Abstract
Piwi-interacting RNA (piRNA) are small RNA abundant in the germline across animal species. In fruit flies and mice, piRNA have been implicated in maintenance of genomic integrity by transposable elements silencing. Outside of the germline, piRNA have only been found in fruit fly ovarian follicle cells. Previous studies have further reported presence of multiple piRNA-like small RNA (pilRNA) in fly heads and a small number of pilRNA have been reported in mouse tissues and in human NK cells. Here, we analyze high-throughput small RNA sequencing data in more than 130 fruit fly, mouse and rhesus macaque samples. The results show widespread presence of pilRNA, displaying all known characteristics of piRNA in multiple somatic tissues of these three species. In mouse pancreas and macaque epididymis, pilRNA abundance was compatible with piRNA abundance in the germline. Using in situ hybridizations, we further demonstrate pilRNA co-localization with mRNA expression of Piwi-family genes in all macaque tissues. Further, using western blot, we have shown the expression of Miwi protein in mouse pancreas. These findings indicate that piRNA-like molecules might play important roles outside of the germline.
INTRODUCTION
Typical piRNA are 24–32 nt in length (1,2–9). Primary piRNA have been suggested to originate via fragmentation of longer transcripts and, therefore, can be traced to discrete genomic locations, or piRNA clusters, which show strong strand specificity (1,7,10,11). Another known property of primary piRNA is marked 5′ uridine bias (1,7,10,11). Secondary piRNA are generated through an amplification mechanism, known as the ping-pong mechanism, which requires base pairing between the first 10 nt of the primary piRNA and target transcripts (1,7,8,10). Thus, presence of the ping-pong mechanism can be deduced through an excess of small RNA with 10 nt long complementary regions (1,7,8–10).
In Drosophila, piRNA have been described to be present in embryos and the germline (1,8,12), as well as in a specific type of somatic tissue: ovarian follicle cells (13,14). Fly germline piRNA include both primary and secondary piRNA, require the presence of three Piwi-family proteins; Piwi, Aub and Ago3, and are mainly derived from transposable elements (1,8). Presence of these piRNA has been shown to be essential for transposable element silencing (1). piRNA found in the fly somatic ovarian follicle cells and a derived cell line, ovarian somatic sheet (OSS) cells, predominantly include primary piRNA that originate from discrete genomic loci (11,13,14–16). These piRNA are associated with one Piwi-family protein (Piwi) and are overrepresented in both transposable elements and certain 3′-UTR regions (11,13,14–16). Functionally, these piRNA play important roles in suppressing transposons in the ovarian somatic cells and are involved in suppression of transposable element transfer into the female germline (13,14,17,18).
In mammals, piRNA are particularly abundant in the male germline. Unlike Drosophila piRNA, piRNA present in adult mammalian testes tend to originate from unannotated genomic regions devoid of transposable elements and other repeats (4,5). The functional significance of piRNA originating outside of transposable elements, if any, is presently unknown.
While most reports provide no indications of piRNA and Piwi-family proteins presence outside of the mammalian and fly germlines, or outside fly ovarian follicle cells (1,3–5), one study cloned and amplified several dozens of piRNA-like small RNA (pilRNA) from a variety of mouse tissues (19). More recently, a specific pilRNA implicated in transcriptional silencing of Ig-like receptors was described in human Natural Killer (NK) cells (20). Finally, thousands of pilRNA, displaying known piRNA sequence features, including presence of 2′-O-methyl group at the 3′-end, have been reported to be present in Drosophila heads (21).
MATERIALS AND METHODS
Sample preparation, tissues
All rhesus macaque samples were obtained from the Suzhou Experimental Animal Center (Suzhou, China). All tissues were dissected and frozen in liquid nitrogen within 20 min after death. For small RNA library construction, we isolated total RNA from testes, epididymis, prostate, seminal vesicles and brain cortex (prefrontal cortex, corresponding to Brodmann area 10) of one adult rhesus macaque individual (9 years 104 days) (see Supplementary Table S6 for sample characteristics).
Adult male mice were purchased from the Animal Center of the Chinese Academy of Sciences (Shanghai, China). Experiments were conducted according to a protocol approved by the Institute Animal Care Committee. The protocol conforms to internationally accepted guidelines for the human care and use of laboratory animals.
Mouse epididymis and pancreas samples were obtained from male mice after they were sacrificed and the samples were frozen immediately in liquid nitrogen.
Total RNA was firstly isolated using the Trizol (Invitrogen, USA) protocol. In addition, low molecular weight RNA with length ranging from 20 to 40 nt were isolated, ligated to the adapters, amplified and sequenced following the Small RNA Preparation Protocol (IL, USA), with no modifications.
Sample preparation, specific cell types
Preparation of specific cell type samples was done independently from the preparation of other tissue samples. Laser Capture Microdissection (LCM) was used to dissect two types of epididymis cells: principal/basal cells (22,23), which contained pilRNA according to in situ hybridization results and, peritubular tissue, which did not contain pilRNA, from four adult rhesus macaque epididymis samples (Supplementary Table S6). Specifically, frozen rhesus macaque epididymis tissue was embedded in OCT. Tissue sections (6–8 µm thick) were stained using the Arcturus HistoGene Frozen Section Staining Kit (Molecular Devices) according to the manufacturer's instructions, with no modifications. For separating the principal/basal cell and peritubular tissue, the Laser Capture Microdissection System Arcturus, Veritas Microdissection System (Molecular Devices) was used with the following settings: 30–40 µm laser spot size, 88–95 mW power and 2500–3800 ms pulse duration. Groups of cells to be collected were localized and marked under the microscope and then transferred into the provided cap (Arcturus, Molecular Devices). Total RNA was extracted from the cell obtained from LCM, using the RNAqueous®-Micro Kit (Ambion) following the manufacturer's instructions and with no modifications. The Small RNA Preparation Protocol (IL, USA) was applied to the LCM dissected samples with modifications, at only one step, such that prior to low molecular weight RNA isolation, the total RNA from four rhesus individuals aged between 8 and 10 years was combined in equal amounts.
All small RNA sequencing data (five macaque tissue samples, two macaque epididymis LCM-dissected samples and one mouse epididymis sample) are deposited to GEO under accession number GSE27369.
Small RNA mapping
We mapped the small RNA sequencing data as described elsewhere (24). Briefly, for each rhesus macaque sequencing data set, all multiple reads representing one sequence were collapsed and the adapter sequence at the 3′-end of the sequence reads was trimmed using the custom trimming procedure. The resulting unique sequences (sequence tags) were mapped to the rhesus macaque genome (MGSC Merged/rheMac2) using Bowtie (25). We allowed one mismatch for the sequences with length >25 nt and required perfect match for the shorter sequences. According to these criteria, 58–69% of all reads (∼7–9 million reads) could be mapped to the rhesus macaque genome of each sample (Supplementary Table S7).
