structures in the final set | 15 |
| X-PLOR energy (kcal/mol) | |
| ENOE | 3.78 ± 1.81 |
| Ecdih | 0.10 ± 0.12 |
| Ebond + Eangle + Eimproper | 172.75 ± 8.30 |
| Eelec | 0.85 ± 0.23 |
| EVDW | 37.18 ± 3.91 |
| ENCS | 0.42 ± 0.22 |
| NOE violations | |
| Number >0.5 Å | none |
| r.m.sa deviation (Å) | 0.017 |
| Deviation from idealized covalent geometry | |
| Angle (°) | 0.65 ± 0.02 |
| Impropers (°) | 0.36 ± 0.02 |
| Bonds (Å) | 0.005 |
| Mean global r.m.s deviation (Å) | |
| Backbone (N,Cα,C') | |
| Residues (secondary structure) | 0.68 ± 0.18 |
| Residues (3–47) | 0.89 ± 0.23 |
| Heavy atoms | |
| Residues (secondary structure) | 1.40 ± 0.31 |
| Residues (3–47) | 1.72 ± 0.39 |
| Ramachandran datab | |
| Residues in most favored regions (%) | 70.2 |
| Residues in allowed regions (%) | 21.3 |
| Residues in generously allowed regions (%) | 8.5 |
| Residues in disallowed regions (%) | 0 |
aRoot mean square.
bThese were calculated using the PROCHECK program.
NMR titration analysis
DNA used for NMR titration was chemically synthesized and gel-purified. 1H-15N HSQC spectra of Sso7c4 for monitoring protein chemical-shift changes at the initial concentration of 1.0 mM dissolved in 20 mM potassium phosphate buffer, pH 5.0, containing 100 mM NaCl were recorded at 300 K by adding increasing amounts of double-stranded 8-mer (5′-CGCTATAGCG-3′) and 16-mer (5′-CGCGTACGCGTACGCGTA-3′) DNAs.
The 12-mer DNA sequence (5′-CAATCCGGATTG-3′) was used to analyze chemical-shift changes for the DNA. One-dimensional spectra of DNA in the same condition as for the HSQC titration experiment, mentioned above, were recorded by adding increasing amounts of Sso7c4. For proton assignment of DNA, DQF-COSY, TOCSY and NOESY spectra were used.
Molecular docking model of Sso7c4–DNA complex
To understand more clearly the binding mechanism and test whether chemical-shift changes and the structure of Sso7c4 are complementary to the DNA structure, we used InsightII to build a model of the complex using a bent dsDNA from the partial nucleosome structure. The DNA-binding surface of the Sso7c4 dimer was docked manually into the major groove with particular attention paid to matching the pattern of chemical shift changes, sharp complementarity, absence of steric clashes (especially with the protein backbone) and favorable electrostatics, so that the molecular 2-fold axis of the dimer was aligned with a local dyad of dsDNA. Finally, we performed a brief minimization to remove steric clashes between the protein side chains and DNA. Minimization was calculated by use of the DISCOVER program (Accelrys, San Diego, CA, USA) with CVFF force-field parameters.
