Abstract
In budding yeast, the eukaryotic initiator protein ORC (origin recognition complex) binds to a bipartite sequence consisting of an 11 bp ACS element and an adjacent B1 element. However, the genome contains many more matches to this consensus than actually bind ORC or function as origins in vivo. Although ORC-dependent loading of the replicative MCM helicase at origins is enhanced by a distal B2 element, less is known about this element. Here, we analyzed four highly active origins (ARS309, ARS319, ARS606 and ARS607) by linker scanning mutagenesis and found that sequences adjacent to the ACS contributed substantially to origin activity and ORC binding. Using the sequences of four additional B2 elements we generated a B2 multiple sequence alignment and identified a shared, degenerate 8 bp sequence that was enriched within 228 known origins. In addition, our high-resolution analysis revealed that not all origins exist within nucleosome free regions: a class of Sir2-regulated origins has a stably positioned nucleosome overlapping or near B2. This study illustrates the conserved yet flexible nature of yeast origin architecture to promote ORC binding and origin activity, and helps explain why a strong match to the ORC binding site is insufficient to identify origins within the genome.
INTRODUCTION
In eukaryotes, the origin recognition complex (ORC) binds to chromosomes and determines the positions of replication origins, the physical sites where DNA replication initiates [reviewed by (1,2)]. In G1-phase, ORC directs the assembly of a pre-replicative complex ‘pre-RC’ that culminates in loading the replicative MCM helicase onto DNA [reviewed by (3)]. During S-phase, modification of the pre-RC leads to activation of the MCM helicase and the initiation of DNA replication (origin firing). Thus in all eukaryotes ORC and MCM chromosome binding are fundamental to defining replication origins. However, with the exception of budding yeast origins, the specific DNA sequences and/or chromatin structure that determine replication origins in other eukaryotes are unclear. Further insights on the mechanism of pre-RC assembly will be significantly aided by the defined and relatively simple origin structure of Saccharomyces cerevisiae, as outlined below.
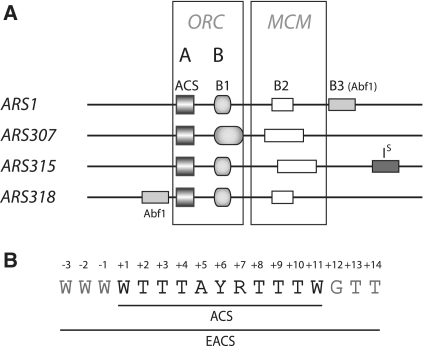
In budding yeast, origins are located within ARS (autonomously replicating sequence) elements, which are defined DNA sequences of ∼150 bp. Although ARSs can vary in the exact sequences that promote activity, focused studies of several ARS elements (4–10) indicate that they share modular A, B1 and B2 elements [Figure 1A, reviewed by (11)]. The A and B1 elements together serve as a bipartite DNA binding site for yeast ORC (12,13). The A element is an essential feature of ARS elements that contains the 11 bp ACS (ARS consensus sequence), a degenerate AT-rich sequence that is present in all origins (Figure 1B). However, functional ACSs may contain one or more mismatches to this sequence. Comparative sequence analysis of many ARS elements subsequently revealed three additional conserved nucleotides on each side of the ACS (14). Although the 3 bp 5′ and 3′ to the ACS (WWW and GTT) are less well conserved than the core ACS, sequence alignments of multiple S. cerevisiae ARSs showed that these nucleotides are preferred in budding yeast (7,15) and in the sensu stricto Saccharomyces species (16). This 17 bp EACS (extended ACS, Figure 1B) is a better predictor of the functional ACS when multiple close matches to the ACS are present within an ARS fragment (14). However, there is little experimental evidence demonstrating that the additional 6 bp in the EACS are important for ARS activity and/or ORC binding.
Figure 1.
(A) The organization of various yeast ARS elements is shown with the ACS, B1, B2 and B3 element, Abf1 binding site and an inhibitory element, IS. The ACS and B1 element comprise the ORC-binding site. B2 is thought to be a loading site for the MCM helicase. (B) Sequence of the 11 bp ACS and the EACS is indicated. ‘W’ denotes A or T, ‘Y’ denotes C or T, ‘R’ denotes G or A.
Although the ORC binding site (A + B1) is necessary for origin function, it is not sufficient. Additional elements are known that further stimulate or inhibit origin activity (7,17–19). In particular, the B2 element is proposed to function as a preferred loading site for the MCM helicase (20,21) but its distance relative to the ORC binding site is not fixed (5,7,8,10,22). Although a study of synthetic B2 elements at ARS1 indicated that this is not simply a region of helical instability (20), to date too few chromosomal B2 elements have been functionally defined to determine a useful consensus or motif. The apparent plasticity of the B2 element and its variable positioning within ARS elements has likely limited efforts to define origins in the yeast genome based on sequence conservation alone.
In this study, we systematically determined the structure of four efficient DNA replication origins (ARS309, ARS319, ARS606 and ARS607) and incorporated these functional data into sequence alignment and nucleosome positioning analyses to gain a more precise view of yeast origin structure. A previous report analyzed stimulatory sequences flanking the ACS of ARS607 (23). The EACS sequence was particularly important for the ARS activity of ARS309, ARS606 and ARS607 since mutations within the 6 bp EACS nucleotides flanking the ACS substantially decreased ARS activity. In addition, linker mutations between the EACS and B1 significantly decreased ARS activity at ARS309, ARS606 and ARS607 but not if the EACS was made closer to the consensus. These data suggest that ORC-EACS nucleotide contacts are often critical for ARS activity and ORC binding to the ARS, which we show directly for ARS606 and ARS607. Our functional data thus explain the sequence conservation of the EACS nucleotides. In addition, our new high-resolution data allowed us to define B2 elements comprehensively enough to reveal a core B2 element motif. This consensus B2 element is significantly enriched (P ≤ 1 × 10−5) in a set of 228 phylogenetically conserved ARSs (16). Finally, we used recently generated genome-wide nucleosome maps to examine nucleosome positioning surrounding the nine ARS elements that have been defined at high resolution by linker scan analyses. This revealed that ARS606, an ARS negatively regulated by the Sir2 histone deacetylase, contains a stably positioned nucleosome overlapping its B1 and B2 elements, similar to the Sir2-regulated ARSs, ARS305 and ARS315 (22). In contrast, these functional components exist within nucleosome free regions (NFRs) in the Sir2-independent ARSs. In summary, these analyses reveal the complexity and flexibility of the yeast ORC binding site and ARS elements in general, and indicate why matches to the previously defined bi-partite ORC binding site are imperfect predictors of ARS elements. Furthermore, we provide evidence for how variations in local chromatin structure might be used to differentially regulate individual origins.