We downloaded 57 mouse small RNA sequencing data sets, from GSE12757 (7), GSE17319 (26), GSE19172 (16), GSE7414 (10), GSE16023 (27) and GSE21630 (28), and 60 Drosophila melanogaster small RNA sequencing data sets from GSE11624 (29), GSE12462 (30), GSE11019 (21), GSE10794 (31), GSE11086 (32), GSE7448 (33), GSE18806 (30), GSE6734 (1) and GSE15378 (11) (Supplementary Table S13). Downloaded mouse data and small RNA sequences from mouse epididymis, as well as downloaded fly data, were processed and mapped to the mouse genome (NCBI37/mm9) and the D. melanogaster genome (BDGP R5/dm3), respectively, using the above described procedure.
Rhesus macaque genome annotation
The rhesus macaque genome (MGSC Merged/rheMac2) was downloaded from UCSC (34). The rhesus macaque transposon/repeat annotation was downloaded from UCSC rhesus RepeatMasker track (34). We classified LINE, SINE, LTR and DNA as ‘transposons' and the rest of the repetitive sequence types, such as simple repeats, as ‘other repeats'
The rhesus macaque protein-coding genes and non-coding RNA (ncRNA) annotation was based on reciprocal liftOver of the genomic coordinates of the corresponding human annotation. Specifically, to obtain rhesus macaque protein-coding gene annotation, we downloaded human protein-coding gene annotation from Ensembl (version 57) (35). From here, we obtained the genomic coordinates of exons (5′-UTR, coding region, 3′-UTR), introns and gene boundaries in the rhesus macaque genome using reciprocal liftOver with default parameters and requiring the length of the matched sequence to be >70 and <130% of the query sequence length (34). Take human region A as one example. For human region A, using default parameters and requiring the length of the ortholog region to be between 70% and 130% of the length of region A, we first used liftOver to find region A's ortholog region (region B) in the macaque genome. We then used the same settings to find region B's ortholog region (region C), in the human genome. Finally, we require regions A and C to be located within 100 nt from one another in the human genome. To annotate rhesus macaque miRNA, we downloaded human miRNA annotation from miRBase (Version 12) (36–38) and used reciprocal liftOver, with the same settings used for protein-coding gene annotation, to obtain the corresponding coordinates in the rhesus macaque genome. The miRNA expression quantification method is described elsewhere (24). The miRNA expression level from five macaque tissues of this study measured by Illumina sequencing platform are listed in Supplementary Table S14. For other ncRNA (rRNA, tRNA, snoRNA, snRNA, scRNA and misc-RNA), we created a combined human ncRNA annotation by taking the union of ncRNA annotations downloaded from UCSC (34), Refseq (39) and Ensembl (35). We then used reciprocal liftOver, with the same settings used for protein-coding gene annotation, to obtain the corresponding coordinates in the rhesus macaque genome.
Mouse and fly genome annotation
The mouse genome (NCBI37/mm9) and the fly genome (BDGP R5/dm3) were downloaded from UCSC (34). Mouse and fly transposon/repeat annotations were downloaded from UCSC mouse and fly RepeatMasker track (34). For mouse annotation, we classified LINE, SINE, LTR and DNA as ‘transposons' and the rest of the repetitive sequence types, such as simple repeats, as ‘other repeats'. Mouse protein-coding gene annotation was downloaded from Ensembl (version 57) (35). Fly protein-coding gene annotation was downloaded from UCSC FlyBase Genes track (34). Mouse and fly miRNA annotations were downloaded from miRBase (Version 14) (36–38). For other mouse ncRNA (rRNA, tRNA, snoRNA, snRNA and misc-RNA), we created a combined mouse ncRNA annotation by taking the union of ncRNA annotations downloaded from Refseq (39) and Ensembl (35). For other fly ncRNA (rRNA, tRNA, snoRNA, snRNA, ncRNA), we created a combined fly ncRNA annotation by taking the union of ncRNA annotations downloaded from UCSC flyBaseNoncoding Track (39) and Ensembl (35).
Small RNA sequence analysis
For each sample, we first removed all sequences mapped to annotated ncRNA regions defined using the methods described above. The length and the 5′-nt distributions of all mapped reads and sequence tags (sequence reads after collapsing identical sequences) are shown in Figures 1A, B, 2A, B, 3A and B, the first two columns of Supplementary Figure S3. The length and the 5′- nt distributions of sequence reads and tags, excluding those mapped to known ncRNA, are shown in Figures 1C, D, 2C, D, 3C, D, the third and the fourth columns of Supplementary Figure S3. The remaining small RNA sequences with length distributions of between 24 and 32 nt (pilRNA) were used in further analyses. Specifically, we tested these small RNA with respect to: length distribution, 5′-nt bias, genomic context, clustering along the genome and presence of sequence features compatible with the ping-pong biogenesis model.
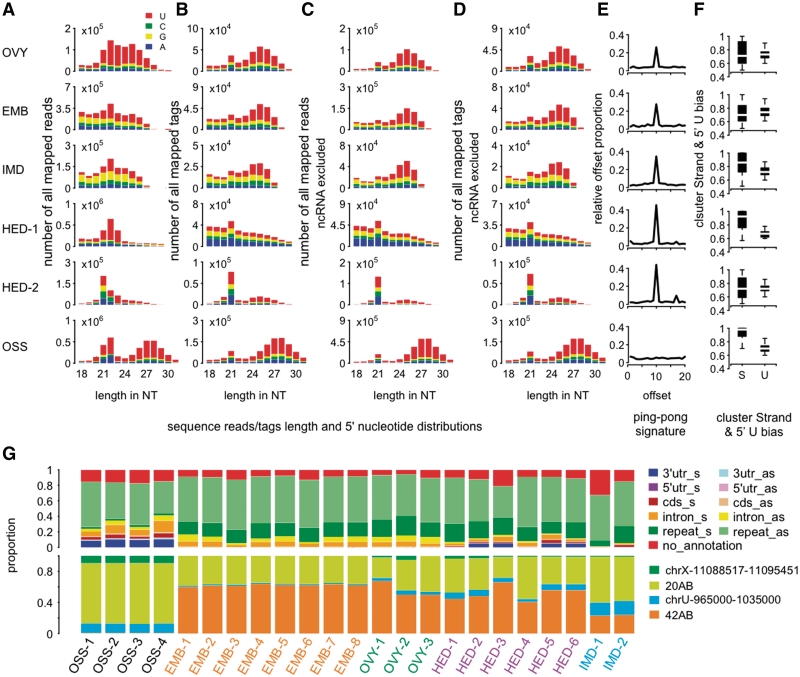
Figure 1.
