Abstract
Homologous recombination is essential for the preservation of genome stability, thereby preventing cancer. The recombination protein RAD51 drives DNA strand exchange, which requires the assembly, rearrangement and disassembly of a RAD51 filament on DNA, coupled to ATP binding and hydrolysis. This process is facilitated and controlled by recombination mediators and accessory factors. Here, we have employed a range of single molecule techniques to determine the influence of the C-terminal RAD51 interaction domain (CTRD) of the breast cancer tumor suppressor BRCA2 on intrinsic aspects of RAD51-DNA interactions. We show that at high concentration the CTRD entangles RAD51 filaments and reduces RAD51 filament formation in a concentration dependent manner. It does not affect the rate of filament disassembly measured as the loss of fluorescent signal due to intrinsic RAD51 protein dissociation from double-stranded DNA (dsDNA). We conclude that, outside the context of the full-length protein, the CTRD does not reduce RAD51 dissociation kinetics, but instead hinders filament formation on dsDNA. The CTRDs mode of action is most likely sequestration of multiple RAD51 molecules thereby rendering them inactive for filament formation on dsDNA.
INTRODUCTION
Double-stranded DNA breaks (DSBs) are severe lesions that can result in chromosomal rearrangements leading to cellular senescence or the onset and progression of cancers (1,2). In S and G2 phases of the cell cycle, homologous recombination (HR) provides a pathway for the faithful repair of such lesions (3). HR can be divided in three steps: pre-synapsis, synapsis and post-synapsis (4). Pre-synapsis consist of the recognition of a DSB, and the resection of one strand leaving tailed DNA that is bound by RAD51 to form a nucleoprotein filament (5,6). During synapsis this nucleoprotein filament interacts with the intact sister chromatid (homology search), and strand invasion occurs at the site of homology resulting in joint molecule formation and strand exchange. Post-synapsis is defined as the events leading to the recovery of lost information by polymerase-mediated DNA resynthesis and the resolution of branched DNA structures resulting in two intact chromatids (7,8).
Biochemical assays have defined RAD51 recombinase as the catalyst of synapsis. It forms a helical filament on single-stranded DNA (ssDNA) that can drive the DNA transactions required for HR in vitro. The RAD51 nucleoprotein filament will invade a homologous double-stranded DNA (dsDNA) and forms a joint molecule. To catalyze strand exchange, the RAD51 nucleoprotein filament undergoes dynamic rearrangements that result in DNA strands exchanging base-paired partners leading to the formation of a filament on heteroduplex DNA—one strand being the invading ssDNA while the other is the complementary strand of the template DNA. The next step in repairing a DNA break requires the invading strand to prime DNA extension by a polymerase. This stage requires RAD51 removal from the heteroduplex DNA (9–11), and these dissociation reactions have been addressed in previous single molecule studies (12–15).
The actions of RAD51 are facilitated by recombination mediators and controlled by accessory factors (16,17). One such mediator is the BRCA2 protein which is essential for efficient HR in mammalian cells (16–18). The role that BRCA2 plays in maintaining genome stability has been attributed to its ability to interact directly with RAD51 (19,20). Biochemical studies with BRCA2 peptides have defined interactions with RAD51 via a series of eight BRC repeats and a C-terminal domain, designated C-terminal RAD51 interaction domain [CTRD; previously referred to as TR2 (21,22)]. This domain contains a cyclin-dependent kinase phosphorylation site that modulates RAD51 binding (21,23). The BRC domains, best characterized by BRC4, which has the highest affinity for RAD51, interact with RAD51 at the polymerization domain and can thereby disrupt RAD51 filaments at concentrations equal to or higher than the RAD51 concentration (22–25). The CTRD of BRCA2 interacts with multimeric RAD51 and can inhibit the destabilization of RAD51 filaments caused by the BRC repeats (22,24,25). In avian cells the CTRD of BRCA2 influences the persistence of RAD51 in local nuclear accumulations in a manner consistent with stabilizing RAD51 filaments (26). To determine whether the CTRD of BRCA2 might provide an effect opposite to the BRC repeats, by helping to stabilize RAD51 filaments on dsDNA, we used an array of single-molecule techniques together with ensemble studies. First, we visualized the effect of the CTRD on RAD51 dissociation from dsDNA. We observed individual filaments formed with fluorescent RAD51 and quantified their disassembly in time by fluorescent microscopy in a microfluidic flow system (12). We then analyzed the effect of the CTRD on RAD51 nucleoprotein filament structure by optical fluorescence microscopy, by scanning force microscopy imaging (27), and a combination of both techniques (28). We then analyzed the effect of the CTRD on RAD51 filament assembly on dsDNA, following the extension of individual dsDNA molecules by RAD51 binding, in magnetic tweezers (13,29).
MATERIALS AND METHODS
Protein production
Wild-type and variants of human RAD51 were purified by similar procedures, essentially as previously described (12). Briefly, RAD51 was expressed in bacteria, precipitated by ammonium sulfate, subsequently dialyzed and further purified by Heparin, gel filtration and anion exchange chromatography. The CTRD peptides were as described previously (21,22).
Protein labeling
RAD51 was Alexa Fluor 488 labeled on a specific cysteine residue using maleimidine chemistry and checked for activity as previously described (12). The degree of labeling was 0.75 fluorophores per RAD51.