RESULTS AND DISCUSSION
Sso7c4 family is a new member of small DNA-binding proteins in the archaeal kingdom
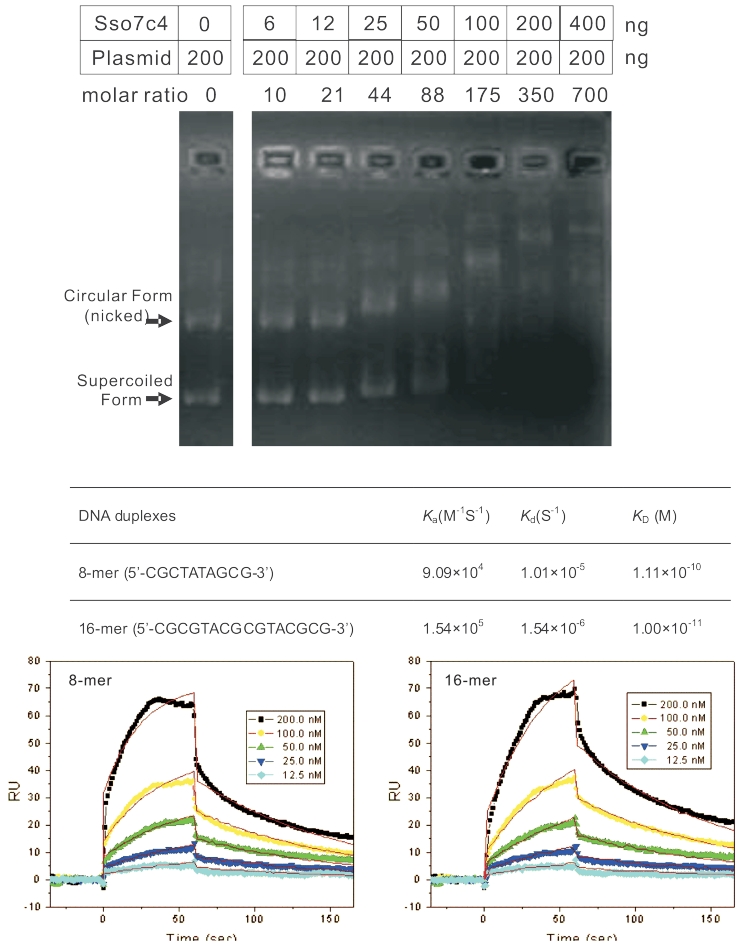
Several sequenced archaeal genomes have homologous proteins of Sso7c4 (Figure 1) belonging to two groups consisting of ∼55 and ∼75 amino acids. BLAST search against the protein structure database revealed similar folds to SpoVT-AbrB superfamily with low identity. To establish a functional role for Sso7c4, we first measured its binding affinities with dsDNA by gel electrophoretic retardation analysis. Binding to dsDNA was observed with presence of at least a 10:1 molar ratio of protein to plasmid (5711 bp) (Figure 2A). The maximal value of the molar ratio of protein to plasmid (700:1) indicates that one protein binds to an ∼8-bp DNA length. Single-stranded DNA and RNA showed little binding affinity to Sso7c4 (data not shown). To evaluate the DNA-binding affinity of Sso7c4, 8 and 16 base-pair duplex DNAs were separately immobilized on BIAcore SA sensorchips, and surface plasmon resonance experiments were performed at 25°C. A typical set of sensorgrams (response units versus time) at different protein concentration for Sso7c4 binding to the 8- and 16-mer duplexes is shown in Figure 2B. Sso7c4 bound rapidly to both 8- and 16-bp duplexes. The association reaction was analyzed by a Langmuir binding isotherm to obtain dissociation constants of 1.01 × 10−5 and 1.54 × 10−6 M to 8- and 16-mer duplexes, respectively.
Figure 2.
(A) Different mixtures of DNA (200 ng) and Sso7c4 (0, 6, 12, 25, 50, 100, 200 and 400 ng) were run on a gel. (B) Surface Plasmon Resonance experiments showing the binding ability of Sso7c4 to duplex DNA. SPR sensorgrams for the binding of Sso7c4 to 8-mer (5′-CGCTATAGCG-3′) (left) and 16-mer (5′-CGCGTACGCGTACGCGTA-3′) (right) duplex DNA in HBS–EP buffer at 25°C. The concentration of the unbound ligand in the flow solution varies from 12.5 nM in the lowest curve to 200 nM. RU denotes response units.
Sso7c4 forms a homodimer in solution
The average T2 relaxation value of Sso7c4 suggested that the protein existed in a form larger than that of the monomer in solution. Size exclusion chromatography showed the presence of a peak of 12 ∼ 18 kDa, which is compatible with the formation of a dimer or higher oligomer. Analytical ultra-centrifugation experiments conclusively showed that Sso7c4 forms a stable dimer at pH 2.0, 5.0 and 9.0 (Supplementary Figure S1).
Spectral analysis and secondary structure determination
Heteronuclear multidimensional NMR techniques were used for resonance assignments (BMRB-17301), sequential connectivities and distance geometry for the development of a structural model. Sso7c4 showed well-resolved proton NMR spectra. However, two peaks (ratio 70:30) appeared in the downfield region (∼9.5–11 ppm) of the 1D spectrum from the indole HN of the single tryptophan, which indicates two conformations of Sso7c4. In addition, two sets of cross-peaks (for simplicity denoted as ‘doublets’ hereafter) were observed for several residues of Sso7c4 in the 2D 1H-15N heteronuclear single quantum coherence (HSQC) spectra (Supplementary Figure S2). The presence of two chemically different species can be ruled out, because MS showed only a single protein species (mass = 6142).