piRNA and pilRNA features in fly tissues. (A) Length distribution of all mapped sequence reads in fly tissues. Here (and further), the labels indicate: OSS, ovarian somatic sheet cells; EMB, embryo; OVY, ovary; HED, head; IMD, imaginal discs. For a full sample description, see Supplementary Table S13. (B) Length distribution of all mapped sequence tags (sequence reads after collapsing identical sequences). (C and D) Length distributions of sequence reads and tags, excluding reads mapped to known ncRNA, respectively. Note that 5′ uridine bias is one known property of primary piRNA. (E) Proportions of complementary small RNA reads (24–32 nt) at different offsets. Excess of complementary reads at offset equal 10 nt indicates the ping-pong model signature. (F) The stand (S) and the 5′ uridine (U) bias of sequence reads' clusters based on 24–32 nt long sequences. (G) The upper panel shows genomic context of small RNA clusters (24–32 nt) identified in five fly tissues using publicly available data sets (11,21,29,30–33). The lower panel shows relative abundance of uniquely mapped small RNA (24–32 nt) among four previously characterized fly TE master loci (11).
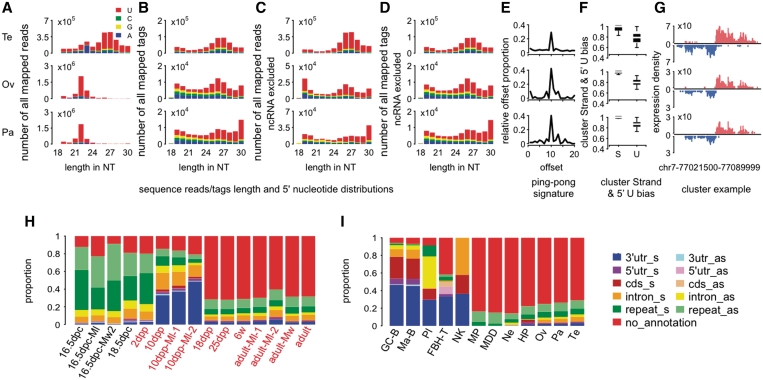
Figure 2.
piRNA and pilRNA features in mouse tissues. (A) Length distribution of all mapped sequence reads in three adult mouse tissues: Te, testes; Ov, ovary; Pa, pancreas. (B) Length distribution of all mapped sequence tags (sequence reads after collapsing identical sequences). (C and D) Length distributions of sequence reads and tags, excluding reads mapped to known ncRNA, respectively. Note that 5′ uridine bias is one known property of primary piRNA. (E) Proportions of complementary small RNA reads (24–32 nt) at different offsets. Excess of complementary reads at offset equal 10 nt indicates the ping-pong model signature. (F) The stand (S) and the 5′ uridine (U) bias of sequence reads' clusters based on 24–32 nt long sequences. (G) Reads coverage of a bidirectional piRNA cluster previously identified in mouse testes (4) in three mouse tissues. Shown are all reads with length 24–32 nt, excluding reads mapped to known ncRNA. The y-axis shows log2-transformed nucleotide coverage normalized by the total number of mapped reads within in a given sample. (H) Genomic context of piRNA clusters found in mouse testes at different age, from embryonic stages, pre-pachytene (10 dpp) and pachytene (18 dpp) stages to adults, based on data from (7,10,16,26,27) (‘Materials and Methods' section). The labels indicate the age of the samples in days post-conception (dpc), days post-partum (dpp) or weeks (w) and piRNA origin: testes tissue (no label) or immunoprecipitations of Mili (Ml), Miwi2 (Mw2) or Miwi (Mw) proteins. For a full sample description, see Supplementary Table S13. (I) Genomic context of pilRNA clusters in adult mouse tissues: GC-B, germinal center B cells; Ma-B, mature B cells; Pl, plasma cells; FH-T, follicular helper T cells; NK, natural killer cells; MP, macrophages; MDD, macrophages-derived dendritic cells; Ne, neutrophils; HP, hematopoietic progenitor cells; Ov, ovaries; Pa, pancreas; Te, testes (28).
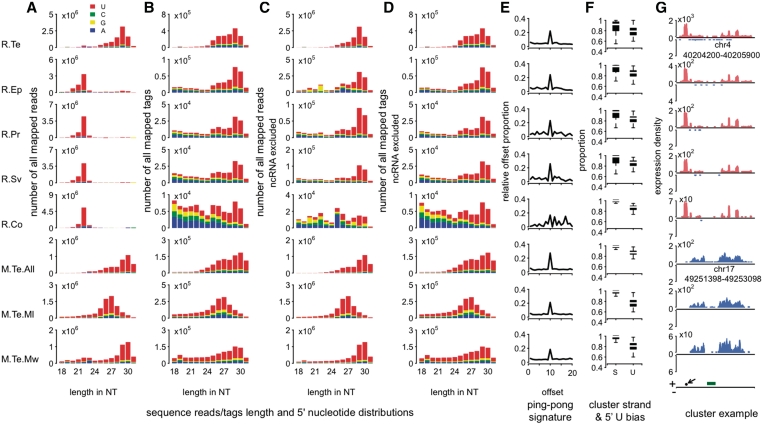
Figure 3.
piRNA and pilRNA features in rhesus macaque tissues. (A) Length distribution of all mapped sequence reads in five adult rhesus macaque tissues and adult mouse testes. The labels indicate: R.Te, macaque testes; R.Ep, macaque epididymis; R.Pr, macaque prostate; R.Sv, macaque seminal vesicles; R. Co, macaque cerebral cortex; M.Te.All, mouse total testes (26); M.Te.ML, mouse testes Mili-immunoprecipitation (16); M.Te.MW, mouse testes Miwi-immunoprecipitation (16) (B) Length distribution of all mapped sequence tags (sequence reads after collapsing identical sequences). (C and D) Length distributions of sequence reads and tags, excluding reads mapped to known ncRNA, respectively. Note that 5′ uridine bias is one known property of primary piRNA. (E) Proportions of complementary small RNA reads (24–32 nt) at different offsets. Excess of complementary reads at offset equal 10 nt indicates the ping-pong model signature. (F) The stand (S) and the 5′ uridine (U) bias of sequence reads' clusters based on 24–32 nt long sequences. (G) Reads coverage of an example piRNA with high coverage in adult macaque testes. Shown are all reads with length 24–32 nt, excluding reads mapped to known ncRNA. The y-axis shows log2-transformed nucleotide coverage normalized by the total number of mapped reads within in a given sample. The bottom of the panel shows annotation of the corresponding rhesus macaque genomic region, dark green—SINE and location of one of the ‘anti-pilRNA’ LNA probes (piRNAa)—arrow. Note that the genomic region orthologous to the macaque small RNA cluster shown in Figure 3G is inverted in the mouse genome. To align the mouse and macaque genomic regions, mouse cluster is shown in reverse orientation with respect to the macaque cluster.
