Abstract
Nucleosomes, the fundamental units of chromatin structure, are regulators and barriers to transcription, replication and repair. Post-translational modifications (PTMs) of the histone proteins within nucleosomes regulate these DNA processes. Histone H3(T118) is a site of phosphorylation [H3(T118ph)] and is implicated in regulation of transcription and DNA repair. We prepared H3(T118ph) by expressed protein ligation and determined its influence on nucleosome dynamics. We find H3(T118ph) reduces DNA–histone binding by 2 kcal/mol, increases nucleosome mobility by 28-fold and increases DNA accessibility near the dyad region by 6-fold. Moreover, H3(T118ph) increases the rate of hMSH2–hMSH6 nucleosome disassembly and enables nucleosome disassembly by the SWI/SNF chromatin remodeler. These studies suggest that H3(T118ph) directly enhances and may reprogram chromatin remodeling reactions.
INTRODUCTION
Histone post-translational modifications (PTMs) play a central role in regulating RNA transcription (1), DNA replication (2) and DNA repair (3). Histone PTMs appear to regulate DNA processing through distinct mechanisms. Histone PTMs, particularly on the disordered histone tails, may indirectly impact nucleosomes by providing binding modules for accessory complexes (4), which in turn alter chromatin structure and function. Histone PTMs may also function to directly influence nucleosome structure and dynamics (5). For example, acetylation of lysine 16 on the H4 histone tail directly reduces higher order chromatin compaction (6), while lysine acetylation in the DNA–histone interface impacts DNA unwrapping (7,8) and nucleosome mobility (9).
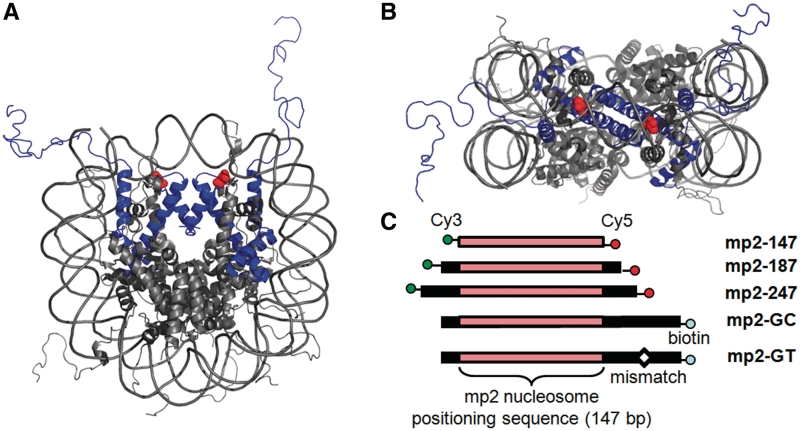
A number of histone phosphorylation sites lie within the DNA–histone interface (10–12). While the impact of histone acetylation and acetylation mimics within the DNA–histone interface is an active area of study, the impact of histone phosphorylation within this nucleosome region remains unexplored. Among the putative histone phosphorylation sites, H3(T118) stands out. The side chain hydroxyl is located within 3Å of the DNA phosphate backbone at the nucleosome dyad (Figure 1A and B). Mutations H3(T118E) and H3(T118A), which are used to mimic constitutively phosphoylated and de-phosphorylated states, respectively, are lethal in haploid yeast. Heteroallelic expression of these mutations with ‘wild-type’ H3 leads to defects in transcriptional regulation and DNA repair (13). These results suggest that the H3(T118) residue, and its apposite modification, is essential for appropriate chromatin metabolism (14).
Figure 1.
Location of H3(T118ph) within the nucleosome structure. (A) and (B) The side and top view of the nucleosome structure (41) with histone H3 in blue and H3(T118) in red. (C) The DNA constructs contain the 147 bp NPS mp2 (25) with no additional DNA (mp2-147), with 20 bp of DNA flanking each side (mp2-187) or with 50 bp flanking each side (mp2-247). Each of these DNA constructs were 5′-labeled with cy3 and cy5 on the left and right ends, respectively. The DNA constructs: mp2-GT and mp2-GC contain 15 bp flanking the left side of the mp2 NPS, 70 bp flanking the right side of the mp2 NPS and a biotin attached to the 5′-end. mp2-GT contains a GT mismatch 35 bp to the right of the mp2 NPS.
H3(T118) was also found to influence the requirement for SWI/SNF chromatin remodeling (15). A Saccharomyces cerevisiae genetic screen identified H3(T118I) as a SWI/SNF independent (Sin) substitution mutation that returned expression from the HO mating-type recombination locus to near wild-type levels in a mutant strain defective in SWI/SNF remodeling (15). A number of studies subsequently showed that nucleosomes containing H3(T118I) exhibit increased mobility (16,17) and accessibility (18). Furthermore, the Sin mutant H4(R45H), which is located near H3(T118) in the nucleosome structure, decreases chromatin higher order structure (19). This suggests alterations at H3(T118) may also impact higher order structure. Taken as a whole, these results suggest that changes at H3(T118) alter nucleosome sliding and chromatin compaction in ways that can compensate for the requirement of SWI/SNF chromatin remodeling.
