Abstract
Tn10/IS10 transposition takes place in the context of a protein–DNA complex called a transpososome. During the reaction, the transpososome undergoes several conformational changes. The host proteins IHF and H-NS, which also are global regulators of gene expression, play important roles in directing these architectural changes. IHF binds tightly to only one of two transposon ends within the transpososome, folding this end into a DNA loop structure. Unfolding this DNA loop is necessary for excising the transposon from flanking donor DNA and preventing integration of the transposon into itself. We show here that efficient DNA loop unfolding relies on the continuity of the flanking donor DNA on the side of the transpososome opposite to the folded transposon end. We also show this same donor DNA is a preferred binding site for H-NS, which promotes opening of the IHF-loop, which is required for productive target interactions. This is counter to the usual mode of H-NS action, which is repressive due to its propensity to coat DNA. The interplay between IHF and H-NS likely serves to couple the rate of transposition to the host cell physiology as both of these proteins are integrated into cellular stress response pathways.
INTRODUCTION
Bacterial chromosomes are associated with a number of small, but abundant, DNA-binding proteins. Together they form a compacted structure visible under the light microscope in appropriately stained cells. This structure, known as the nucleoid, is loosely analogous to the eukaryotic nucleus because it is seen to divide and segregate during cell division. The nucleoid proteins have been described as ‘histone-like’ because of their role in compaction, and because some bind and wrap DNA with low sequence specificity. However, this is probably not a strict analogy because, unlike the majority of bulk histones, bacterial nucleoid proteins have specific and active roles in numerous aspects of DNA metabolism.
The long relationship between transposons and their hosts has provided ample opportunity for mutual adaptation. The RNA interference (RNAi) machinery in eukaryotes is an example of host-mediated control of transposition. However, few transposon-encoded mechanisms have been documented, with the possible exception of the topological selectivity and over-production-inhibition mechanisms in the mariner family elements (1–3). In contrast, there are several well-documented transposon-encoded mechanisms in bacteria, which will be discussed further subsequently. There are also host-mediated mechanisms in the form of some bacterial nucleoid proteins, which have been identified as host factors in different systems. For Tn10/IS10 these are IHF (integration host factor), HU (heat unstable nucleoid protein) and H-NS (histone-like nucleoid structuring protein) (4,5). These proteins are among the most important global regulators in Escherichia coli and have distinct modes of binding DNA (6). H-NS binds to DNA with high A + T content and intrinsic curvature (7,8). IHF has a specific recognition sequence, where it binds and bends the helix in a U-turn (9). HU is closely related to IHF but binds DNA with low sequence specificity and a preference for various types of distortions (10).
H-NS, IHF and HU are each involved in a number of cellular processes including the initiation of DNA replication and the control of transcription. Their activities are mediated directly by binding at specific effector sites, and also indirectly by modulating the level of DNA supercoiling. In Tn10/IS10 transposition the DNA-bending activities of IHF and HU stimulate early steps of the reaction prior to excision of the element (4). After excision, they may also regulate the choice of target site, downregulating the rate of transposition if they remain associated with the transpososome. By contrast, the effects of H-NS on Tn10/IS10 transposition are entirely stimulatory, promoting productive target interactions by opposing the effects of IHF and HU (5,11).
H-NS has a propensity to polymerize on intrinsically curved DNA with a high A + T content (7,8). More recently, a high-affinity consensus sequence has been recognized. This is proposed to act as a nucleation site for initiation of the H-NS polymer. One of the best-known examples of genetic regulation by H-NS is in the repression of the cryptic bgl operon in E. coli. Curiously, transposon insertions upstream of the gene not only relieve repression, but also restore the operon’s sensitivity to regulation by its substrate and cyclic adenosine monophosphate (AMP; 12–14). The explanation is that the transposon insertion physically separates the promoter from the upstream H-NS nucleation sites. This example is reasonably characteristic of H-NS, whose action is predominantly repressive, mediated by the occluding effects of its filament. It is therefore surprising to find that H-NS stimulates Tn10/IS10 transposition by unfolding the transpososome and promoting target capture. This is not unprecedented as H-NS binds to the open complex of RNA polymerase at some promoters [(8) and references therein]. However, its effect on RNA polymerase is repressive, and the activation of Tn10 transposition by H-NS remains a highly unusual example.
METHODS
Chemicals and oligonucleotides were from Sigma. Enzymes were from NEB. Radio-nucleotides were from Amersham Biosciences.
Proteins and DNA substrates
IS10 transposase, IHF and H-NS were expressed and purified as described previously (15–17). The expression plasmids were as follows: wild-type transposase was from pRC60, which contains the transposase gene on an NdeI–BamHI fragment cloned into pET11a; IHF was expressed from pRC188, which is identical to pPR204 obtained from Phoebe Rice. This plasmid contains the IHF operon cloned downstream of the bacteriophage T7 promoter in pET27b (Novagen).
The IS10 outside end substrates encode the terminal inverted repeat and the adjacent IHF-binding site, which extends to bp +42 of the transposon. The inside end of the transposon was a modified derivative, in which the DNA between bp +19 and +47 is replaced by a tandem repeat of the bases 5′-CTGA. This is referred to as an ‘even-end’ because of the evenness of the hydroxyl radical footprint produced (18).