MATERIALS AND METHODS
Yeast strains and plasmid construction
Plasmid loss assays were performed in W303-1 A (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100) (24). Yeast transformation and culturing were done according to standard methods. Strains were propagated in YPD or synthetic complete medium (SCM) (7). The plasmids containing wild-type chromosome III/VI ARS elements were constructed to replace ARS1 within the pARS1-WT (CEN4 URA3) backbone (5), and were described previously (22). A series of 7 bp SalI linkers and other point mutations were then introduced into pARS309, pARS319, pARS603 and pARS606 by QuikChange® Site-Directed Mutagenesis (Agilent Technologies).
Plasmid stability assay
This assay was performed as described previously (7). Plasmids were transformed into W303-1 A, plated onto SCM-Ura plates and incubated for 3–4 days at 30°C. At least six colonies for each plasmid were streak purified on SCM-Ura plates before inoculating into SCM-Ura broth for ∼18–24 h at 25°C. These overnight cultures were further diluted 1:2000 into SCM and cultured for another 24 h at 25°C where they underwent ∼10 cell doublings. The total number of cells with and without plasmids was determined by plating onto SCM and SCM-Ura plates after overnight growth in selective and non-selective medium. Loss rates per generation (%) were then calculated as described (25) and reported with standard errors of the mean (SEM). Deviations from the wild-type loss rate of <50% were not significant due to the nature of the assay.
Electrophoretic mobility shift assay
ORC was purified from baculovirus-infected Sf9 cells as described (26). The ORC-ARS EMSA were performed as described (27) with some modifications. Biotin-labeled specific DNA probes (102 bp) of ARS1, ARS606 and ARS607 were generated by PCR using the primers 5′-biotin- AAAGCCAAATGATTTAGCAT-3′/5′-GTGCACTTGCCTGCAGGCCT-3′ for ARS1, 5′-biotin-CATCCTCAATCATCAATTAA-3′/5′-GGGCGCTTTTTTTGTGACAT-3′ for ARS606, 5′-biotin- ACACTACATTCGCTAAATTAC-3′/5′-TGGTCACTCCAGATCTAGTTT-3′ for ARS607 (IDT). A typical binding reaction of 20 µl was prepared on ice containing 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 5 mM MgCl2, 2 mM ATP (pH 7), 1 µg/ml of bovine serum albumin, 6.5% v/v glycerol, 0.125 µg/ml of poly (dI•dC) competitor, 5 fmol of biotin DNA probes and the specified amounts of purified yeast ORC. Binding reactions were then incubated at 25°C for 20 min in an Eppendorf PCR cycler and separated on a 3.5% of polyacrylamide mini-gel (29:1) using 0.5× TBE as running buffer. The run time was ∼60 min at 4°C with a constant 100 V. Biotin-labeled DNA fragments were transferred to a Biotrans nylon membrane (ICN) and cross-linked at 120 mJ/cm2 (Stratagene UV 1800) before detection using a LightShift® Chemiluminescent EMSA Kit (Pierce).
Alignment of B2 elements and statistical analysis
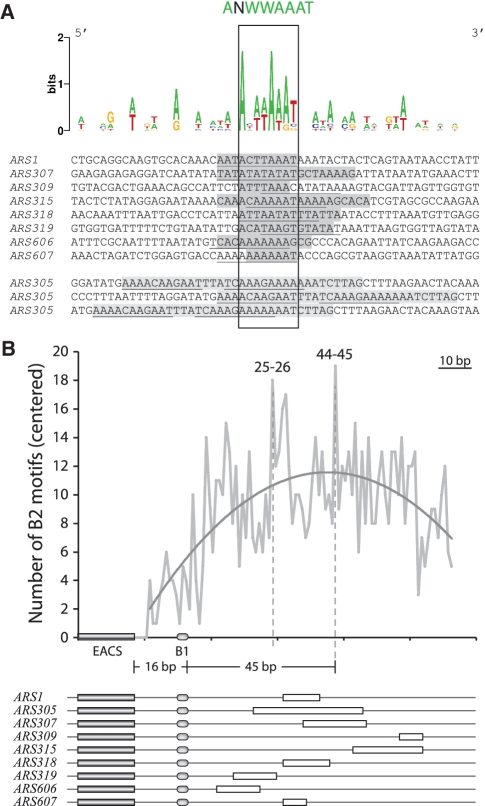
Initially we used Clustal W2 (28) to align 8 B2 sequences and viewed the alignment using Jalview 2.3. Because of the small sample size and short sequences, especially for ARS309 and ARS607, Clustal W2 didn’t give us a centered alignment that covered all 8 B2 sequences, i.e. the B2 sequences for ARS309 and ARS607 were off-center and only had a two-nucleotide overlap with each other. As a result, we manually modified the alignment by centering ARS309 and ARS607 and then sliding these sequences by 1 bp increments to determine the optimal alignment. The resulting figure of all 8 B2 elements was generated using WebLogo (http://weblogo.berkeley.edu/). Close matches to this sequence are also present within the B2 element of ARS305 (Figure 6A).
Figure 6.
Multiple sequence alignment of eight B2 elements. (A) A gray box indicates each B2 element determined by mutational analysis. The WebLogo diagram above the alignment relates to the frequency of each particular nucleotide in the alignment. The common sequence ANWWAAAT is found in all B2 elements with at most one mismatch. The ARS305 B2 element, which spans a broader area and is less important than at other ARSs, is shown at the bottom with three close matches to ANWWAAAT. (B) The number of B2 motifs (identities plus motifs with one mismatch at positions 6, 7 or 8) in the 228 phylogenetically conserved S. cerevisiae ARSs are plotted relative to distance from the EACS in base pairs. There were clear maxima present between 25–26 and 44–45 bp from the B1 element (WTW). The average (centered) distance of all B2 consensus matches in this set was 38 bp distal to the B1 (WTW) element. The positions of the EACS, B1 and B2 elements are shown below for the known ARSs.