Here we have examined the influence of H3(T118) phosphorylation [H3(T118ph)] on nucleosome dynamics and remodeling in vitro. We prepared H3(T118ph) by expressed protein ligation (EPL; 19) and found that H3(T118ph) dramatically decreases DNA–octamer binding, increases nucleosome sliding and increases DNA accessibility near the nucleosome dyad. Recently, we reported that the DNA mismatch recognition complex, hMSH2–hMSH6, disassembles nucleosomes near a mismatch (21). We find that H3(T118ph) enhances hMSH2–hMSH6 induced nucleosome disassembly by 25-fold. Moreover, H3(T118ph) enables nucleosome disassembly by the SWI/SNF chromatin remodeler. Our results suggest that H3(T118ph) may function to destabilize nucleosomes in vivo by regulating their mobility, disassembly and remodeling.
MATERIALS AND METHODS
Nucleosome preparation
Nucleosomes were prepared by salt dialysis with purified histone octamer and DNA, and purified by sucrose gradient centrifugation (9). The addition of 0.5 mM MgCl2 was required to maintain stability during the purification of H3(T118ph) nucleosomes. H3(T118ph) was prepared by EPL as previously described (9). The peptide synthesized with phosphothreonine was prepared using standard Fmoc protocols; C110 was introduced as thiazolidine. Wild-type histones and H3(T118E) were prepared by recombinant expression in Escherichia coli and purified as previously described (22). The DNA constructs, mp2-147, mp2-187, mp2-247, mp2-GC and mp2-GT were prepared and purified as previously described (9,21).
Nucleosome dynamics analyses
Competitive reconstitutions were performed as previously described (9) at 12, 25 and 33°C. Thermal shifting experiments were done with 50 nM nucleosomes in 20 mM Tris (pH 8.0), 0.5 mM MgCl2 at 53 and 37°C as previously described (9). Electrophoretic mobility shift assays (EMSAs) were used to determine the fraction of each nucleosome position within either mp2-187 and mp2-247 DNAs over 90 min. We used EMSA conditions (16), such that the nucleosome gel mobility increases as nucleosomes reposition from the central to the end positions. In addition, we confirmed nucleosome positions with ExoIII mapping (Supplementary Figures S3–S6) as previously described (9).
Restriction enzyme (RE) accessibility studies were carried out at 37°C (HindIII, HaeIII, HhaI, PmlI) or 65°C (TaqαI) with 1 nM nucleosomes in the recommended RE buffer (New England Biolabs) as previously described (9). Briefly, this method determines the nucleosomal DNA site accessibility equilibrium, Keq, which is defined as the equilibrium between nucleosome states that expose the DNA site for RE binding and nucleosome states that protect the DNA site from RE binding. We carry out RE digestions in the rapid pre-equilibrium regime such that the observed rate of DNA cleavage is proportional to Keq. The digestions were quantified by PAGE and analyzed using Image Quant (Molecular Dynamics) software. The background for each band of interest was subtracted using the local pixel median of a box enclosing the band. The digestions were fit to a single exponential to determine the initial digestion rate. An initial drop in undigested DNA between the 0 and 1 min time point is due to (i) a small fraction of DNA that copurifies with the mononucleosomes (Supplementary Figure S1C) and (ii) a fraction of nucleosomes that dissociates during the rapid mixing. We therefore neglect this initial time point as has been done in previous studies (23,24). The fraction of nucleosomal DNA cleaved was limited over the 30-min digestion, as is typical for RE studies of nucleosome site accessibility, because the absolute Keq is between 10−3 and 10−6 (23). Since nucleosome dynamics measurements were carried out at different temperatures (Supplementary Table S1), results from experiments done at different temperatures are only qualitatively compared.
DNase I footprinting
DNase I sensitive sites within the nucleosome were determined by mild DNase I digestion. Reactions were carried out in an initial volume of 50 µl with 10 nM nucleosomes and 22 U/ml DNase I (Invitrogen) 20 mM Tris pH 8, 0.5 mM MgCl2 at 16°C to prevent H3(T118ph) nucleosome disassembly in the low salt buffer required for DNase I digestion (data not shown). Each time point was acquired by quenching the reaction with a final concentration of 5 mM EDTA, 1 mg/ml of proteinase K and 0.02% SDS, and then analyzed by denaturing PAGE as described for ExoIII mapping (See Methods in Supplementary Data).
hMSH2–hMSH6 nucleosome remodeling assay
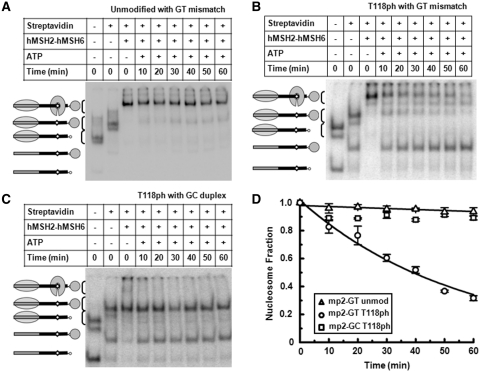
Nucleosome disassembly reactions were carried out at 37°C as previously described (21) with 0.25 nM of H3(T118ph) nucleosomes reconstituted with the mp2-GT nucleosome positioning sequence (NPS) and 5 nM of unlabled unmodified nucleosomes reconstituted with the mp2-147 DNA molecule. Kinetic analysis was performed by staggering the time ATP was added. The fraction of disassembled nucleosome was analyzed by gel shifts on polyacrylamide gels as previously described (21). A small fraction of free DNA appears in initial time points because (i) a small amount of naked DNA co-purifies with nucleosomes and (ii) a fraction of nucleosomes falls apart during the rapid mixing of the nucleosomes. To control for this, we do not include the zero time point in the exponential decay fit. The GC duplex control experiments show no change in the fraction of H3(T118ph) containing nucleosomes from the second to the last time point (Figure 7C). This demonstrates that nucleosome disassembly is dependent on hMSH2–hMSH6 as we previously reported (21).