The transposon end fragments were prepared as follows: pRC850 was digested with XbaI and AccI to yield an inside end with 40 bp of flanking DNA. pRC98 was digested with BamHI and AccI to yield an outside end fragment with 40-bp flanking DNA and an 87-bp transposon arm. The pre-cleaved outside and inside end fragments were produced by digesting pRC35 and pRC99 with PvuII + BstEII and PvuII + AccI, respectively. The inside end fragments with short flanking DNA were produced in the following way: 3 bp by digesting pRC847 with AfeI; 7 bp by digesting pRC850 with EcoRI; 11 bp by digesting pRC850 with EcoRI. Following digestion, the linearized plasmids were blunt ended by treatment with mung bean nuclease or by filling in with the Klenow enzyme, as appropriate. The linearized plasmids were next digested with AccI to release the transposon end fragment, which was radioactively labeled by filling in using the Klenow enzyme as described below.
Inside end fragments containing uracil substitutions on the non-transferred strand were produced by polymerase chain reaction (PCR) using pRC100 as template and the following forward primers: U2, 5′-aggaattcgatatcacactcagUgCTGATGAATCCCC; U4, 5′-aggaattcgatatcacactcUgcgCTG; U6, 5′-aggaattcgatatcacacUcagcgC; control fragment, 5′-aggaattcgatatcacactcagcgCTGATGAATCCCC. Uppercase letters indicate the transposon end. Lowercase letters indicate the flanking DNA. The reverse primer was 5′-CGCGTAATACGACTCACTATAGG. The PCR products were digested with AccI to provide a 5′-overhang for labeling. The uracil substitutions were converted into single nucleoside gaps by treatment with Endonuclease VIII and Uracil–DNA Glycosylase (UDG) according to the protocols supplied by NEB. After treatment, the abasic sites were stabilized by incubating in 100 mM sodium borohydride (NaBH4) buffer on ice for 30 min.
When DNA fragments were to be labeled they were treated with the Exo- Klenow enzyme in the presence of a single 32P-labeled dNTP. The labeled fragments were purified by electrophoresis on a TAE-buffered 5% polyacrylamide gel, and recovered by the crush and soak method as described previously (19,20).
PEC assembly and unfolding
Transpososomes were assembled and visualized using the electrophoretic mobility shift assay (EMSA) as described (12,15). The standard reaction was 20 µl and contained 50 fmol of radioactively labeled transposon end, 20 fmol transposase and 300 fmol IHF. Mixed complexes were assembled by incubating 50 fmol of the labeled inside end with 200 fmol of the unlabeled outside end. IHF was the standard 300 fmol, but transposase was increased to 100 fmol. Transposase was added last and the reactions were incubated at room temperature for 1 h before the addition of CaCl2, heparin and/or H-NS, as required. Complexes were visualized using the EMSA and were quantified using a Fuji phosphorimager (20).
Missing nucleosides interference assay
Labeled transposon end fragments were treated with hydroxyl radicals (21), precipitated with ethanol and resuspended in TE buffer. Transpososomes were assembled and then purified using the EMSA. The DNA was recovered by the crush-and-soak method, denatured and the footprint was displayed on a DNA sequencing gel. The footprint was recorded using a phosphoimager and analyzed using the NIH Image program.
RESULTS
Experimental system
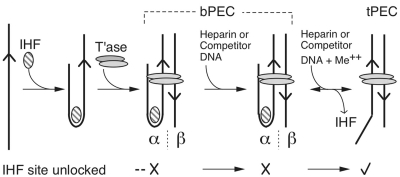
IS10 is flanked by a pair of inverted repeats, which are defined as ‘inside’ and ‘outside’ ends according to their arrangement in Tn10. The key difference between the ends is that the outside end is flanked by a consensus-binding site for IHF. During transposition, the transposon ends are synapsed together by a dimer of transposase into a paired ends complex (PEC), also known as the transpososome. Assembly of the complex is stimulated by IHF binding, particularly in the absence of negative supercoiling. Previously we developed a molecular model of the IHF-folded arm of IS10 by combining the structure of the related Tn5 transpososome with that for the IHF co-crystal. The 180° bend in the DNA imposed by IHF provides a set of ‘subterminal’ transposase contacts, located distal to the IHF-binding site (Figure 1). The contacts in the model fit well with the experimentally determined hydroxyl radical protection pattern and together with several other lines of evidence support the model as an accurate representation of the general shape of the Tn10 transpososome (18).
Figure 1.
Assembly of the Tn10 transpososome. The assembly and unfolding of the Tn10/IS10 transpososome (18,29). IHF binds specifically to the outside-end of Tn10 and activates assembly of the transpososome. IHF remains locked in position on the α side of the complex until released by the addition of divalent metal ion. Further details are given in the text. Arrowhead, transposon end; hatched oval, IHF; grey ovals, transposase (T’ase); tPEC, top-PEC; Me++, divalent metal ion; IE, inside end; *, radioactive label.
The IHF-binding site imparts the transpososome with structural asymmetry (Figure 1). The two sides of the complex are defined as alpha (α) and beta (β), corresponding to the outside and inside ends, respectively (22,23). On the α side of the complex, IHF is locked in position, probably by the subterminal contacts, until its release as the catalytic metal ion enters into the reaction (22). We refer to the IHF-bound and unbound forms of the complex as bottom-PEC (bPEC) and top-PEC (tPEC), respectively, because of their behavior in an EMSA (20). The slower mobility of the tPEC reflects the extended conformation of the transposon arms, rather than the total molecular mass of the complex, which is of course lower than the bPEC. Once the α side of the complex has been unlocked, IHF is free to associate or dissociate, according to the prevailing concentration.