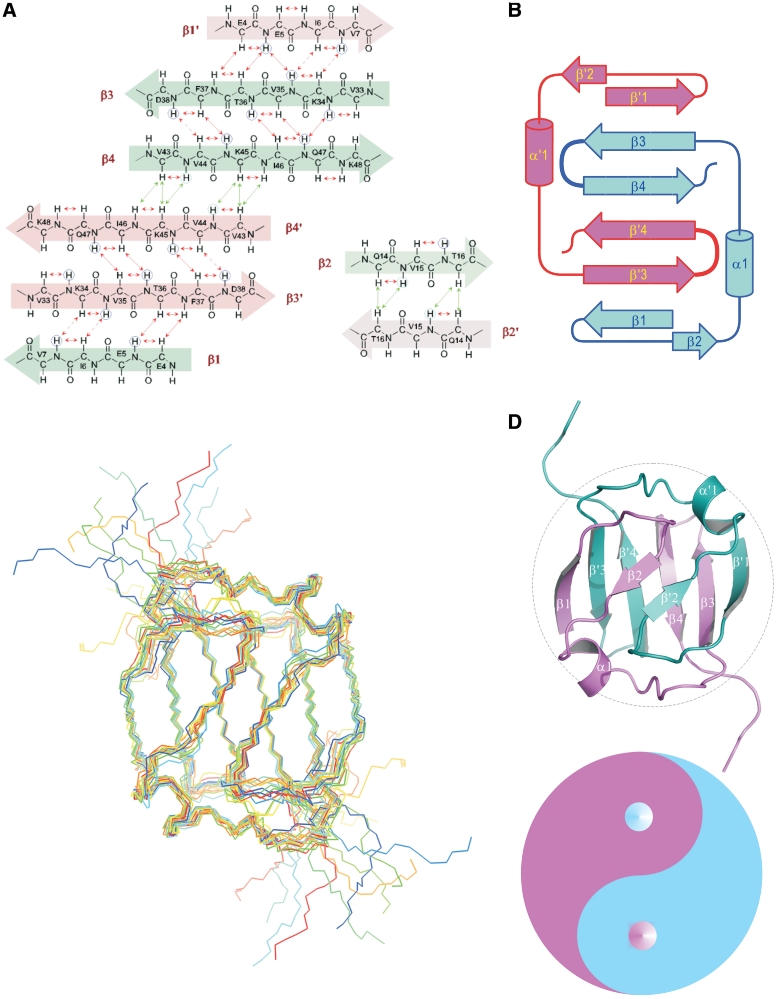
Two amide proton resonances were observed for each the following 11 residues: Glu4, Leu32, Glu39, Ser40, Glu41, Gly42, Gln47, Leu48, Leu49, Trp53 and Lys54. Two resonances were also observed for several side-chain protons of these residues. These residues scattered around the C-terminal end and loop 3, which suggests the presence of two conformers of the C-terminal tail due to the cis and trans forms of Pro52 According to the observed NOEs between Glu51 Hα and Pro52 Hδ, the major conformer could be identified as the trans form of Pro52. Analysis of Hα chemical shifts with chemical shift index (Supplementary Figure S3) and nuclear Overhauser enhancement (NOE) connectivities (Figure 3A) indicated that the folding topology of the Sso7c4 monomer contains four β strands and one α helix (Figure 3B). Strong, completely unambiguous NOEs could be identified between the solvent exchange-protected amide protons of Val43 and Lys45 located in β4 and β4′ (primes indicate the second monomer in the dimer). These protons are too far (>7 Å) from each other in the monomeric structure, but are close enough in a dimer. The β4 strand shows several NOE connectivities that are best explained by an intermolecular antiparallel β-sheet contact with its symmetry-related equivalent, β4′. In addition, unambiguous strong NOEs were observed due to the intermolecular short distance of side-chain protons between Phe25 and Val44, Ile46 and Leu48, as well as Tyr13 and Ile27 (Supplementary Figure S4). This observation enabled the development of a low-resolution model of the dimeric Sso7c4 topology. Further refinement of the monomeric structure contributed to the identification of 66 (of the 124 total NOEs defining the dimeric interface) inter-subunit NOEs (Table 1). These NOEs were excluded from the monomer structure calculation (Figure 3A).
Figure 3.