Figure 7.
H3(T118ph) facilitates nucleosome disassembly by hMSH2–hMSH6. (A–C) EMSA of unmodified nucleosomes adjacent to a GT mismatch, H3(T118ph) nucleosomes adjacent to a GT mismatch and H3(T118ph) nucleosomes without a DNA mismatch disassembled by hMSH2–hMSH6, respectively. Lane 1: sucrose gradient purified H3(T118ph)-containing nucleosomes, Lane 2: H3(T118ph) nucleosomes bound by streptavidin, Lane 3: H3(T118ph) nucleosomes bound by streptavidin and MSH2–MSH6, Lanes 4–9: kinetic analysis of streptavidin-bound H3(T118ph) nucleosome disassembly by hMSH2–hMSH6 in the presence of 1 mM ATP. (D) The fraction of unmodified mp2-GT NPS nucleosomes (triangle), H3(T118ph) mp2-GT NPS nucleosomes (circles) and H3(T118ph) mp2-GC NPS nucleosomes (squares) verses time in the presence of hMSH2–hMSH6 (250 nM) and ATP (1 mM). The error bars were determined from the standard deviation of at least three separate experiments. The fraction of nucleosomes versus time were fit excluding the zero time point to a single exponential decay, Ae-t/τ with τGT•H3(T118ph) = 55 ± 4 min and τGT•H3 = 1400 ± 400 min. We interpret the negative decay time of H3(T118ph) mp2-GC NPS nucleosomes to imply that no nucleosomes were disassembled within the uncertainty of the measurement.
SWI/SNF nucleosome binding and remodeling assays
SWI/SNF binding reactions were performed in 7 µl with 28 nM of unmodified H3(T118ph) nucleosomes with mp2-187 and 3.5–56 nM of SWI/SNF at 30°C for 30 min. Half of the binding reactions were examined by 4% (79:1 acrylamide:bis-acrylamide) native PAGE in 0.5× TBE at 200 V for 4 h in 4°C. The remaining half was cooled down to 18°C and ATP was added to 800 µM final concentration. The remodeling reactions were incubated at 18°C to slow the reaction such that it could be monitored over 30 min and stopped with 1 µl of a 1:1 mix of 10 mg/ml salmon sperm DNA and 10 mM γ-thio-ATP as previously described (25). For gel shift analysis of the remodeling reactions, 4 µl samples were loaded on a 4% (35:1 acrylamide: bis-acrylamide) native PAGE in 0.5× TBE at 200 V for 4 h in 4°C.
RESULTS
Constructing defined nucleosomes containing H3(T118ph)
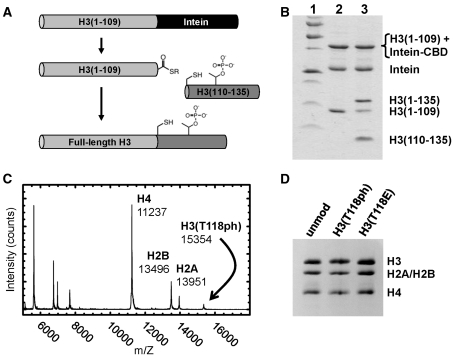
We examined the consequences of H3(T118ph) (Figure 1) by semisynthetic construction of this modified histone using EPL (9). We introduced a single phosphothreonine by ligation of a synthetic modified peptide to an unmodified recombinant protein thioester. We used the native cysteine at residue 110 as a ligation site to generate the native protein sequence and then refolded the H3(T118ph) protein with recombinant H2A, H2B and H4 into histone octamers (Figure 2). These purified octamers were used for nucleosome reconstitutions (Supplementary Figure S1) with DNA molecules containing a NPS (Figure 1C) (26). Mononucleosomes were then purified by sucrose gradient centrifugation (Supplementary Figure S1).
Figure 2.
Preparation of histone octamer containing H3(T118ph). (A) Scheme for the synthesis of H3(T118ph). A fusion of truncated H3(1-109) with Intein-CBD was expressed as a recombinant protein and then refolded by dialysis into high salt buffer. Thiolysis was initiated with the addition of 100 mM MESNA, which generated H3(1-109) with a thioester terminus. This was then ligated to a peptide containing the phosphothreonine in 6 M urea, 1 M NaCl, 50 mM HEPES (pH 7.5), 1 mM EDTA, 20 mM tris(2-carboxyethyl)phosphine and the product purified by ion-exchange chromatography. (B) SDS–PAGE analysis of the thiolysis and ligation. Lane 1: molecular weight standard, Lane 2: the generation of H3(1-109) following thiolysis and Lane 3: the generation of H3(T118ph) by ligation of H3(110-135) to H3(1-109). (C) MALDI-TOF MS analysis of purified histone octamer containing H3(T118ph). (D) SDS–PAGE analysis of histone octamers containing unmodified H3, H3(T118ph) and H3(T118E).