Sequence length distributions were compared with piRNA sequence length distributions found in three mouse piRNA data sets: adult mouse Mili-IP (16), adult mouse Miwi-IP (16) and 16.5dpc mouse Miwi-2 IP (7) (Figure 3 and Supplementary Figure S5).
The 5′-nt bias was measured as a percentage of sequences with a certain nucleotide at the 5′-end among all pilRNA. The significance of a 5′-nt bias was tested by comparing it to the genome average, using a binomial test. For pilRNA clusters, the 5′-nt bias was estimated, following the criteria described below, as a percentage of sequences with a certain nucleotide at the 5′-end among all pilRNA that clustered along the genome. As these criteria include minimal percentage of small RNA with 5′ uridine, the nucleotide bias within clusters is not an independent indicator of piRNA-like properties.
To investigate whether pilRNA cluster along the genome, we used a 3 kb long sliding window with 0.5 kb steps to identify regions containing at least 30 uniquely mapped small RNA sequences and showing at least 60% 5′ uridine bias. For the mouse small RNA data sets [GSE21630 (28)] and for all but 4 of the OSS samples for the fly small RNA data sets, we required 10 unique mapped sequences instead of 30, due to relatively low sequencing coverage. Identified windows located within 1 kb distance from each other were further merged. To identify numbers of windows that expected to be found by chance, we randomized small RNA locations along the genome and repeated the analysis with the same requirements 1000 times.
According to the suggested piRNA biogenesis model, the ping-pong model, there are two groups of piRNA populations: primary piRNA and secondary piRNA (1,7,8). Primary piRNAs have 5′ bias for uridine and do not have nucleotide bias at position 10. Secondary piRNAs, generated by primary piRNA guided cleavage, have a bias for adenine at position 10 and do not have 5′ bias. Based on this knowledge, we tested the ping-pong biogenesis model by checking the complementarity, at different offsets, between putative primary piRNA and putative secondary piRNA. Here, small RNA sequences (24–32 nt), excluding ncRNA, were divided into two categories: (i) putative primary piRNA—small RNA that have U as the first nucleotide and (ii) putative secondary piRNA—the rest of the sequences. Sequences that have both U at position 1 and A at position 10 were excluded from the putative primary piRNA group because these sequences could not be assigned to one of the groups unambiguously. Next, we investigated the ping-pong model signature by checking the complementarity, again at different offsets, between putative primary piRNA and putative secondary piRNA that originated from the opposite strand (of the primary piRNA) and that were within 20 nt from the 5′-end of the primary piRNA. The relative complementary rate at each offset value was calculated by normalizing by the sum of complementary rate at each offset. We then determined the signal to noise ratio (SNR) for each offset position, calculated as a ratio of complementary rate at a given offset to the mean value of complementary rates at the other 19 offset positions. If the SNR at position 10 had the highest SNR value and was >4, and the ratio between SNR at the 10th position and the second highest SNR was >3, we classified this tissue as showing the ping-pong amplification model signature. We further investigated the ping-pong model signature based on small RNA sequences (24–32 nt, excluding ncRNA) originating within identified clusters, using the same procedure (Supplementary Tables S1, S4, S8). If the ping-pong model signature based either on small RNA sequences (24–32 nt, excluding ncRNA) or small RNA sequences (24–32 nt, excluding ncRNA) originated from identified small RNA clusters fit the above-mentioned criteria, we classified the tissue as showing the ping-pong amplification model signature. In addition, we investigated the ping-pong signature using the method described in (14). This approaches produced similar results (Supplementary Figure S2). To estimate pilRNA strand specificity, we first calculated total pilRNA reads from each strand of one cluster and, further, calculated the ratio between the reads from one strand that has more pilRNA inside, to total pilRNA reads of the cluster.
To investigate the genomic context, pilRNA located within clusters were classified according to their genomic localization: repeat regions, exon regions (including 5′-UTR, coding region and 3′-UTR), intron regions and no annotation regions. Annotation was done hierarchically in the listed order. A small RNA was assigned to a given annotation category if at least 1 nt mapped within the corresponding genomic region. The relative percentage of small RNA in each annotation category was compared to the percentage occupied by each category in the rhesus macaque genome according to the above-mentioned hierarchical order, using a binomial test. We note that because repeat regions had the highest annotation priority, our results might be biased toward over-annotation of small RNA within repeat regions.
To measure four previous TE master loci activity (11) in fly OSS, embryo, ovary, head and imaginal discs, we collected pilRNA mapped within each TE master loci, and used both uniquely mapped reads and weighted reads (reads divided by mapped loci number) to estimate the activities (Figure 1G and Supplementary Table S2).
Small RNA cluster expression correlation
To calculate the correlation in the expression of small RNA (piRNA and pilRNA) clusters between rhesus macaque testes and rhesus macaque somatic tissues, we defined cluster loci based on small RNA found in rhesus macaque testes. All small RNA clusters identified independently in somatic tissues overlapped with clusters with small RNA clusters found in testes. To calculate the correlation in the expression of small RNA clusters between fly ovaries or mouse testes and other fly or mouse tissues, respectively, for each comparison, we considered the union of small RNA clusters found in the two tissues. In comparison of small RNA cluster expression between mouse and rhesus macaque epididymis, reciprocal liftOver was used to find corresponding small RNA cluster regions in the other species. The expression correlation was calculated based on the union of small RNA clusters found in mouse and rhesus macaque epididymis. In comparison of pilRNA cluster expression between stained and unstained cells dissected from rhesus macaque epididymis by LCM, we took a union of small RNA clusters identified in these two samples. The control clusters were required to contain at least one uniquely mapped sequence and show <40% 5′ U-bias in the two samples.
Comparison of rhesus macaque testes small RNA clusters with human and mouse known piRNA
To compare rhesus macaque testes, small RNA clusters with known human testes piRNA clusters, we downloaded human piRNA cluster coordinates from (3) and mapped rhesus macaque small RNA clusters to the human genome using reciprocal liftOver.
