Abstract
CD166/ALCAM plays an important role in tumor aggression and progression as well as protecting cancer cells against apoptosis and autophagy. However, the mechanism by which pro-cell death signals control CD166 expression remains unclear. Here we show that following serum deprivation (SD), upregulation of CD166 protein is shorter than that of CD166 mRNA. Molecular analysis revealed both CD166 and miR-9-1 as two novel NF-κB target genes in hepatoma cells. In vivo activation and translocation of the NF-κB P50/P65 hetero-dimer into the nucleus following the phosphorylation and accompanied degradation of its inhibitor, IκBα, contributes to efficient transcription of both genes following SD. We show that following serum starvation, delayed up-regulation of miR-9 represses translation of CD166 protein through its target sites in the 3′-UTR of CD166 mRNA. We also propose that miR-9 promotes cell migration largely due to inhibition of CD166. Collectively, the study elucidates a novel negative auto-regulatory loop in which NF-κB mediates differential regulation of CD166 after SD.
INTRODUCTION
CD166, also known as activated leukocyte cell adhesion molecule (ALCAM) or MEMD, is a 105-kDa transmembrane glycoprotein of the immunoglobin superfamily (1) mapped to human chromosome 3q13 (2). Its expression has been described in subsets of cells involved in dynamic growth and migration, including developing neuronal cells, hematopoietic cells (3), endothelial cells during embryogenesis, lymphoid and myeloid cells, fibroblasts, hepatocytes, pancreatic accinar and islet cells and bone marrow stromal cells (4). CD166 was studied in malignant melanoma, where it seems to be significantly correlated with cancer progression and distinguishes the invasive and metastasizing vertical growth phase of melanoma from its non-invasive, radial growth phase (5–7). Additionally, CD166 expression was altered in prostate and breast cancer carcinoma tissue (8,9). In particular, upregulation of CD166 mRNA and protein levels were found in prostate cancer compared with adjacent normal tissue, with the exception of downregulation in some high-grade tumors (8). Recently, a novel soluble isoform of CD166 (sCD166), produced via alternative splicing, was isolated (10). sCD166 demonstrated an CD166-independent effect in endothelial cell assays as well as a regulatory effect on CD166 function. CD166–CD166 interactions are crucial to the survival and primary site maintenance of cancer cells (11). Additionally, CD166 gene silencing in breast cancer cells decreased the concentration of BCL-2 and increased levels of apoptosis (PARP, active caspase7) (12) and autophagy (MAP1LC3, Beclin1) markers (13), therefore, CD166 may also play an important role in protecting cancer cells against apoptosis and autophagy.
Given that CD166 modulates many cellular functions, it can be hypothesized that aberrant CD166 expression may be responsible for the development of human cancer. CD166 could represent a novel therapeutic target, as the underlying mechanism of CD166-mediated carcinogenesis has been progressively elucidated. However, the exact regulation of CD166 has yet to be well-described, especially the mechanism by which pro-cell death signals control CD166 expression.
In the present study, we observed that CD166 mRNA is greatly upregulated in hepatoma cell lines after serum deprivation (SD), a well-known condition which inhibits cell growth and migration and leads to either apoptosis (14,15) or autophagy (16,17). However, up-regulation of CD166 protein is not as prolonged as that of mRNA. The aim of the present study was to define the mechanism responsible for the induction of CD166 after SD and provide a basic model to aid future studies.
MATERIALS AND METHODS
Materials
Human hepatoma cell lines, HepG2, GQY-7701 and Bel-7402 were maintained in Dulbecco's modified Eagle's medium (DMEM) with or without fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA). The following primary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA): Brg I (SC-10768), RNA polymerase II (SC-9001), CD166/ALCAM (SC-25624); β-actin (SC-130656); NF-κB P65 (SC-372x), NF-κB P52 (SC-848x), NF-κB P50 (SC-7178x), c-Rel (SC-71x), Rel B (SC-48366x), TFIIAα (SC-134080) and IκBα (SC-847). The anti-phospho-IκBα antibody (2859) was from Cell Signaling Technology (Boston, MA, USA). NF-κB inhibitor BAY 11-7082, inhibitor of translation, cycloheximide (CHX) and bacterial lipopolysaccharides (LPS, L2880) were purchased from Sigma (St Louis, MO, USA). Tumor necrosis factor alpha (TNFα, 210-TA) was purchased from R&D systems (Minneapolis, MN, USA). All primers and probes are available in Supplementary Data.
Cell culture
To examine the effects of NF-κB on CD166 and miR-9 expression, cells were pretreated with 100 μM BAY 11–7082 for 1 h and then stimulated with SD for another 24 h. LPS and TNFα were used at concentrations of 100 ng ml−1 and 10 ng ml−1, respectively. Cells were transfected with miR-9 mimics (50–70 nM), miR-9 inhibitor (50–70 nM), siRNA (50 nM) against P50, P65, Brg I, Brm or negative control scramble RNA (GenePharma Co., Ltd, Shanghai China) using Lipofectamine 2000 (Invitrogen Corp, Carlsbad, CA, USA) according to the manufacturer's instructions. Certification of interference effects by siRNA is available in Supplementary Figure S1. Pre-incubation with 10 mM CHX was performed for 30 min, followed by serum-deprivation treatment and further incubation for 1 day in the continued presence of CHX.
5′-RLM-RACE
The GeneRacer system (Invitrogen), based on RNA ligase-mediated and oligo-capping rapid amplification of cDNA, was carried out according to the manufacturer's instructions. CD166-specific primers were used for amplification by PCR using the LaTaq GC-Rich PCR system (Takara, Inc., Dalian, China). The PCR products were then cloned into pGEM-T vector (Promega, Madison, WI, USA) and sequenced.
Chromatin accessibility analysis
Accessibility of DNA to digestion with DNase I or restriction enzymes (all from Takara, except Dde I and Mae III from New England Biolabs, Beverly, MA, USA) analyzed using chromatin accessibility by real-time PCR (CHART-PCR) as described previously (18–20). After purification, 0.1 μg DNA from nuclease-digested or non-digested control cells was used in semi-quantitative PCR.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) assays were performed according to the manufacturer's instructions (Active Motif, Carlsbad, CA, USA). Briefly, 2 × 106 cells were fixed with 1% formaldehyde, washed with cold phosphate-buffered saline and lysed in buffer. Nuclei were sonicated to shear DNA, and the lysates were pelleted and precleared. The protein–DNA complexes were incubated with 4 μg antibodies overnight and then incubated with protein A beads followed by elution in 1% SDS/0.1 M NaHCO3 and cross-links were reversed at 65°C. At last, DNA was subjected to semi-quantitative PCR analysis after recovered.
Reverse transcription PCR
Total RNA was extracted by Trizol reagent (Invitrogen). Reaction mixture (20 μl) containing 1 μg total RNA was reverse transcribed to cDNA using PrimeScript RT-polymerase (Takara). Quantitative PCR was performed on the cDNA using primers specific for CD166, P50, P65, Brg I, Brm, pri-miR-9-1 and pri-miR-9-2. Specifically, Stem-loop reverse transcription (RT)–PCR for mature miR-9 was performed as described previously 21). RNA input was normalized to the level of β-actin, both for genes and micro RNA (miRNA) analysis (22). All reactions were carried out using SYBR Green Mix (Takara), and the PCR conditions for quantitative RT–PCR were as follows: activation of enzyme at 94°C for 5 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 20 s. qRT–PCR was carried out using a Mx3000P real-time PCR System (Stratagene, La Jolla, CA, USA). The data from qRT–PCR were analyzed by the ΔCt method as previously described (20,23).