H3(T118ph) reduces DNA–histone binding free energy
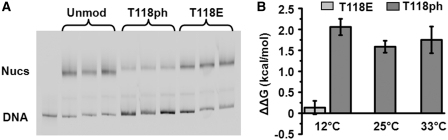
The influence of H3(T118ph) on the free energy for nucleosome formation, a measure of nucleosome stability, was examined by competitive reconstitutions (9,27). H3(T118ph) reduced the nucleosome formation free energy by 2.1 ± 0.2, 1.6 ± 0.2 and 1.7 ± 0.3 kcal/mol at 12, 25 and 33°C, respectively (Figure 3). These results indicate that the probability for nucleosome formation is reduced by ∼16-fold at physiological temperatures. In contrast, the H3(T118E) amino acid substitution, which adds a negative charge and is often used to mimic phosphothreonine (14), has no influence on the nucleosome formation free energy (Figure 3). Together these findings suggest that the precise moiety introduced by the phosphorylation of H3(T118) is more important than simply the presence of negative charge for the reduction in the DNA–histone binding free energy.
Figure 3.
H3(T118ph) reduces the free energy of nucleosome formation. (A) EMSA of competitive reconstitutions with unmodified, H3(T118ph) and H3(T118E) histone octamers with the mp2-147 NPS in triplicate. The ratio of the nucleosome band to the DNA band is used to determine the equilibrium constant, Keq, of each octamer, Keq. (B) The difference in nucleosome formation free energy, ΔΔG, between H3(T118ph) or H3(T118E) containing histone octamer and unmodified octamer at 12, 25 and 33°C. The change in the nucleosome formation free energy was determined from ΔΔG = kBT ln(Keq), where kB is the Boltzmann constant and T = 285, 298 and 306K.
H3(T118ph) dramatically enhances nucleosome mobility
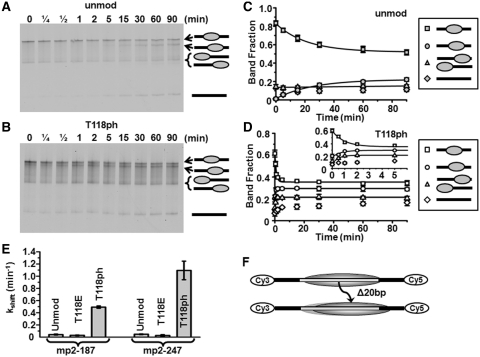
The reduction in nucleosome formation free energy could result in increased nucleosome mobility as has been reported for histone Sin mutants including H3(T118I) (16,17), and nucleosomes acetylated at the dyad axis (9). We examined the effect of H3(T118ph) on nucleosome mobility by heating nucleosomes to 53°C and then measuring the kinetic evolution of nucleosome positions by electrophoretic mobility shift analysis (EMSA) (16) within the mp2-187 and the mp2-247 DNAs (NPS; Figures 1C, 4 and Supplementary Figure S2). We found that centrally positioned unmodified nucleosomes repositioned within mp2-247 with a characteristic decay time (τH3 = 22 ± 3 min; Figure 4A, C and E), while centrally positioned H3(T118ph) containing nucleosomes repositioned significantly faster (τH3(T118ph) = 0.8 ± 0.2 min; Figure 4B, D and E). In contrast, nucleosomes containing H3(T118E) repositioned slower than unmodified nucleosomes (τH3(T118E) = 34 ± 6 min; Figure 4E). We confirmed that the nucleosomes retained their 147 bp footprint with exonuclease III mapping before and after repositioning for each of the histone octamers studied (Supplementary Figures S3–S6). We also investigated the influence of H3(T118ph) on nucleosome mobility at the physiological temperature of 37°C. We find that H3(T118ph) allows for 20 bp repositioning with a decay of τH3(T118ph),37C = 40 ± 10 min, while unmodified nucleosome repositioning was undetectable (Supplementary Figure S7). These results indicate that the histone octamer within a repositioned nucleosome remains intact and that the reduction in nucleosome formation free energy increases nucleosome mobility by 28 ± 8 times. The increase in nucleosome mobility appears to facilitate thermally induced repositioning at physiological temperatures.
Figure 4.
H3(T118ph) increases the rate of nucleosome mobility. (A and B) Cy5 fluorescence images of thermally (53°C) induced repositioning visualized by EMSA of unmodified and H3(T118ph) mp2-247 NPS nucleosomes, respectively. The top band is centrally positioned nucleosomes, the second band is nucleosomes shifted toward the Cy5 end from the center by 20 bp, the third band is nucleosomes positioned at the Cy3 or Cy5 end of the DNA and the bottom band is naked DNA (diagramed on right, See Supplementary Figures S3 and S4 for mapping of nucleosome positions). (C and D) Quantification of the fraction of center positioned nucleosomes (squares), 20 bp shifted nucleosomes (circles), end positioned nucleosomes (triangles) and naked DNA (diamonds). The error bars were determined from the standard deviation of each time point from at least three separate experiments. The kinetic evolution of each nucleosome position was fit to a single exponential decay. The inset shows the first 6 min of plot (D). (E) Summary of the characteristic decay times for repositioning of the central nucleosome position within the mp2-187 NPS (Supplementary Figure S3) and mp2-247 NPS. (F) Diagram of the change in nucleosome positions at 53°C.