Budding yeast sequences 100 bp distal to the proACS in the 228 phylogenetically conserved ARS elements (16) were compiled in Supplementary Table S1, and then searched for the presence of the B2 consensus sequence: ANWWAAAT in the forward direction. Although T is often present in last position (75%) it can also be a C or G, so we searched using an N at this position. Seventy four percent of the 228 ARS sequences contained a match to this string. The statistical significance of the proportion of B2 consensus matches (ANWWAAAN) in the 228 ARS elements was estimated by first identifying all potential ACS sequences in the S. cerevisiae genome allowing a 1 bp mismatch to the 11 bp ACS. This yielded 13 978 ACS-like sequences (7). We then created 10 000 random sets containing 228 members: each set contained 228 × 100-mer sequences using the 100-mer sequences directly 3′ to each ACS-like sequence, i.e. oriented as in the genome. For each of the 10 000 sets, the proportion of the 228 sequences containing ANWWAAAN was calculated as above. The P-value was found to be <1 × 10−5 on the basis of this permutation testing. The maximum occurrence of the ANWWAAAN motif in any of the random 228 × 100mer sets was 31.3% implying that the P-value is likely to be considerably <1 × 10−5. The location of potential B2 consensus sequences within the 228 ARS elements was performed using ‘DNA Pattern Find’ in the Sequence Manipulation Suite: (http://www.ualberta.ca/∼stothard/javascript/dna_pattern.html). The forward matches allowing a mismatch at positions 6, 7, or 8 are plotted in Figure 6B relative to the positions of known B2 elements.
RESULTS
A goal of this study was to closely examine sequences spanning the ORC binding site to discover additional nucleotides that influence ARS activity and/or ORC binding. We also wanted to define additional B2 elements with the goal of discovering a consensus motif that would enhance the ability to predict functional origins with yeast genomes based on DNA sequence. We therefore constructed an ordered series of substitution mutations spanning four highly efficient ARSs to identify base pairs that impact ARS activity (Figures 2–4, Supplementary Figure S1). Mutant plasmids that failed to transform yeast revealed essential ARS sequences. To define important ARS sequences, mutant plasmids that transformed yeast were quantified for their loss rates in the absence of plasmid selection, which is a measure of ARS efficiency.
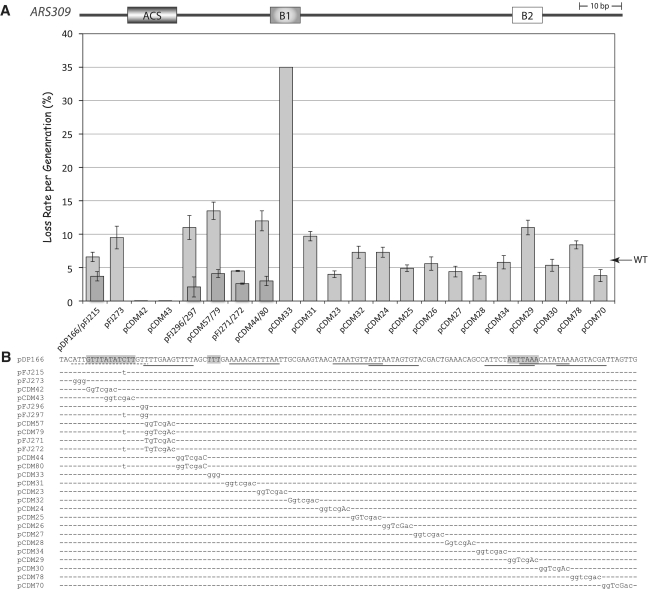
Figure 2.
Linker scan analysis of ARS309. (A) Plasmid loss rates per generation (±SEM) are plotted for an ARS CEN plasmid contain wild-type ARS309 (pDP166 and indicated by an arrow) versus particular mutant versions of pDP166 containing GGTCGAC linker substitutions and additional mutations. Where two plasmids are indicated in one column, the darker gray bar indicates the loss rate of the second plasmid. (B) Detailed view of mutations and plasmid names. The wild-type ARS309 sequence is indicated above. Shaded boxes indicate the position of the ACS, B1 and B2 elements (from left to right). Partial 8 of 11 or better matches to the ACS are underlined. The dotted line beneath the functional ACS indicates the EACS.
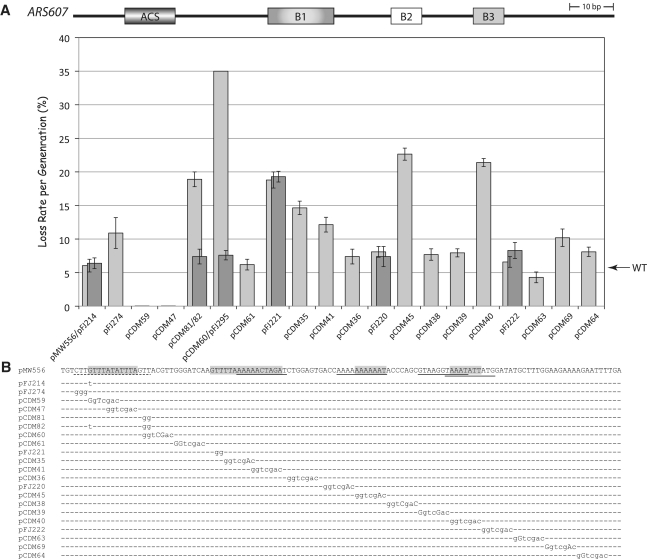
Figure 3.
Linker scan analysis of ARS607. (A) Plasmid loss rates per generation (±SEM) are plotted for wild-type ARS607 (pMW556) and mutant versions of pMW556 containing GGTCGAC linker substitutions and additional mutations. Where two plasmids are indicated in one column, the darker gray bar indicates the loss rate of the second plasmid. (B) Detailed view of wild-type ARS607 sequence and mutant derivatives, marked as in 2B with additional shading of B3 element.
Figure 4.
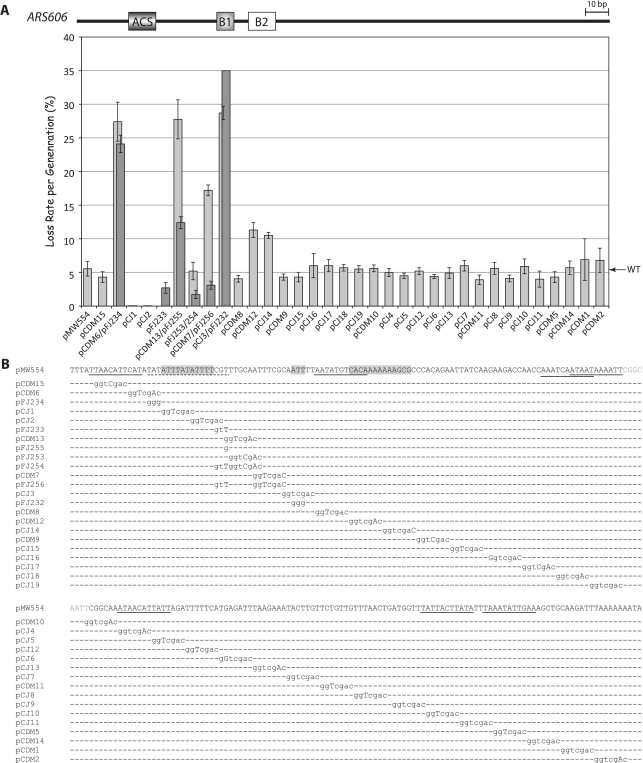
Linker scan analysis of ARS606. (A) Plasmid loss rates per generation (±SEM) are plotted for wild-type ARS606 (pMW554) and mutant versions of pMW554 containing GGTCGAC linker substitutions and additional mutations. Where two plasmids are indicated in one column, the darker gray bar indicates the loss rate of the second plasmid. (B) Detailed view of wild-type ARS606 sequence and mutant derivatives, marked as in 2B.