The structural asymmetry of the bPEC has functional consequences. Cleavage of the α or β transposon end yields the α- and β-single end break complexes (αSEB and βSEB), respectively. However, the initiation of cleavage is biased toward the α side of the complex (24). This appears to be the preferred reaction pathway because the αSEB unfolds rapidly and goes on to complete cleavage, producing the double end break (DEB) complex. Provided that IHF has dissociated, the top-DEB (tDEB) will quickly establish target interactions and perform the integration step of the reaction. If IHF remains associated with the complex it blocks intermolecular target interactions and promotes suicidal autointegration events (4). This is the fate of the βSEB, which unfolds poorly (24). The bottom-DEB (bDEB) is likewise unable to unfold. It therefore appears that unfolding requires the presence of the β flanking DNA. We will present experiments to support this view and further show that the β flanking DNA is likely the site at which H-NS acts to mediate unfolding.
Short β side flanking DNA inhibits unfolding
We monitored the unfolding of the IS10 transpososome using an established EMSA (24). Briefly, transposase and IHF are incubated with linear fragments of DNA encoding the inside and outside ends of the transposon. The inside end, which has a radioactive label, is recruited into the developing transpososome by an IHF-bound outside end (Figure 1). The inside end lacks an IHF-binding site and cannot form a PEC on its own. The outside end can form PEC on its own, but these complexes lack a radioactive label and are therefore invisible in the EMSA. The assay therefore detects only mixed complexes between outside and inside transposon ends.
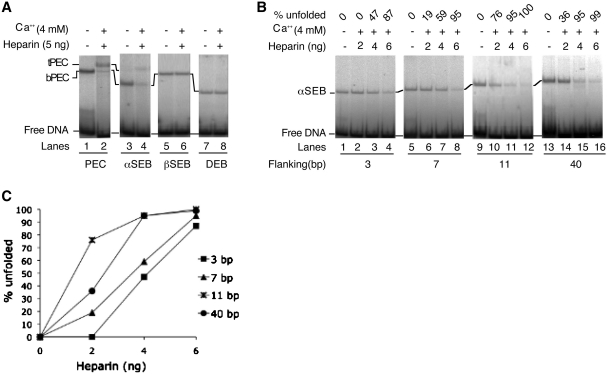
Although Ca2+ does not support any of the catalytic steps in IS10 transposition, it acts as an analog of the catalytic metal ion in several other respects (22). This allows us to monitor the metal-ion-dependent unfolding of the transpososome without the added complication of the ongoing cleavage and integration reactions. When the bPEC was treated with Ca2+ and heparin, it was converted to a tPEC (Figure 2A, lanes 1 and 2). This occurs because Ca2+ unlocks the IHF-binding site on the α side of the complex, while the heparin acts like competitor DNA and sequesters any IHF that dissociates spontaneously from the complex (25). The amount of tPEC produced by unfolding was significantly less than the starting amount of bPEC (compare lanes 1 and 2). This is because the unfolded transpososomes are less stable and some is lost during electrophoresis. This is known because the bPEC can be made to reappear after unfolding if the IHF is added back (20).
Figure 2.
Unfolding of the single end break complexes. Transpososomes were assembled by mixing appropriate combinations of un-cleaved and pre-cleaved transposon ends. Unfolding was initiated by the addition of Ca2+, an analog of the catalytic metal ion, and heparin, which sequesters any IHF that dissociates from the complex. Unfolding was analyzed using the EMSA and gels were recorded using a phosphoimager. (A) The bPEC and αSEB unfold in the presence of Ca2+ and heparin. The tPEC and the tSEB are relatively unstable during electrophoresis and the most accurate measure of unfolding is given by the disappearance of the bottom complexes (see text for details). The βSEB and the DEB are resistant to unfolding. The un-cleaved transposon end had 40 bp of flanking DNA. (B) The αSEB was assembled using un-cleaved transposon ends with progressively shorter flanking DNA. (C) Unfolding of the complexes in part B were quantified using a phosphoimager. The amount of αSEB present in the absence of Ca2+ and heparin was defined as 100%. If half of this was lost after heparin treatment, this would be given as 50% unfolding.
The two structural isomers of the SEB complex, the αSEB and βSEB, were prepared by cleaving the respective transposon ends with a restriction endonuclease prior to the assembly of the complex (Figure 2A). We refer to this technique as pre-cleavage of the transposon end (20,24). Note that the αSEB, in which the IHF-bound transposon end lacks flanking DNA, migrates faster than the βSEB (for further details see Figure 3 in reference 24). Presumably, the βSEB complex is retarded in the gel because its conformation is more extended. When the αSEB was treated with Ca2+ and heparin there was a drastic reduction in the amount of αSEB (lane 4), indicating that it had unfolded. However, only a very faint band was detected at the position expected for the unfolded tSEB. This is because the tSEB is even more unstable during electrophoresis than the tPEC. However, we know that the tSEB is in fact present before electrophoresis because integration products are detected if the reaction is supplemented with Mg2+ and target DNA (24).