Topology and NMR structure of the dimeric Sso7c4 protein. (A) The proton nuclear Overhauser enhancement (NOE) networks of the swapped six-stranded and the short two-stranded antiparallel β-sheets of Sso7c4 are defined from the NOEs and amide exchange rate. Long-range NOEs between β-strands are indicated by double arrows. The amide protons with very slow exchange rates are circled. (B) Topology diagram of the Sso7c4 structure showing the connectivity between strands in two β-sheets. (C) NMR ensemble of the selected structures is shown with a backbone chain. (D) The individual secondary structure elements are indicated in the ribbon diagram. The two monomers are colored differently for clarity. Association of the monomers is through two β-sheets; in each sheet, two strands derive from one monomer and the first strand forms the other monomer. In addition, another short β-sheet is constructed by the β2 strand from each monomer. These arrangements form a strand-switched dimer interface. The architecture of homodimeric Sso7c4 is presented as the Chinese traditional ‘Tai-Chi’ symbol. The separation line (also called ‘Yin-Yang’ diameter) formed by two semicircles of the ‘Tai-Chi’ symbol generate more binary interaction than does the linear diameter.
Sso7c4 possesses the swapped β-loop-β ‘Tai-Chi’ fold
The solution structure of Sso7c4 is represented by an ensemble of 10 calculated structures selected on the basis of lowest residual restraint violations (Figure 3C). Sso7c4 possesses a homodimeric fold in which the two monomers form an extensive hydrophobic dimer interface that buries ∼4370Å2 of the surface area. Each monomer forms a β1-β2-α1-β3-β4 topology without a hydrophobic core on its own and represents only one-half of a typical protein domain. The swapped-β1 strand forms an inter-molecular anti-parallel sheet with the β3′ strand. A complete domain structure is formed by the association of two chains, thus creating a hydrophobic core between two four-stranded β-sheets. This hydrophobic core consists exclusively of short aliphatic residues.
The dimer interface is largely composed of hydrophobic residues located in loop 1 (Y13) and loop 2, as well as the head of the hairpin (F37, V44, I46, I48 and F25). The double-layered architecture of the dimeric Sso7c4 is composed of two anti-parallel β3-β4 hairpins at the bottom and two swapped-β-loop-β motifs on top. The two β-loop-β motifs are present in a symmetric manner as in the Chinese ‘Tai Chi’ symbol (also called ‘Yin Yang’) (Figure 3D). CD analysis showed Sso7c4 has the melting transition with a Tm value of 60°C in the presence of 1.5 M GdnHCl at pH 5.0. In contrast, in the absence of GdnHCl, the Tm value of Sso7c4 could not be determined since it did not reach a completely denatured state even at near 100°C (Supplementary Figure S5).
Analysis by DALI (24) revealed that Sso7c4 shares significant structural homology to the N-terminal DNA-binding domain of AbrB (AbrB-N) from Bacillus subtilis (25–27) with a rmsd of 3.47 Å between them. Additional homologs of Sso7c4 and AbrB-N were found for N-terminal domain of another DNA binding protein MazE (28) from E. coli. Interestingly, Sso7c4 shares <30% identities with AbrB-N and MazE-N but a common fold with swapped homodimeric scaffold (Supplementary Figure S6). Since there are few consensus residues between Sso7c4 and the two protein family members for DNA binding, the further NMR experiments were carried out for DNA-binding studies of Sso7c4.
Mapping the DNA-binding surface of Sso7c4
The DNA-binding mode of Sso7c4 was analyzed by NMR analysis of the 15N-enriched protein in the presence of the two DNA oligomers. The NMR spectra of the Sso7c4–DNA complex were well dispersed, thus enabling the assignment of the protein backbone resonance (Figure 4A and B), although the quality was not sufficient for structure determination. A large number of resonances exhibited broadening, presumably as a result of conformational exchange that is intermediate on the chemical-shift time-scale.
Figure 4.