Multiple ARSs depend on nucleotides within the EACS and between the ACS and B1 element for robust activity
ARS309 has been shown to contain an unusual 9 of 11 match to the ACS, gTTTATATcTT (14). Its ACS has a ‘C’ at +9 in place of the highly conserved ‘T’, which is unusual since substitutions at this position in ARS1 or ARS307 inactivate or severely impair ARS activity (29,30). ARS309, however, contains an exact match to the additional 6 bp of the EACS. We postulated that the EACS nucleotides were optimized in ARS309 and therefore compensated for the weak core ACS element in this ARS. To test this idea, we constructed an ordered series of SalI linker substitution mutations within ARS309 using the sequence GGTCGAC (Figure 2). These plasmids were scored for the ability to transform wild-type yeast at high frequency and second, for their loss rates in the absence of plasmid selection.
The loss rate for the wild-type ARS309 plasmid was 6% per generation (Figure 2). As expected, two mutations that disrupted the 11 bp core ACS failed to transform yeast at high frequency, confirming that the unusual ACS was essential for ARS activity (14). Mutation of the 5′-‘WWW’ EACS nucleotides ATT to GGG (Figure 2, pFJ273) increased the loss rate to 10% indicating that these EACS nucleotides were important for activity. Similarly, a linker mutation that altered the 3′-EACS nucleotides ‘GTT’ to GTG (Figure 2, pCDM57) increased plasmid loss rates to 13%. This effect on ARS activity was likely due to the GTG change since a similar linker mutation that only differed by retaining the GTT sequence (Figure 2, pFJ271), gave a wild-type phenotype.
Unexpectedly, mutation of nucleotides between the EACS and B1 element (detailed below) also reduced ARS activity, indicated by the increased plasmid loss rate (Figure 2, pCDM44). This was surprising since these nucleotides have been shown to be relatively unimportant for the function of other ARSs (5,7,8,10,22). However, one hypothesis was that nucleotides between the ACS and B1 compensate for the suboptimal ARS309 ACS. To test this idea, we changed the +9 nucleotide within the ACS to a consensus ‘T’ and measured loss rates of ARS309 containing the 10 of 11 ACS (gTTTATATTTT) with or without the linker mutation between the EACS and B1 element. ARS309 containing the 10 of 11 ACS (Figure 2, pFJ215) had a loss rate of ∼3%, which was significantly better than wild-type indicating that this mutation made ARS309 a better ARS. Furthermore the same linker substitution between the ACS and B1 that reduced wild-type ARS309 activity (Figure 2, pCDM80) had no effect on ARS309 containing a 10 of 11 ACS (∼3% loss rate) as predicted. Similarly, mutation of the EACS nucleotides ‘GTT’ to GTG or GGG (Figure 2, pCDM79 and pFJ297) had no effect on ARS309 activity in the 10 of 11 ACS context (∼3% loss rate). Thus, a suboptimal ACS made ARS309 more reliant on both the EACS nucleotides and those between the EACS and B1 elements, raising the possibility that ORC-nucleotide contacts spanning its binding site are somewhat flexible.
Analysis of ARS607 produced similar results to that of ARS309. The ACS of ARS607 matches 10 of 11 ACS consensus nucleotides gTTTATATTTA (9) and 15 of 17 EACS nucleotides (Figure 3). Mutating the ‘WWW’ EACS nucleotides CTT to GGG (Figure 3, pFJ274) increased the plasmid loss rate 2-fold. However, a linker mutation that changed the ‘GTT’ EACS nucleotides to GGG (Figure 3, pCDM60, GTTACGTT > GggtCGac) resulted in very high plasmid loss rates (>35% per generation). A GG substitution of just the +13 and +14 EACS nucleotides from GTT to GGG (Figure 3, pCDM81) increased the plasmid loss rate 3-fold, indicating that the 3′-EACS nucleotides are very important for ARS function. In contrast, this same GTT to GGG mutation in the context of a perfect 11 of 11 match to the ACS (Figure 3, pCDM82) generated only a weak phenotype. Thus, although the +1 nucleotide of the ACS consensus is not highly conserved (7,15,16), a consensus nucleotide at the +1 position significantly compensates for mismatches in the 3′-EACS nucleotides. Thus as for ARS309, a hierarchy of nucleotides exists within the EACS: a better match to the ACS can compensate for substitutions within the EACS nucleotides.
ARS606 contains a perfect match to the ACS ATTTATATTTT (9) and matches 15 of 17 EACS positions, differing from the consensus only at the +12 and +13 positions. Nonetheless, ARS606 activity was also significantly decreased by mutations in the EACS. A 7 bp linker substitution immediately preceding the ACS (Figure 4, pCDM6) significantly decreased ARS stability from the wild-type, i.e. the plasmid loss rate increased from 5% to 27%. A triple TAT to GGG substitution at the ‘WWW’ EACS residues (Figure 4, pFJ234) gave a similar high rate of plasmid loss (25%) indicating that these three nucleotides were very important for ARS606 activity. Similarly, the SalI linker mutation that changed the ‘GTT’ EACS residues from CGT to CGG (Figure 4, pCDM13) had a high loss rate of 27%—but the SalI linker displaced one bp 3′ to the EACS that did not alter EACS residues (Figure 4, pFJ253) had a loss rate equivalent to the wild-type (5%). A single G substitution at position +14 (Figure 4, pFJ255) increased the plasmid loss rate 2-fold. Thus, individual EACS nucleotides are important for ARS606 activity. In contrast, mutation of residues +12 and +13 to give a perfect match to the EACS TATATTTATATTTGTT (Figure 4, pFJ233), improved ARS606 activity, evidenced by a 2-fold decrease in the plasmid loss rate compared to the wild-type. As for ARS309, mutation of nucleotides between the EACS and B1 resulted in a high plasmid loss rate of 17% (Figure 4, pCDM7). However, this same linker mutation had no effect on ARS606 stability within the context of a perfect EACS (Figure 4, compare pFJ233 and pFJ256). Therefore, EACS nucleotides are generally very important for ARS activity, as are nucleotides between the EACS and B1 element when the EACS sequence is suboptimal. For ARS309 and ARS607, mismatches in the 11 bp ACS sequence can be compensated by consensus EACS nucleotides or nucleotides between the EACS and B1 element.