Surface tethering and visualization of fluorescent filaments in buffer
Flow cells, constructed from #1 glass cover slips (Menzel-Glaser) separated by a double-layer Parafilm in a custom holder, were prepared as follows: Neutravidin (Pierce) was introduced at 1 mg/ml and allowed to interact for 30 min. After removal of excess Neutravidin, the flow cell was washed and blocked with 2 mg/ml acetylated BSA, 2 mg/ml α-casein, 10 mM DTT, 50 mM Tris–HCl (pH 7.5), 30 mM KCl and 10% glycerol. λ phage dsDNA was biotinylated at one end and labeled with digoxygenin at the other by annealing biotinylated oligonucleotides to CosL (5′-P-AAG TCG CCG CCC dR-bio dR-bio-bioTEG-3′) and to CosR (5′-P-GGG CGG CGdRDig CCT CGG CGC CCG GCC GCG dTDigAA ACG CGG CCG GGC GCC GG-3′) as described (30). Filaments were assembled in a 20 μl reaction mixture containing 91 pM (molecules) of λ phage dsDNA, 1.4 μM RAD51, 50 mM Tris–HCl (pH 7.5), 1 mM ATP, 2 mM CaCl2, 1 mM DTT and 60 mM KCl. Reaction mixtures were incubated at 37°C for 30 min. When indicated 4.8 nM CTRD peptide was added and the reaction mixtures incubated for another 15 min (RAD51:CTRD of 291:1). Alternatively, CTRD and RAD51 were incubated for 15 min before addition of the other reaction components. All reaction mixtures were diluted by addition of 380 μl CaCl2/ATP buffer [50 mM Tris–HCl (pH 7.5), 1 mM ATP, 2 mM CaCl2, 1 mM DTT and 60 mM KCl], and injected into a flow cell. For the CTRD reactions the CaCl2/ATP buffer also included 4.8 nM CTRD, changing the RAD51:CTRD ratio to 15:1. The flow was stopped for ∼10 min to allow interaction of the biotynilated DNA with the Neutravidin surface. Unbound filaments were flushed by flow of CaCl2/ATP buffer, including 4.8 nM CTRD if present in assembly reaction. Dissociation was triggered by switching to Mg2+/ATP buffer [50 mM Tris–HCl (pH 7.5), 1 mM ATP, 10 mM MgCl2, 1 mM DTT and 60 mM KCl]. Hydrodynamic flow was controlled with a precision pump (Harvard Apparatus). Dynamic visualization of filaments was performed with a Nikon 60× or 100× (NA 1.45) TIRF objective in a Nikon TE2000U inverted microscope equipped with a Cascade 512B CCD camera (Princeton Instruments) driven by Metamorph software (Molecular Devices). Excitation was performed with a mercury arc lamp. Intensity measurements were obtained defining regions of interest around the construct contour length in all planes, tracking them over time and correcting for background. The data were plotted using Origin software.
Combined fluorescence and scanning force microscopy
RAD51 filaments were assembled as described above. After 30 min incubation at 37°C, the CTRD or a phosphorylated CTRD (P-CTRD) was added to a final concentration of 45 nM (RAD51:CTRD of 31:1). Reaction mixtures were diluted 10-fold in 50 mM Tris–HCl (pH 7.5), 1 mM ATP, 2 mM CaCl2, 1 mM DTT, 60 mM KCl and 45 nM CTRD for flow cell experiments, changing the RAD51:CTRD to 1.5:1. Ten microliter aliquots of the dilution were supplemented with 3 pM red fluorescent (580/605) 40 nm diameter beads (FluoSpheres® microspheres from Invitrogen) and deposited on a freshly cleaved Mica. Fluorescent images were obtained with the set-up described above and correlated with topographic images obtained with a NanoWizard II scanner (JPK instruments) as described (28).
Electrophoretic mobility shift assays
Reactions were performed in a final volume of 20 μl containing 5′-end Alexa Fluor 532-labeled 66 bp dsDNA (31) at 66 nM (nucleotides), 1 μM RAD51, 50 mM Tris–HCl (pH 7.5), 1 mM DTT, 60 mM KCl, 2 mM CaCl2 and 1 mM ATP (32). After 5 min incubation at 37°C, the CTRD peptide was added to the indicated concentrations in a 2 µl volume and incubations were continued for a further 15 min at 37°C. Reaction mixtures were then supplemented with 3 μl 30% glycerol and 24 μl were fractionated by 0.8% agarose gel electrophoresis in 0.5× Tris–Borate for 2.5 h at 60 V at 4°C. Gels were analyzed using a T9600 typhoon scanner exciting the dye-coupled DNA with a 532 nm laser and detecting emission intensity using a the 555 nm BP 20 filter at 800 V PMT, 3 mm focal plane. Images obtained were analyzed with ImageQuant version 5.2 (Molecular Dynamics) or Image J.
Scanning force microscopy
Nucleoprotein filaments were formed in 10 µl reaction mixtures containing 7.5 µM 3-kb dsDNA (concentration in bp), 2.5 µM human RAD51, 25 mM HEPES–KOH (pH 7.5), 5 mM CaCl2, 2 mM ATP and 30 mM KCl. Reaction mixtures were incubated at 37°C for 1 h and then placed on ice. When indicated, 500 nM CTRD was added to RAD51 for 15 min at 37°C before addition of the other components (RAD51:TR2 = 5:1). Aliquots of the reaction mixtures were then diluted 15-fold in 10 mM HEPES–KOH (pH 7.5) and 10 mM MgCl2 and deposited on freshly cleaved mica, scanned and analyzed as previously described (27).