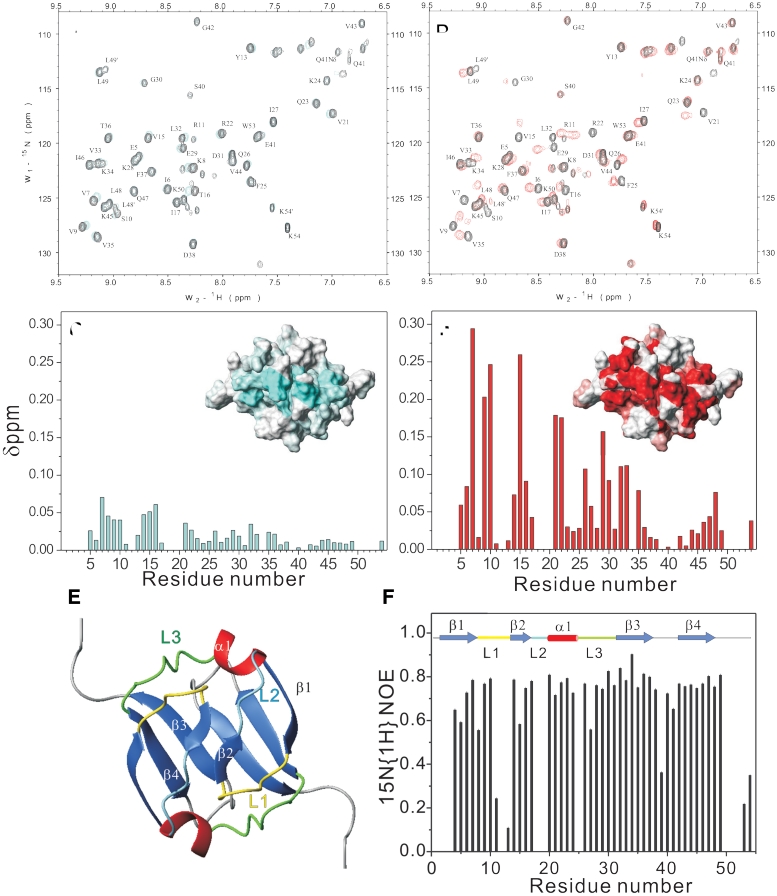
Mapping the DNA-recognition region of Sso7c4. (A and B) overlay of 1H-15N HSQC spectra of Sso7c4 (black) and Sso7c4/8-mer DNA complex (cyan) and Sso7c4/16-mer DNA complex (red). (C and D) combined chemical shift change (Δδ) for Sso7c4 with the addition of 8-mer DNA in (C) and 16-mer in (D). The inset figures in (C) and (D) indicate the mapping of the duplex DNA interacting with the Sso7c4 surface. (E) Ribbon diagram of Sso7c4 is shown in the same orientation as the inset surface figures in (C) and (D). (F) Hetero-nuclear NOE results with secondary structure elements color-coded as in (E).
The DNA-binding surface was mapped by comparing the backbone chemical shifts of Sso7c4 alone with those of the Sso7c4–DNA complex. The regions of the protein showing the largest chemical-shift difference are from loop 1 (I6 to S10), strand 2 (Q14 to T16), helix 1 (V21, R22) and loop 3 (Q26 to V33) (Figure 2D). Because of broad or disappearing signals, determination of the Kd and the stoichiometry of the complex was difficult. The binding of protein to DNA is in intermediate to fast exchange on the NMR time scale as judged by the 1H-15N HSQC broadened cross-peaks. The 1H and 15N chemical-shift changes for backbone amides are shown in Figure 4C and D, and they define the DNA-binding surface (inset for each Figure 4C and D).
Dynamic properties of Sso7c4
The presence of backbone amide proton resonances in a 1H-15N HSQC spectrum after exchange of Sso7c4 in deuterated NMR buffer for 24 h defined residues 43 and 45′ (and 43′ to 45) as stable intermolecular hydrogen-bonded partners at the dimer interface. The steady-state heteronuclear 15N-1H-NOEs for most of the Sso7c4 backbone amides are >0.7, which indicates that the backbone of Sso7c4 is well ordered in solution (Figure 4E and F). The concerned residues (V43 to L48) of the β4 strand show the rigid properties associated with the stable dimeric interface. Related inter- (V44 to F25′ and I46′) and intra-molecular residues (F25 to I46 and L48) for forming the hydrophobic core also show little mobility. The residues in the loop L1 (K8-Q14) with higher mobility are in good agreement with its DNA-binding-regulatory role. In contrast, loop L3 (Q26-L32) with lower mobility is relatively rigid for stabilizing part of the dimer interface.