To check how many rhesus macaque small RNA clusters (piRNA and/or pilRNA) could be supported by known mouse piRNA, we mapped rhesus macaque clusters to the mouse genome (mm9) using reciprocal liftOver. Then we checked mouse piRNA density within each liftovered cluster region using piRNA sequences from published Piwi-family proteins immunoprecipitation studies (7,10,16,26,27). To simulate mouse piRNA density expected by chance, we randomly chose start coordinates of the clusters, while keeping the length consistent, 1000 times.
Measure the expression level of four previous piRNA candidates using Deep sequencing data
We measured expression levels of four specific piRNA, used as piRNA markers across tissues in previous studies (4,5), in deep sequencing data sets from mouse testes, ovary (28) and epididymis. Specifically, for each piRNA candidate, small RNA sequence reads that mapped within 50 nt upstream and downstream of the genomic location of the piRNA sequences were summed (Supplementary Table S12).
Expression patterns of piRNA-like molecules in five rhesus macaque tissues using in situ hybridizations
We designed three LNA-probes (Supplementary Table S10) corresponding to highly expressed small RNA sequences. Hybridizations were performed as described in (40). Briefly, tissue sections were collected on Superfrost/plus slides (Fisher). After washing in two changes of excess PBS, sections were acetylated with 0.1 M triethanolamine/0.25% acetic anhydride for 10 min and then incubated in humidified bioassay trays for prehybridization at 50°C (20–25°C below the Tm of the probe) for 4 h in hybridization buffer [5×SSC/l× Denhardt's solution/5 mM EDTA/0.1% Tween/0.1% DHAPS/50% deionized formamide/0.1 mg/ml Heparin and 0.3 mg/ml yeast tRNA] (40,41). This was followed by an over-night hybridization step using a DIG-labeled LNA oligonucleotide probe complementary to the target piRNA. Sections were rinsed at a temperature below 50°C and washed twice in 2×SSC and 3 times in 0.2×SSC. The non-specific binding could be reduced by more stringent washing conditions or by an intense RNase A digestion (10 mg/ml). An in situ signal was detected by incubation with alkaline phosphatase (AP)-conjugated anti-DIG antibody, using NBT/BCIP as substrate.
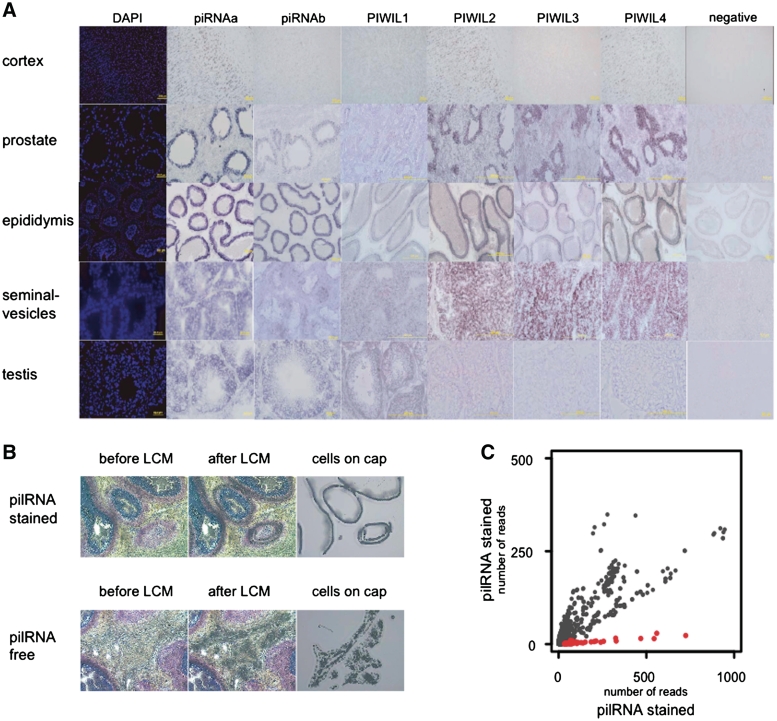
Figure 4A and Supplementary Figures S9–S14 show in situ hybridizations with two different ‘anti-pilRNA’ LNA probes (piRNAa and piRNAb) and four probes corresponding to rhesus macaque PIWI transcripts (PIWIL1-4) in adult rhesus macaque tissues. The negative controls shown were subjected to the same hybridization and staining procedure, as the test samples and included alkaline phosphatase (AP)-conjugated anti-DIG antibody, but omitted specific LNA probes. A negative control shown in Supplementary Figure S12B included scrambled LNA probe in addition to alkaline phosphatase (AP)-conjugated anti-DIG antibody.
Figure 4.
Localization of pilRNA and Piwi-family mRNA expression in macaque tissues. (A) In situ hybridizations with two different ‘anti-pilRNA’ LNA probes (piRNAa and piRNAb) and probes against four rhesus macaque PIWI transcripts (PIWIL1-4) in adult rhesus macaque tissues. Negative control shows tissue staining after identical hybridization and staining procedure, omitting specific probes. (B) Rhesus macaque epididymis tissue before and after LCM of principal/basal cells (pilRNA stained) and peritubular tissue (pilRNA free). (C) Small RNA sequencing coverage of pilRNA clusters (red) and background expression clusters (gray) (‘Materials and Methods' section) in rhesus macaque LCM-purified principal/basal cells (pilRNA stained) and peritubular tissue (pilRNA free).
Western blot
For protein extracts used in the western blots, mouse tissues were lysed in RIPA buffer, using Polytron tissue homogenizer. Rabbit anti-Miwi antibody raised against the peptide VIRVPAPCQYAHKLAFLVGQ (amino acids 826–845) and mouse anti-actin antibody (Sigma) were used for immunoblotting as described in (42).
Ethics statement
All rhesus macaques and mice used in this study suffered sudden deaths for reasons other than their participation in this study and without any relation to the tissue used. Biomedical Research Ethics Committee of Shanghai Institutes for Biological Sciences completed the review of the use and care of the animals in the research project (approval ID: ER-SIBS-260802 P).