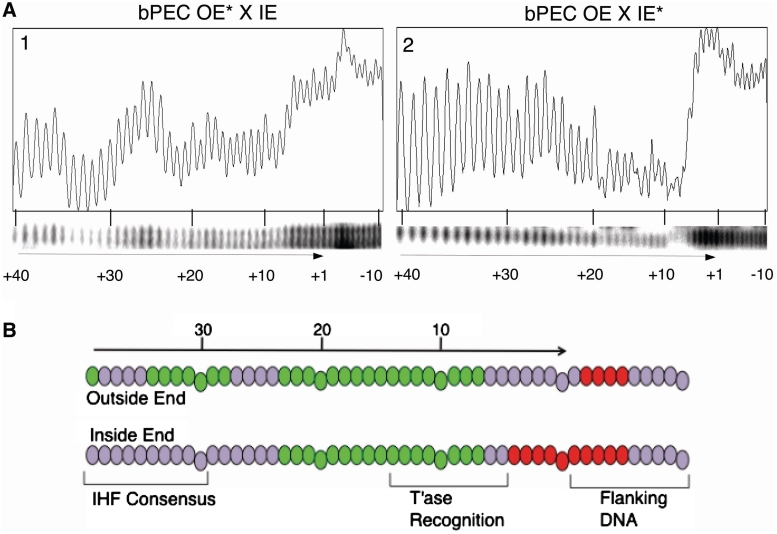
Figure 3.
Missing nucleotide interference assay. (A) The outside end (OE) and inside end (IE) of the transposon were treated with hydroxyl radicals, which attack at random positions, eliminating a base and breaking the phosphodiester backbone. The extent of treatment was limited so that there was on average less than one hit per DNA fragment. The fragments were subsequently used to assemble bPEC, which was purified using the standard EMSA. The radioactively labeled DNA was recovered from the gel, denatured and analyzed on a DNA sequencing gel to reveal the patterns of interference and enhancement. Under-represented positions are those at which the missing base interferes with assembly of the complex. Over-represented positions are those at which the presence of the base itself is unimportant, but where the presence of the single-stranded gap enhances assembly of the complex. (B) The interference patterns in (A) are illustrated. The DNA base pairs are represented as ovals, with depletions in green and enhancements in red. Genetic analysis previously identified the residues between bp +6 and +13 as the primary determinants of the transposase binding site.
When the βSEB and the bDEB were challenged with Ca2+ to unlock the α side of the complex and with heparin to sequester the IHF, they were resistant to unfolding (Figure 2A, lanes 5–8). As mentioned above, this suggests that unfolding requires the β flanking DNA. To explore this issue further, the αSEB was assembled using DNA fragments with progressively shorter segments of β flanking DNA (Figure 2B). After assembly, the complexes were titrated with increasing concentrations of heparin. The disappearance of the complex was used to quantify the extent of unfolding (Figure 2C). In this experiment, the fragment with the longest flanking DNA (40 bp) is identical to that shown in part B of the figure, and 95% of the αSEB unfolded when 4 ng of heparin was added (Figure 2B, lane 15). Unfolding was similar with 11 bp of flanking DNA, but the efficiency dropped significantly when it was reduced to 7 bp or 3 bp (Figure 2B and C).
Distortion of the α and β flanking DNA segments
Hydroxyl radical footprinting previously revealed no significant difference between the protein–DNA interactions in or around the flanking DNA on the α and β sides of the complex, which correspond to the outside and inside ends of the transposon, respectively (18). Nevertheless, there are clear functional differences between the two types of end, most notably that the inside end needs an IHF-bound outside end partner during transpososome assembly. Unfolding of the αSEB and βSEB suggest that the conformation of the flanking DNA on either side of the complex may be a significant determinant of these differences. This idea is supported by the observation that the DEB complex assembles more efficiently and is more stable than the PEC, suggesting that flanking DNA may in some way hinder transpososome assembly (20). This could arise from the energetic cost associated with a hitherto unrecognized distortion of the flanking DNA. We therefore performed a missing nucleoside interference assay to probe the contribution of each end during assembly of the complex.
Outside and inside ends were treated with hydroxylradicals to delete single nucleosides and subsequently used to form bPECs. The PECs were then purified using the standard EMSA. The DNA was recovered and displayed on a DNA sequencing gel to yield the interference footprints (Figure 3). Missing nucleosides can affect assembly of a protein–DNA complex in two ways. Positions at which a given nucleoside is required for assembly of the complex are depleted in the footprint. In contrast, enhancements appear at positions where specific contacts are unimportant, but where the increased flexibility of the DNA, due to the loss of base stacking interactions, overcomes a limitation on assembly of the complex. Both types of signature are detected in the transpososome, with clear differences between each side of the complex.
The interference patterns for the transposon ends on both sides of the complex are depleted between residues +23 and +7, which correspond to the transposase binding sites (Figure 3). On the outside end of the transposon, which lies on the α side of the complex, the IHF-binding site is depleted from bp +34 to +28 (Figure 3). There is also an enhancement between bp −2 and −5 in the flanking DNA. The interference pattern on the β side of the complex is significantly different. The depletion of the transposase-binding site between bp +23 and +7 is followed immediately by enhancement between bp +5 and −5. The distortion of the DNA on either side of the terminal nucleotides therefore appears to be greater on the β side of the complex.