Construction of plasmids and mutagenesis
PCR was performed using sets of primers specific for the CD166 and pri-miR-9-1 promoters, of which the forward primers were Mlu I-site-linked and the reverse primer Xho I-site-linked. These PCR products were cloned into the pGL3-Basic vector (Promega), yielding the promoter reporter plasmids. CD166 3′-UTR, and flanking sequences around pre-miRNA-9-1, which containing miR-9 putative target sites were amplified and then inserted into the Xho I/Not I sites of the psiCHECK2 vector (promega) as previously reported (20). For overexpression studies, the complete ORF sequences of P65, P50, P52, c-Rel and Rel B were cloned into pcDNA3.1(+) expression vector (Invitrogen). Verification of these vectors is seen in Supplementary Figure S1. Deletion constructs were generated by a site-directed mutagenesis kit (Takara). All the vectors have been verified via sequencing by Invitrogen Inc (Shanghai, China).
Luciferase assay
Promoter and miRNA sensor reporter assays were performed as described previously (20). Luciferase activity was measured as chemiluminescence in a luminometer (PerkinElmer Life Sciences, Boston, MA, USA) using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed using the light shift kit (Pierce, Rockford, IL, USA). Five micrograms of nuclear extract proteins [prepared by a kit from Active Motif and quantized by BCA protein quantification kit (Pierce)] were incubated in 20 μl of reaction mixture with or without excess of DNA competitors on ice for 10 min followed by the addition of the biotin labeled probes (synthesized and 5′ labeled by Sangon Inc, Shanghai, China). For super-shift assays, 5 μg antibodies were added to the reaction mixture 10 min before the addition of the probe. All DNA–protein complexes were resolved by electrophoresis on 5% native polyacrylamide and trans-membrane to Immobilon-Ny+ (Millipore, Billerica, MA, USA).
Western-blot analysis
To determine protein expression levels, we prepared total-cell extracts (by RIPA buffer, Beyotime Inc, Haimen, China) and fractions from either cytoplasm or nucleus (extracted by a kit from Active Motif sequentially). The analysis was carried out by ECL western blotting (Promega) according to the manufacturer's recommendations.
Cell migration assay
To carry out the wound healing assay, the cells were plated onto 24-well plate. The monolayer HepG2 cells were then scratched manually with a plastic pipette tip, and after being washed with PBS, the wounded monolayers of the cells were transfected with either siRNA or pcDNA 3.1 vector in serum free opti-MEM (Gibco) for 5 h. Afterwards, transfection medium was replaced, and cells were allowed to heal for 24–30 h in DMEM containing 10% FBS. To perform a rescue experiment, 1 μg pcDNA3.1 or 50 nM siRNA targeting CD166 was co-transfected with 70 nM either miR-9 mimics or miR-9 inhibitor into HepG2 cells.
To measure the cell migration activity, trans-well assays were done using Corning 8.0-μm Transwell® cell culture inserts (Corning, NY, USA). Membranes were coated with purified fibronectin (Sigma) at a concentration of 10 μg ml−1. After transfection, either siRNA or expression vectors in opti-MEM medium for 5 h, cells were collected and resuspended in serum-free DMEM medium containing 0.1% bovine serum albumin. Subsequently, cells (4 × 105 ml−1) were seeded in trans-well chambers. After 24 h incubation, the cells on the upper surface of the filter were completely wiped away with a cotton swab. The cells on the lower surfaces of the membrane were fixed with 100% methanol, stained with Giemsa solution and counted under a microscope.
Statistical analysis
All data were expressed as the mean ± SD. The statistical significance of differences between values was evaluated by the Student's t-test.
RESULTS
CD166 is upregulated after SD
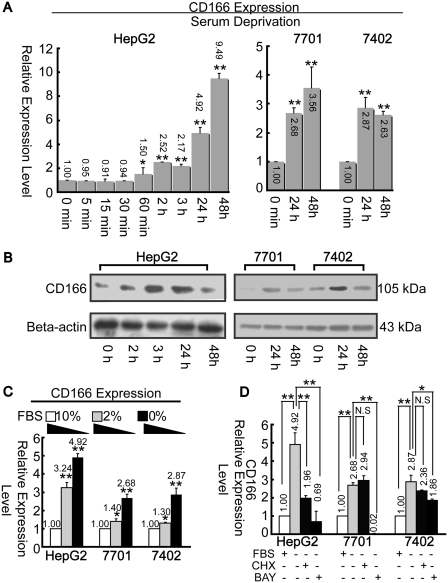
CD166 mRNA expression levels were upregulated following SD in both HepG2 and GQY-7701 cells; however, CD166 mRNA levels decreased after 48 h (compared to that of 24 h) in Bel-7402 cells (Figure 1A). The expression pattern of CD166 protein was similar with that of mRNA, but declined remarkably 48 h after induction in all three cell lines (Figure 1B). We then discovered that CD166 expression is negatively associated with serum concentration (Figure 1C). CHX, a protein synthesis inhibitor (23,24), was applied to determine whether SD-induced upregulation of CD166 is dependent upon translation. Unlike in GQY-7701 and Bel-7402 cells, pre-treatment with CHX in HepG2 cells significantly inhibits the starvation-induced effects as analyzed by real-time PCR (Figure 1D).
Figure 1.
SD induces upregulation of CD166. (A) Change in CD166 mRNA level under SD during the whole detection period as indicated was quantified by real-time PCR. *P < 0.05 and **P < 0.01 versus ‘0 min’ point. (B) Alteration of CD166 protein level by SD was detected by western-blotting assays. (C) CD166 mRNA expression levels were measured at different serum (FBS) concentration as indicated. *P < 0.05 and **P < 0.01 versus ‘10%’ point. (D) Cells were stimulated with BAY 11-7082 (100 μM) or CHX (10 mM) in combination with SD for 24 h, then total RNA was extracted for CD166 detection. N.S., non-significant; *P < 0.05 and **P < 0.01 level using t-test. β-actin was treated as internal control both for real-time PCR and western blotting.
A κB motif acts as an enhancer to CD166 gene
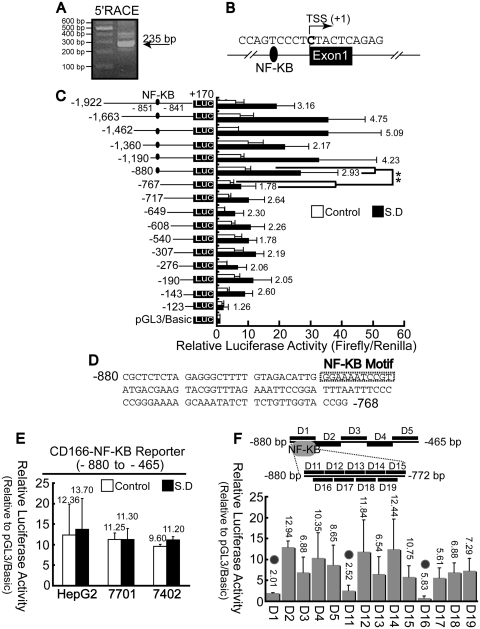
The transcription start site (TSS) of the human CD166 gene was identified by 5′ RACE analysis and predicted to be ∼235-bp upstream from our gene-specific nested primer (Figure 2A). Sequencing of PCR products indicated that the first base was cytosine (C) (Figure 2B), which is 35-bp downstream of the first base provided by National Center for Biotechnology Information (NCBI Gene ID: 214).
Figure 2.
Analysis of CD166 promoter activity. (A) 5′RACE was performed using the reverse 5′RACE nested primer. (B) Mapping the TSS of CD166 gene in hepatoma cells. (C) HepG2 cells were transiently co-transfected with reporter plasmids containing truncated versions of the promoter region of the CD166 gene, as indicated, and pRL-TK. **P < 0.01 level using t-test. (D) The κB motif presents in the promoter region predicated by web software TFSEARCH and MatInspector, as indicated by a rectangle box. (E) CD166 NF-κB reporter is defined as the promoter reporter containing the minified upstream region (from −880 to −465), which the κB motif located in, and whose promoter activities were measured before and after SD. (F) Scanning deletion analysis of the CD166 NF-κB reporter. As shown on the upper panel, a series of deletions were made within the fragment from −880 to −465 of the NF-κB reporter. D1, D11 and D16 overlap the possible κB motif are indicted by gray circle in the lower panel. Firefly luciferase activity was normalized to renilla-luciferase activity, and the relative luciferase activities are presented as fold increase over the promoter-less pGL3 basic vector.