DNA constructs for magnetic tweezers
A 7.3-kb dsDNA construct was prepared as described previously (33).
Magnetic tweezers assay
The magnetic tweezers set-up used in these experiments was described previously (34). By using image processing, 10-nm position accuracy of the bead was obtained in all three dimensions. To exclude the effect of thermal drift, all positions were measured relative to beads fixed to the bottom of the flow cell. DNA constructs carrying a magnetic bead at one end were anchored to the bottom of a flow cell. Experiments were started by addition of RAD51 or RAD51 pre-incubated with the CTRD at the indicated concentrations, in 50 mM NaCl, 50 mM Tris–HCl, pH 7.5, 2 mM CaCl2, 1 mM ATP. All measurements were carried out at 25°C.
The nucleation rate dependence on the CTRD concentrations was fitted with a model that takes RAD51 nucleation into account. RAD51 nucleation requires multiple RAD51 monomers according to nRAD51 + DNA → (RAD51)n–DNA, where values of n are reported between 2.7 and 4.3, indicating that nucleation requires 3–5 RAD51 monomers (13,15). This model leads to a power-law dependence of the nucleation rate according to Rnuc ∝ [RAD51]n. We propose a model where the CTRD can interact with multiple RAD51 monomers to reduce the concentration of active RAD51 according to [RAD51]active = [RAD51] − i[CTRD], where i is the number of RAD51 monomers that a CTRD peptide binds.
RESULTS
The initial aim was to measure the dissociation kinetics of RAD51 in the absence and presence of the CTRD of BRCA2 to determine whether the CTRD played a role in filament stabilization.
Filament dissociation
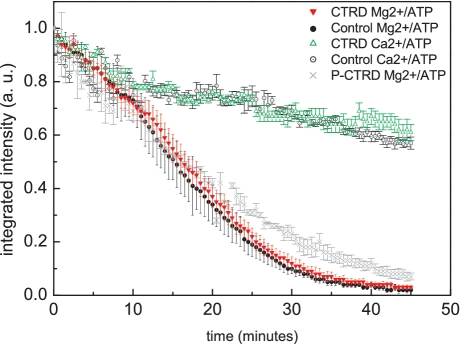
RAD51 filaments were formed by incubating Alexa Fluor 488 labeled protein with 48-kb λ phage dsDNA molecules in the presence of ATP and CaCl2, conditions that stabilize RAD51 nucleoprotein filaments (12,27,32). The λ phage dsDNA molecules were biotinylated at one end in order to tether the nucleoprotein filaments on the neutravidin-coated surface of a flow cell, where they were extended by buffer flow and observed in a fluorescent microscope. ATPase dependent dissociation of RAD51 from dsDNA was measured as a loss of fluorescent signal over time, corrected for background. Control traces in Ca2+/ATP buffer in absence or presence of a synthetic CTRD peptide (Figure 1, black hollow circles and green triangles, respectively) showed that the RAD51 complex was stable over time and that the CTRD did not affect the photostability of the fluorophore. Switching buffer to one containing ATP and MgCl2 triggered RAD51 dissociation from dsDNA (12,14). Stabilization of RAD51 filaments by the CTRD was expected to slow the kinetics for RAD51 dissociation (23). As a reference, RAD51 filaments were assembled in absence of the CTRD and RAD51 filament disassembly was measured over time (Figure 1, black solid circles). When the CTRD was added at a ratio of RAD51:CTRD = 15:1 to pre-formed filaments, there was no detectable effect of the CTRD on RAD51 dissociation from dsDNA (Figure 1, red solid triangles). The dissociation data overlap which showed that the rate is the same and implies that there is no difference in the way dissociation takes place. A phosphorylated CTRD peptide (P-CTRD), which does not interact with RAD51, also did not affect the rate of RAD51 dissociation significantly (Figure 1, grey crosses).
Figure 1.
The effect of the CTRD on RAD51 disassembly from individual dsDNA molecules in a flow cell. RAD51 disassembly from dsDNA was measured as loss of fluorescence signal. The CTRD did not significantly influence the dissociation rate when ATP hydrolysis was triggered by Mg2+/ATP. The Ca2+/ATP control filaments were also not affected by the presence of the CTRD. The non-interacting P-CTRD also did not influence the dissociation rate significantly. The error bars represent the standard error.