Transpososome unfolding requires continuity of the flanking DNA
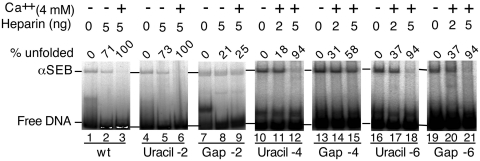
Distortion of the flanking DNA will place the transpososome in a state of tension. We therefore wondered whether such tension could have a role in unfolding of the αSEB complex. Based on the results from the missing nucleoside experiment (Figure 3), we reasoned that the loss of stacking interactions associated with a missing base would prevent the build up of tension in the transpososome and may confer an unfolding defect. We therefore developed a technique to introduce missing nucleosides at specific positions in the DNA flanking the inside end of the transposon. We began by using PCR primers containing uracil residues introduced at positions −2, −4 and −6 in the flanking DNA. The uracil-substituted inside ends were then treated with DNA uracil glycosylase and endonuclease VIII. Together these enzymes remove the uracil base and cleave the phosphodiester backbone. The resulting lesion is very similar to the missing nucleosides generated by hydroxyl radical treatment.
The uracil-substituted inside ends, together with their gapped derivatives, were used to assembly αSEB complexes (Figure 4). The uracil-substituted ends behaved similarly to wild type, unfolding in the presence of Ca2+ and heparin (lanes 3, 6, 12 and 18). However, the complexes with gaps at positions −2 and −4 failed to unfold properly (compare lane 6 with 9 and 12 with 15). By contrast, the αSEB with the gap at position −6 unfolded normally (compare lanes 18 and 21). These results suggest that continuity of the flanking DNA is important out as far as position −4, but not as far as position −6. This is consistent with the enhancements in the missing nucleoside footprints, which extended as far as position −5 (Figure 3).
Figure 4.
Single-strand gaps in the flanking DNA inhibit unfolding. The αSEB complex was assembled from a mixture containing a pre-cleaved outside end with an un-cleaved inside end. The inside end, which is always on the β side of the transpososome, had a uracil residue in the flanking DNA at position –2, –4 or –6 of the non-transferred strand. Treatment of the uracil-containing transposon end with DNA uracil glycosylase and endonuclease VIII prior to transpososome assembly eliminated the uracil base and cleaved the phosphodiester backbone. This lesion is very similar to that caused by hydroxyl radicals in the interference assay presented in Figure 3. Unfolding was initiated by the addition of Ca2+ and heparin as previously described. Unfolding was analyzed using the EMSA and gels were recorded and quantified using a phosphoimager.
Transpososome unfolding by H-NS
H-NS mutant strains of E. coli support much lower rates of transposition than wild-type strains (5). Although H-NS mutants are highly pleiotropic, the protein was found to interact directly with the transpososome to promote and maintain unfolding of the transpososome (11,26). These interactions are mediated by contacts with the transposase and multiple sites on the DNA, including the flanking segment(s). We therefore investigated whether H-NS could distinguish between the two structural isomers of the SEB complex.
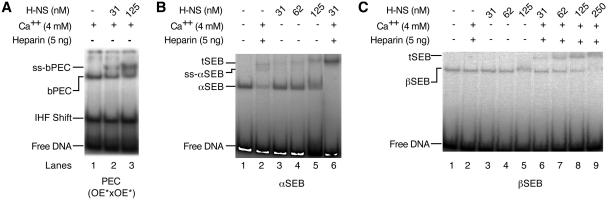
When the bPEC was titrated with H-NS in the absence of heparin, a partial super-shift was detected (ss-bPEC, Figure 5A). IHF remains bound to the transpososome in the absence of heparin and remains unaffected by the highest H-NS concentrations used in these assays, as evidenced by the presence of the IHF-shifted transposon ends (Figure 5A). The super-shift is caused by H-NS binding to the IHF-folded transpososome (11).
Figure 5.
H-NS promotes the unfolding of the transpososome. Transpososomes were assembled by mixing appropriate combinations of un-cleaved and pre-cleaved transposon ends. Unfolding was initiated by the addition of Ca2+ and heparin. The complexes were analyzed using the EMSA and gels were recorded using a phosphoimager. SS is the super-shifted complex that forms when H-NS binds to the bPEC or the bottom αSEB. The tSEB is not usually detected in the EMSA, but is stabilized by H-NS binding. Note that the βSEB is closer to the tSEB and the super-shifted complex because it migrates more slowly than the αSEB. For further details of the relative mobility of the αSEB and βSEB see Figures 2A and 3 in ref. 24. The DNA sequences flanking the outside and inside transposon ends were identical out as far as bp –12. (A) Unfolding of the PEC. (B) Unfolding of the αSEB. (C) Unfolding of the βSEB.
When the αSEB was unlocked by Ca2+ treatment in the presence of heparin, most of it unfolded (Figure 5B, compare lanes 1 and 2). As before, very little tSEB was detected owing to the instability of the unfolded complexes during electrophoresis (see Figure 2A for a previous example, and the associated text for the explanation). The αSEB complex was next titrated with H-NS in the absence of heparin (Figure 5B, lanes 3–5). A band appeared fractionally above the position of the tPEC and increased in abundance with increasing H-NS concentration. This band corresponds to the super-shift caused by H-NS binding to the folded SEB complex, and is analogous to that shown in Figure 5A. When the αSEB was unfolded by treatment with heparin and Ca2+ in the presence of H-NS, an abundant super-shifted complex was detected (Figure 5B, lane 6). This represents the tSEB, which is now detected in the EMSA owing to the stabilizing effects of H-NS binding (27). Notice also that H-NS completes the partial unfolding of the αSEB that occurred in response to treatment with heparin and Ca2+ alone (compare lanes 2 and 6). H-NS therefore does more than stabilize the tSEB: it actively unfolds the αSEB, helping to dissociate IHF.
