Abstract
Bacterial single-stranded (ss) DNA-binding proteins (SSBs) bind and protect ssDNA intermediates formed during cellular DNA replication, recombination and repair reactions. SSBs also form complexes with an array of genome maintenance enzymes via their conserved C-terminal tail (SSB-Ct) elements. In many cases, complex formation with SSB stimulates the biochemical activities of its protein partners. Here, we investigate the mechanism by which Escherichia coli SSB stimulates hydrolysis of ssDNA by Exonuclease I (ExoI). Steady-state kinetic experiments show that SSB stimulates ExoI activity through effects on both apparent kcat and Km. SSB variant proteins with altered SSB-Ct sequences either stimulate more modestly or inhibit ExoI hydrolysis of ssDNA due to increases in the apparent Michaelis constant, highlighting a role for protein complex formation in ExoI substrate binding. Consistent with a model in which SSB stabilizes ExoI substrate binding and melts secondary structures that could impede ExoI processivity, the specific activity of a fusion protein in which ExoI is tethered to SSB is nearly equivalent to that of SSB-stimulated ExoI. Taken together, these studies delineate stimulatory roles for SSB in which protein interactions and ssDNA binding are both important for maximal activity of its protein partners.
INTRODUCTION
The double-stranded structure of genomic DNA is a remarkably stable storage form for cellular information. However, this structure must be separated into single-stranded (ss) DNA to allow enzymes that catalyze DNA replication, recombination or repair reactions access to genomic information. To prevent spontaneous reannealing and to protect exposed ssDNA from damage, cells utilize specialized ssDNA-binding proteins (SSBs) to bind unwound ssDNA (1–3). As such, genome maintenance enzymes have adapted to utilize SSB/ssDNA nucleoprotein substrates rather than free ssDNA in cells. Understanding the structural and biochemical basis underlying enzymatic processing of SSB/ssDNA substrates is critical for developing molecular models of cellular genome maintenance.
Eubacterial SSBs generally function as homotetramers in which each monomer contains both DNA-binding and protein-interaction elements (1–3). The DNA-binding domain, which also mediates tetramer formation, is composed of an N-terminal oligonucleotide/oligosaccharide-binding (OB) fold. DNA wraps around the SSB OB domains in a high-affinity, sequence-independent manner (4). In spite of the stability of SSB/ssDNA complexes, SSB tetramers are surprisingly dynamic, moving freely along the DNA to which they bind (5). Outside of eubacterial SSBs, bacteriophage and eukaryotic SSBs can have very different quaternary structures but still utilize OB folds to bind DNA. For example, the bacteriophage T4 SSB (gene 32 protein, gp32) functions as a monomer with a single OB domain (6) and eukaryotic Replication Protein A (the major nuclear eukaryotic SSB) functions as a heterotrimer that contains six OB folds (7).
The protein-binding activities of eubacterial SSBs are mediated by C-terminal tail (SSB-Ct) segments located on each monomer in the protein (3). This sequence includes six evolutionarily conserved residues (Asp–Asp–Asp–Ile–Pro–Phe) that form a docking site used for interactions with at least a dozen different DNA replication, recombination and repair proteins. Consistent with the importance of SSB/protein interactions, mutations within the SSB-Ct sequence are highly detrimental to cell growth. One such mutation in Escherichia coli (ssb113) changes the penultimate Pro of the SSB-Ct sequence to a Ser (8). This mutation confers temperature-sensitive lethality by producing an SSB variant that retains the ability to bind ssDNA but that cannot support DNA replication at non-permissive temperatures and is hypersensitive to DNA damage even under permissive conditions (8–13). The SSB113 protein is defective for binding several proteins, including Exonuclease I (ExoI) (14,15), RecQ DNA helicase (16,17), PriA DNA helicase (18), Topoisomerase III (19), RecG DNA helicase (20), DNA polymerase V (21) and the χ subunit of DNA polymerase III (22–24). Additionally, altering the C-terminal Phe residue from E. coli SSB to Cys disrupts heterologous protein interaction and is lethal in E. coli (14). Among the proteins that bind SSB, structural descriptions of the SSB-Ct binding sites on ExoI and RecQ have identified conserved electrostatic features that mediate binding to SSB (15,17). These include a hydrophobic pocket and basic residue that, respectively, accommodate the side chain and α-carboxyl group of the C-terminal Phe residue, and a ‘basic ridge’ element that is thought to bind the SSB-Ct Asp residues.