In contrast to the previous ARSs, ARS319 contains a nearly perfect match (16 of 17) to the EACS (7). Therefore, we predicted that the EACS and sequences between the A and B1 elements would be less important. The wild-type ARS319 plasmid was lost at 4% per generation and linker mutations within the ACS, resulted in plasmids that failed to transform yeast (Supplementary Figure S1). Mutation of the ‘WWW’ EACS sequence TAT to GGG (Supplementary Figure S1, pFJ275) increased the ARS319 plasmid loss rate 2-fold. Mutation of the 3′-‘GTT’ EACS sequence GGT to GGG or mutations between the EACS and B1 element had little effect on ARS stability (Supplementary Figure S1). Thus, ARS319 differed considerably from the previous three ARS elements in its dependence on nucleotides surrounding the ACS and B1 elements (summarized in Table 1). In summary, these data reveal a previously unrecognized flexibility and hierarchy of nucleotides within the ORC recognition elements.
Table 1.
Summary of loss rates for EACS mutants
| EACS | WWW | GTT | ||
|---|---|---|---|---|
| WWW-WTTTAYRTTTW-GTT | WT (%) | → GGG (%) | → GGG (%) | |
| ARS309 | ATT-gTTTATATcTT-GTT | 6 | 10 | 11 |
| ARS319 | TAT-TTTTATGTTTA-GgT | 4 | 8 | 6 |
| ARS606 | TAT-ATTTATATTTT-cgT | 5 | 25 | 27 |
| ARS607 | cTT-gTTTATATTTA-GTT | 6 | 11 | 19 |
The B1 element is largely defined by consensus WTW nucleotides
The B1 element core sequence contains WTW nucleotides from 17 to 19 base pairs 3′ to the T-rich strand of the ACS (7,31). Mutation of these nucleotides in ARS309 (Figure 2, pCDM33, TTT to GGG) and in ARS606 (Figure 4, pFJ232, ATT to GGG) resulted in a plasmid loss rate of >35% per generation, indicating that these residues are very important for ARS309 and ARS606 function. Within ARS607, mutation of the WTW nucleotides GTT to GGG (Figure 3, pFJ221) also caused a large (4-fold) increase in the plasmid loss rate to 19%. However, the following two linker mutations in ARS607 (Figure 3, pCDM35/pCDM41) also significantly increased plasmid loss rates to 15 and 12%, indicating that the B1 element extends 3′ to the WTW motif. The B1 element also extends beyond the WTW nucleotides in ARS307 (8,10). This reliance on residues 3′ to WTW might reflect that both of these ARSs have an imperfect match to the WTW consensus. Mutation in the WTW B1 motif of ARS319 (Supplementary Figure S1, pFJ90, ATT to GGG) increased the plasmid loss rate more modestly, from 4% to ∼10%. This effect is similar to that previously reported for an ATT to AGG mutation in ARS319 (7). Therefore as predicted based on analysis of previous ARSs (5,7,12,13,22), the WTW motif largely defines the B1 element of ARS309, ARS606 and ARS319 but nucleotides directly 3′ to WTW are important for ARS607 activity. Notably as well, the relative importance of the B1 element correlates with the importance the EACS nucleotides in each ARS.
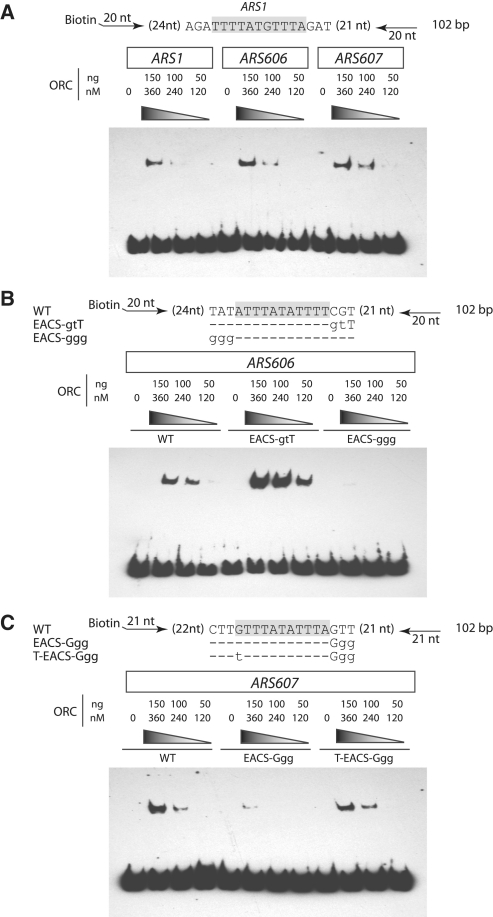
ORC binding to ARS606 and ARS607 depends strongly on EACS residues
Since ARS606 and ARS607 activities exhibited a strong dependence on EACS residues, we examined whether ORC binding was sensitive to changes in EACS residues using electrophoretic mobility shift assays. We used ARS1 DNA as a control since this is an efficient ARS that has been extensively examined in vivo and in vitro (5,30). ARS606 and ARS607 are highly efficient origins in our plasmid ARS assay and on the chromosome (32,33) and ORC bound to both ARSs with a similar affinity as to ARS1 (Figure 5A). Compared to the wild-type ARS606 DNA probe, ORC binding was substantially diminished to the ARS606 probe that mutated the −1 to −3 ‘WWW’ EACS nucleotides TAT to GGG (Figure 5B). This result paralleled the very high loss rates for the ARS606 plasmid containing this same GGG mutation (Figure 4, pFJ234). In contrast, ORC bound better to ARS606 when it contained a perfect match to the EACS, TATATTTATATTTGTT. We saw a similar pattern at ARS607, which has a 15 of 17 match to the EACS. ORC bound poorly to ARS607 with a GG mutation in the 3′-EACS, CTTGTTTATATTTGGG (Figure 5C). However, changing the +1 nt of the ARS607 ACS to a consensus ‘T’ restored ORC binding even in the presence of the GG EACS mutation, CTTTTTTATATTTGGG. The ORC binding data correlated perfectly with the ARS607 activity in vivo (Figure 3, pCDM81 versus pCDM82). Therefore, the EACS nucleotides significantly influenced ORC binding at ARS606 and ARS607 in a manner that paralleled their effects on ARS activity in vivo (Figures 3 and 4). This strongly supports the hypothesis, also based on the ARS309 mutational data (Figure 2), that nucleotides outside the previously defined core elements of the ORC binding site significantly influence ORC binding.
Figure 5.