When the βSEB was titrated with H-NS in the absence of Ca2+ and heparin, it was unaffected, although the band did become slightly fuzzy at the highest concentration (Figure 5C, lane5). This likely indicates that H-NS was loosely associated with the complex, but dissociated soon after the start of the electrophoresis. This is similar to the behavior of the folded αSEB (Figure 5, compare lane 5 in parts B and C). However, with the βSEB no super-shifted bands were detected in the absence of Ca2+ and heparin, even at the highest H-NS concentration (Figure 5C, lanes 3–5). H-NS interactions with the folded βSEB are therefore significantly weaker than with the folded αSEB and bPEC, which are both super-shifted. This suggests that H-NS interacts strongly with the flanking DNA on the β side of the complex and weakly, or not at all, with the α side flanking DNA.
Finally, we titrated the βSEB with H-NS in the presence of Ca2+ and heparin (Figure 5C, lanes 6–9). At the lowest concentration of H-NS tested (31 nM) very little of the complex unfolded. This contrasts with the αSEB, which unfolded completely at this concentration (compare lane 6 in parts B and C). At higher H-NS concentrations, the βSEB unfolded progressively, until almost none remained at 250 nM. This was accompanied by the appearance of the tSEB, which is stabilized by H-NS binding.
DISCUSSION
Tn10/IS10 transposition is strongly dependent on supercoiling or IHF. These factors are interchangeable and seem to function by helping the α transposon end to wrap round the transposase dimer during transpososome assembly (4,18). However, if IHF remains associated with the transpososome after cleavage of the transposon ends it inhibits intermolecular target interactions and promotes a suicidal autointegration reaction, which may serve to protect the host from an excessively high rate of transposition (4). H-NS opposes the effect of IHF by unfolding the transpososome and promoting intermolecular integration (11,28). Results presented here add to our understanding of the determinants involved in architectural changes in the Tn10/IS10 transpososome that take place over the course of the transposition reaction. First, we have shown that the length and continuity of the flanking donor DNA associated with the β end of the transpososome contributes strongly to unfolding the α end in the context of the SEB complex. Second, we have shown that in the precursor to the SEB complex (i.e. the PEC) the DNA structure of the β end is more distorted than that of the α end. We have also added to our understanding of how H-NS promotes Tn10/IS10 transposition. H-NS binds preferentially to the αSEB complex to promote SEB unfolding, although at high concentration it also unfolds the βSEB, which is otherwise the least productive of the transposition intermediates.
Several unique characteristics of the Tn10/IS10 system have allowed us to address the nature and consequences of the conformational changes at different stages of the reaction (23,24,29,30). The most important experimental advantage of Tn10/IS10 is that the requirement for IHF binding to one of the transposon ends allowed opposite ends to be distinguished experimentally, which in turn revealed the differences between the α and β sides of the complex (22). By contrast, in the related Tn5/IS50 system IHF is not involved in the assembly of the transpososome. Notwithstanding, there is strong evidence that the molecular mechanisms of Tn10 and Tn5 are almost identical (30,31). It is therefore possible that the asymmetry of the intermediates in Tn5 has simply gone unnoticed because the ends cannot be distinguished experimentally.
The Tn10 molecular spring and the mechanism of H-NS action
The molecular spring model for Tn10 transposition proposed that IHF acted as an architectural catalyst: after promoting the assembly of a folded complex it is actively ejected by a conformational change in transposase (4). In this model, force is provided by the transposase, while the DNA provides the elastic component of the spring through which it is transmitted. At the time, it was assumed that the IHF-folded arm of the transposon provided the DNA component of the spring. However, the 180° bend introduced by IHF binding breaks the stacking interactions, which are the main sources of DNA’s stiffness and elasticity. Where then may the molecular spring be located? The present results suggest that the spring may reside in a small region at the tip of the β transposon end and the first few bases of the flanking DNA. This region was enhanced in the missing nucleoside interference assay, indicating that it becomes bent during the assembly of the complex (Figure 3). The rational for this inference is that the missing nucleoside at these positions lowers the energy required to deform the DNA, which is preferentially assembled into the complex. The importance of the flanking DNA was also revealed by the site-specific single nucleoside gaps introduced on the β side of the complex. Although transposon ends with gaps at positions −2 and −4 were assembled efficiently, they were more difficult to unfold than their un-gapped counterparts (Figure 4).
Earlier studies performed with the outside-by-outside end PEC revealed that flanking donor DNA provides key binding determinants for H-NS in the folded transpososome (bPEC) (26). In an OP-Cu footprint H-NS protected 15–20 bp into the flanking donor DNA as well as both an expanded zone of protection and hypersensitivity at the transposon–donor junction. The latter is consistent with H-NS binding within the flanking donor DNA immediately adjacent the transposon–donor junction. In the present work, we show that H-NS preferentially binds the β-side flanking DNA, which is present in the αSEB and appears to contain the most severe distortion over the transposon–donor junction. The implication of these observations is that H-NS binding to the β end of the transpososome may contribute to either loading the molecular spring or triggering the spring resulting in IHF release. Further testing of this idea will require localizing H-NS within the αSEB and this has not yet been attempted.