H3(T118ph) increases DNA accessibility near the nucleosome dyad
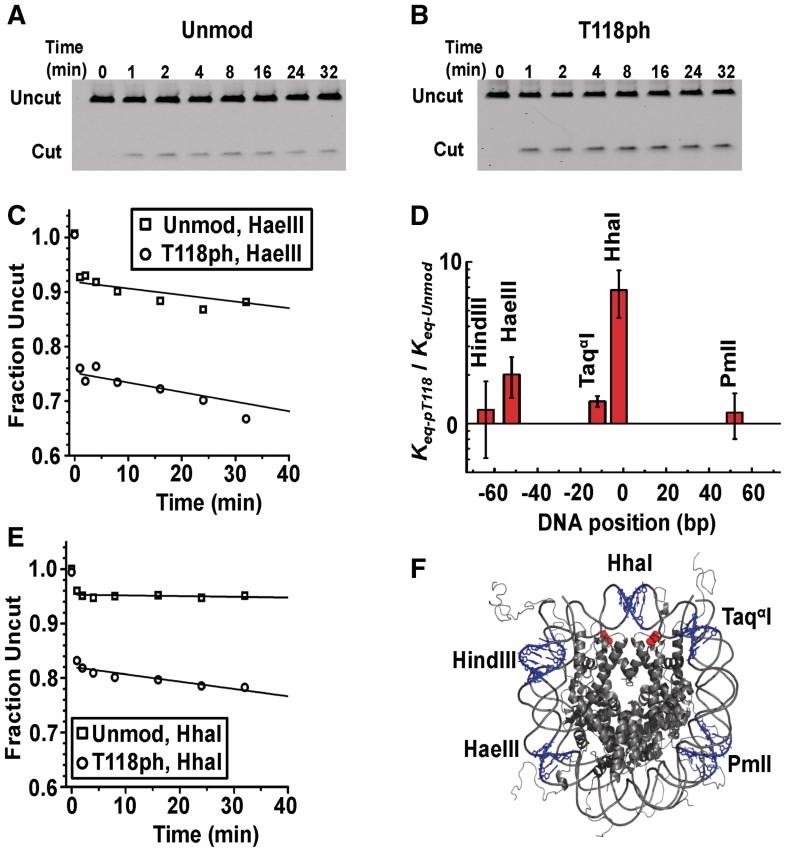
To determine whether the H3(T118ph) influences nucleosomal DNA accessiblity, we quantified the DNA site exposure equilibrium of nucleosomes containing H3(T118ph) relative to unmodified nucleosomes with RE digestions (24,28). We define the DNA site exposure equilibrium, Keq, as the equilibrium between nucleosome states that are accessible for RE binding and nucleosome states that are inaccessible to RE binding. Four of the five restriction sites displayed no significant increase in site exposure equilibrium in nucleosomes containing H3(T118ph). However, the HhaI site near the dyad symmetry axis of the nucleosome displayed a 6-fold increase in site exposure (Figure 5). This suggests that H3(T118ph) increases DNA site accessibility only near the dyad symmetry axis.
Figure 5.
H3(T118ph) increases DNA accessibility near the dyad symmetry axis. (A and B) PAGE analysis of unmodified and H3(T118ph) nucleosomes, respectively, digested with HhaI. The lanes are labeled with the digestion quench time in minutes. (C) and (D) Plots of the fraction of HaeIII and HhaI undigested DNA, respectively. Digestions of unmodified (squares) and H3(T118ph) (circles) nucleosomes were fit with a single exponential decay. (E) Plot of the site accessibility of H3(T118ph) nucleosomes, Keq-pT118 relative to the site accessibility of unmodified nucleosomes, Keq-unmod, for five separate RE sites. The error bars were determined from the standard deviation of at least three separate experiments. (F) The nucleosomes crystal structure (40) with the five RE sites is shown in blue and the two H3(T118) residues are shown in red.
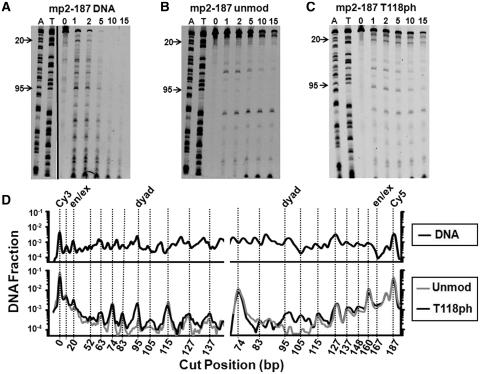
We also investigated the influence of H3(T118ph) on DNA accessibility with DNase I footprinting. After analyzing DNase I digestion of nucleosomes reconstituted with mp2-187 positioning DNA by denaturing PAGE (Supplementary Figure S8), we determined the line trace of the 2 min lane for both the Cy5 (Figure 6A, B and C) and Cy3 images, representing detection of both the top (Cy3) and bottom (Cy5) strands of mp2-187. We find that nucleosomes with and without H3(T118ph) protect the central 147 bp of mp2-187 from DNase I cleavage and display the periodic 10 bp cleavage pattern that is typical of nucleosome footprinting by DNase I.
Figure 6.