To examine the transcriptional activity of the sequences at the 5′-end of the CD166 gene, various fragments of the proximal sequence were cloned upstream of the firefly luciferase reporter gene. A 5′ deletion series with a fixed 3′-end at the +170 position (relative to the TSS) were generated through PCR amplification. All the constructs (containing −880/−768 fragment) tested were capable of inducing a more significant increase in luciferase activity compared to those which lacked this region (Figure 2C). This suggests that the most important 113 nucleotides of the CD166 gene contain the enhancer element capable of inducing CD166 expression.
We then performed a prediction using the software MatInspector and TFSEARCH, which revealed a κB motif (from −851 to −841) within this region (Figure 2D). To confirm the role of this 113-bp fragment in transcriptional activity, minified sequences between −880 and −465 were cloned into pGL3/Basic vector, yielding the CD166 NF-κB reporter. Results indicated this isolated fragment had promoter activity; however, no significant changes in luciferase activity were detected before or after SD (Figure 2E).
To further test the role of this κB motif, we generated a series of deletions within the CD166 NF-κB reporter construct. The individual deletions in D1, D11 and D16 (which disrupts the kB motif) all reduced promoter activity significantly, whereas no disruption in promoter activity was observed in the other constructs (Figure 2F). This demonstrates that this κB motif may act as an enhancer necessary for increasing transcriptional activity.
Recruitment of P50/P65 to the CD166 promoter
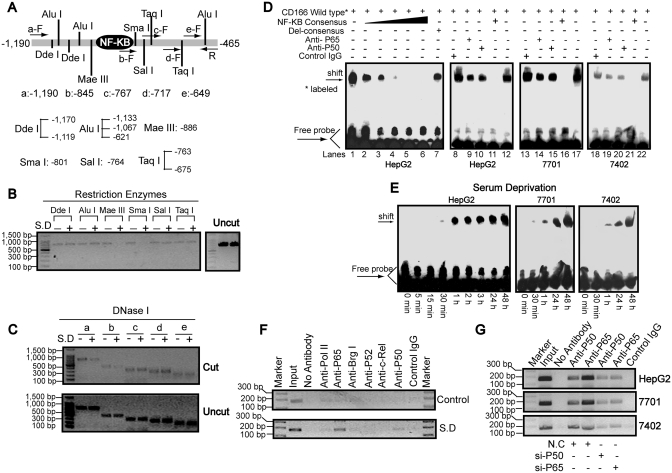
To analyze the locations and boundaries of chromatin changes around κB motif after SD, we designed five forward primers (labeled a through e) within the −1190 to −465 for Chart-PCR assays (Figure 3A). Nuclei isolated from HepG2 cells before and after SD were subjected to limited digestion with a panel of restriction enzymes and DNase I. When the nuclei were incubated with Mae III and Sma I, starvation-dependent restriction enzyme cleavages were apparent. In contrast, such cleavages were not observed for other restriction enzyme sites, including Dde I, Alu I, Sal I and Taq I (Figure 3B). In addition, stronger cleavages by DNase I were only detected when using primers a and b (Figure 3C), suggesting the region from c to e does not cover the starvation-response element. Because digestion with enzymes with cut sites just upstream (Alu I/-1067) and downstream (Sal I/-764) of the regions remain unchanged, we mapped the boundaries undergoing chromatin alteration following serum starvation to between −1067 and −764 bp, in which the κB motif is located.
Figure 3.
Binding of NF-κB to the CD166 promoter. (A) Schematic representation of the restriction enzyme positions and primers used to map the serum starvation-response site from −1190 to −465 (relative to the TSS) region by chart-PCR assays. (B) Nuclei of HepG2 cells before or after SD for 1 day were purified and digested with 5–15 U of different restriction enzymes, as indicated, per 100 μl reaction system for 30 min. Purified DNA treated with or without enzymes (uncut) was semi-quantitated using primer sets a–F and R after 30 cycles of PCR reactions. (C) Purified DNA from HepG2 cells before or after SD for 1 day treated with (cut) or without (uncut) 3 U of DNase I for 5 min at 37°C was semi-quantitated using primer sets R and a–e, which were the same as indicated in Figure 3A after 29 PCR cycles. (D) Nuclear extracts from three different hepatoma cell lines treated with SD for 1 day were assayed by EMSA using biotin-labeled oligonucleotide probes (from −858 to −834) and NF-κB consensus probes. Shifted bands were verified by addition 5 μg either nonspecific control IgG or specific antibodies against P65 or P50, as indicated. (E) EMSA assays were performed using nuclear extracts from cells by treatment of SD for indicated time and wild-type probes (from −858 to −834). (F) ChIP analysis was performed to qualitatively confirm the interaction of NF-κB components, Pol II and Brg I with the promoter in vivo. As the negative controls, the protein–DNA complexes were incubated without antibodies or with non-specific control IgG. The input DNA represents 1/20th of the starting material. (G) In vivo interactions of NF-κB with CD166 promoter after SD upon suppression of P50/P65 protein by specific siRNA were determined by ChIP assay.
To test DNA–protein interactions, we performed EMSA using the DNA sequence from −858 to −834 bearing the κB motif and nuclear extracts from cells 1 day after SD. As illustrated, one strong DNA–protein complex was detected when wild type probes (Figure 3D, lanes 1, 12, 17 and 22) were used. Then, a series of fold excess (10,20,30,40,50) unlabeled NF-κB consensus sequence (lanes 2–6) was used to compete with the complexes. We observed that the 40-fold excess of competitor (lane 5) is enough to compete away the complexes, possibly by sequestering the available transcription factors present in the nuclear extract. In contrast, the 50-fold excess of unlabeled mutated consensus oligonucleotide bearing the disrupted κB could not compete for protein binding capacity (lane 7). To verify if NF-κB is involved in the formation of these protein–DNA complexes, antibodies specific against P50 and P65 were used. As shown [lanes 9 and 10 (HepG2), 14 and 15 (QGY-7701), 19 and 20 (Bel-7402)], although no super-shifted bands were detected, the complexes were greatly abrogated by either anti-P50 or anti-P65, similar to previously reports (25–27); however, non-specific control IgG (lanes 8, 13 and 18) had no competed effects. We also detected that the amount of DNA–protein complex began to increase around 30 min following SD and remained high afterwards (Figure 3E), suggesting the induction of the NF-κB binding capacity.
Next, ChIP assays were performed to examine whether NF-κB components (P50, P65, P52 and c-Rel), RNA polymerase II (Pol II) and key factor of SWI/SNF complexes, Brg I interact with promoter in vivo before and after SD. After 1 day of serum starvation, anti-P65 and anti-P50 antibodies specifically enriched the region, which contains the κB motif. However, no inducible enrichment was detected for other factors (Figure 3F). We also detected P50/P65-specific siRNA could reduce P50/P65 loading onto the promoter, compared to non-specific control siRNA (N.C) (Figure 3G). The above results suggest recruitment of P50/P65 to the CD166 promoter after SD.