Confirming CTRD interaction with RAD51–DNA complexes
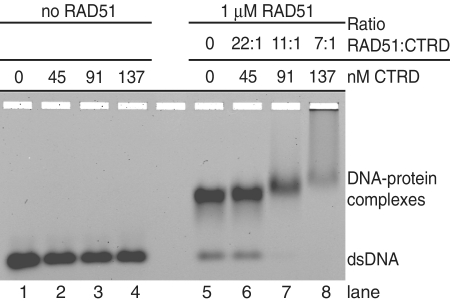
To demonstrate that the CTRD peptide can interact with RAD51–dsDNA complexes under our reaction conditions, we employed electrophoretic mobility shift assays (EMSA). Using conditions identical to those in the flow cell before switching to Mg2+/ATP buffer (i.e. ATPase suppressing Ca2+/ATP buffer) complex formation between RAD51–dsDNA and the CTRD was assessed using agarose gels (Figure 2). In the flow cell experiment the ratio RAD51:CTRD was ∼15:1. Therefore the amount of the CTRD was varied in a range encompassing the ratio in the flow cell. A higher amount of RAD51 was needed to fully shift the short dsDNA (see Supplementary Figure S1) and therefore the CTRD concentrations were adjusted to keep the ratios of RAD51:CTRD similar. The CTRD concentrations used gave RAD51:CTRD ratios of 22:1 (45 nM CTRD), 11:1 (91 nM CTRD) and 7:1 (137 nM CTRD). Figure 2 shows a representative gel where titration of the CTRD had a minor effect on dsDNA migration in the controls (lanes 2–4), but showed clear interaction with the RAD51–dsDNA complexes by ‘super-shifting’ them in a concentration dependent manner (lanes 6–8). To show specificity, the EMSA assay was also performed with P-CTRD, which includes a phopsphate group at the cyclin-dependent kinase phosphorylation site (S3291E), that does not interact with RAD51 (21).This non-interacting P-CTRD failed to form ternary complexes with the RAD51–dsDNA complex (Supplementary Figure S2). The assays confirm that the CTRD interacts with RAD51–dsDNA complexes at a similar ratio of RAD51:CTRD and in the same buffer conditions that were used in the flow cell.
Figure 2.
CTRD interaction with RAD51 filaments. Electrophoretic mobility shift assays (EMSA) were preformed with fluorescently end-labeled dsDNA and RAD51 (1 µM), CTRD or a combination of both using agarose gels to separate unbound from bound DNA. Reaction mixtures contained the indicated concentrations of the CTRD. Interaction of the CTRD with the RAD51–dsDNA complexes is evident in lanes 6–8 as ‘super-shifting’ of the complex in a CTRD concentration dependent manner.
CTRD-induced entanglement of RAD51–DNA complexes
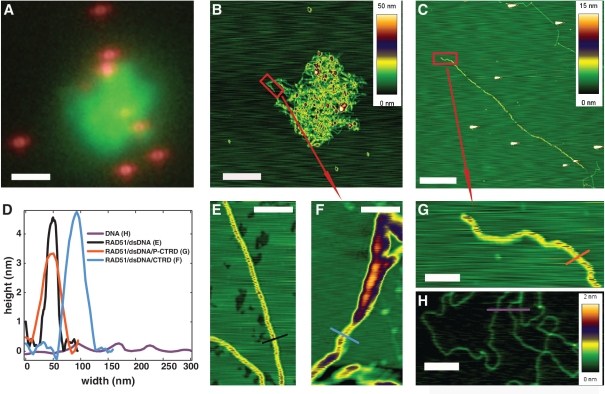
Previous biochemical assays indicate that the BRCA2 CTRD stabilizes RAD51 filaments (23). These experiments, however, utilized excess of the CTRD with respect to RAD51. To approach a comparable ratio of RAD51:CTRD, we increased the CTRD concentration 10-fold with respect to the conditions used for the experiments shown in Figure 1. However, distinctly separated, flow-stretched filaments were not observed, even at a RAD51:CTRD ratio of 1.5:1. Instead round, bright fluorescent signals were observed that presumably represented CTRD-induced entanglement of RAD51–dsDNA complexes (data not shown). A portion of these RAD51/CTRD–dsDNA complexes was deposited for imaging using combined fluorescence and scanning force microscopy (SFM). Fluorescence detection allows locating and identifying the labeled RAD51 and SFM provides nm resolution structural information of the protein–DNA complexes (Figure 3). The fluorescent image (Figure 3A) shows an overlay of a representative complex of RAD51 protein and fluorescent polystyrene beads. The polystyrene beads serve to align the combined fluorescent picture (Figure 3A) with the topographic image (Figure 3B) obtained by SFM (28). Analysis of the SFM image (Figure 3F) revealed that it was indeed an entanglement of (partial) RAD51 filaments since the height of the structure corresponded to single RAD51 filaments on dsDNA, while unbound dsDNA was also observed (compare Figure 3, panel F with E and H, respectively; see Figure 3D for height traces of the corresponding cross sections). Entanglement was due to the presence of the CTRD because in a control experiment using the same concentration of non-interacting P-CTRD no aggregation was observed (Figure 3C). Instead regular RAD51 filaments on dsDNA could be seen (compare Figure 3G with E, corresponding height traces in Figure 3D). In flow cell experiments without CTRD ∼15 ± 6 filaments on average could be seen in a 40 by 40 µm field (n = 95). However, at CTRD concentrations above 45 nM ∼1.5 ± 0.7 intense, round fluorescent signals were observed in the same size field (n = 9). Since the same amount of DNA and RAD51 were used in both filament formation reactions this suggests that one such aggregate consists on average of ∼10 filaments.
Figure 3.