RESULTS AND DISCUSSION
Small RNA in fly tissues
To assess presence of piRNA-like small RNA in D. melanogaster tissues, we analyzed publicly available small RNA data sets from embryo, ovary, head, imaginal discs and OSS cells generated using high-throughput sequencing (11,16,21,29–33). Embryo, ovary and OSS cells were previously shown to contain piRNA (1,11,16,29,32). Indeed, we observed large numbers of sequences with length 24–32 nt, an interval corresponding to the piRNA length, in small RNA data from these three tissues (1,11,16,29). Besides length, these small RNA showed other known properties of fly piRNA, such as 5′ uridine bias, clustering along the genome and preferential localization in the antisense repeat regions (54–65%) (Figure 1A–D, F and G). Further, small RNA present in embryo and ovaries showed clear signature of the ping-pong biogenesis model and low piRNA cluster strand specificity (Figure 1E and F; Supplementary Figures S1 and S2). In contrast, in agreement with previous reports (11), OSS cells did not show any detectable ping-pong model signature and largely contained strand-specific piRNA clusters (Figure 1E and F; Supplementary Figures S1 and S2). Thus, overall analysis of high-throughput sequencing data from fly embryo, ovary and OSS cells has allowed us to ‘rediscover’ piRNA in these tissues by identifying small RNA that display all previously reported piRNA sequence features.
Notably, applying the same analysis to the fly head and imaginal disc data sets, we also identified a distinct fraction of small RNA, with length distribution of 24–32 nt, displaying piRNA sequence features. Presence of these piRNA-like small RNA (pilRNA) was not affected by exclusion of sequences originating from known non-coding RNA (ncRNA) (Figure 1C and D). Further, similar to piRNA, head and imaginal disc pilRNA showed 5′ uridine bias, clustering along the genome and preferential localization in the antisense repeat regions (Figure 1 and Supplementary Tables S1–S3). More than 55% of pilRNA clusters found in both head and imaginal discs overlapped with previously reported ovarian piRNA clusters, while <10% would be expected to overlap by chance (P < 0.0001). Further, pilRNA cluster coverage in fly head and imaginal discs correlated well with piRNA cluster coverage in fly ovaries (R > 0.7, P < 0.0001).
Both head and imaginal disc pilRNA showed a distinct ping-pong model signature and low cluster strand specificity, characteristic to fly germline piRNA (Figure 1E and F; Supplementary Figures S1 and S2; Supplementary Tables S1–3). Further, as is the case for fly germline piRNA (11), head and imaginal disc pilRNA were enriched in 42AB genomic region and were underrepresented in 20AB/flamenco locus (Figure 1G and Supplementary Table S2). Finally, in agreement with previous observations (21), the proportion of pilRNA detected in fly heads increased substantially after enrichment for small RNA containing 2′-O-methyl group at its 3′-end, a known piRNA sequence feature (43–46), during small RNA library preparation. This enrichment could be reproduced in our analysis in three independent small RNA data sets (21,30) (Supplementary Figure S3).
Thus, our analysis confirmed previously reported presence of pilRNA in fly heads and showed presence of pilRNA in fly imaginal discs. In both tissues, the proportion of pilRNA, defined as small RNA with length between 24 and 32 nt after exclusion of known ncRNA, was lower than in tissues previously shown to contain piRNA: embryo, ovary or OSS cells. Specifically, average proportions of pilRNA among all mapped sequences were 2 and 14% in heads and imaginal discs and 44, 34 and 59% in embryo, ovary and OSS cells, respectively. In the fly head samples, this proportion increased to an average of 10% after enrichment for RNA containing 2′-O-methyl group. Despite smaller numbers, however, pilRNA present in fly heads and imaginal discs showed all known sequence properties of fly germline piRNA, as well as significant overlap of cluster locations and significant correlation of the cluster coverage. It was surprising that pilRNA present in fly somatic tissues did not resemble piRNA found in ovarian follicle cells, another somatic tissue, but showed all features of piRNA found in the germline. Without proof of direct association between these small RNA and Piwi proteins, however, they cannot be classified as piRNA. Still, the fact that substantial numbers of small RNA displaying piRNA features are present in fly somatic tissues might, by itself, be worthwhile noting.
Small RNA in mouse tissues
To investigate pilRNA presence in mammalian tissues, we took advantage of a published data set, which contains small RNA sequences from 11 mouse tissues and 31 immune system cell types (28). In agreement with previous reports (28), in these data, we detected abundant piRNA presence in testes and moderate numbers of piRNA in ovaries. In addition to germline, we found small RNA showing distinct sequence features of mammalian piRNA in 17 out of the 40 mouse somatic tissues and cell types (‘Materials and Methods' section, Supplementary Table S4 and S5). Specifically, in addition to universal piRNA features, such as length distribution between 24 and 32 nt, 5′ uridine bias and clustering along the genome, we observed other sequence features typical to the mammalian piRNA: strand specificity of the clusters, under-representation in repetitive element regions and compatibility with the ping-pong piRNA biogenesis model (‘Materials and Methods' section, Supplementary Figure S2 and Supplementary Table S4 and S5). While in some tissues not all of these features could be detected due to relatively small pilRNA cluster numbers, they were apparent in tissues with substantial pilRNA cluster numbers, such as in the pancreas (Figures 2 and Supplementary Figure S2; Supplementary Table S4 and S5).
With respect to genomic context and ping-pong amplification features of identified pilRNA, mouse tissues could be separated into two types: resembling the germline and resembling the OSS. Pancreas, hematopoietic progenitor cells, neutrophils, macrophages and macrophage-derived dendritic cells contained pilRNA clusters preferentially located in unannotated genomic regions and vastly overlapping with piRNA clusters identified in mouse testes using piRNA co-precipitating with PIWI proteins (average overlap 97.2%) (Figure 2I and Supplementary Table S4). In contrast, pilRNA found in germinal center B cells, mature B cells, plasma cells, follicular helper T cells and NK cells showed bias toward the 3′-UTR sense strand regions and overlapped only weakly with piRNA clusters found in mouse testes (average overlap 21.4%) (Figure 2I and Supplementary Table S4). The piRNA bias toward the 3′-UTRs was previously described for mouse pre-pachytene testes (Figure 2H) and Drosophila OSS cells (Figure 1G) (10,16). Notably, in both cases only one of the Piwi-family proteins (Mili or Piwi, respectively) was expressed (10,16). Based on these observations, we could speculate that the difference between the two pilRNA types could be caused by expression of different sets of Piwi-family proteins in these tissues.
Interestingly, pilRNA presence in B cells appeared to require an intact activation-induced cell death mechanism, as none—or very few—pilRNA could be found in germinal center and mature B cells, in the BclXL mouse strain (Supplementary Table S4). While apoptosis might lead to generation of small RNA through mRNA degradation, experimental procedure used for small RNA library preparation should not result in biased pilRNA distribution along transcripts in germinal center and mature B cells in the wild type mice. Our observation, that pilRNA, found in these cells show preferential localization in 3′-UTR is not compatible with a simple mRNA degradation scenario (‘Materials and Methods' section and Supplementary Table S4).