H3(T118ph) increases DNase I accessibility near the dyad symmetry axis. (A) Cy5 fluorescence image of a denaturing PAGE analysis of a DNase I digestion with naked mp2-187 DNA. The first two lanes are A and T ladders of the mp2-187 DNA molecule. Lanes 3 through 8 are labeled with the time in min. (B) Cy5 fluorescence image of a denaturing PAGE analysis of a DNase digestion with unmodified nucleosomes containing the mp2-187 DNA molecule. The labels 20 and 95 denote cleavage at the 20th and 95th bp positions within the mp2-187 DNA molecule, which are located in the nucleosome DNA entry–exit and dyad regions, respectively. (C) Cy5 image of a denaturing PAGE analysis of a DNase I digestion with H3(T118ph) containing nucleosomes. (D) Line trace of the 2 min DNase I digestion lane from images A–C. The location of a peak in each trace corresponding to a DNase I cut position is calibrated using the A and T ladders. Each trace is background corrected and normalized by the intensity of the uncut DNA in the 0 min lane. The decrease in digestion of unmod and T118ph nucleosomes with respect to naked DNA from 167–140 bp is due to the ∼147 bp footprint of the nucleosome. Both unmodified and H3(T118ph) containing nucleosomes exhibit a similar 10 n phasing in digestion intensity except at the dyad (95 bp) where H3(T118ph) containing nucleosomes are ∼10 times more sensitive than unmodified nucleosome to DNase I cleavage.
The DNase I cleavage is enhanced near the nucleosome dyad at the 95th bp of both the top (Cy3) and bottom (Cy5) DNA strands of mp2-187 for H3(T118ph) containing nucleosomes relative to unmodified nucleosomes (Figure 6D). This result independently confirms the RE measurement that DNA accessibility is increased near the dyad symmetry axis region by H3(T118ph). The region near the dyad symmetry axis typically contains the least accessible nucleosomal DNA with a site exposure equilibrium constant of ∼10−5 (23,24). While the 6-fold increase in the site accessibility by H3(T188ph) is significant, the overall site accessibility equilibrium constant remains much less than naked DNA. The enhanced accessibility could be due to alterations in DNA unwrapping where rare unwrapping fluctuations that extend close to the nucleosome dyad more often continue to unwrap past the dyad when H3(T118) is phosphorylated. Alternatively, H3(T118ph) could increase the probability that a DNA bulge forms near the dyad (29) or that the DNA near the dyad slips off the side of the histone octamer. Each of these models would only increase accessibility near the dyad region without increasing site accessibility closer to the DNA entry–exit region.
H3(T118ph) also enhanced DNase I cleavage in the DNA entry–exit region at the 20th bp of mp2-187 relative to unmodified nucleosomes (Figure 6). However, the DNase I cleavage site near the opposite entry–exit region at the 160th bp of mp2-187, is not altered by H3(T118ph) (Figures 6D and S8). This is consistent with our results from Exo III nucleosome mapping (Supplementary Figures S5 and S6), which indicate that a fraction of nucleosomes is positioned off the mp2 sequence toward the Cy5 labeled end of mp2-187.
Nucleosome disassembly by hMSH2–hMSH6 is enhanced by H3(T118ph)
H3(T118ph) appears to be important for transcriptional regulation, resistance to DNA damage and replication fidelity (13). Recent studies from our group have demonstrated that the DNA mismatch repair recognition complex hMSH2–hMSH6 disassembles nucleosomes by passively unwrapping and/or repositioning nucleosomes (21). This activity is enhanced 5-fold by acetylation of histone H3 near the nucleosome dyad at lysine 115 and 122 [H3(K115ac,K122ac)] (21). H3(T118ph) is located between these two acetylation PTMs in the dyad region, which suggested that phosphorylation might also influence hMSH2–hMSH6 induced nucleosome disassembly.
We initially examined the influence of H3(T118ph) on hMSH2–hMSH6 nucleosome disassembly using the 5S rDNA NPS for comparison to acetylated nucleosomes (21) and found the disassembly kinetics to be too rapid for quantification by gel shift analysis (data not shown). Using the significantly more stable 601 NPS derivative mp2, we found that H3(T118ph) nucleosomes were disassembled by hMSH2–hMSH6 25 ± 7 times faster (t1/2•H3(T118ph) = 55 ± 4 min) than unmodified nucleosomes (t1/2•H3 = 1400 ± 400 min (Figure 7). This increase in rate is nearly identical to the increase in mobility induced by H3(T118ph) containing nucleosomes using an identical NPS substrate (Figure 4).
Previously, we proposed two mechanisms by which hMSH2–hMSH6 might disassemble nucleosomes (21). The first model is that hMSH2–hMSH6 traps DNA unwrapping fluctuations from the entry–exit region toward the dyad such that the nucleosome spontaneously disassembles. The second model is that hMSH2–hMSH6 traps fluctuations in nucleosome position such that the histone octamer slides off the end of the DNA. Our findings that H3(T118ph) enhances RE and DNase I accessibility near the dyad are consistent with increased DNA unwrapping in the nucleosome dyad region. This could enhance nucleosome disassembly by increasing the proportion of unwrapping fluctuations that extend past the dyad, which appears to significantly destabilize the nucleosome (30). We also observed that H3(T118ph) enhances thermally induced nucleosome repositioning. This enhanced sliding could facilitate nucleosome disassembly within the framework of the nucleosome sliding model. These models are nonexclusive, so each could be contributing to the H3(T118ph) enhancement of nucleosome disassembly by hMSH2–hMSH6.