Sso7c4 binds to the major groove of duplex DNA
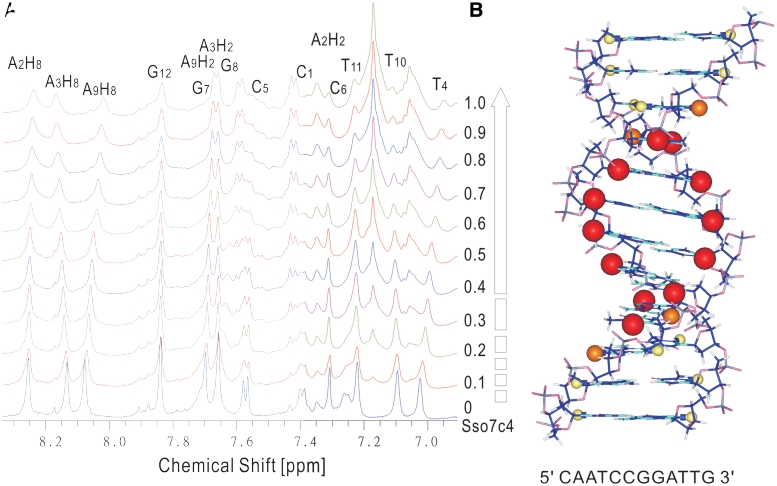
The 1H NMR resonances of the free 12-mer dsDNA (5′-CAATCCGGATTG-3′) were assigned according to the previous study (29) and confirmed from a combination of NOESY, TOCSY and DQF-COSY. The NMR spectra of 12-mer DNA at various ratios of added Sso7c4 are shown in Figure 5A. Only one set of 12-mer resonances were observed upon addition of protein at all molar ratios of protein to DNA. This indicates a fast-exchange (on the NMR time scale) between the free DNA and the protein-bound forms. Titration of DNA with Sso7c4 results in continuous and small perturbations along the entire DNA sequence. This finding is consistent with a non-specific interaction with multiple binding sites on DNA. Furthermore, the strongest chemical-shift perturbations of the major groove purine H8 and pyrimidine H6 resonances were observed for the middle 8 bp (A3H8, T4H6, C5, C6, G7H8, G8H8, A9H8, T10H6) of the 12-mer DNA. However, the minor groove A2H2, A3H2 and A9H2 resonances show only small chemical shift changes. The broaden peaks of DNA protons at higher protein–DNA molar ratio are due to the slower tumbling rate for the protein–DNA complex. From chemical shift analysis of the DNA fragment, the protein–DNA binding interactions appear to be in the major groove. The observed site size of 8 for Sso7c4 binding may be due to a more complete major groove of 12-mer dsDNA located on the central 8 bp. Interestingly, the size of the major perturbed DNA region matches not only that of the mapped surface of Sso7c4 involved in DNA binding, but also the molar ratio (one protein to ∼8 bp) from the gel retardation experiment.
Figure 5.
Sso7c4 recognizes the major groove of duplex DNA. (A) One-dimensional spectra of 12-mer DNA (5′-CAATCCGGATTG-3′) show the ring proton peaks perturbed with various concentrations of Sso7c4. (B) Duplex DNA model based on chemical-shift perturbation observed in DNA. Chemical-shift perturbations of DNA proton are proportional to the sphere radii and colored red, ΔδDNA > 0.050; orange, 0.050 ≥ ΔδDNA ≥ 0.025; and yellow, ΔδDNA ≤ 0.025.