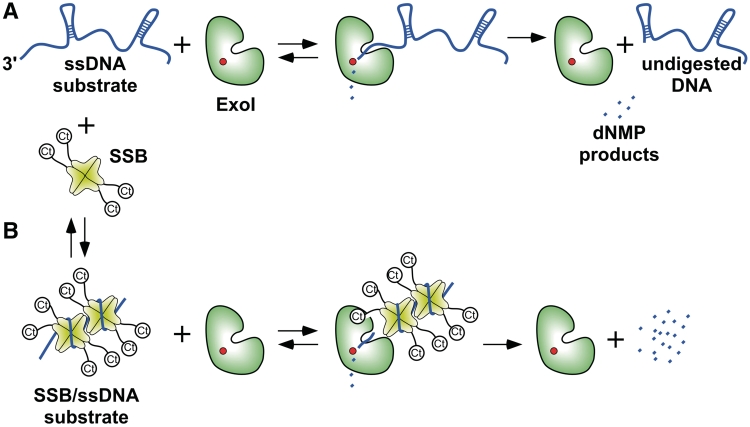
ExoI has emerged as a model for studying the roles of SSB in stimulating biochemical activity of SSB-associated enzymes. Escherichia coli ExoI degrades homopolymeric ssDNA in a highly processive fashion; however, its activity on heteropolymeric DNA is less processive, which has been attributed to ExoI dissociation from DNA upon encountering duplex structures (25–28). ExoI nuclease activity is stimulated by SSB in a manner that requires physical association between the two proteins (14,15,29–32). Disruption of the ExoI/SSB interaction by sequence changes in either protein or by peptide or small-molecule inhibitors that compete with SSB for binding ExoI reduces nuclease activity on SSB/ssDNA substrates to its SSB-free level (15,31,32). Interestingly, steady-state kinetic studies with peptide and small-molecule competitors have shown that the inhibitor effects arise from increases in Km and not effects on substrate turnover, suggesting that complex formation with SSB stimulates ExoI activity by improving its association with SSB/ssDNA and/or its retention on such substrates (31,32). However, given that ExoI is exclusively active on ssDNA substrates (25–28), a role for SSB in stimulating its nuclease activity by preventing formation of inhibitory secondary structures in the DNA is likely to be important as well. Resolution of the contributions of SSB DNA binding and protein binding to the stimulation of ExoI activity would help elucidate the mechanisms by which SSB stimulates enzyme activity on SSB/ssDNA substrates.
We have investigated the mechanism of SSB stimulation of ExoI using steady-state kinetic experiments. SSB stimulates ExoI activity through effects on both the apparent kcat and Km. Relative to SSB/ssDNA, ExoI hydrolysis of SSB113/ssDNA substrates occurs with an elevated Km, app, whereas hydrolysis of ssDNA in complex with SSBΔC8, which removes the SSB-Ct sequence, is strongly inhibited. These observations are consistent with a role for SSB/ExoI protein interaction in stabilizing ExoI substrate binding. Bacteriophage T4 gp32, which lacks the SSB-Ct, can stimulate ExoI nuclease activity by increasing kcat, app; however, a hybrid protein in which the SSB-Ct is appended onto gp32 stimulates ExoI to a greater extent by reducing Km, app relative to the wild-type gp32/ssDNA substrate. These data lead to a model in which SSB mediates ExoI binding to SSB/ssDNA substrates and melts secondary structures that could impede ExoI processivity. Consistent with this hypothesis, the specific activity of a fusion protein in which ExoI is tethered to SSB is nearly equivalent to that of SSB-stimulated ExoI. Taken together, these studies delineate stimulatory roles for SSB in which both protein binding and ssDNA binding are important for maximal activity of its protein partners.