H3(T118ph) enables nucleosome disassembly during the SWI/SNF remodeling reaction
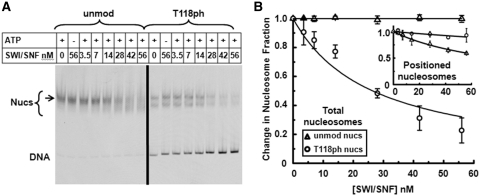
The report that H3(T118A) and H3(T118E) impact transcriptional regulation (13) and our observation that H3(T118ph) enhances nucleosome sliding and reduces DNA–histone binding suggests that this modification could influence chromatin remodeling. Moreover, the H3(T118) residue was found to contribute to the Sin phenotype (15). We examined the influence of H3(T118ph) on chromatin remodeling by the purified SWI/SNF complex (30). SWI/SNF binds unmodified and H3(T118ph) containing nucleosomes similarly in the absence of ATP (Supplementary Figure S9). In the presence of ATP, SWI/SNF repositioned unmodified nucleosomes without inducing nucleosome disassembly as previously observed (25,32) (Figure 8). In contrast, SWI/SNF remodeling of H3(T118ph) containing nucleosomes resulted in a dramatic increase in free DNA (Figure 8), suggesting that SWI/SNF directly disassembles H3(T118ph) containing nucleosomes. This is consistent with our other studies that show modifications at the nucleosome dyad enhance the dissociation or disassembly of partially wrapped nucleosomes (21).
Figure 8.
H3(T118ph) enables nucleosome disassembly by the SWI/SNF remodeling complex. (A) EMSA of ATP-dependent chromatin remodeling with unmodified and H3(T118ph) nucleosomes in the presence of increasing concentrations of SWI/SNF. Lanes are labeled with the nanomolar (nM) concentration of SWI/SNF. Each reaction was incubated for 30 min with or without 1 mM ATP. The bracket indicates the range of nucleosome electrophoretic mobility and the arrow indicates the location of central positioned nucleosomes. (B) The change in the fraction of total unmodified (triangles) or H3(T118ph) (circles) nucleosomes versus SWI/SNF concentration. The error bars were determined from the standard deviation of three separate experiments. The total unmodified nucleosome fractions and the total H3(T118ph) nucleosome fractions were each fit to: nucleosome fraction = 1-[SWI/SNF]/(K½+[SWI/SNF], where K½ is the concentration at which half of the nucleosomes are disassembled. We find that there is not a decrease in the fraction of unmodified nucleosomes (K½, unmod total > 2 × 10−5 nM), while K½,H3(T118ph) total = 26 ± 4 nM. (B, inset) The change in the fraction of centrally positioned unmodified (triangles) and H3(T118ph) (circles) nucleosomes versus SWI/SNF concentration. We fit the faction of positioned nucleosomes to the equation used to fit the total nucleosome fraction. We find H3(T118ph) decreases by 6-fold the fraction of positioned nucleosomes (K½, unmod positioned = 80 ± 3 nM and K½,H3(T118ph) positioned = 500 ± 100 nM). The values were determined from the ratio of the top band (arrow) to the entire nucleosome band (bracket) and normalized by the fraction of positioned nucleosomes at zero concentration of SWI/SNF.
We also analyzed the influence of H3(T118ph) on repositioning of intact nucleosomes. Interestingly, while the free DNA band increased and the total nucleosome content decreased, the initial positions within the nucleosome fraction remained, indicating that H3(T118ph) reduced the repositioning of nucleosomes to other DNA sites by SWI/SNF by 6-fold (Figure 8B). However, H3(T118ph) nucleosomes are already distributed to two separate positions in the absence of SWI/SNF (Supplementary Figures S1 and S2), yet retain a 147 bp DNA footprint (Supplementary Figures S5 and S6). We cannot rule out the possibility that the two dominant nucleosome positions are in dynamic equilibrium and that SWI/SNF repositions the nucleosomes rapidly between these two positions.
There are two nonexclusive mechanisms by which H3(T118ph) might enable nucleosome disassembly by SWI/SNF in our experiments. The first model is that SWI/SNF allows the histone octamer to directly disassociate from the DNA. This model relies on our observation that H3(T118ph) significantly reduces the DNA–histone binding free energy, which combined with the 50 bp of DNA that are unwrapped from the histone octamer by SWI/SNF could directly release the histone octamer. Alternatively, H3(T118ph) could allow SWI/SNF to slide the histone octamer off the end of the DNA. This model is consistent with our observation that H3(T118ph) dramatically enhances the rate of nucleosome mobility. Since DNA ends are rare in vivo, this alternate model would be consistent with the conclusion that H3(T118ph) increases SWI/SNF repositioning in vivo.