Contribution of P50/P65 to upregulation of CD166
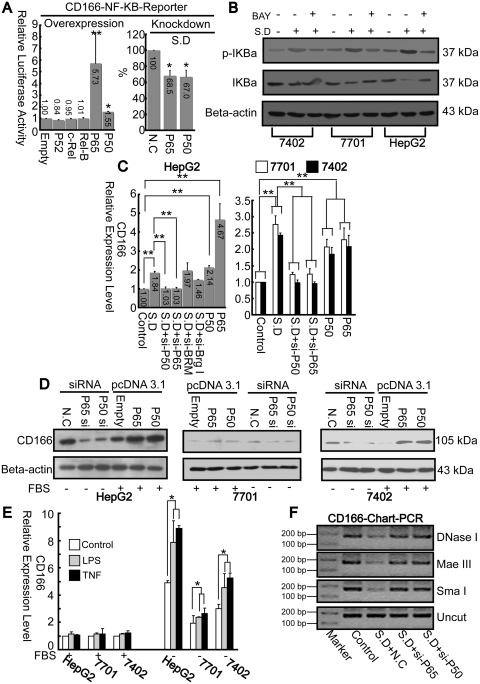
Next, we determined whether deregulation of NF-κB components affect promoter activity. In HepG2 cells, promoter activities from the NF-κB reporter increased only after transfection of P50/P65 expression vectors. However, no significant changes were detected after overexpression P52, c-Rel or Rel B (Figure 4A, left panel). Additionally, promoter activities from the same vector could be reduced after using siRNA against P50/P65 (Figure 4A, right panel).
Figure 4.
NF-κB contributes to upregulation of CD166. (A) Promoter activities from CD166 NF-κB reporter in HepG2 cells were measured after overexpression of NF-κB components, as indicated, with FBS condition, *P < 0.05 and **P < 0.01 versus ‘Empty’ or knockdown either P65 or P50 using specific siRNA under SD, *P < 0.05, versus ‘NC’. (B) Effects of SD with or without BAY 11–7082 (100 μM) treatment on the amount and phosphorylation of IκBα protein. β-actin was treated as internal control. (C) Total RNA from cells was extracted after 12 h treatment with either siRNA or pcDNA3.1, as indicated. Then CD166 mRNA levels were measured using real-time PCR. **P < 0.01 level using t-test. (D) CD166 protein levels from cells under similar treatment as described in (C) were assayed by western blotting. (E) Effects of LPS or TNFα on CD166 expression before or after SD. (F) Open chromatin accessibility status of HepG2 cells under different treatment as indicated was measured via chart-PCR (29 reaction cycles) using either DNase I, Mae III or Sma I by same primer sets used in Figure 3F.
BAY 11–7082, a known pharmacological inhibitor of IκBα phosphorylation, was used to further define the critical role of NF-κB on CD166 expression. As indicated in Figure 1D, BAY 11–7082 can block the starvation-induced upregulation of CD166 mRNA in all three cell lines. To rule-out the possible off target effects of this inhibitor, we monitored phosphorylation and degradation of IκBα by western blotting. As expected, SD efficiently promotes phosphorylation of IκBα. Subsequent degradation of the IκBα protein was most obvious in HepG2 cells. On the contrary, BAY 11-7082 could reduce SD-induced effects on IκBα (Figure 4B).
Then we tested the effect of P50/P65 and SWI/SNF complexes on endogenous CD166 expression. Starvation-induced upregulation of CD166 was attenuated to basal level after knockdown of P50/P65; however, CD166 levels were not affected by knockdown of either Brg I or BRM, two key factors of SWI/SNF complexes. The enhanced effects of overexpression of P50/P65 on CD166 mRNA expression were also observed, which further confirms the importance of P50/P65 on transactivaion of CD166 gene (Figure 4C). Similarly, the P50/P65 effect on CD166 expression was also seen at protein level (Figure 4D). Interestingly, when evaluating the LPS or TNFα-induced effects on CD166 expression, no major changes were observed when cells were cultured with FBS, yet both two inducers could quantitatively enhance SD-promoted effects (Figure 4E), suggesting activation of NF-κB by these inducers alone is not sufficient to activate CD166 expression, but may synergize the impact of SD.
As chromatin structure represents the ultimate integration site of both environmental and differential inputs which determine gene expression, we wanted to explore whether P50/P65 directly affects chromatin accessibility after SD. Chart-PCR was performed using restriction enzymes (Mae III and Sma I) and DNase I. As expected, the bands amplified from intact DNA isolated from P50/P65 siRNA-treated cells after digestion with restriction enzymes were more pronounced than those transfected with non-specific control siRNA (Figure 4F). Overall, P50/P65 upregulates CD166 expression may due to enhancing open chromatin accessibility around the κB motif independent of SWI/SNF complexes.
Activation and translocation of P50/P65 induced by SD
To verify whether expression of P50 or P65 has effects on CD166 expression under SD, both real-time PCR and western blotting assays were used. In HepG2 cells, the P65 mRNA began to increase 24 h after SD (Supplementary Figure S2A), decreased rapidly after re-addition of serum in 30 min, and remained at low level throughout the detection period (Supplementary Figure S2D). For protein, total P65 induced gradually after induction (Supplementary Figure S2B), whereas reduced slowly after re-addition of serum (Supplementary Figure S2D). With difference to HepG2 cells, no obvious change of P65 protein level was detected after SD in other two cell lines, albeit P65 mRNA decreased at 48 h compared to that of 24 h in QGY-7701 cells (Supplementary Figure S2A and B). We also detected that P50 (both for mRNA and total protein) remained almost unchanged in all three cell lines (Supplementary Figure S2C). In addition, no alteration of P50 (both for mRNA and total protein) was detected after re-addition of serum in HepG2 cells (Supplementary Figure S2D).
To further analyze the response of NF-κB to SD, P50/P65 translocation was evaluated. SD-stimulated P50/P65 translocation to the nucleus, whereas cytoplasmic P50/P65 levels were reduced after SD in all three cell lines (Supplementary Figure S2B and C), which is consistent with nuclear translocation of a finite cytosolic pool of these proteins. Specifically, it was also found that SD resulted in accumulation of cytoplasmic P65 during the early phase of stimulation (at least for the first 3 h) in HepG2 cells, and eventually led to translocation of all these induced P65 into the nucleus (Supplementary Figure S2B). We next examined the effect of serum re-addition on the amount of P50/P65 in HepG2 cells. As shown in Supplementary Figure S2D, serum re-addition decreased the amount of nuclear P50/P65, whereas increased that of cytoplasmic (albeit both cytoplasmic and total P65 finally returned to the basal level), suggesting the nuclear export of these proteins after removal of the stimulation.
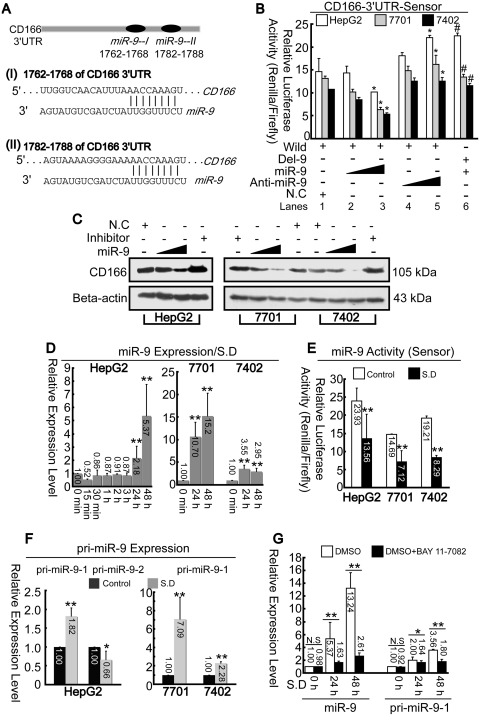
Suppression CD166 protein by miR-9
Unlike mRNA (Figure 1A), CD166 protein was markedly reduced 48 h after SD compared to that of 24 h in all three cell lines (Figure 1B). To investigate these results, we explored whether protein synthesis of CD166 was affected 24 h after stimulation. As shown in Figure 5A, the 3′-UTR of CD166 contains two adjacent elements complementary to miR-9 seed regions predicted by Targetscan online software. To verify, miR-9 can genuinely affect CD166 expression, sensors were constructed (described in the ‘Materials and Methods’ section). As indicated in Figure 5B, luciferase assays revealed that overexpression of miR-9 could reduce the renilla-luciferase activities from the wild sensor (lanes 3 versus 1). To inhibit the function of miR-9, 2′-O-methyle antisense inhibitory oligoribonucleotides targeted toward miR-9 (50–70 nM, lanes 4 and 5) were co-transfected, and it was observed that relative-luciferase activities were upregulated compared to that of control (lane 5 versus 1). To further confirm these putative target sites of miR-9, a deletion assay was performed. Luciferase assays indicated that overexpression of miR-9 had no effect on deleted 3′-UTR compared to the wild type (lane 6 versus 3). Instead, the increased luciferase activity that was observed may have resulted from a failure of endogenous miR-9's effect to the sensor (lane 6 versus 1). It was also confirmed that CD166 protein levels could be suppressed by miR-9. Transfection with anti-miR-9 blocked miR-9 function, thus resulting in enhanced CD166 protein level (Figure 5C).