MATERIALS AND METHODS
Peptides and proteins
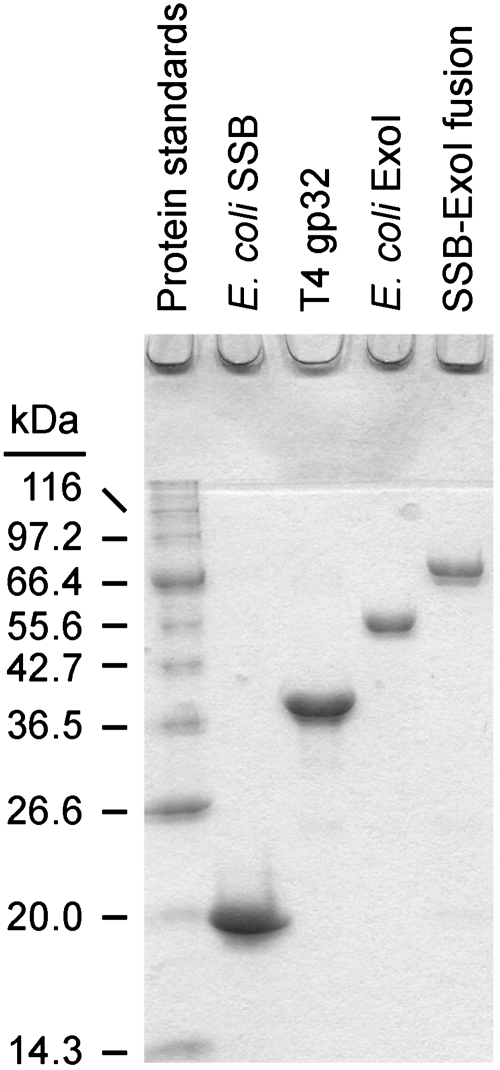
SSB-Ct (Trp-Met-Asp-Phe-Asp-Asp-Asp-Ile-Pro-Phe), F-SSB-Ct (SSB-Ct with an N-terminal fluoresceine moiety) and F-SSB113 (Pro-to-Ser variant of F-SSB-Ct) peptides were prepared as described earlier (15). Escherichia coli ExoI and SSB were purified as described earlier (15) (Figure 1). pDL002 is a T7 overexpression plasmid that encodes an SSB–ExoI fusion protein in which E. coli SSB is tethered directly to the N-terminus of E. coli ExoI. No linker is used in the fusion protein. Expression and purification of the SSB–ExoI fusion protein were carried out as previously described for ExoI (15) (Figure 1). The SSB–ExoI fusion eluted as an oligomeric species of between 2 and 4 protomers per complex in size exclusion chromatography experiments. pET15b-gp32 is a T7 overexpression plasmid that encodes the bacteriophage T4 gp32 protein with a N-terminal hexa-histidine purification epitope (Figure 1), and pET15b-gp32-SSB-Ct is a derivative of pET15b-gp32 that encodes a gp32 variant protein in which the C-terminal residues of T4 gp32 have been changed to encode the final nine residues of E. coli SSB (changing the sequence from Asp–Leu–Asp–Asp–Leu–Leu–Asn–Asp–Leu to Met–Asp–Phe–Asp–Asp–Asp–Ile–Pro–Phe).
Figure 1.
Coomassie-stained samples of purified proteins used in this study resolved by SDS–polyacrylamide gel electrophoresis.
BL21(DE3) E. coli transformed pET15b-gp32 or pET15b-gp32-SSB-Ct were grown at 37°C in Luria-Bertani medium (33) supplemented with 100 µg ml−1 ampicillin. Cells at early logarithmic phase (OD600 of 0.3–0.4) were induced to overexpress T4 gp32 or gp32-SSB-Ct by the addition of 1 mM isopropyl β-d-thiogalactopyranoside and were harvested by centrifugation after an additional 3 h of growth. Cells were resuspended in lysis buffer (20 mM Tris–HCl, pH 8.0, 250 mM NaCl, 20 mM imidazole, 1 mM EDTA, 1 mM phenylmethanesulfonylfluoride, 10% glycerol) and lysed by sonication on ice. Soluble lysates were incubated with nickel–NTA agarose beads (Qiagen). The beads were washed with lysis buffer and bound protein was eluted by the addition of 20 mM Tris–HCl, pH 8.0, 250 mM NaCl, 200 mM imidazole, 1 mM EDTA, 1 mM phenylmethanesulfonylfluoride, 10% glycerol. The eluent was dialyzed against low salt buffer (20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 10% glycerol), bound to a heparin column and eluted with a salt gradient. Fractions containing protein were pooled, concentrated to ∼2 ml and further purified through a Sephacryl S-300 size exclusion column (Pharmarcia) equilibrated in 20 mM Tris–HCl, pH 8.0, 0.5 M NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 10% glycerol. Protein fractions were pooled, concentrated and stored at 4°C (Figure 1). Protein and peptide concentrations were determined by measuring their A280 in 6.0 M Guanidine–HCl (34) [ε280 nm = 73 570 M−1 cm−1 (ExoI), 27 880 M−1 cm−1 (SSB, SSB113 and SSBΔC8, as monomers), 38 690 M−1 cm−1 (gp32 and gp32-SSB-Ct), 100 850 (ExoI-SSB, as monomer) and 5690 M−1 cm−1 (SSB-Ct peptide)]. All SSB and gp32 proteins and variants (except for the SSB–ExoI fusion protein) were free of contaminating nuclease activity.