CTRD-induced RAD51 filament entanglement. Fluorescent (A) and SFM (B, C, E) images of RAD51 filaments on phage λ dsDNA. Filaments were either assembled in the presence of 45 nM CTRD (A, B, F), 45 nM of phosphorylated control peptide P-CTRD (C and G) or without peptide (E). (A) Image of combined fluorescence signals from RAD51 and fluorosphere markers. The bright green object in the center of the image in panel A was scanned for nanometer topography. (B) The topographic image reveals entangled RAD51 filaments. (F) A higher resolution picture of the red boxed area in panel B shows stretches of single filament (cyan cross section) and partial filaments revealing DNA stretches not bound by RAD51. (D) Plot of height and width of the cross sections taken in scan F (blue trace) and G (red trace) comparing them to control depositions of RAD51 filaments in absence of peptide (panel E, black trace) and dsDNA alone (panel H, magenta trace). Images A–C are 10 by 10 µm, the white bar is 2 µm. Images E–H are 1 by 0.5 µm, the white bar 0.2 µm. The height in the topography scans is indicated with color as shown by the scale bars to the right of each image. The height scale in panels E–G corresponds to the one in panel C.
Filaments formed in the presence of CTRD are qualitatively distinct
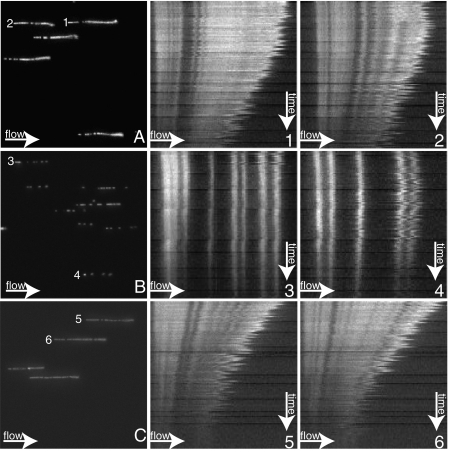
In the RAD51 dissociation experiments in the flow cell (such as plotted in Figure 1) the CTRD was added after RAD51 filament formation and incubated for an additional 15 min at 37°C. Examples of such filaments are shown in the upper left picture in Figure 4A. These filaments appear similar as filaments with RAD51 alone (12); the fluorescence covers most of the DNA contour with some gaps. In previous experiments the CTRD influenced filaments when pre-incubated with RAD51 for 15 min at 37°C before addition of DNA (21,23). Therefore we prepared filaments after pre-incubation of the CTRD with RAD51. Strikingly, RAD51 filaments were distinct from the filaments formed in the absence of the CTRD (compare Figure 4B to A). The filaments formed after pre-incubation appeared to be less complete, consisting of shorter filament patches covering less DNA (Figure 4B). Thus a change in the order of addition revealed that a pre-incubation of the CTRD with RAD51 is interfering with extensive filament formation. The presence of the CTRD during filament formation results in incomplete, ‘patchy’ filaments, which could not be analyzed reliably in dissociation experiments. Therefore we decided to analyze these filaments using SFM.
Figure 4.
Appearance of RAD51 filaments in flow cell experiments. RAD51 nucleoprotein filaments (A–C) and their corresponding kymographs showing disassembly over time after ATP hydrolysis has been triggered (A 1/2, B 3/4, C 5/6). Fluorescent signal was acquired as described previously for flow cell experiments (12). The images on the left (A–C) show 40 by 40 µm images of RAD51 filaments on dsDNA inside the flow cell before ATP hydrolysis was triggered. Adjacent to images A, B and C are the example kymographs of the filaments numbered accordingly. The kymographs are pictures horizontally displaying a filament, with its anchor point on the left, and vertically displaying disassembly of RAD51 over time. Each pixel line towards the bottom of the kymograph represents a 20 s step for a total time of 45 min. The upper panels show the typical appearance of RAD51 filaments. The middle panels show filaments formed after pre-incubation of RAD51 and the CTRD peptide. These filaments appear patchy as evidenced by stretches of bare DNA and less protein bound per DNA molecule. The lower panels show filaments formed after pre-incubation with the phosphorylated control peptide (P-CTRD). These filaments appear similar to the ones without peptide as they are not patchy.
SFM analysis of filament length in presence of the CTRD
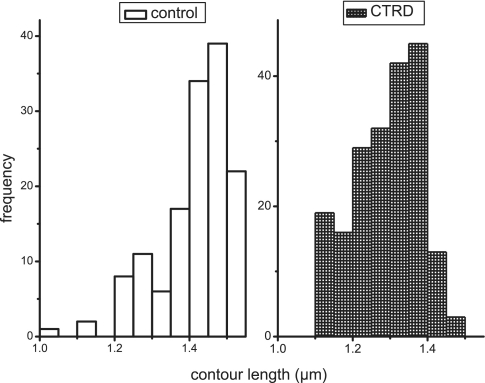
To assess the effect of the CTRD on filament formation and structure, nucleoprotein filaments were formed after pre-incubation of RAD51 and the CTRD, at a ratio of 5:1, on 3-kb dsDNA were analyzed by SFM (27). The images did not show a strong difference in appearance of the control filaments and those formed in the presence of the CTRD (data not shown). As RAD51 polymerization onto DNA stretches it ∼50% over regular B-form length, we compared the contour lengths of RAD51 filaments without and with the CTRD (RAD51:CTRD = 5:1). The average elongation of filaments was not much decreased by the presence of the CTRD (1.4 ± 0.1 µm versus 1.29 ± 0.09 µm). However, the distribution of filament lengths was clearly skewed towards shorter filaments in the presence of the CTRD (Figure 5). A reduction in contour lengths could be attributed to reduced RAD51 polymerization onto DNA. Filament formation is a dynamic process consisting of nucleation and extension (growth) and the CTRD could specifically influence one or the other of these processes.
Figure 5.