Linking Tn10 transposition to cellular growth conditions
IHF and H-NS are almost universally present in the eubacteria. Moreover, they are both global regulators of gene expression and as such are integrated into many different regulatory pathways (7,32). Accordingly, their involvement in Tn10 transposition connects Tn10 to a network of other interactions. These will modulate the rate of transposition depending on the prevailing conditions, and provide for an integrated physiological response. Evidence that Tn10 transposition is sensitive to growth conditions and/or cellular stress includes the following: (1) Tn10 transposition is much more sensitive to the H-NS status of the cell when transposition is assayed under conditions where colonies are subject to gradual nutrient deprivation compared to when they are propagated in rich media (5); (2) Over-expression of H-NS, which triggers premature entry into a state resembling stationary phase (33), increases the frequency of Tn10 transposition (11); (3) UV-irradiation has been reported to increase IS10 transposition (34).
Since IHF and H-NS act in opposition to each other in the unfolding of the transpososome, factors influencing H-NS and IHF expression would be expected to affect the frequency of Tn10/IS10 transposition. As previously discussed, both IHF and H-NS are highly expressed proteins and have comparable binding affinities for at least some forms of the Tn10 transpososome (27,28). However, IHF protein levels increase in stationary versus exponential growth (35), whereas H-NS protein levels are relatively constant (36). This might therefore limit Tn10 transposition in stationary phase. On the other hand, the activity of H-NS (with regard to DNA binding) is sensitive to temperature (37) and levels of DNA supercoiling (38). Accordingly, changes in either of these parameters might alter the balance of IHF and H-NS binding within the transpososome and thereby impact the transposition frequency.
IHF/H-NS competition in other transposition systems
The DNA distortions associated with transpososome assembly in other systems may also provide targets for H-NS binding. For example, the rate of Tn5 transposition is depressed in strains lacking the H-NS protein, even though this element probably lacks significant IHF-binding sites (39). Tn5 does, however, have a binding site for FIS, a protein that is functionally related to IHF (40). Interestingly, FIS and H-NS have been shown to work in opposition in the regulation of gene expression (41). In contrast to Tn5, there are other transposons that do contain IHF-binding sites within their end sequences. Examples from the transposition literature include IS1, γδ and ISV-A1 (42–44). In the case of IS1, IHF has been shown to bind the end sequences, and IHF mutant E. coli cells exhibit reduced levels of transposition (42). However, the mechanism for IHF function in this system has not been defined.
Notably, the absence of IHF (or FIS) binding sites within a transposon end does not preclude H-NS from regulating bacterial transposition reactions by modulating the structure of transposition intermediates. We have shown here that the distortion of the flanking DNA provides an attractive target for H-NS binding. Furthermore, the DNA folding and looping in the Tn10/IS10 system are not entirely dependent on IHF, which can be substituted by supercoiling (4). In this case, DNA looping is an inherent property of the superhelix. It is therefore possible that H-NS can play an important role as an architectural catalyst in transposition reactions that do not employ IHF for transpososome assembly and development.
FUNDING
Funding for open access charge: The Wellcome Trust (to R.M.C.); and by a Canadian Institutes of Health Research grant (MOP 11281 to D.B.H.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Claeys Bouuaert C, Liu D, Chalmers R. A simple topological filter in a eukaryotic transposon as a mechanism to suppress genome instability. Mol. Cell. Biol. 2011;31:317–327. doi: 10.1128/MCB.01066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claeys Bouuaert C, Chalmers R. Transposition of the human Hsmar1 transposon: rate-limiting steps and the importance of the flanking TA dinucleotide in second strand cleavage. Nucleic Acids Res. 2010;38:190–202. doi: 10.1093/nar/gkp891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson H, Chalmers R. Delivering the goods: viral and non-viral gene therapy systems and the inherent limits on cargo DNA and internal sequences. Genetica. 2010;135:485–489. doi: 10.1007/s10709-009-9434-3. [DOI] [PubMed] [Google Scholar]
- 4.Chalmers R, Guhathakurta A, Benjamin H, Kleckner N. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell. 1998;93:897–908. doi: 10.1016/s0092-8674(00)81449-x. [DOI] [PubMed] [Google Scholar]
- 5.Swingle B, O'Carroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 2004;52:1055–1067. doi: 10.1111/j.1365-2958.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 6.Dorman CJ. Nucleoid-associated proteins and bacterial physiology. Adv. Appl. Microbiol. 2009;67:47–64. doi: 10.1016/S0065-2164(08)01002-2. [DOI] [PubMed] [Google Scholar]
- 7.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 8.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice PA. Making DNA do a U-turn: IHF and related proteins. Curr. Opin. Struct. Biol. 1997;7:86–93. doi: 10.1016/s0959-440x(97)80011-5. [DOI] [PubMed] [Google Scholar]
- 10.Swinger KK, Rice PA. Structure-based analysis of HU-DNA binding. J. Mol. Biol. 2007;365:1005–1016. doi: 10.1016/j.jmb.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward CM, Wardle SJ, Singh RK, Haniford DB. The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. Mol. Microbiol. 2007;64:1000–1013. doi: 10.1111/j.1365-2958.2007.05708.x. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds AE, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 13.Dole S, Nagarajavel V, Schnetz K. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 2004;52:589–600. doi: 10.1111/j.1365-2958.2004.04001.x. [DOI] [PubMed] [Google Scholar]