A model of DNA–Sso7c4 complex
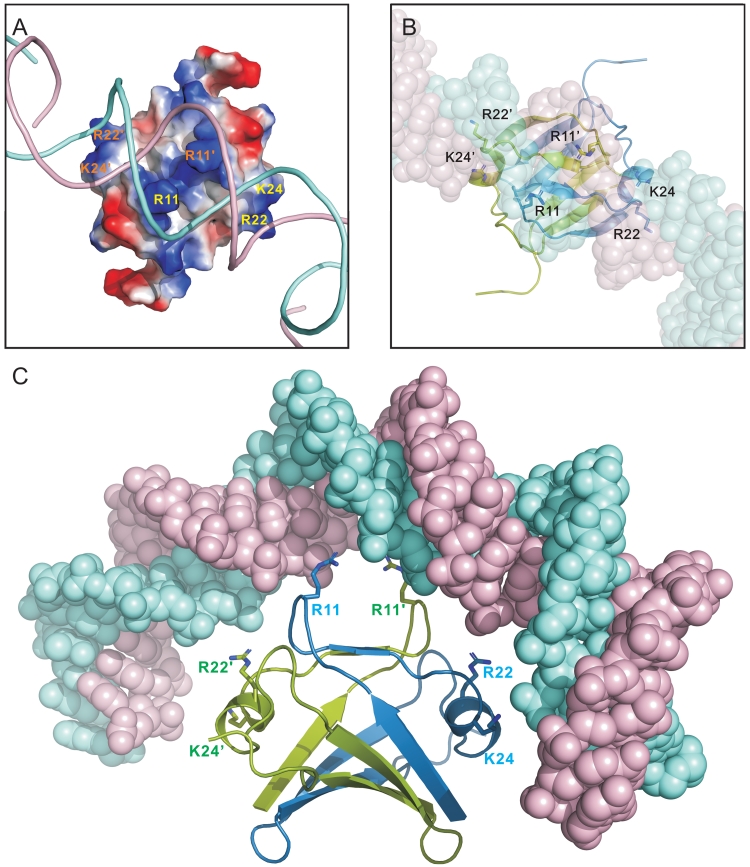
Our data showed loop 1, β2, α1 and loop 3 of Sso7c4 involved in DNA binding in the major groove. The protein surface for DNA binding was mapped by chemical-shift perturbation and consisted of considerable positive electrostatic characters. Charged residues in Sso7c4 are grouped into well-defined clusters on the protein surface (Figure 6A). The main basic patch comprises residues R11, K20, R22 and K24, and the side chain amides N12, Q14, Q23 and Q26, thus defining a putative DNA-binding site. R11, N12 and R22 are highly conserved in the archaeal homologs. The Sso7c4 dimer may present two DNA-binding loops (L1 and L2) for non-specific DNA binding. Compared to the similar fold of AbrB-N, DNA binding related conserved residues, Asp11, Arg15 and Arg 23 on AbrB-N are Arg 11, Gln14 and Arg22, respectively, on the relative position of Sso7c4. This suggests that Sso7c4 may bind DNA with similar mode but different DNA sequences from AbrB-N. In addition, our NMR perturbation with Sso7c4 upon dsDNA supported the DNA major groove binding model for the ApoVT-AbrB protein family. Thus, a plausible model of Sso7c4 binding to DNA was constructed (Figure 6) with the dyad of the DNA duplex aligned with that of the Sso7c4 dimer in the DNA major groove.
Figure 6.
Model of Sso7c4 interaction with a bent DNA. (A) Putative DNA-binding site of Sso7c4. Conserved residues appear on the same face corresponding to NMR chemical-shift perturbation data. The transformed surface is complementary to the bent DNA in charge and shape. (B) The DNA-binding surface of the Sso7c4 dimer was docked manually into the major groove so that the molecular 2-fold axis of the dimer was aligned with a local dyad of dsDNA. (C) The side view of Sso7c4 binding to bent DNA relates to the top view of that in (B) by 90° rotation around the horizontal axis.
Figure 6C shows the complex model with surface potential of protein and ribbon of DNA. In this model, residues in the centre of this site are crucial for the ability of Sso7c4 to interact with different nucleotide sequences in the major groove. In Figure 6B and C, the optimal fitting of DNA to the dimeric structure of Sso7c4 involved a bent DNA. This model is supported by the matching of the positively charged clustered surface of Sso7c4 with the negative charge of DNA.
In addition, NMR dynamics data suggested that K8, R11, N12, Y13 and R15 (located in L1) have high mobility. The flexible L1 suggests that DNA binding depends on some level of structural plasticity in Sso7c4, which allows for the recognition of multiple unrelated DNA target substrates. The flexibilities of these residues may be a contributing factor in the ability of Sso7c4 to kink DNA. These residues are highly conserved across species and should be important for the function of the Sso7c4 family (Figure 1).
Although the function of the Sso7c4 protein is unknown, the Sso7c4-related fold is found in many chromatin-associated proteins. Sso7c4 appears to be a small archaeal protein that binds to dsDNA without sequence specificity. The Sso7c4/bent DNA model suggests that Sso7c4 could be another significant member of chromatin proteins.
ACCESSION NUMBER
PDB code: 2L66.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council (NSC 97-2113-M-002-005-MY2 to C.-H.H. and NSC 97-9112-B-001-035-B4 to A.H.-J.W., in part); Academia Sinica, Taiwan (in part). Funding for open access charges: Academia Sinica.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The NMR spectra were obtained at the High-Field Biomacromolecular NMR Core Facility for Proteomic Research, National Research Program for Genomic Medicine, Taiwan.