Thus, our analysis of published small RNA sequencing data showed widespread presence of pilRNA across mouse somatic tissues. In most of these tissues, the total proportions of pilRNA were small and the proportions of pilRNA that fell into clusters were yet smaller: ∼0.6% on average. In pancreas, however, pilRNA presence was substantial −3.5% of the total mapped reads. Notably, the proportion of total reads occupied by piRNA in mouse ovaries, defined using the same criteria, was 3%. Furthermore, 76% of all pancreas pilRNA reads mapped within pilRNA clusters. Similarly, 82 and 83% of piRNA reads mapped within clusters in ovaries and testis, respectively. Importantly, western blot results confirmed Miwi protein expression in mouse pancreas (Supplementary Figure S4)
Again, as in case of fly pilRNA, mouse pilRNA found in our study cannot be classified as piRNA until their association with Piwi proteins is shown. Still, substantial presence of pilRNA in some somatic tissues, such as in the pancreas, might indicate their potential functional importance.
Small RNA in rhesus macaque tissues
We next tested for presence of pilRNA in somatic tissues in a mammalian species closely related to humans—rhesus macaque. To do so, we sequenced small RNA with a size range of 20–40 nt in adult rhesus macaque testes, epididymis, prostate, seminal vesicles and cerebral cortex (prefrontal cortex, corresponding to Brodmann area 10) using Illumina sequencing platform (‘Materials and Methods' section and Supplementary Table S6). For each tissue, we obtained 10 266 594—13 972 125 reads, 58–69% of which could be mapped to the rhesus macaque genome (‘Materials and Methods' section and Supplementary Table S7).
In all tissues except testes, the majority of reads (75–95%) were 21–23 nt in length and mapped within the annotated miRNA precursor regions (‘Materials and Methods' section). In testes, in agreement with the previous studies (3,26), the majority of the reads (92%) had length of 24–32 nt, corresponding to the length of piRNA (Supplementary Figure S5) and showed all other known piRNA sequence features (‘Materials and Methods' section; Supplementary Tables S8 and S9; Figure 3). Expression of small RNA found in macaque testes overlapped well with expression of mouse piRNA identified by co-precipitation with Piwi proteins (7,10,16): out of 1548 macaque clusters with orthologs in the mouse genome, 71% contain 5 or more mouse piRNA, while <8% would be expected by chance (P < 0.0001). Similarly, 92% of the annotated human piRNA clusters (3) overlap with macaque clusters. Taken together, these features indicate that the vast majority of analyzed small RNA sequences with length distribution between 24 and 32 nt correspond to genuine testes piRNA.
Unexpectedly, in all other macaque tissues studied, we found small RNA with the same sequence properties. Such properties included length distribution, 5′ uridine bias, genomic localization and clustering, cluster strand specificity and ping-pong model mechanism features (Figure 3; Supplementary Figures S2 and S6; Supplementary Tables S8 and S9). Compared to testes, these pilRNA occupy a much smaller proportion of all mapped sequence reads: 1–15% versus 92% in testes (Supplementary Table S8). Consequently, only a few pilRNA clusters (58–248) can be identified in somatic tissues (Supplementary Tables S8 and S9). All pilRNA clusters found in somatic tissues overlapped with macaque testes' piRNA clusters and cluster read coverage correlated well among tissues (R > 0.90, P < 10−15) (Supplementary Figure S7).
To further investigate pilRNA expression in somatic tissues, we designed ‘anti-pilRNA’ LNA probes complementary to small RNA sequences highly expressed in two piRNA clusters in macaque testes (Figure 3; Supplementary Table S10; ‘Materials and Methods' section). In mice, one of the orthologous clusters was shown to generate large numbers of piRNA in adults, but not in prenatal or 10 day-old animals (7,10) (Supplementary Figure S8 and Supplementary Table S11). Accordingly, we found few small RNA originating from this genomic region in immature rhesus macaque testes (Supplementary Figure S8 and Supplementary Table S11). For all LNA probes, we observed strong in situ hybridization patterns in adult macaque testes (Figure 4A and Supplementary Figure S9). Notably, we also observed clear hybridization signals in the four somatic tissues, most of them localized within specific cell types (Figures 4A and Supplementary Figures S10–S16). In the epididymis and the prostate, the hybridization signal was located in the principal and basal cells of epithelia surrounding the ducts, but not in the peritubular tissue or spermatozoa (Supplementary Figures S10–S12). In the cerebral cortex, the hybridization signal showed a pattern resembling a neuronal body distribution (Supplementary Figure S14). These hybridization patterns were consistent among different ‘anti-pilRNA’ LNA probes and were reproduced in another adult macaque individual (Supplementary Figures S15 and 16).
Presence of piRNA requires expression of Piwi-family proteins. In the absence of suitable antibodies, we checked mRNA expression of the four macaque PIWI proteins, (PIWIL1-4), by in situ hybridization. We found that PIWIL mRNA were expressed both in testes and in somatic tissues. Most importantly, in all cases PIWIL mRNA expression co-localized with hybridization signals from ‘anti-pilRNA’ LNA probes (Figure 4A). While in testes PIWIL1 mRNA had the strongest signal, in somatic tissues PIWIL2 and PIWIL4 mRNA signals were the strongest ones. Although not quantitative, this observation is consistent with previous reports of PIWIL4 mRNA expression across human somatic tissues (47) and indicates that pilRNA might be associated with a subset of PIWI proteins.
The LNA-probes used in our study cannot discriminate between small and long transcripts. To test whether hybridization signal comes from pilRNA, rather than long transcripts, we isolated and sequenced small RNA from cells with and without LNA hybridization signal. Cells stained after LNA-probes hybridization should contain pilRNA, while unstained cells should not. Using laser capture microdissection (LCM), we dissected principal and basal cells that had a clear LNA hybridization signal and peritubular tissue that had no signal, from epididymis of four adult rhesus macaques (Supplementary Table S6; Figure 4A and B; Supplementary Figure S11). We indeed found that the principal and basal cells contained 30 times more pilRNA reads than the peritubular tissue. In contrast, expression of other small RNA did not differ between these cell types (Figure 4C). Furthermore, expression of pilRNA in the principal/basal cells correlated well with expression of pilRNA found in the total epididymis (R = 0.85, P < 2.2 e-16). The residual presence of pilRNA in the peritubular tissue, detected by sequencing, might be caused by their incomplete separation from the epithelial cells in the LCM procedure. Based on these results, we can conclude that principal/basal cells of rhesus macaque epididymis indeed contained pilRNA, while the peritubular tissue did not. This result could not be explained by spermatozoa contamination, as LCM-dissected cells did not contain detectable spermatozoa (Figure 4B) and, according to in situ hybridization results, spermatozoa did not contain detectable levels of pilRNA (Figure 4A and Supplementary Figure S11).