Exonuclease assays
Nuclease assays were carried out as described previously (15,31). Briefly, a 1369-bp region of a pET-derived plasmid was amplified by PCR in the presence of [α-32P] dTTP to generate internally radiolabeled double-stranded DNA. PCR products were purified by sequential size-exclusion spin columns and boiled for 5 min followed by incubation on ice to produce ssDNA substrate. For specific activity measurements, substrate [1.1 µM (nucleotides)] was mixed with 0–1000 nM SSB (or SSB variant) in Exo buffer (20 mM Tris–HCl, pH 8.0, 10% glycerol, 3 mM MgCl2, and 0.1 g l−1 bovine serum albumin) and incubated for 10 min. In some experiments, SSB-Ct peptide (0–100 µM) was included. Reactions were initiated by the addition of ExoI diluted in Exo buffer (30–100 pM final enzyme concentration) and incubated at 37°C. At several time points, reaction aliquots were quenched by adding EDTA to 0.05 M; remaining substrate was precipitated by adding Torula RNA to 1 g l−1 and perchloric acid to 0.25 N (final concentrations) followed by incubation on ice. Precipitated substrate was pelleted and 80% of the supernatant was removed for liquid scintillation counting to determine the acid-soluble counts per minute (cpm). Reaction velocities were determined by measuring the time-dependent generation of acid-soluble cpm using 3–4 linear time points (<40% of the substrate was hydrolyzed in all time points). One unit of exonuclease activity is the amount of enzyme required to generate 1 µmol of acid-soluble products per min at 37°C; specific enzyme activity is the units per mg of protein. Data are presented as the mean of three measurements with error bars depicting one standard deviation of the mean.
For Michaelis–Menten kinetic measurements, DNA substrate concentrations were varied from 0.5 to 160 µM (nucleotides) and the SSB concentration was altered to maintain a constant ratio of 200 nM SSB (monomers)/1.1 µM (nucleotides) DNA. This ratio is sufficient to saturate the ssDNA with SSB. Saturation kinetic data were fitted using GraphPad Prism as described earlier (31). Km and vmax values were fitted using the equation:
| (1) |
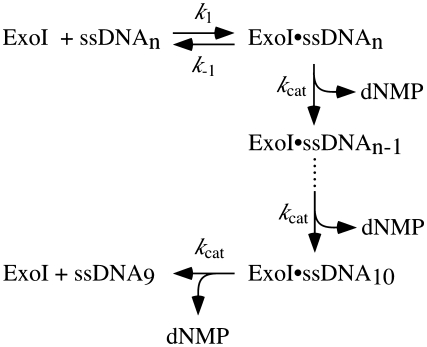
where vmax is the maximal velocity, [S] is ssDNA concentration (in nucleotides) and Km is the [S] required for v to equal one half of vmax. kcat values, which were determined by dividing vmax by [ExoI], were interpreted using equations derived for kinetics of processive ExoI activity by Brody et al. (35), which is described in the following kinetic Scheme 1.
Scheme 1.
Kinetic model of Exonuclease I-catalyzed ssDNA hydrolysis.
In this scheme, k1 is the rate constant for ExoI–substrate association, k−1 is the rate constant for ExoI–substrate dissociation, kcat is the rate constant for nucleotide hydrolysis and n is the number of nucleotides in the ssDNA substrate. Dissociation of ExoI from homopolymeric ssDNA once hydrolysis has begun is only significant when the length of available ssDNA is 10 nt or less (35). In this scheme,
| (2) |
Given the heterogeneous structural nature of the DNA substrate used in this analysis, we use the terms Km, app and kcat, app to describe measured kinetics parameters. Assays were performed with several independently generated substrate preparations.
For processivity experiments, substrate amplification was carried out as described above except using unlabeled dTTP. The DNA was heat denatured, quick-cooled on ice and labeled with 32P on 5′- or 3′-ends using polynucleotide kinase or terminal transferase, respectively, according to the manufacturer's protocol (New England Biolabs). The products were purified as above. Reaction conditions were as described above, with SSB, SSBΔC8, SSB113 or gp32 present at 1.8 µM (monomers), ssDNA at 10 µM (nucleotides) and ExoI at 1 nM. Reactions were quenched at 0, 5, 10, 15 or 20 min and acid-soluble products measured as above.
Peptide binding isotherms
Peptide binding assays were carried out as described previously (15). Briefly, ExoI at 0.1–10 000 nM was incubated with 10 nM F-SSB-Ct or F-SSB113 in 20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 4% (v/v) glycerol, 1 mM 2-mercaptoethanol and 0.1 g l−1 bovine serum albumin at room temperature for 10 min. Fluorescence anisotropy was measured at 25°C with 490 nm excitation and 535 nm emission wavelengths for three replicates; the average anisotropy value was plotted with 1 SD of the mean shown as error. Apparent dissociation constants were the [ExoI] needed for 50% binding saturation.