REFERENCES
- 1.Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 2.White MF, Bell SD. Holding it together: chromatin in the Archaea. Trends Genet. 2002;18:621–626. doi: 10.1016/s0168-9525(02)02808-1. [DOI] [PubMed] [Google Scholar]
- 3.Baumann H, Knapp S, Lundback T, Ladenstein R, Hard T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat. Struct. Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 4.Robinson H, Gao YG, McCrary BS, Edmondson SP, Shriver JW, Wang AH. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 5.Gao YG, Su SY, Robinson H, Padmanabhan S, Lim L, McCrary BS, Edmondson SP, Shriver JW, Wang AH. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Struct. Biol. 1998;5:782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- 6.Agback P, Baumann H, Knapp S, Ladenstein R, Hard T. Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nat. Struct. Biol. 1998;5:579–584. doi: 10.1038/836. [DOI] [PubMed] [Google Scholar]
- 7.Forterre P, Confalonieri F, Knapp S. Identification of the gene encoding archeal-specific DNA-binding proteins of the Sac10b family. Mol. Microbiol. 1999;32:669–670. doi: 10.1046/j.1365-2958.1999.01366.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Chen LR, Zhou XE, Wang Y, Kahsai MA, Clark AT, Edmondson SP, Liu ZJ, Rose JP, Wang BC, et al. The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod. J. Mol. Biol. 2004;341:73–91. doi: 10.1016/j.jmb.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 9.Kahsai MA, Vogler B, Clark AT, Edmondson SP, Shriver JW. Solution structure, stability, and flexibility of Sso10a: a hyperthermophile coiled-coil DNA-binding protein. Biochemistry. 2005;44:2822–2832. doi: 10.1021/bi047669t. [DOI] [PubMed] [Google Scholar]
- 10.Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 11.Wardleworth BN, Russell RJ, Bell SD, Taylor GL, White MF. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou CC, Lin TW, Chen CY, Wang AH. Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms. J. Bacteriol. 2003;185:4066–4073. doi: 10.1128/JB.185.14.4066-4073.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cam K, Bejar S, Gil D, Bouche JP. Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 1988;16:6327–6338. doi: 10.1093/nar/16.14.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck von Bodman S, Hayman GT, Farrand SK. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl Acad. Sci. USA. 1992;89:643–647. doi: 10.1073/pnas.89.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the Methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 16.Oppermann UC, Knapp S, Bonetto V, Ladenstein R, Jornvall H. Isolation and structure of repressor-like proteins from the archaeon Sulfolobus solfataricus. Co-purification of RNase A with Sso7c. FEBS Lett. 1998;432:141–144. doi: 10.1016/s0014-5793(98)00848-5. [DOI] [PubMed] [Google Scholar]
- 17.Wuthrich K. NMR of Protein and Nucleic Acids. New York: Wiley Interscience; 1986. [Google Scholar]
- 18.Bax A, Vuister GW, Grzesiek S, Delaglio F, Wang AC, Tschudin R, Zhu G. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 19.Hsu CH, Liao YD, Pan YR, Chen LW, Wu SH, Leu YJ, Chen C. Solution structure of the cytotoxic RNase 4 from oocytes of bullfrog Rana catesbeiana. J. Mol. Biol. 2003;326:1189–1201. doi: 10.1016/s0022-2836(02)01472-9. [DOI] [PubMed] [Google Scholar]
- 20.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 21.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 24.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 25.Vaughn JL, Feher V, Naylor S, Strauch MA, Cavanagh J. Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator abrB. Nat. Struct. Biol. 2000;7:1139–1146. doi: 10.1038/81999. [DOI] [PubMed] [Google Scholar]
- 26.Coles M, Djuranovic S, Soding J, Frickey T, Koretke K, Truffault V, Martin J, Lupas AN. AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure. 2005;13:919–928. doi: 10.1016/j.str.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Bobay BG, Andreeva A, Mueller GA, Cavanagh J, Murzin AG. Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett. 2005;579:5669–5674. doi: 10.1016/j.febslet.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 28.Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J. Biol. Chem. 2001;276:5975–5984. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- 29.Collins JG. Assignment of the non-exchangeable proton resonances and solution conformation of the dodecanucleotide-d(CAATCCGGATTG) Biochem. Int. 1988;16:819–828. [PubMed] [Google Scholar]
- 30.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.