ORC EMSA on ARS606 and ARS607. (A) Sequence of the ARS1 EACS is shown and positions of oligonucleotides for amplifying biotinylated wild-type ARS1 probe (102 bp). Purified ORC was incubated with the indicated DNAs (described in ‘Materials and Methods’ section), electrophoresed on a 3.5% polyacrylamide gel, transferred to nitrocellulose and then probed with a streptavidin-HRP-conjugated antibody. The ARS606 (B) and ARS607 (C) EACSs are shown together with two mutant derivatives in each EACS sequence. Plasmids containing the wild-type EACS or the mutant derivates were used as PCR templates to generate biotinylated probes (102 bp). EMSA performed as in (A).
Mapping B2 elements and identification of a shared B2 consensus sequence
B2 elements were initially identified as ∼11–18 bp sequences that enhanced ARS function and mapped variable distances immediately downstream of the B1 element in ARS1 and ARS307 (Figure 1). The B2 elements from these ARSs were interchangeable (8,10) suggesting that they shared a common (but unknown) function. B2 sequences were subsequently mapped in ARS305, ARS315 and ARS318 (7,22), however, no sequence conservation has been identified among the five known B2 elements. Importantly, it is now thought that B2 sequences enhance MCM loading or activity based on genetic analyses of several ARSs (20,21) and the fact that B2 overlaps the MCM loading site (34).
To gain a deeper insight into the nature of the B2 element, we mapped four additional B2 elements in ARS309, ARS319, ARS606 and ARS607 using linker substitution mutations. B2 elements are defined as regions downstream and proximal to the B1 element, which enhance ARS activity ∼2–3-fold. For ARS309, a single linker substitution (Figure 2, pCDM29) defined the B2 element, which is positioned similarly to B2 in ARS315 (22) but is more distal than for the other ARS elements in this study. The B2 elements in ARS319 and ARS606 were each defined by two mutations (Supplementary Figure S1, pFJ218 and pCDM21; Figure 4, pCDM12 and pCJ14) suggesting that they might be broader. A single linker substitution 3′ to B1 in ARS607 (Figure 3, pCDM45) defined its B2 element. Interestingly, yet another sequence beginning 14 bp further downstream of B2 defined an important ‘B3’ element in ARS607 (Figure 3, pCDM40). This B3 element may bind a transcription factor that enhances ARS activity, but it is not an Abf1 binding site as at ARS1. Thus ARS309, ARS319 and ARS606 had an A–B1–B2 structure similar to ARS307. ARS607 more closely resembled ARS1 since it also had a B3 element.
It is not known if a consensus sequence exists for B2 elements. Because we now had eight different functionally defined B2 elements we generated a B2 alignment and identified a shared sequence (see ‘Materials and Methods’ section). This analysis identified an optimal consensus centered within the B2 elements that was AT-rich: ANWWAAAT (Figure 6A). Each of the B2 elements contained this sequence with at most one mismatch at position 6, 7 or 8. We used the sequence ‘ANWWAAAN’ (since the T was not invariant) to search 100 bp downstream of the ACS in the 228 phylogenetically conserved ARS elements in budding yeast [Supplementary Table S1; (16)]. We found this exact sequence at least once in 74% of these ARSs. However, this sequence was much less represented in the 100 bp 3′ to the ∼14 000 ACS-like sequences (7) in the budding yeast genome (P-value ≤ 1 × 10−5), the vast majority of which are not functional ARSs. There is therefore a very strong preference for this B2 consensus sequence downstream of the ORC binding site, with an average distance centered at 38 bp 3′ to B1 (Figure 6B). A caveat to the statistical analysis is that the ‘B region’ is A-rich in multiple ARSs (31). The region downstream of B1 in the 228 phylogenetically conserved ARSs is also A-rich (Supplementary Figure S11). Therefore, any conserved B2 sequence will likely be A-rich and more represented downstream of bona fide ARSs than in the genome as a whole. Nonetheless, our GC-rich linker scan analysis of multiple ARSs indicates that the A-rich character is not important except at the B2 elements we experimentally identified.
Partial ACS matches are not required for B2 activity
The determination of additional B2 sequences also allowed us to test the importance of partial 11 bp ACS matches overlapping B2. Inverted, (non-functional) ACSs often map downstream of the ACS and overlap the B2 element, however the significance of this placement is unknown. Linker scan mutations in this study can be used to determine the importance of the partial ACS match to B2 activity. This is most readily apparent from an analysis of ARS309 and ARS607 (Figures 2 and 3). ARS309 contains a 10 of 11 ACS match partially overlapping the B2 element we defined: ATTTAAACATAtAA (B2 is in italics and the inverted ACS is underlined). Mutating the bona fide ACS at ARS309 allowed ORC to footprint this 10 of 11 ACS overlapping B2 but the ARS was inactive nonetheless (14). Importantly, our linker scan mutation 3′ to the ARS309 B2 element substantially disrupts this 10 of 11 sequence ATTTAAAggTcgAc (giving a 6 of 11 ACS) but this mutant retains wild-type ARS activity (Figure 2, pCDM30). Similarly, a 9 of 11 ACS match overlaps the B2 element at ARS607, AAAAaAaAAAT. Mutation of this sequence to cgAcaAaAAAT (resulting in a 6 of 11 ACS match) also retains wild-type ARS activity (Figure 3, pFJ220). Analysis of these two ARSs therefore suggest that close matches to the 11 bp ACS might be co-incidental to some more important underlying sequence conservation within the B2 element.
Nucleosomes surrounding ARS309 and ARS606 overlap replicator elements
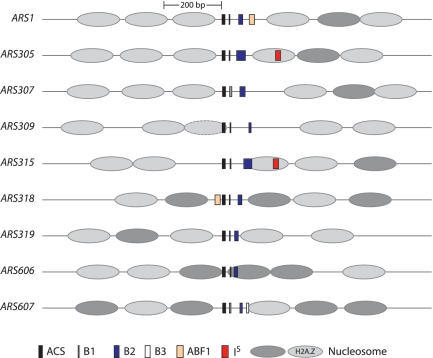
ARSs generally exist within NFRs in budding yeast (35–37) and recently, local nucleosome structure has been implicated in helping to define functional ARSs (36,38,39). Since many studies have determined the positions of stable nucleosomes throughout the genome (35–37,40–44), we took advantage of our detailed ARS structural data to investigate stable nucleosome positions surrounding the functional components of ARS309, ARS319, ARS606 and ARS607. In Figure 7, we utilized a published composite view of six genome-wide nucleosome-positioning studies to include the consensus nucleosome position (45). Obviously, there were some small base pair variations in the exact positioning of the same nucleosome between studies, but this did not have an impact our conclusions.
Figure 7.