DISCUSSION
We find H3(T118ph) dramatically reduces the DNA–histone binding free energy and increases nucleosome mobility by ∼25 times. Furthermore, we observe a significant increase in DNA accessibility near the modification site in the nucleosome dyad. These results indicate that H3(T118ph), which is located in the DNA–histone interface near the dyad symmetry axis, directly impacts nucleosome dynamics. The side chain hydroxyl of H3(T118) is within 3 Å of the DNA phosphate backbone (17). The phosphorylation of H3(T118) would place significant negative charge in close proximity to the DNA phosphate backbone. One possibility is that electrostatic repulsion between the phosphothreonine and the DNA phosphate backbone is responsible for the observed alterations in nucleosome properties. However, the H3(T118E) substitution also introduces a negative charge, but does not reduce the DNA–histone binding free energy or nucleosome mobility (Figures 3 and 4). Moreover, the H3(T118I) Sin mutant induces similar but less pronounced alterations to nucleosome mobility (16,17) and DNA accessibility (18) compared to the H3(T118ph). The substitution of a threonine with an isoleucine replaces a hydrophilic residue with a hydrophobic residue and slightly increases steric bulk. The crystal structure of nucleosomes containing H3(T118I) and the much bulkier substitution H3(T118H) illustrate a distortion of the nucleosome DNA and a reduction in the number of hydrogen bonds in the nucleosome dyad region (17). Taken as a whole these observations suggest that specific properties of H3(T118ph) are responsible for the dramatic alterations in nucleosome properties and that it is the combination of sterics and the precise properties of the negatively charged phosphate that significantly disrupts the DNA–histone interactions. Furthermore, the fact that H3(T118E) does not reproduce the effects of H3(T118ph) but that both H3(T118E) and H3(T118A) are lethal in budding yeast is consistent with the notion that switching between the phosphorylated and unphosphorylated states of H3(T118) is required for yeast viability. Finally, since H3(T118ph) is observed at low levels (10) and dramatically perturbs nucleosome dynamics, it might be anticipated that this histone PTM would occur only transiently and be tightly regulated.
SWI/SNF appears to remodel nucleosomes by binding the histone octamer face creating a ∼50 bp DNA bulge near the entry–exit region of the nucleosome that propagates to the opposite entry–exit region, ultimately resulting in nucleosome sliding without disassembly (33,34). The presence of histone chaperones or adjacent nucleosomes appears to facilitate histone octamer release (35–38). Our observation that H3(T118ph) enables nucleosome disassembly by SWI/SNF suggests that H3(T118ph) may intrinsically regulate the outcome of a SWI/SNF remodeling reaction. These results are consistent with the hypothesis that the H3(T118ph) PTM may target the SWI/SNF chromatin disassembly reaction to distinct chromatin sites.
The H3(T45) (11,12) and H4(S47) (10) histone residues within the DNA–histone interface of the nucleosome are also known to be phosphorylated. H3(T45ph) is important for apoptosis (12) and replication (11), while H3(S47ph) appears to play a role in transcriptional silencing and DNA repair (13). H4(T80) is suspected to be phosphorylated since its location in the L2 loop of H4 is structurally similar to the L2 loop of H3 where H3(T118) is located (39), phosphorylation is observed in that sequence region by MS analysis (10), and mutations near the L2 loop of H4 eliminate silencing within rDNA, telomeres and the mating locus of yeast (40). Our studies suggest that these additional histone phosphorylations within the nucleosome DNA–histone interface could function to influence nucleosome dynamics, facilitate nucleosome disassembly and regulate or target chromatin-remodeling mechanisms. Further studies will be required to fully enumerate the biophysical effects of these PTMs on nucleosome dynamics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Heart Association Predoctoral Fellowship 0815460D (to J.A.N.); NSF CAREER MCB0845695 (to J.J.O.); a Career Award in the Basic Biomedical Sciences from the Burroughs Wellcome Fund (to M.G.P.); NIH CA067007 and GM080176 (to R.F.); NIH GM048413 (to B.B.) and NIH GM083055 (to M.G.P. and J.J.O.) Funding for open access charge: NIH GM083055.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jonathan Widom and Karolin Luger for the X. laevis histone vectors. The authors are grateful to Karin Musier-Forsyth for access to a Typhoon Trio fluorescence scanner and a fluorescence plate reader and Dongping Zhong for access to a FPLC.
REFERENCES
- 1.Suganuma T, Workman JL. Crosstalk among Hhstone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 2.Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 6.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 7.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of fully synthetic histone h3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J. Mol. Biol. 2011;408:187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 2009;284:23312–23321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 11.Baker SP, Phillips J, Anderson S, Qiu Q, Shabanowitz J, Smith MM, Yates JR, 3rd, Hunt DF, Grant PA. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 2010;12:294–298. doi: 10.1038/ncb2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurd PJ, Bannister AJ, Halls K, Dawson MA, Vermeulen M, Olsen JV, Ismail H, Somers J, Mann M, Owen-Hughes T, et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J. Biol. Chem. 2009;284:16575–16583. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 16.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. EMBO J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein–DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurumizaka H, Wolffe AP. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 1997;17:6953–6969. doi: 10.1128/mcb.17.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn PJ, Crowley KA, Carruthers LM, Hansen JC, Peterson CL. The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat. Struct. Biol. 2002;9:167–171. doi: 10.1038/nsb762. [DOI] [PubMed] [Google Scholar]
- 20.Shogren-Knaak MA, Fry CJ, Peterson CL. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J. Biol. Chem. 2003;278:15744–15748. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 21.Javaid S, Manohar M, Punja N, Mooney AM, Ottesen JJ, Poirier MG, Fishel R. Nucleosome remodeling by hMSH2–hMSH6. Mol. Cell. 2009;36:1086–1094. doi: 10.1016/j.molcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Met. Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 23.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 24.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 25.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 26.Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J. Mol. Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 28.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 29.Schiessel H, Widom J, Bruinsma RF, Gelbart WM. Polymer reptation and nucleosome repositioning. Phy. Rev. Lett. 2001;86:4414–4417. doi: 10.1103/PhysRevLett.86.4414. [DOI] [PubMed] [Google Scholar]
- 30.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone–DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. et Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF–nucleosome complex. Mol. Cell. Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl Acad. Sci. USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 38.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 40.Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat. Gen. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 41.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.