Contour length of RAD51 filaments formed in the presence and absence of the CTRD. The contour length of RAD51 filaments, from images obtained by SFM, were measured and plotted in histograms for control filaments (left) and filaments formed in the presence of the CTRD (right). The y-axis indicates the number of filaments, while the contour length in micrometer with a binning step of 50 nm is plotted along the x-axis. The left panel displays the histogram for the control filaments; the right panel for filaments formed after pre-incubation of RAD51 with CTRD at a ratio of 5:1.
The CTRD decreases RAD51 filament assembly rate in magnetic tweezers measurements
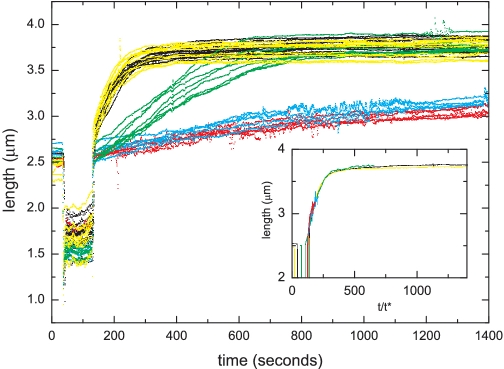
RAD51 filament assembly was followed on individual dsDNA molecules (7.3 kb) in magnetic tweezers (13,33). The DNA molecules were tethered between a magnetic bead and the surface of a flow cell. A force of 7 ± 2 pN was applied by a pair of magnets and video microscopy was used to measure the end-to-end distance of the DNA tether. Due to the known extension of dsDNA upon RAD51 binding an increase in tether length can be directly correlated to the polymerization of RAD51 onto dsDNA. Assembly of a RAD51 filament in the absence of the CTRD resulted in a DNA tether length increase from 2.54 ± 0.04 μm to 3.76 ± 0.07 μm corresponding to a 1.48-fold extension (Figure 6, black curves), similar to previous observations (13). RAD51 filament assembly was, however, very sensitive to pre-incubation of RAD51 with the CTRD in a concentration dependent manner. At a ratio of RAD51:CTRD = 12.5:1 filament assembly was already markedly reduced (Figure 6, cyan curves).
Figure 6.
RAD51 filament assembly in real time. RAD51 filament assembly was measured as changes in the length of a DNA, tethered in magnetic tweezers, over time. Different CTRD concentrations are indicated by color: [CTRD] = 0 (black), [CTRD] = 2 nM (yellow), [CTRD] = 6 nM (green), [CTRD] = 12 nM (cyan) and [CTRD] = 60 nM (red). Filament assembly was measured on several individual DNA molecules (n = 5–12) for each CTRD concentration. RAD51 and the CTRD were pre-incubated for 15 min at 37°C. RAD51 concentration was constant at 150 nM in all experiments. The inset shows the average growth profile for each concentration of the CTRD rescaled by t/t*, where t* = 1/γ and γ is the nucleation rate determined from fitting to Monte Carlo simulations.
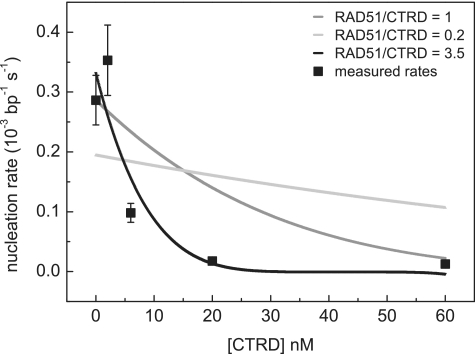
The assembly profiles obtained by magnetic tweezers experiments were analyzed by fitting to Monte Carlo simulations, assuming a binding size of 3 bp per RAD51 monomer and filament nucleation and extension by RAD51 pentamers as described previously (13,29,33). Nucleation is the dominant step in filament formation. For the simulations the extension rate was linked to the nucleation rate by setting the co-operativity number ω to a value of 100 in line with results of fits with cooperativity as a free independent fit parameter and previous results (29). The Monte Carlo fitting procedure yielded nucleation rates of RAD51 at different CTRD concentrations (Figure 7). The validity of this approach for extracting filament assembly rates was demonstrated by plotting averaged assembly curves for each CTRD concentration against a rescaled time axis (inset, Figure 6). The time axis was normalized based on rates found after fitting to Monte Carlo simulations. After normalizing, the filament assembly profiles at different CTRD concentrations overlapped (inset, Figure 6), confirming that the shape of the assembly curves is similar and that these curves differ in only one parameter, namely the assembly rate. Filament assembly was strongly decreased by the presence of the CTRD in a concentration dependent manner. The influence of the CTRD on filament assembly rates indicated that the amount of RAD51 available for filament assembly was decreased in the presence of the CTRD. When RAD51 nucleation rates are plotted against CTRD concentration (Figure 7), the power-law shape of the fit indicates that the CTRD interacts with multimeric RAD51, calculated to be on average three to four RAD51 monomers (see ‘Materials and Methods’ section) consistent with previous observations (23,25).
Figure 7.