Figure 5.
Upregulation of miR-9 leads to down-regulation of CD166. (A) CD166 3′-UTR contains two miR-9 target sites predicted by Targetscan online software. (B) psiCHECK2 vectors containing CD166 3′-UTR with (wild) or without (Del-9) predicted miR-9-binding sites were co-transfected with miR-9 (50–70 nM) or miR-9 inhibitor (50–70 nM). Luciferase assays were performed 1 day post-transfection. *P < 0.05 versus ‘Wild + NC’, and #P < 0.05 versus ‘Wild + miR-9 (higher dose)’. (C) SD-treated cells were transfected with or without miR-9 (50–70 nM) or anti-miR-9 (70 nM). CD166 was then detected by western blotting 1 day post-transfection. (D) Total RNA from hepatoma cells were detected for miR-9 after SD at different time point as indicated by real-time PCR. **P < 0.01 versus ‘0 min’ point. (E) Activities of miR-9 before and after SD were measured via dual-luciferase assays. (F) Primary miRNAs (pri-miR-9-1 and pri-miR-9-2) expression levels were measured before and after SD by real-time PCR. (G) Expression levels of mature miR-9 and its precursor pri-miR-9-1 were detected with incubation of DMSO or BAY 11-7082 under SD at different time points as indicated in HepG2 cells. Renilla-luciferase activities were normalized to Firefly luciferase activities (B and E).
Next we evaluated both expression and activity of miR-9 before and after SD. Results indicated that miR-9 began to upregulate 24 h following SD (Figure 5D). To verify induction of miR-9 activity, sensors were constructed as described previously (20). Reduction of renilla-luciferase activity suggests induction of miRNAs activity. As expected, the activities from miR-9 sensors decreased significantly after SD (Figure 5E). Mature miR-9 can be generated from its precursor pri-miR-9-1 and -2 in human genome, therefore it is important to clarify which precursor is associated with upregulation of miR-9. As shown in Figure 5F, only pri-miR-9-1 increased significantly after SD while pri-miR-9-2 decreased. We also confirmed that induction of both mature miR-9 and pri-miR-9-1 by starvation could be blocked by NF-κB inhibitor, BAY 11-7082 (Figure 5G).
Taken together, starvation induced up-regulation of miR-9 may be associated with induction of pri-miR-9-1 via NF-κB. Additionally, miR-9 may have a negative role in controlling CD166 protein level.
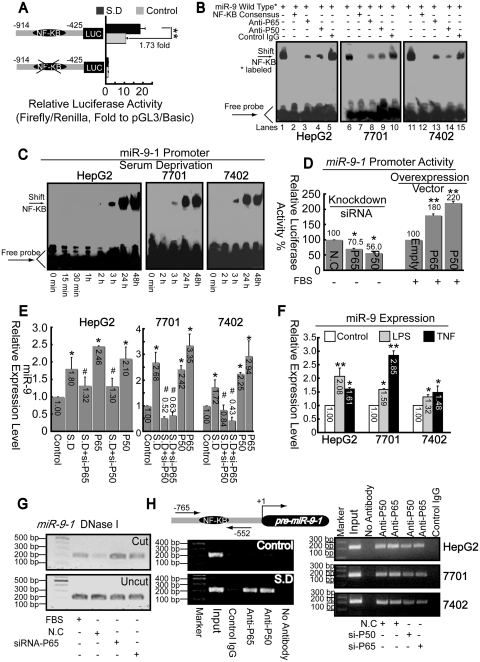
Activation of pri-miR-9-1 promoter via P50/P65
To confirm whether sequences upstream of pre-miR-9-1 had promoter capacity, fragments encompassing from −914 to −425 (relative to the first base of pre-miR-9-1) with or without the putative κB motif (from −693 to −681) were cloned into the pGL3/Basic vector. As shown in Figure 6A, after SD, the promoter activity can be significantly induced by 1.73-fold compared to the basal level. Also, deletion of the κB motif resulted in almost total loss of its promoter activity.
Figure 6.
P50/P65 mediates up-regulation of miR-9 via interaction with pri-miR-9-1 promoter. (A) Promoter activities of fragment from −914 to −425 relative to pre-miR-9-1 with or without putative κB motif were detected before and after SD in HepG2 cells. **P < 0.01. (B) Nuclear extracts from cells 1 day after SD were assayed by EMSA using biotin-labeled oligonucleotide probes (from −700 to −673) and NF-κB consensus probes. Shifted bands were verified by addition of 5 μg either non-specific control IgG or specific antibodies against P65 or P50, as indicated. (C) EMSA assays were performed using nuclear extracts from three hepatoma cell lines after SD for indicated time and wild-type probes (from −700 to −673). (D) Promoter activities from miR-9-1 reporter were measured after overexpression of NF-κB components, as indicated, with FBS condition or knockdown either P65 or P50 using specific siRNA under SD. *P < 0.05, and **P < 0.01 versus the control (NC or empty). (E) Total RNA from cells was extracted after treatment with siRNA or pcDNA3.1 vector targeting either P50 or P65. Then miR-9 levels were measured using real-time PCR. *P < 0.05, versus the control and #P < 0.05 versus the SD. (F) Effects of LPS or TNFα on miR-9 expression. *P < 0.05, and **P < 0.01 versus the control. (G) Open-chromatin-accessibility status around the κB motif under different treatment as indicated in HepG2 cells was measured via Chart-PCR (31 reaction cycles) using DNase I by same primer sets used in Figure 6H. (H) ChIP analysis was performed to qualitatively confirm the interaction of NF-κB components, P50 and P65 with the miR-9-1 promoter in vivo (left panel). Right panel, in vivo interactions of NF-κB with pri-miR-9-1 promoter after SD upon suppression of P50/P65 protein by specific siRNA were determined by ChIP assay again.
EMSA was then performed to identify the DNA sequence from −700 to −673, which overlaps κB motif, has protein-binding capacity. The probes which interacted with nuclear extracts from hepatoma cells 1 day after SD lead to generation of a single band (Figure 6B, lanes 1, 6 and 11), which could be competed by NF-κB consensus probe (lanes 2, 7 and 12). Super-shift assays using antibodies specific against either P50 or P65 resulted in disruption of the band, whereas non-specific control IgG had no significant effects (similar with results illustrated in Figure 3D). Then we investigated the influence of starvation on DNA–protein complex formation at this promoter region. After treatment, the amount of DNA–protein complex increased progressively (began at 3 h) in a time-dependent manner (Figure 6C).