A previous study, which used the northern blot hybridization technique with specific probes, did not detect presence of piRNA in mouse epididymis (5). To test weather piRNA are indeed absent in this tissue, we sequenced small RNA from mouse epididymis, following the same procedure (‘Materials and Methods' section). As in macaques, mouse epididymis contained small RNA with length 24–32 nt that formed pilRNA clusters. These pilRNA clusters largely overlapped with piRNA clusters identified in mouse testes (96%, P < 0.0001) and contained sequences showing a clear ping–ping amplification signature, as well as other piRNA features (Supplementary Table S4 and ‘Materials and Methods' section). Furthermore, epididymis pilRNA coverage across the orthologous clusters was largely conserved between mice and macaques (R = 0.57, P < 10−13). Thus, presence of pilRNA in epididymis could be observed in both mice and macaques.
Why, then, were mouse epididymal pilRNA not detected previously? Using our sequence data, we tested the abundance of small RNA complementary to the four piRNA-specific probes used in the previous studies in order to detect piRNA expression in mouse epididymis and ovaries (4,5). We found that levels of these specific small RNA were, on average, 75-fold lower in ovaries and epididymis compared to testes (Supplementary Figure S17; Supplementary Table S12; ‘Materials and Methods' section). Thus, it is possible that the chosen probes did not provide enough sensitivity. Indeed, while no hybridization signal using these probes was detected in both epididymis and ovaries (4,5), piRNA presence in mouse ovaries has since been described (7,28,48,49).
Taken together, our results demonstrate that small RNA sharing sequence features of piRNA are widespread among somatic tissues in flies, mice and macaques. Notably, in all three species, pilRNA found in somatic tissues, with possible exception of several mouse cell types, resembled germline piRNA, rather than piRNA present in fly ovarian somatic cells. Even though our study did not test association between pilRNA and Piwi proteins, presence of pilRNA across animal somatic tissues is intriguing. In some tissues, this presence was quite substantial: in macaque epididymis pilRNA constituted 15% of the total mapped reads and in mouse pancreas −3.5%. While 3.5% is not a large proportion, it was greater than the proportion of total reads (3%) occupied by piRNA in mouse ovaries. Furthermore, our in situ hybridization results indicated that pilRNA expression in somatic tissues tends to be localized to specific cell types. Thus, tissues containing low overall pilRNA amounts might still have high pilRNA concentrations in discrete cells. Presence of pilRNA in somatic tissues across a wide range of animal species, from flies to macaques, indicates their potential functional significance. Based on their sequence features and genomic origins, pilRNA closely resemble the germline piRNA from the corresponding species, rather than piRNA found in the fly ovarian somatic cells. It is, therefore, conceivable that functions of pilRNA in somatic tissues are similar to functions of piRNA in the germline. Further studies are needed to explore the full scope of pilRNA in animal somatic tissues, their association with Piwi proteins, as well as the functional roles they play.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Ministry of Science and Technology of the People's Republic of China (grant numbers 2007CB947004 and 2006CB910700); Chinese Academy of Sciences (grant numbers KSCX2-YW-R-094 and KSCX2-YW-R-251); Shanghai Institutes for Biological Sciences (grant number 2008KIT104); Max Planck Society; Bundesministerum fuer Bildung und Forschung.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Suzhou Drug Safety Evaluation and Research Center and C.Lian, H.Cai and X.Zheng in particular for providing the macaque samples; Professor Yong Liu and Dr Liu Yang for providing mouse pancreas and 832/3 cell line; W. Tang for enabling us to do LCM experiments; J. Dent for editing the manuscript; E. Lizano and F. Xue for assistance and all members of the Comparative Biology Group in Shanghai for helpful discussions and suggestions.
REFERENCES
- 1.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 2.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell. Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 4.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 5.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 7.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 9.Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 16.Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr. Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalvet F, Teysset L, Terzian C, Prud'homme N, Santamaria P, Bucheton A, Pelisson A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18:2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ro S, Park C, Song R, Nguyen D, Jin J, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA. 2007;13:1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting Edge: KIR Antisense Transcripts Are Processed into a 28-Base PIWI-Like RNA in Human NK Cells. J. Immunol. 185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki Y, Suzuki K, Matusik RJ, Obinata M, Orgebin-Crist MC. Immortalized epididymal cell lines from transgenic mice overexpressing temperature-sensitive simian virus 40 large T-antigen gene. J. Androl. 2002;23:854–869. [PubMed] [Google Scholar]
- 23.Zhu CF, Liu Q, Zhang L, Yuan HX, Zhen W, Zhang JS, Chen ZJ, Hall SH, French FS, Zhang YL. RNase9, an androgen-dependent member of the RNase A family, is specifically expressed in the rat epididymis. Biol. Reprod. 2007;76:63–73. doi: 10.1095/biolreprod.106.054635. [DOI] [PubMed] [Google Scholar]
- 24.Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 28.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic. Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucl. Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silahtaroglu AN, Nolting D, Dyrskjot L, Berezikov E, Moller M, Tommerup N, Kauppinen S. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 41.Jankowsky JL, Derrick BE, Patterson PH. Cytokine responses to LTP induction in the rat hippocampus: a comparison of in vitro and in vivo techniques. Learn. Mem. 2000;7:400–412. doi: 10.1101/lm.32600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu M, Medvedev S, Yang J, Hecht NB. MIWI-independent small RNAs (MSY-RNAs) bind to the RNA-binding protein, MSY2, in male germ cells. Proc. Natl Acad. Sci. USA. 2009;106:12371–12376. doi: 10.1073/pnas.0903944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 44.Kirino Y, Mourelatos Z. 2′-O-methyl modification in mouse piRNAs and its methylase. Nucleic Acids Symp. Ser. 2007:417–418. doi: 10.1093/nass/nrm209. [DOI] [PubMed] [Google Scholar]
- 45.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2'-O-methylated at their 3' termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 46.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugimoto K, Kage H, Aki N, Sano A, Kitagawa H, Nagase T, Yatomi Y, Ohishi N, Takai D. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem. Biophys. Res. Commun. 2007;359:497–502. doi: 10.1016/j.bbrc.2007.05.136. [DOI] [PubMed] [Google Scholar]
- 48.Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, Chen ZJ, Milosavljevic A, Marra MA, Rajkovic A. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol. Hum. Reprod. 16:463–471. doi: 10.1093/molehr/gaq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ro S, Song R, Park C, Zheng H, Sanders KM, Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.