Effect of the CTRD on filament assembly rate. The nucleation rates of RAD51 filaments were extracted from fitting Monte Carlo simulations to the data from Figure 6 at a constant cooperativity number of ω = 100. Error bars indicate the standard deviations in the nucleation rates found by fitting the assembly profiles of Figure 6 at each CTRD concentration. A model used to fit nucleation rates (see ‘Materials and Methods’ section) resulted in a best fit for the number of RAD51 monomers interacting with CTRD of 3.5 ± 0.4 (black trace). For comparison a model assuming a CTRD interaction with monomeric RAD51 (dark grey trace), and a model assuming an interaction of monomeric RAD51 with multiple CTRDs (light gray trace) is shown.
DISCUSSION
Here we have addressed the effect of the CTRD domain of BRCA2 on the key entity in HR, the RAD51 filament. RAD51 nucleoprotein filaments are dynamic structures undergoing constant rearrangements coupled to ATP hydrolysis. Either suppressing ATP hydrolysis or mechanical protein–protein interactions could stabilize RAD51 filaments. The CTRD of BRCA2 was proposed to stabilize RAD51 filaments by stabilization of protein–protein interactions, acting as a bridge over the RAD51 monomer interface in one model (23,25). In the context of full-length BRCA2, the CTRD domain is expected to have a role modulating RAD51 activity based on observations that phosphorylation of S3291 of CTRD by CDKs abolishes RAD51 binding (21,26). However, we observe no effect of the isolated CTRD domain on filament disassembly rates by direct visualization of RAD51 dissociation from λ-phage dsDNA. Although this result suggests the model based on CTRD bridging RAD51 monomers in a filament to prevent access of BRC-repeats might not hold up it is not contradictory to previous work showing that RAD51 filament disassembly by BRC-repeat peptides is inhibited by the CTRD (22–25). That observation could also be due to CTRD induced RAD51 filament entanglement or aggregation as discussed below. Direct stabilization by altering inherent RAD51 dissociation was not previously tested (22–25). Since RAD51 dissociation in our set-up is dependent on ATP hydrolysis (12,14), we can conclude that the presence of the CTRD (at a ratio of RAD51:CTRD = 15:1) does not suppress the ATPase activity of RAD51, nor is its interaction with the filament strong enough to slow dissociation in a flow cell set-up.
The absence of an effect of the CTRD peptide on RAD51 filament stability could have been due to absence of interaction with the filaments formed in our conditions. Although the amount of peptide available precluded adding it to the buffer flow during dissociation the persistence of CTRD-induced filament entanglement in the same flow conditions argues that the peptide does not simply dissociate. The EMSA experiments show that the CTRD interacts with RAD51–dsDNA complexes in a concentration-dependent manner in the same starting reaction conditions used in the flow cell i.e. Ca2+/ATP. Because these gel assays required RAD51 at higher concentrations than in the flow cell, the concentration of the CTRD was also adjusted so that the ratio of RAD51:CTRD was 22:1, 11:1 and 7:1, encompassing the 15:1 ratio used in the flow cell. Thus, while the CTRD interacts with RAD51 filaments, it does not influence filament disassembly rates, suggesting that filament stability is not affected.
Several lines of evidence indicate that at higher concentration, or ratios to RAD51, CTRD causes entanglement of RAD51 filaments. Flow cell experiments were attempted at a higher protein concentration, such as required for the EMSA. At CTRD concentration of 45 nM, the lowest concentration showing an effect in the EMSA and corresponding to a final ratio of RAD51:CTRD of 1.5:1 in the flow cell, filaments could not be analyzed by flow stretching. The observation of round, intense, fluorescent signals, which sometimes untangled upon ATP hydrolysis, indicated that the long RAD51 filaments on λ-phage dsDNA may have aggregated in these conditions. Combined fluorescence and SFM imaging revealed that the large, brightly fluorescent objects are indeed CTRD-induced entangled RAD51 filaments that include stretches of naked DNA (Figure 3). This aggregation of RAD51–DNA complexes at higher CTRD concentration would have a detrimental effect on interactions of other proteins with the filament structure. Indeed, CTRD-induced filament aggregation has also been noted in other studies (23). This could explain the previously observed protective effect of the CTRD on filaments exposed to high concentrations of BRC4 that will otherwise disrupt filaments (23,25). The correlation between stronger CTRD:RAD51 interaction and longer persistence of local nuclear accumulations of RAD51 is also consistent with CTRD-induced filament aggregation or entanglement (26).
By contrast, the CTRD–RAD51 interaction did inhibit filament assembly. Interaction of RAD51 and the CTRD before adding dsDNA had a strong effect on filament appearance, even at a final CTRD concentration of ∼5 nM (ratio RAD51:CTRD = 291:1). The nucleoprotein filament formed had more frequent and larger gaps of naked DNA between protein-bound patches (compare Figure 4 upper and lower panel), an appearance we refer to as ‘patchy’. Furthermore, the concentrations used here were equivalent to the ones in the disassembly experiments, indicating that the lack of an effect on filament disassembly in the flow cell was not due to concentrations of the CTRD insufficient for interaction with RAD51. RAD51filaments, formed on 3 kbp-long dsDNA, in the presence of the CTRD are also somewhat shorter than control filaments (Figure 5). The average DNA extension by filament pre-incubated with the CTRD is only ∼70% that of the control filaments (29 versus 40%). In the magnetic tweezers assay, RAD51 filament assembly was very sensitive to pre-incubation with the CTRD. Rates progressively decreased with increasing CTRD concentration (Figure 6). The shape of the assembly curves reflects the interplay between nucleation and extension events in RAD51 DNA-binding to assemble a filament (13). The average assembly curves for each CTRD concentration have the same shape, implying that the CTRD has not changed the way RAD51 extends dsDNA as reflected in the ratio of nucleation to extension. These data all support the idea that the CTRD inhibits RAD51 filament assembly by effectively reducing the active RAD51 concentration.