Schematic diagram of nine ARS elements whose detailed structures have been determined by linker scan analysis (5,7,8,10,22,52). The functional components of each ARS and the positions of nearby stably positioned nucleosomes, determined from a composite of six genome-wide positioning studies (45) are indicated as in the key. Details of nucleosome positioning surrounding each ARS from this and other data sets are presented in Supplementary Figures S2–S10.
ORC and Abf1 have been shown to exclude nucleosomes within ARS1 (46) and this activity likely occurs at other ARSs (36,38), although Abf1 is not associated with most ARSs. ARS319 and ARS607 conform to this general designation since they exist within nucleosome free regions (Figure 7). However, ARS309, which has a significant mismatch to the ACS, has a nucleosome overlapping the ACS + B1 region. This nucleosome is not strongly positioned over the ACS in the consensus view (45) and is displaced further 5′ in others (36,45). This variability may reflect suboptimal ORC binding at ARS309 in vivo. However, all the origins examined by high-resolution linker scanning have a stable nucleosome within ∼200 bp 5′ to the ACS, and this is an H2A.Z containing nucleosome at six of nine origins examined here (Figure 7). ARS606, in contrast has a stable nucleosome immediately 5′ to the ACS plus a stable nucleosome overlapping the B1 and B2 elements. The Sir2 histone deacetylase inhibits pre-RC formation at ARS606, as it does at ARS305 and ARS315 (7). It is noteworthy that the three Sir2-sensitive origins analyzed to date have a stably positioned nucleosome directly adjacent to or overlapping the B2 region, i.e. the putative MCM loading site. For ARS606, this nucleosome extends into the B1 region, perhaps because the B1 and B2 elements are closely positioned in this ARS.
DISCUSSION
In this study, we determined the structure of four efficient chromosome III and VI ARS elements at nucleotide resolution. This analysis revealed that EACS nucleotides surrounding the 11 bp ACS sequence are important for ARS activity (Table 1), even when the ARS contains a perfect match to the 11 bp ACS. For ARS309, ARS606 and ARS607, mutation of EACS significantly residues decreased ARS stability. At the biochemical level, this suggested that the EACS nucleotides contributed significantly to ORC binding and in vitro analysis of ARS606 and ARS607 supported this idea. Using an electrophoretic mobility shift assay, ORC bound poorly to its binding site in ARS606 and ARS607 when mutations in the EACS nucleotides were present. In contrast, ORC bound significantly better to ARS606 if it contained a perfect ‘GTT’ EACS sequence at positions +12 to +14 (Figure 5). Therefore, EACS nucleotides can significantly alter ORC binding to ARS elements.
We note that our analysis of ARS607 differs somewhat from a previous linker scan analysis of ARS607, which did not identify the ‘WTW’ nucleotides (GTT) within the B1 element as important for ARS function (23). Instead, the previous study found three regions downstream of WTW that stimulated ARS activity: the first corresponds to the extended B1 region we identified; however, mutations within the second region gave a wild-type phenotype in our hands; both studies revealed a short 7 bp A-rich stretch (the B2 element) within ARS607. Given the established functional importance of the WTW nucleotides (5,7,8,10), the B1 element for ARS607 likely compromises the WTW nucleotides and nucleotides immediately downstream (as shown in Figure 3).
ARS309 has a suboptimal ACS (9 of 11 match) and is unusual in that it contains a T > C transition mutation at a highly conserved nucleotide (+9) within the ACS (14). In spite of this, ARS309 is an efficient chromosomal origin that fires in 90% of cell cycles (47) and was maintained efficiently as a plasmid ARS (Figure 2). ARS309 was sensitive to linker scan mutations in the EACS but also to mutations between the EACS and the B1 element. However these same mutations had no effect if the ARS309 ACS was changed to contain a consensus ‘T’ at +9. This suggests that nucleotides between the bi-partite ORC binding site (ACS + B1) contribute to ORC binding as well. Alternatively, these nucleotides could contribute to subsequent steps in pre-RC assembly such as Cdc6 binding (48). ARS309 activity was exceptionally dependent on the WTW nucleotides within B1. Mutating these residues resulted in a high plasmid loss rate that was not measurable. This is also likely due to the suboptimal ACS that makes ORC binding to ARS309 very dependent on its B1 contacts, unlike for instance, the situation for the ARS319 origin, which has a 16 of 17 bp match to the EACS (Supplementary Figure S1). Taken together, our mutational data show that additional nucleotides spanning the ACS + B1 ORC binding site not only contribute to origin efficiency, but in some highly efficient origins are actually critical for ARS activity. Footprinting and cross-linking studies have determined that ORC protects a region spanning the EACS and B1 element at particular origins and have shown that nucleotides within the ACS, B1 element and immediately 3′ to the T-rich strand of the EACS crosslink to ORC subunits (12,13,30,49). Taken together with our data, it therefore seems likely that multiple nucleotide-ORC contacts in the 33 bp window from the EACS to the WTW element contribute significantly to ORC-DNA binding at some ARSs.
A recent report integrates multiple genome-wide nucleosome mapping studies into a consensus view of stable nucleosome positions in the budding yeast (45). Several studies have also examined the locations of yeast ARS elements relative to known stably positioned nucleosomes (35–38). This has led to the view that yeast ARSs generally fall within NFRs and that nucleosome positioning also depends upon ORC binding. We utilized the nucleosome data to position nucleosomes surrounding the particular yeast ARS elements for which detailed maps are available (Figure 7). First, we made the unexpected observation that ARS309 had a positioned nucleosome overlapping the A element. The nucleosome that overlaps the ARS309 ACS is not a strongly positioned nucleosome in asynchronous cells implying that it is only positioned here in a fraction of the cells or in all cells at a particular cell-cycle stage (36,44,45). Therefore, ARS309 might maintain its strong chromosomal replicator activity by binding ORC during G1 and early S-phase but not at other cell-cycle stages.
Also of interest, we found that ARS606 had a strongly positioned nucleosome overlapping the B1 and B2 elements that are 3′ to the T-rich strand of the ACS. This finding is interesting since we previously identified ARS606 as an origin negatively regulated by the Sir2 histone deacetylase (22). In the absence of Sir2 multiple ARSs throughout the genome (including ARS606) have increased activity and load the MCM helicase even when pre-RC assembly is severely compromised, as occurs in a temperature sensitive cdc6 mutant (22,25). Two Sir2-regulated ARSs, ARS305 and ARS315, have inhibitory DNA sequence elements that map 3′ to their B2 elements and these ‘IS’ elements map within stably positioned nucleosomes that either overlap or are adjacent to the B2 element (22). Although ARS606 lacks an inhibitory site 3′ to the B2 element (Figures 4 and 7), it does have a stably positioned nucleosome within the MCM loading site. However, five of six origins not regulated by Sir2 do not have a nucleosome close to B2. ARS319 has a nucleosome close to its B2 element, similar to ARS305, however ARS319 is a sub-telomeric ARS that is likely also negatively regulated by Sir2 on the chromosome (50,51). To date therefore, a nucleosome overlapping or quite near the B2 sequence correlates with negative regulation by Sir2, which may or may not coincide with an inhibitory DNA sequence element. This suggests that Sir2 regulates a subset of origins through this nucleosome per se and not through a particular inhibitory DNA sequence. For ARS305 and ARS315, the IS element may only be needed to help position this nucleosome.