To confirm P50/P65 has genuine effects on miR-9-1, we used gain–loss of function experiments. Overexpression of either P50 or P65 could enhance both expression and promoter activities of miR-9, but in contrast, knockdown by siRNA had the opposite effects (Figure 6D and E). As miR-9 is previously reported to be upregulated by both LPS and TNFα in human polymorphonuclear neutrophils (PMN) and monocytes (46), we analyzed whether these two stimuli exert analogous function on miR-9 in hepatoma cells. As shown in Figure 6F, both LPS and TNF could induce miR-9 expression in all three cell lines, suggesting the regulatory mechanism may be similar. Additionally, siRNA against P50/P65 reduced open chromatin accessibility induced by SD, around the κB motif of pri-miR-9-1 promoter, as indicated in Figure 6G. Furthermore, we observed that P50/P65 is recruited to κB motif of pri-miR-9-1 promoter 24 h after SD by ChIP assays (Figure 6H, left panel). It is also confirmed that P50/P65-specific siRNA decreased P50/P65 loading onto the promoter, compared to non-specific control siRNA (Figure 6H, right panel). In summary, NF-κB P50/P65 plays a similar role in activation of both CD166 and pri-miR-9-1 promoters.
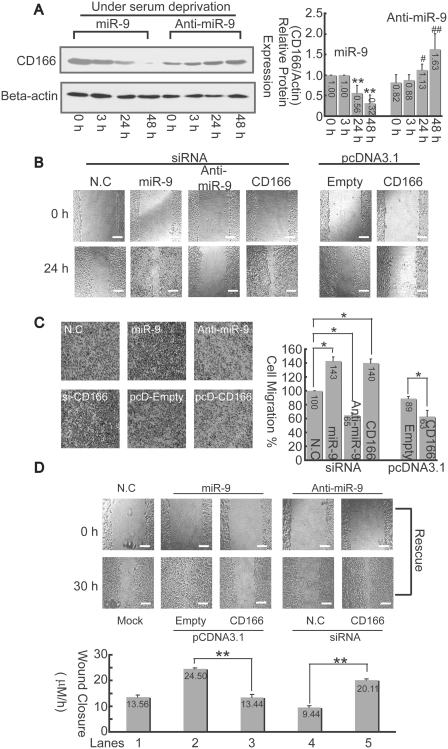
The miR-9 promotes cell migration via inhibitory to CD166
As shown in Figure 7A, the CD166 protein in HepG2 cells transfected with miR-9 mimics (70 nM) was gradually downregulated under SD while miR-9 inhibitor (70 nM) has the opposite effects. As Masedunskasa et al. (28) reported that CD166 markedly inhibited migration of THP1 and HL60 monocytes. Then we explored whether similar effects of the CD166 interaction with miR-9 in controlling cell migration also exist in hepatoma cells. It was observed that overexpression of miR-9 significantly stimulated cell migration, while anti-miR-9 suppressed such effect. Furthermore, knockdown of CD166 by RNA interference promotes migration, which resembles that of miR-9, whereas overexpression of CD166 has the opposite effects (Figure 7B).
Figure 7.
miR-9 promotes cell migration via inhibition to CD166. (A) Changes of CD166 protein in HepG2 cells were detected by western-blotting assays after transfection with 70 nM either miR-9 mimics or miR-9 inhibitor under SD at different time point as indicated (left panel). The relative CD166 protein expression levels were quantitated, and the intensities of the bands of CD166 were normalized to those of β-actin (right panel). *P < 0.05, and **P < 0.01 versus the ‘miR-9 treated-0 h’; #P < 0.05 and ##P < 0.01 versus the ‘anti-miR-9 treated 0 h’. (B) Cells were wounded before transfection with either siRNA or expression vectors as indicated. Then, the culture was continued for 24 h. The bar, 200 μm. (C) Cell-migration assay was carried out with trans-well culture chambers. Transfected with either siRNA or pcDNA3.1 expression vectors as described in (B), 4 × 104 HepG2 cells were seeded into the upper wells. After 24 h, the cells migrating to the lower surface of the membrane were examined (original magnification 40×). Five different areas of migrated cells were counted for each data point (n = 5), and the counts were averaged (right panel). The data is presented relative to cells transfected with negative control siRNA, which is normalized to 100%. The asterisk indicates a significant difference (*P < 0.05). (D) CD166 reverse miR-9 promoted cell-migration activity. Either miR-9 mimics or inhibitor were co-transfected with CD166 expression vector or siRNA into HepG2 cells for 30 h. Afterwards, average rates of wound closure were calculated from three independent experiments. The bar, 200 μm.
To further confirm the above results (Figure 7B), the trans-well approach was used. Consistently, the results indicated that cell migration can be enhanced by either overexpression of miR-9 or knockdown of CD166, while migration can be suppressed by either inhibition of miR-9 or overexpression of CD166 (Figure 7C). To rule out the possibility that miR-9-promoted cell migration through inhibition of other genes, rescue experiments were performed. The results showed that overexpression of CD166 reduced the effects induced by miR-9 (Figure 7D, lane 3 versus 2), whereas knockdown of CD166 rescued inhibitory effects by anti-miR-9 (lane 5 versus 4). All the results suggest that miR-9 promoted cell migration is largely via inhibition of its genuine target, CD166 in hepatoma cells.
DISCUSSION
In our study, we illuminated the mechanism by which SD mediates differential regulation of CD166 expression on the grounds that (i) activation and translocation of P50/P65 into the nucleus occurs after phosphorylation of IκBα (Figure 4B, Supplementary Figure S2B and C); (ii) recruitment of P50/P65 to both CD166 and pri-miR-9-1 promoters, which in turn enhances open chromatin accessibility (Figures 3, 4 and 6); and (iii) suppression of CD166 translation by miR-9 results in decreasing CD166 protein levels later after induction (Figures 1B, 5A–C and 7A).
Biological systems in cells ultimately need to be explained in terms of the activity, regulation and modification of proteins. The ubiquitous occurrence of post-transcriptional regulation makes mRNA an imperfect proxy for such information (29), which is also supported by our observation that induction of CD166 protein and mRNA is inconsistent (Figure 1A and B). miRNAs are endogenous ∼22-nt non-coding RNAs, whose action is to specify translational repression without affecting the levels of the targeted mRNA (30–32). By luciferase and western-blotting assays, we confirmed that miR-9 can genuinely affect CD166 expression (Figure 5A–C and 7A). Inhibition of CD166 by miR-9 was also verified when cell migration assays were utilized (Figure 7B–D). Moreover, we detected that miR-9 was not up-regulated until 24 h after induction (Figure 5D), which may explain why CD166 protein levels decreased at 48 h compared to that of 24 h (Figure 1B).
Using BAY 11-7082, which decreases NF-κB activation (33), we found both CD166 mRNA (Figure 1D) and miR-9 (Figure 5G) were greatly inhibited. Also, both endogenous expression levels and promoter activities were associated with the amounts of P50 and P65 (Figure 4A, C and D and 6D and E). Furthermore, both in vitro and in vivo experiments revealed P50/P65 hetero-dimer is recruited to the promoters of both CD166 and miR-9-1 genes once serum is deprived (Figures 3 and 6). Consistent with previous report that miR-9 could be induced by LPS and TNFα (34), we also verified the same effects by these two typical NF-κB inducers in hepatoma cells (Figure 6F). In contrast, LPS and TNFα only upregulated endogenous CD166 expression after serum withdrawal (Figure 4E). Further results, in which CD166 expression was correlated with NF-κB-binding activity at its promoter (Figure 1A and 3E), indicate that the concentration of activated NF-κB must exceed a critical threshold before transcription of CD166 initiates. We propose here that the amounts of activated NF-κB induced by LPS or TNFα fail to reach this threshold; however, synergistic effects occur by additional activated NF-κB once transcription of CD166 is fully initiated by SD. Additionally, some other proteins may also aid NF-κB mediated transactivation of endogenous CD166. In this study, we observed a great decreasing activity in promoter without −880 to −768 (which contains a κB motif) both before and after SD (Figure 2C). Although the sequence from −880 to −465 has an obvious promoter activity, no inducible effects were detected after induction (Figure 2E). Such results may due to the loss of the region from −143 to −124, which is also responsive to SD (Figure 2C), albeit no valuable information has been obtained from online prediction software, we hypothesize this region may interact with other proteins, which assists recruitment of P50/P65 to the CD166 promoter.