Alternatively, the CTRD could specifically cap nascent filament patches and block further extension. This would produce filaments with different appearance, shorter protein patches with a length correlating to the CTRD concentration, and would require more nucleation events to cover the DNA. However, filaments formed after pre-incubating RAD51 and the CTRD at a 5:1 ratio appeared mostly regular, without obvious gaps or multiple kinks that would indicate reduced extension events. Additionally, filament capping by the CTRD is expected to inhibit dissociation and to change the ratio of nucleation to extension in filament assembly kinetics. None of these effects were observed, thus the CTRD interaction with RAD51 apparently does not cap filaments.
The RAD51 assembly reactions show a power-law dependence of the nucleation rate on CTRD concentration. This was used to determine the stoichiometry of RAD51 affected by the CTRD. Assuming a nucleation unit of 5 RAD51 monomers (13,29) the power-law fit had a minimum for least squares error i = 3.5 ± 0.4, indicating that one CTRD reacts on average with three to four RAD51 monomers. This is in accordance with the observation that the CTRD binds multimeric RAD51 in solution (23,25) and suggests that the mode of the CTRD action in this experiment is to sequester RAD51 from filament assembly.
Inactivation of three to four RAD51 monomers for filament formation by CTRD binding can explain altered filament appearance as well as reduced assembly rates. The alternative model, that extending RAD51 filament patches are capped in RecX-like fashion is unlikely (35–37). Overall, our data are in accordance with recent work on the full-length BRCA2 (38–40) and previous studies with BRC peptides (22–25,41–43). Those studies show that BRCA2 and its fragments interact with RAD51–dsDNA complexes and that BRCA2 modulates RAD51 DNA-binding such that filaments preferentially form on ssDNA over dsDNA. Here we show that not only BRC repeats (23,41–43) but also the CTRD of BRCA2 interferes with RAD51 filament formation on dsDNA. These two peptide portions of BRCA2 differ in details of how they affect RAD51 filaments. Stimulation of RAD51 binding to ssDNA specifically occurs when ATP hydrolysis is possible (41,42). In contrast, the effect we report of the CTRD on RAD51 filaments does not require ATP hydrolysis, as reactions occur in conditions where RAD51′s ATPase activity is suppressed. The interaction stoichiometry of the two domains is also different. The BRC repeat forms a complex with RAD51 at a 1:1 ratio (24,44,45). The CTRD interacts with RAD51 multimers (23,24), and we show it affects filaments formation at substoichiometric amounts. How these different interaction phenomena are manifested in the context of full-length BRCA2 is an important question that can now be addressed. The CTRD domain could work in concert with the BRC domains modulating DNA binding towards ssDNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants from the Netherlands Organization for Scientific Research TOP (to R.K.); VICI 700.56.441 (to C.W.); the Netherlands Genomics Initiative; The European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement No. HEALTH-F2-2010-259893; National Cancer Instititute, USA (SBDR 5PO1CA092584); Association for International Cancer Research (09-0633 to M.M); Agence Nationale de la Recherche (RADORDER to M.M.) and Marie Curie Intra European Fellowship (FP7-221069 to H.S.). Funding for open access charge: Grant funding from Netherlands Organizations for scientific research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Tina Thorslund and Dr Stephen West for the generous gift of CTRD peptides and for insightful comments on the manuscript.
REFERENCES
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 3.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 4.Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair. 2004;3:827–833. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. EMBO J. 2010;29:2864–2874. doi: 10.1038/emboj.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rass U, Compton SA, Matos J, Singleton MR, Ip SC, Blanco MG, Griffith JD, West SC. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010;24:1559–1569. doi: 10.1101/gad.585310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svendsen JM, Harper JW. GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 2010;24:521–536. doi: 10.1101/gad.1903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Heyer WD. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–646. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol. Cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 13.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl Acad. Sci. USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 17.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 18.Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 19.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 20.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 21.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 22.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl Acad. Sci. USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 24.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc. Natl Acad. Sci. USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayoub N, Rajendra E, Su X, Jeyasekharan AD, Mahen R, Venkitaraman AR. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr. Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez H, Kanaar R, Wyman C. Molecular recognition of DNA-protein complexes: a straightforward method combining scanning force and fluorescence microscopy. Ultramicroscopy. 2010;110:844–851. doi: 10.1016/j.ultramic.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Heijden T, Dekker C. Monte carlo simulations of protein assembly, disassembly, and linear motion on DNA. Biophys. J. 2008;95:4560–4569. doi: 10.1529/biophysj.108.135061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graneli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 31.van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res. 2009;37:1580–1588. doi: 10.1093/nar/gkn1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Loenhout MT, van der Heijden T, Kanaar R, Wyman C, Dekker C. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37:4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 36.Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. A RecA filament capping mechanism for RecX protein. Mol. Cell. 2004;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 37.Ragone S, Maman JD, Furnham N, Pellegrini L. Structural basis for inhibition of homologous recombination by the RecX protein. EMBO J. 2008;27:2259–2269. doi: 10.1038/emboj.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl Acad. Sci. USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 45.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.