RESULTS
Based on several lines of evidence, it has been proposed that SSB stimulates ExoI nuclease activity in two ways: by melting out secondary structures that impede ExoI activity and by stabilizing ExoI binding to ssDNA substrates with which SSB forms a nucleoprotein complex (15,29,31,32). To test this model, a steady-state kinetic approach examining E. coli SSB stimulation of the nuclease activity of E. coli ExoI was undertaken. Substrates that included free ssDNA or ssDNA in complex with saturating amounts of different SSBs were tested as substrates for E. coli ExoI and steady-state kinetic parameters were derived from fits of the data.
Escherichia coli SSB stimulation of ExoI activity
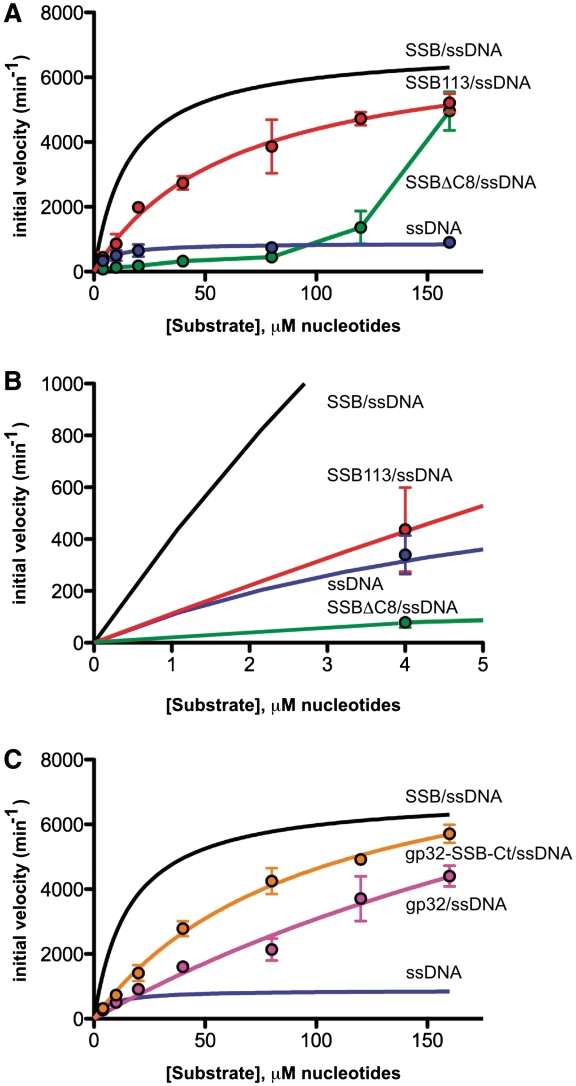
Comparing the hydrolytic activity of E. coli ExoI on free ssDNA and SSB/ssDNA identified a striking difference in kcat, app between the two substrates (Figure 2A and Table 1). The kcat, app for ExoI-catalyzed hydrolysis of free ssDNA was 880 ± 70 min−1, whereas the kcat, app with the same ssDNA substrate saturated with SSB was 6930 ± 260 min−1 (31). Interestingly, the kcat, app for ExoI-catalyzed hydrolysis of SSB/ssDNA is similar to that observed previously with long homopolymeric ssDNA substrates that lack secondary structure [10 400–16 500 min−1 (35,36)], which could indicate that the kcat, app difference between free heteropolymeric ssDNA and SSB/ssDNA arises from secondary structure melting by SSB producing more optimal substrates for ExoI. Consistent with this model, earlier experiments have demonstrated that apparent ExoI hydrolysis rates with homopolymeric ssDNA are reduced by annealing complementary ssDNA to the substrate (27,28).
Figure 2.
Saturation kinetics of ExoI nuclease activity on ssDNA nucleoprotein substrates. (A) Initial ExoI DNA hydrolysis velocities are plotted as a function of ssDNA (blue), SSBΔC8/ssDNA (green) or SSB113/ssDNA (red) concentrations. The saturation kinetic plot for ExoI with SSB/ssDNA measured under identical conditions (31) is shown (black line) for comparative purposes. Data points are reported as the mean of three independent measurements with error bars as 1 SD of the mean. Lines depict fits of the data to Michaelis–Menten kinetics. (B) Close-up of (A) to demonstrate the effects of each SSB variant at low substrate concentrations. Data points are reported as the mean of three independent measurements with error bars as 1 SD of the mean. (C) Initial ExoI DNA hydrolysis velocities are plotted as a function of gp32/ssDNA (magenta) or gp32-SSB-Ct/ssDNA (orange) concentrations. The saturation kinetic plots for ExoI with SSB/ssDNA measured (31) (black line) or ssDNA [from (A)] (blue line) are shown for comparative purposes. Data points are reported as the mean of three independent measurements with error bars as 1 SD of the mean. Lines depict fits of the data to Michaelis–Menten kinetics.