Lastly, we utilized our data on the functional components of ARS309, ARS319, ARS606 and ARS607 to analyze sequence properties of the B2 element in more detail. The biochemical function of B2 is unknown but it could facilitate MCM loading at origins as suggested by genetic analysis of ARS1 (20,21). Alternatively, one or both single-strands of B2 might productively contact the replicative helicase following origin unwinding. Multiple sequences were evaluated for B2 activity within the context of ARS1 leading to the conclusions that the B2 element is A-rich and that active B2 elements do not correlate with helical instability (20). Also this study found that both functional and non-functional B2 elements could overlap partial matches to the inverted ACS. Since the poor ACS sequences (<9 bp matches) are most likely incapable of binding ORC (20), this questions their functional relationship to the ORC binding site. Our data strongly suggest that the overlapping ACS is co-incidental to B2 function: linker scan mutations that decreased the overlapping match to the ACS within the ARS607 and ARS309 B2 elements retained wild-type ARS activity.
We identified a shared 8 bp degenerate sequence that was present in the 8 B2 elements mapped to date: ANWWAAAT. All known B2 elements matched seven or eight nucleotides in this consensus, which was present in ∼90% of 228 conserved ARS elements allowing one mismatch at positions 6, 7 or 8 (Figure 6B). Interestingly, one version of the shared B2 sequence (AYATAAAW) exactly matches eight base pairs of the ACS: specifically, the reverse complement of AYATAAAW matches positions +1 to +8 of the T-rich sequence of the ACS, WTTTAYRT. This could possibly explain the frequent co-occurrence of B2 elements and the inverted ACS, especially since S. cerevisiae has an AT-rich genome that would favor A/T base pairs at the additional three ACS positions from +9 to +11. It is important to point out however, that this 8 bp sequence was not present in several synthetic B2 sequences that functioned well as ARS1 B2 elements (20). Therefore, the presence of this B2 consensus within naturally occurring B2 elements may depend on the context of the ARS and/or may speak to the evolution of this sequence within the S. cerevisiae genome. Clearly, biochemical characterization is required to define its precise role in DNA replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation (MCB-0950464 to M.W.); National Institutes of Health (R01 GM56890 to C.A.F.). Funding for open access charge: Van Andel Institute.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 2.DePamphilis ML. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene. 2003;310:1–15. doi: 10.1016/s0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- 3.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Broach JR, Li YY, Feldman J, Jayaram M, Abraham J, Nasmyth KA, Hicks JB. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harbor Symp. Quant. Biol. 1983;47:1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- 5.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 6.Palzkill TG, Oliver SG, Newlon CS. DNA sequence analysis of ARS elements from chromosome III of Saccharomyces cerevisiae: identification of a new conserved sequence. Nucleic Acids Res. 1986;14:6247–6264. doi: 10.1093/nar/14.15.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang F, Theis JF, Miller J, Nieduszynski CA, Newlon CS, Weinreich M. Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site. Mol. Cell. Biol. 2008;28:5071–5081. doi: 10.1128/MCB.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol. Cell. Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirahige K, Iwasaki T, Rashid MB, Ogasawara N, Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theis JF, Newlon CS. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol. Cell. Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newlon CS, Theis JF. The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- 12.Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl Acad. Sci. USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowley A, Cocker JH, Harwood J, Diffley JF. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theis JF, Newlon CS. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl Acad. Sci. USA. 1997;94:10786–10791. doi: 10.1073/pnas.94.20.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W, Aparicio JG, Aparicio OM, Tavare S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieduszynski CA, Knox Y, Donaldson AD. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006;20:1874–1879. doi: 10.1101/gad.385306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang VK, Donato JJ, Chan CS, Tye BK. Mcm1 promotes replication initiation by binding specific elements at replication origins. Mol. Cell. Biol. 2004;24:6514–6524. doi: 10.1128/MCB.24.14.6514-6524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irlbacher H, Franke J, Manke T, Vingron M, Ehrenhofer-Murray AE. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 2005;19:1811–1822. doi: 10.1101/gad.334805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker SS, Francesconi SC, Eisenberg S. A DNA replication enhancer in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1990;87:4665–4669. doi: 10.1073/pnas.87.12.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilmes GM, Bell SP. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc. Natl Acad. Sci. USA. 2002;99:101–106. doi: 10.1073/pnas.012578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crampton A, Chang F, Pappas DL, Jr, Frisch RL, Weinreich M. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol. Cell. 2008;30:156–166. doi: 10.1016/j.molcel.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Rashid MB, Shirahige K, Ogasawara N, Yoshikawa H. Anatomy of the stimulative sequences flanking the ARS consensus sequence of chromosome VI in Saccharomyces cerevisiae. Gene. 1994;150:213–220. doi: 10.1016/0378-1119(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 24.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 25.Pappas DL, Jr, Frisch R, Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 27.Bolon YT, Bielinsky AK. The spatial arrangement of ORC binding modules determines the functionality of replication origins in budding yeast. Nucleic Acids Res. 2006;34:5069–5080. doi: 10.1093/nar/gkl661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Van Houten JV, Newlon CS. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol. Cell. Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 31.Breier AM, Chatterji S, Cozzarelli NR. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 2004;5:R22. doi: 10.1186/gb-2004-5-4-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman KL, Brewer BJ, Fangman WL. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]
- 34.Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 35.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 36.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berbenetz NM, Nislow C, Brown GW. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 2010;6:e1001092. doi: 10.1371/journal.pgen.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller P, Park S, Shor E, Huebert DJ, Warren CL, Ansari AZ, Weinreich M, Eaton ML, MacAlpine DM, Fox CA. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 2010;24:1418–1433. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 41.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 44.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 45.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 47.Poloumienko A, Dershowitz A, De J, Newlon CS. Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell. 2001;12:3317–3327. doi: 10.1091/mbc.12.11.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cosgrove AJ, Nieduszynski CA, Donaldson AD. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 2002;16:2485–2490. doi: 10.1101/gad.231602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang RY, Kowalski D. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.