- 14.Sankar TS, Neelakanta G, Sangal V, Plum G, Achtman M, Schnetz K. Fate of the H-NS-repressed bgl operon in evolution of Escherichia coli. PLoS Genet. 2009;5:e1000405. doi: 10.1371/journal.pgen.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalmers RM, Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J. Biol. Chem. 1994;269:8029–8035. [PubMed] [Google Scholar]
- 16.Lynch TW, Read EK, Mattis AN, Gardner JF, Rice PA. Integration host factor: putting a twist on protein–DNA recognition. J. Mol. Biol. 2003;330:493–502. doi: 10.1016/s0022-2836(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 17.Cusick ME, Belfort M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 1998;28:847–857. doi: 10.1046/j.1365-2958.1998.00848.x. [DOI] [PubMed] [Google Scholar]
- 18.Crellin P, Chalmers R. Protein–DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J. 2001;20:3882–3891. doi: 10.1093/emboj/20.14.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bischerour J, Lu C, Roth DB, Chalmers R. Base flipping in V(D)J recombination: insights into the mechanism of hairpin formation, the 12/23 rule, and the coordination of double-strand breaks. Mol. Cell. Biol. 2009;29:5889–5899. doi: 10.1128/MCB.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai J, Chalmers RM, Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schickor P, Heumann H. In: DNA-Protein Interactions: Principles and Protocols. Kneale GG, editor. New Jersey: Humana Press; 1994. pp. 21–32. [Google Scholar]
- 22.Sewitz S, Crellin P, Chalmers R. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 2003;31:5868–5876. doi: 10.1093/nar/gkg797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crellin P, Sewitz S, Chalmers R. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell. 2004;13:537–547. doi: 10.1016/s1097-2765(04)00052-8. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Crellin P, Chalmers R. Cyclic changes in the affinity of protein-DNA interactions drive the progression and regulate the outcome of the Tn10 transposition reaction. Nucleic Acids Res. 2005;33:1982–1992. doi: 10.1093/nar/gki348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allingham JS, Wardle SJ, Haniford DB. Determinants for hairpin formation in Tn10 transposition. EMBO J. 2001;20:2931–2942. doi: 10.1093/emboj/20.11.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wardle SJ, O'Carroll M, Derbyshire KM, Haniford DB. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 2005;19:2224–2235. doi: 10.1101/gad.1338905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wardle SJ, Chan A, Haniford DB. H-NS binds with high affinity to the Tn10 transpososome and promotes transpososome stabilization. Nucleic Acids Res. 2009;37:6148–6160. doi: 10.1093/nar/gkp672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh RK, Liburd J, Wardle SJ, Haniford DB. The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. J. Mol. Biol. 2008;376:950–962. doi: 10.1016/j.jmb.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Liu D, Sewitz S, Crellin P, Chalmers R. Functional coupling between the two active sites during Tn 10 transposition buffers the mutation of sequences critical for DNA hairpin processing. Mol. Microbiol. 2006;62:1522–1533. doi: 10.1111/j.1365-2958.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- 30.Bischerour J, Chalmers R. Base flipping in Tn10 transposition: an active flip and capture mechanism. PLoS ONE. 2009;4:e6201. doi: 10.1371/journal.pone.0006201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bischerour J, Chalmers R. Base-flipping dynamics in a DNA hairpin processing reaction. Nucleic Acids Res. 2007;35:2584–2595. doi: 10.1093/nar/gkm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman DI. Integration host factor: a protein for all reasons. Cell. 1988;55:545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 33.McGovern V, Higgins NP, Chiz RS, Jaworski A. H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie. 1994;76:1019–1029. doi: 10.1016/0300-9084(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 34.Eichenbaum Z, Livneh Z. UV light induces IS10 transposition in Escherichia coli. Genetics. 1998;149:1173–1181. doi: 10.1093/genetics/149.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aviv M, Giladi H, Schreiber G, Oppenheim AB, Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol. Microbiol. 1994;14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 36.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madrid C, Nieto JM, Paytubi S, Falconi M, Gualerzi CO, Juarez A. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 2002;184:5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tupper AE, Owen-Hughes TA, Ussery DW, Santos DS, Ferguson DJ, Sidebotham JM, Hinton JC, Higgins CF. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 1994;13:258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitfield CR, Wardle SJ, Haniford DB. The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. Nucleic Acids Res. 2009;37:309–321. doi: 10.1093/nar/gkn935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinreich MD, Reznikoff WS. Fis plays a role in Tn5 and IS50 transposition. J. Bacteriol. 1992;174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol. Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 42.Gamas P, Chandler MG, Prentki P, Galas DJ. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J. Mol. Biol. 1987;195:261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- 43.Wiater LA, Grindley ND. Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. EMBO J. 1988;7:1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolmasky ME, Crosa JH. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposonlike structure. Plasmid. 1995;33:180–190. doi: 10.1006/plas.1995.1019. [DOI] [PubMed] [Google Scholar]