Table 1.
Steady-state kinetic parameters
| Exonuclease substrate |
||||||
|---|---|---|---|---|---|---|
| ssDNA | SSB/ssDNAa | SSB113/ssDNA | SSBΔC8/ssDNA | gp32/ssDNA | gp32-SSB-Ct/ssDNA | |
| Km, app (µM, nucleotides) | 7 ± 2 | 16 ± 2 | 63 ± 10 | ND | 380 ± 200 | 100 ± 12 |
| kcat, app (min−1) | 880 ± 70 | 6930 ± 260 | 7160 ± 500 | ND | 14 900 ± 5,800 | 9200 ± 540 |
| kcat, app/Km, app | 130 ± 40 | 430 ± 60 | 110 ± 20 | ND | 40 ± 20 | 90 ± 10 |
aValues are from Ref. (31). ND, not determinable.
In contrast to the kcat, app enhancement, there was little or no difference between the Km, app values for the hydrolysis of ssDNA and SSB/ssDNA substrates (Table 1). Comparison of Km, app values between these two substrates could be difficult to interpret, however, since the Michaels constants are sensitive to several rate constants and to the length of available ssDNA [see Equation (1)]. Since SSB and ExoI directly interact with one another, the impact of protein interaction from that of SSB/ssDNA interactions cannot be distinguished in the measured kinetic parameters. To better compare Km, app values between related substrates, the effects of two SSB variant proteins with reduced ExoI binding affinity (SSB113 and SSBΔC8) on ExoI activity were measured.
Comparison of ExoI activity on SSB/ssDNA and SSB113/ssDNA substrates revealed a difference in Km, app values (Figure 2A and Table 1). SSB113, which has a Pro-to-Ser substitution in the SSB-Ct sequence, has a dramatically reduced ExoI binding affinity but has wild-type ssDNA binding properties (8,14,15,31). The Km, app for ExoI-catalyzed hydrolysis of the SSB113/ssDNA substrate was 4-fold higher than SSB/ssDNA but the kcat, app values were indistinguishable between the two substrates (Table 1). Since the kcat, app value is constant between the two substrates, the Km, app differences must arise from changes in the binding or dissociation kinetics of ExoI/SSB113/ssDNA complex formation [see Equation (1)]. Interestingly, the catalytic efficiency of ExoI hydrolysis of SSB113/ssDNA and naked ssDNA, as expressed by kcat, app/Km, app, were essentially identical; only the SSB/ssDNA substrate was measurably elevated (Table 1). These data, therefore, support a model in which the SSB-Ct provides a site for binding ExoI to SSB/ssDNA substrates.
The SSBΔC8/ssDNA substrate, in which the entire ExoI-binding site on SSB (14,15) is removed, is a markedly inferior substrate relative to all others tested (Figure 2A). ExoI nuclease activity with low SSBΔC8/ssDNA substrate concentrations was greatly retarded relative to the activity with ssDNA, SSB/ssDNA or SSB113/ssDNA and increased only in the highest substrate concentrations tested (Figure 2A and 2B). Saturated activity levels could not be measured but assuming that the kcat, app value for ExoI hydrolysis of SSBΔC8/ssDNA is similar to that with other SSB/ssDNA substrates tested, 50% maximal velocity requires ∼140 µM SSBΔC8/ssDNA concentrations. Taken together, these results indicate that SSBΔC8 likely has effects beyond impairment in association with ExoI in the assay. Indeed, E. coli SSB variants that lack the SSB-Ct bind ssDNA with higher affinity and cooperativity than SSB (37–39), which could be related to inhibition of ExoI nuclease activity by SSBΔC8. This possibility is considered in the ‘Discussion’ section.
It is interesting to note that ExoI activity on the SSBΔC8/ssDNA substrate appears to be cooperative. Although it is not clear how this relates to the ExoI association with SSBΔC8/ssDNA, it could possibly reflect physical interactions between multiple ExoI monomers and the SSBΔC8/ssDNA substrate. Since the activity only occurs with very high substrate concentrations, this could be an artifact of our reconstituted biochemical systems rather than an activity that is biologically relevant.
We examined the kinetics of ExoI-catalyzed hydrolysis of each of the substrates at low concentrations to better appreciate the effects that SSB when a limited number of 3′ ssDNA ends are present, as might be found in vivo (Figure 2B). Given an average sized E. coli cell, a single 3′ ssDNA end would provide a substrate concentration of ∼1 nM (3′ ends), which is equivalent to a 1.4 µM (nucleotides) concentration of the substrate used in this study. Interestingly, it is apparent that only SSB significantly stimulates the velocity of the ExoI hydrolysis reaction under this condition; SSB113/ssDNA is hydrolyzed at essentially the same rate as ssDNA alone (Figure 2B). This effect is due ultimately to the Km, app difference between the SSB/ssDNA and SSB133/ssDNA substrates. Since the presence of ssDNA ends is likely to be quite limited under most conditions in E. coli, activities at the lower substrate concentrations shown in Figure 2B may reflect more typical ExoI activity levels and thus highlight the importance of the Km, app difference between SSB/ssDNA and SSB113/ssDNA substrates.