Very recently, King et al. (35) reported another distinct functional κB motif (−643/−634 relative to our TSS, which is 156-bp upstream from the one identified by King et al.) on the endogenous CD166 promoter, which can be induced by overexpression of P65 in melanoma cells lines. However, there seems to be no functional role of this site in hepatoma cells, because the little impacts on promoter activity after deletion the region compassing from −649 to −607 (Figure 2C). Although the presence of κB motif in pri-miR-9-1 promoter has been predicted by bioinformatics previously (36), we confirmed this possibility experimentally for the first time. Thereby, in the present study, we have revealed another two novel NF-κB target genes in hepatoma cells. NF-κB is involved in stimulating cell proliferation, inhibiting apoptosis, and promoting metastasis and angiogenesis (37). More than 180 NF-κB target genes have been proposed and many of them, such as BclII, cyclin D1, matrix metalloproteinase-9 and vascular endothelial growth factor, regulate critical steps of cancer progression (37). Therefore, it is important to point out that CD166 and miR-9 may not be the only downstream targets of NF-κΒ that contribute to development of cancer. Further study is needed to elucidate the genes and signaling pathways that are critical mechanisms of NF-κΒ activity that participate in liver cancer development and progression.
One unexpected finding was that unlike other two cell lines, QGY-7701 and Bel-7402, upregulation of CD166 mRNA following serum starvation could be greatly reduced after treatment of CHX in HepG2 cells (Figure 1D), suggesting dependence of translation. Here, we illustrate that both mRNA and protein levels of P65 can be simultaneously induced in HepG2 cells (Supplementary Figure S2A and B), inhibition of translation, and impaired protein expression of P65, which results in blocking CD166 gene activation. By comparison, no alteration of P65 was detected in QGY-7701 and Bel-7402 cells, which may explain why CHX had no such effect (Figure 1D). Translocation the most highly increased P65 concentration into the nucleus (Supplementary Figure S2B) results in the strongest upregulation of CD166 in HepG2 cells compared to other two cell lines (Figure 1). The patterns of mRNA and protein expression of P65 were not synchronized because mRNA was not upregulated until induction for 24 h, however, the protein began to increase just 2 h after SD. In addition, both P65 mRNA and protein decreased after re-addition of FBS (Supplementary Figure S2D). Thereby, we assume that upregulation of P65 protein after SD may be due to two independent mechanisms; the first one may be transcriptional activation of P65 gene in a slow and moderate manner (however, this activation is unstable and may easily lose its function immediately after re-addition of serum accompanied by the prompt degradation of excess mRNA) and the other is at the post-transcriptional level. As previously reported, NF-κB function is regulated by ubiquitin-mediated proteolysis of its P65 subunit. Upon cytokine treatment, Pin1 binds to the pThr254-Pro motif in P65, resulting in protein stability of P65 and enhanced NF-κB activity (38), and phosphorylation of P65 prevents its degradation. Pim-1 may act as a further kinase responsible for the phosphorylation of P65 (39). Such events may occur in cells under SD, and miRNA may also play a potential role in regulation of P65, yet further studies are required to explore such mechanisms underlying the upregulation of P65 in HepG2 cells.
Some other proteins that are critical for activity of the endogenous CD166 promoter may not be necessary in the transient assay because of the high-plasmid copy number or the aberrant chromatin structure that assembles on transiently transfected plasmids (34,40). By chromatin accessibility assay, we observed the κB motif is in two linker regions, with greater accessibility to restriction enzymes (Mae III and Sma I) and DNase I after SD compare to that at basal level (Figure 3A–C), which is similar with studies described in other promoters (20,23,41). Changes in the accessibility of nucleosome-packaged DNA can be mediated by the SWI/SNF remodeling complex (41–43), but we do not believe the SWI/SNF complex works on the CD166 promoter after SD, because Brg I, a key factor of SWI/SNF does not recruit to sequences around the κB motif (Figure 3F). Moreover, knockdown assays showed that neither BRM nor Brg I has obvious negative effects on starvation-induced upregulation of CD166 expression (Figure 4C). Other mechanisms may involve chromatin remodeling around the κB motif after induction, as reported previously, sequential recruitment of P65, accompanying ordered elevation of the levels of histone H4 and H3 acetylation at the native TGF-beta1 promoter after IL-1beta stimulation (44). Furthermore, NF-κB increases the recruitment of HAT p300 and acetylated histones H3 and H4 of the FasL promoter, thus making more open chromatin accessibility and helping to expose the κB motif (45). P50/P65 binding to the CD166 promoter results in the activation of the promoter and a more open chromatin status, which may be dependent on histone modification, but further studies should be done to clarify the underlying mechanism.
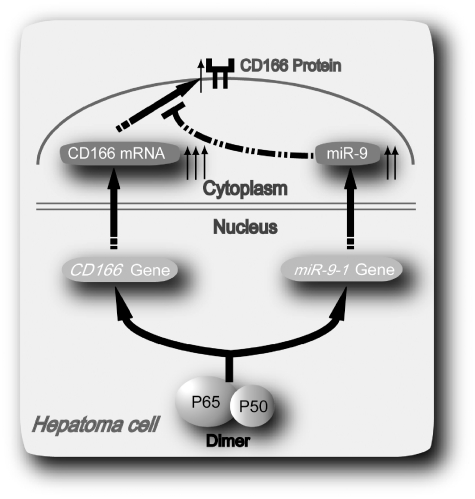
In conclusion, in this work, we demonstrate that a novel negative auto-regulatory loop in which P50/P65 mediates differential regulation of CD166 via interaction with miR-9 after SD in hepatoma cells (Figure 8). Induction of CD166 and miR-9 provides a means to smooth out the fluctuations in gene expression and fine tune the synthesis of protein, thus allowing the specific response to correctly proceed.
Figure 8.
Possible mechanism underlying NF-κB P50/P65 hetero-dimer mediates differential regulation of CD166 expression via interaction with miR-9 under SD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Sciences Foundation of China (grant No. 81071745). Funding for open access charge: Shanghai Ruijin Hospital.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank Prof. Xinmin Zheng for critical comments on the article, and Dr Zanlin Yu and Jennifer Cotton (Department of Cancer Biology, University of Massachusetts Medical School) for carefully editing the language and grammar of this article.
REFERENCES
- 1.Degen WG, van Kempen LC, Gijzen EG, van Groningen JJ, van Kooyk Y, Bloemers HP, Swart GW. MEMD, a new cell adhesion molecule in metastasizing human melanoma cell lines, is identical to ALCAM (activated leukocyte cell adhesion molecule) Am. J. Pathol. 1998;152:805–813. [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen MA, Patel DD, Li X, Modrell B, Malacko AR, Wang WC, Marquardt H, Neubauer M, Pesando JM, Francke U, et al. Cloning, mapping and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J. Exp. Med. 1995;18:2213–2220. doi: 10.1084/jem.181.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohneda O, Ohneda K, Arai F, Lee J, Miyamoto T, Fukushima Y, Dowbenko D, Lasky LA, Suda T. ALCAM (CD166): its role in hematopoetic and endothelial development. Blood. 2001;98:2134–2142. doi: 10.1182/blood.v98.7.2134. [DOI] [PubMed] [Google Scholar]
- 4.Arai F, Ohneda O, Miyamoto T, Zhang XQ, Suda T. Mesenchymal stem wells in perichondrium express Activated Leukocyte Cell Adhesion Molecule and participate in bone marrow formation. J. Exp. Med. 2002;195:1549–1563. doi: 10.1084/jem.20011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Kempen LCLT, Nelissen JMDT, Degen WGJ, Torensma R, Weidle UH, Bloemers HPJ, Figdor CG, Swart GWM. Molecular basis for the homophilic Activated Leukocyte Cell Adhesion Molecule (ALCAM)-ALCAM interaction. J. Biol. Chem. 2001;276:25783–25790. doi: 10.1074/jbc.M011272200. [DOI] [PubMed] [Google Scholar]
- 6.van Kempen LCLT, van den Oord JJ, van Muijen GNP, Weidle UH, Bloemers HPJ, Swart GWM. Activated Leukocyte Cell Adhesion Molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. Am. J. Pathol. 2000;156:769–774. doi: 10.1016/S0002-9440(10)64943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kempen LC, Meier F, Egeblad M, Kersten-Niessen MJF, Garbe C, Weidle UH, van Muijen GNP, Herlyn M, Bloemers HPJ, Swart GWM. Truncation of activated leukocyte cell adhesion molecule: a gateway to melanoma metastasis. J. Invest. Dermatol. 2004;122:1293–1301. doi: 10.1111/j.0022-202X.2004.22531.x. [DOI] [PubMed] [Google Scholar]
- 8.Kristiansen G, Pilarsky C, Wissmann C, Stephan C, Weissbach L, Loy V, Loening S, Dietel M, Rosenthal A. ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate. 2003;54:34–43. doi: 10.1002/pros.10161. [DOI] [PubMed] [Google Scholar]
- 9.King JA, Ofori-Acquah S, Stevens T, Al-Mehdi AB, Fodstad O, Jiang WG. Activated leukocyte cell adhesion molecule in breast cancer: prognostic indicator. Breast Cancer Res. 2004;6:78–87. doi: 10.1186/bcr815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda K, Quertermous T. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J. Biol. Chem. 2004;279:55315–55323. doi: 10.1074/jbc.M407776200. [DOI] [PubMed] [Google Scholar]
- 11.Jezierska A, Matysiak W, Motyl T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Med. Sci. Monit. 2006;12:BR263–BR273. [PubMed] [Google Scholar]
- 12.Scott FL, Deanault JB, Riedl SJ, Shinb H, Renatusc M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved echanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bursh W. The autophagosomal-lysosomal compartment in programmed cell death. Cell. Death. Differ. 2001;8:569–581. doi: 10.1038/sj.cdd.4400852. [DOI] [PubMed] [Google Scholar]
- 14.Zhu W, Chen J, Cong X, Hu S, Chen X. Hypoxia and serum deprivation-induced apoptosis in mesenchymal stem cells. Stem Cell. 2006;24:416–425. doi: 10.1634/stemcells.2005-0121. [DOI] [PubMed] [Google Scholar]
- 15.Wei J, Sun Z, Chen Q, Gu J. Serum deprivation induced apoptosis in macrophage is mediated by autocrine secretion of type I IFNs. Apoptosis. 2006;11:545–554. doi: 10.1007/s10495-006-5146-7. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Liu X, Ni P, Gu Z, Fan Q. SP1 is required for basal activation and chromatin accessibility of CD151 promoter in liver cancer cells. Biochem. Biophys. Res. Commun. 2010;393:291–296. doi: 10.1016/j.bbrc.2010.01.127. [DOI] [PubMed] [Google Scholar]
- 19.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue X, Sun J, Zhang Q, Wang Z, Huang Y, Pan W. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS ONE. 2008;3:e4034. doi: 10.1371/journal.pone.0004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 23.Sun FY, Chen QY, Yang SH, Pan QH, Ma J, Wan Y, Chang CH, Hong A. Remodeling of chromatin structure within the promoter is important for bmp-2-induced fgfr3 expression. Nucleic Acids Res. 2009;37:3897–3391. doi: 10.1093/nar/gkp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 25.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl Acad. Sci. USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simeonova PP, Luster MI. Asbestos induction of nuclear transcription factors and interleukin 8 gene regulation. Am. J. Respir. Cell. Mol. Biol. 1996;15:787–795. doi: 10.1165/ajrcmb.15.6.8969274. [DOI] [PubMed] [Google Scholar]
- 27.Fraser DA, Arora M, Bohlson SS, Lozano E, Tenner AJ. Generation of inhibitory NFkappaB complexes and phosphorylated cAMP response element-binding protein correlates with the anti-inflammatory activity of complement protein C1q in human monocytes. J. Biol. Chem. 2007;282:7360–7367. doi: 10.1074/jbc.M605741200. [DOI] [PubMed] [Google Scholar]
- 28.Masedunskasa A, King JA, Tana F, Cochrana R, Stevens T, Sviridove D, Ofori-Acquaha SF. Activated leukocyte cell adhesion molecule is a component of the endothelial junction involved in transendothelial monocyte migration. FEBS Lett. 2006;580:2637–2645. doi: 10.1016/j.febslet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 29.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 30.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 31.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA–Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao J, Luo XB, Lin HX, Zhang Y, Lu Y, Wang N, Zhang YQ, Yang BF, Wang ZG. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J. Biol. Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 33.Mori N, Yamada Y, Ikeda S, Yamasaki Y, Tsukasaki K, Tanaka Y, Tomonaga M, Yamamoto N, Fujii M. Bay 11-7082 inhibits transcription factor NF-κB and induces apoptosis of HTLV-I-infected T-cell lines and primary adult T-cell leukemia cells. Blood. 2002;100:1828–1834. doi: 10.1182/blood-2002-01-0151. [DOI] [PubMed] [Google Scholar]
- 34.Smith CL, Hager GL. Transcriptional regulation of mammalian genes in vivo. J. Biol. Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 35.King JA, Tan F, Mbeunkui F, Chambers Z, Cantrell S, Chen H, Alvarez D, Shevde LA, Ofori-Acquah SF. Mechanisms of transcriptional regulation and prognostic significance of activated leukocyte cell adhesion molecule in cancer. Mol. Cancer. 2010;9:266. doi: 10.1186/1476-4598-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bazzonia F, Rossatoa M, Fabbrib M, Gaudiosib D, Mirolob M, Morib L, Tamassiaa N, Mantovanib A, Cassatellaa MA, Locatib M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl Acad. Sci. USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 38.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottape R, Yamaoka S, Lu KP. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 39.Nihira K, Ando Y, Yamaguchi T, Kagami Y, Miki Y, Yoshida K. Pim-1 controls NF-κB signalling by stabilizing RelA/p65. Cell. Death Differ. 2010;17:689–698. doi: 10.1038/cdd.2009.174. [DOI] [PubMed] [Google Scholar]
- 40.Brightbill HD, Plevy SE, Modlin RL, Smale ST. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J. Immunol. 2000;164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 41.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 42.Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL. Promoter targeting of chromatin-modifying complexes. Front Biosci. 2001;6:D1054–D1064. doi: 10.2741/hassan. [DOI] [PubMed] [Google Scholar]
- 43.Coisy-Quivy M, Disson O, Roure V, Muchardt C, Blanchard JM, Dantonel JC. Role for Brm in cell growth control. Cancer Res. 2006;66:5069–5076. doi: 10.1158/0008-5472.CAN-05-0596. [DOI] [PubMed] [Google Scholar]
- 44.Lee KY, Ito K, Hayashi R, Jazrawi EP, Barnes PJ, Adcock IM. NF-kappaB and activator protein 1 response elements and the role of histone modifications in IL-1beta-induced TGF-beta1 gene transcription. J. Immunol. 2006;176:603–615. doi: 10.4049/jimmunol.176.1.603. [DOI] [PubMed] [Google Scholar]
- 45.Aurora AB, Biyashev D, Mirochnik Y, Zaichuk TA, Sánchez-Martinez C, Renault MA, Losordo D, Volpert OV. NF-kappaB balances vascular regression and angiogenesis via chromatin remodeling and NFAT displacement. Blood. 2010;116:475–484. doi: 10.1182/blood-2009-07-232132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.