T4 gp32 stimulation of ExoI activity
To complement the experiments described above, the effects of the SSB from bacteriophage T4 (gp32) on E. coli ExoI nuclease activity were measured. T4 gp32 differs from E. coli SSB in two ways: (i) it lacks the conserved SSB-Ct sequence that is required for complex formation with ExoI and (ii) it is a monomer rather than a tetramer (6,40). These differences make gp32 a useful tool for testing the general effects of ssDNA binding in ExoI stimulation by SSB.
ExoI catalyzed hydrolysis of the gp32/ssDNA substrate with a Km, app of 380 ± 200 µM and a kcat, app of 14 900 ± 5800 min−1 (Figure 2C and Table 1). These fitted values are notably less precise than those obtained for ssDNA or SSB/ssDNA since the substrate concentrations for which initial velocities were measured were all below the estimated Km, app. However, they indicate that gp32/ssDNA is a poor substrate compared with SSB/ssDNA in terms of Km, app (∼24-fold difference). In contrast, the kcat, app is similar to that of SSB/ssDNA or SSB113/ssDNA substrates. These data are consistent with SSBs generally increasing kcat, app for ExoI-catalyzed hydrolysis by melting secondary structures in the DNA (an activity present in both E. coli SSB and T4 gp32).
To further test the role of recruitment in ExoI stimulation, a chimeric protein in which the T4 gp32 C-terminus is changed to that of the E. coli SSB-Ct (gp32-SSB-Ct) was created, purified and tested for stimulation of ExoI nuclease activity. ExoI hydrolyzed the gp32-SSB-Ct/ssDNA substrate with a Km, app of 100 ± 12 µM and a kcat, app of 9200 ± 540 min−1 (Figure 2C and Table 1). Relative to the gp32/ssDNA substrate, the Km, app of the ExoI nuclease activity was reduced ∼4-fold. This improvement is remarkably similar to the ∼4-fold lower Km, app for hydrolysis of SSB/ssDNA relative to SSB113/ssDNA (Table 1) and could reflect the extent to which protein interaction-mediated recruitment augments ExoI activity. In contrast, the addition of the SSB-Ct tail to T4 gp32 had little or no effect on kcat, app.
ExoI processivity is impaired on the SSBΔC8/ssDNA substrate
One potential complication in the above kinetic studies is that the presence of different SSB species could influence the processivity of ExoI. To test for this possibility, an assay similar to that developed by Thomas and Olivera (28) was used to measure the relative processivity of ExoI on several different substrates (ssDNA, SSB/ssDNA, SSB113/ssDNA, SSBΔC8/ssDNA and gp32/ssDNA). In this method, ssDNA hydrolysis progress is monitored on substrates with either 3′ or 5′ 32P label. Since ExoI is a 3′–5′ nuclease, processivity is determined by examining the hydrolysis kinetics of the 3′ versus 5′ labels; for a perfectly processive enzyme, these kinetics would be identical.
The most striking effect in this assay was observed with the SSBΔC8/ssDNA substrate (Supplementary Figure S1). Hydrolysis of the SSBΔC8/ssDNA 3′ label was modestly slowed relative to other substrates, whereas hydrolysis of the 5′ label was nearly absent over the time course, consistent with a very poorly processive nuclease activity. Apparent ExoI processivity on SSB/ssDNA, SSB113/ssDNA and gp32/ssDNA was quite similar (Supplementary Figure S1).
Tethering E. coli SSB to ExoI increases specific activity of the nuclease
The kinetic experiments described above suggested that SSB stimulates ExoI nuclease activity by melting out secondary structures in the ssDNA and by stabilizing ExoI complexes with SSB/ssDNA substrates through direct protein–protein interaction. This model predicts that a fusion protein in which SSB is linked to ExoI would have heightened specific activity on ssDNA relative to ExoI alone, particularly at lower substrate concentrations where activity is most limited by ExoI substrate binding. To test this idea, a recombinant fusion protein in which ExoI is tethered to the C-terminus of SSB was created and purified. The nuclease activity of the SSB–ExoI fusion enzyme was then measured and compared with the results from above and from previous experiments (31) carried out under the same conditions. Saturation kinetic experiments with the fusion protein and several SSB/ssDNA substrates failed to reach saturation under the substrate concentrations tested (Supplementary Figure S2). Experiments with the fusion protein were thus limited to comparing its specific nuclease activity to that of ExoI and to examining the effects of SSB on its nuclease activity.
As predicted by the model, the SSB–ExoI fusion protein had a significantly higher specific activity than ExoI with free ssDNA (Table 2). At a 1.1 µM (nucleotides) substrate concentration, the purified ExoI–SSB fusion protein hydrolyzes ssDNA with a specific activity of 16.3 ± 1.5 U mg−1, which is 3.5-fold higher than that of ExoI under the same conditions (15) (Table 2). Interestingly, this higher specific activity for the ExoI–SSB fusion protein was similar to that of ExoI stimulated by the addition of saturating amounts of SSB [20.7 ± 2.9 U mg−1 (15)]. Thus, fusing ExoI with SSB creates an enzyme that appears to mimic SSB-stimulated ExoI.
Table 2.
Specific activities of ExoI and ExoI fusion proteins
| Specific activity of nuclease with free ssDNA or SSB variant/ssDNA substrates (U mg−1) |
||||||
|---|---|---|---|---|---|---|
| ssDNA | SSB/ssDNA | SSB113/ssDNA | SSBΔC8/ssDNA | gp32/ssDNA | gp32-SSB-Ct/ssDNA | |
| ExoI | 4.7 ± 0.4a | 20.7 ± 2.9a | 5.6 ± 1.3a | 0.6 ± 0.1 | 1.0 ± 0.1 | 6.1 ± 4.6 |
| SSB–ExoI | 16.3 ± 1.5 | 29.4 ± 2.1 | 27.4 ± 1.4 | 9.9 ± 1.9 | 12.4 ± 2.4 | 19.7 ± 4.0 |
aValues are from Ref. (15).
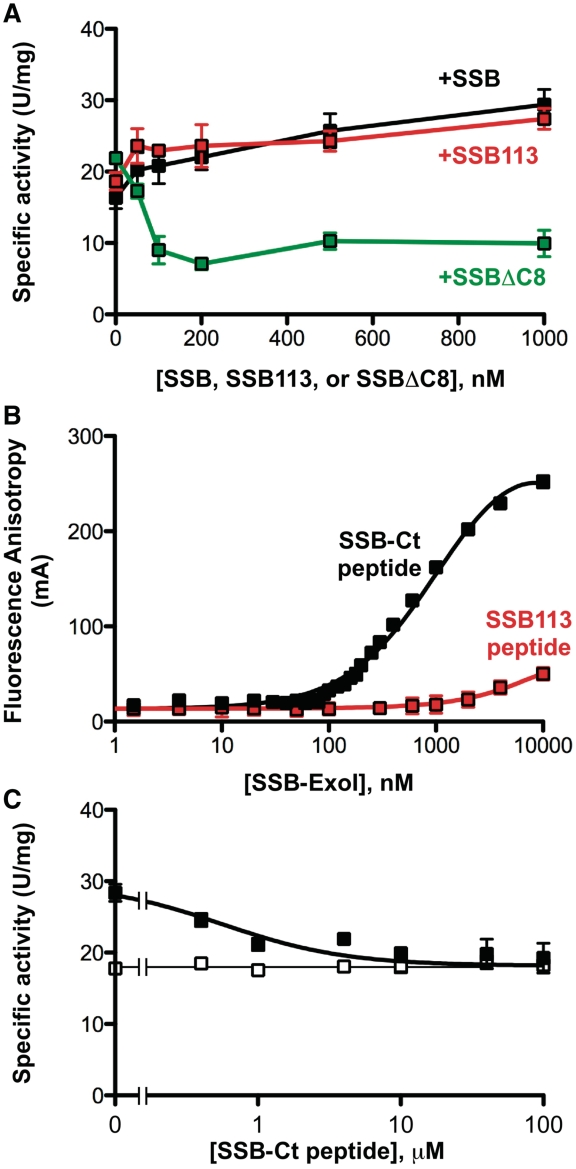
The SSB-dependence of the ExoI–SSB fusion protein activity was tested to determine whether its activity could be stimulated similarly to that of ExoI. The addition of SSB stimulated the activity of the ExoI–SSB fusion protein ∼1.8 fold (Figure 3A and Table 2), which is less than the SSB's 4.4-fold stimulation of ExoI (15). The addition of SSB113 provided a similarly modest 1.7-fold stimulation to the ExoI–SSB fusion protein, whereas SSBΔC8 inhibited activity (Figure 3A and Table 2). Modest stimulation of ExoI by SSB113 (15) and inhibition by SSBΔC8 were also observed (Table 2).
Figure 3.
Nuclease activities of an SSB–ExoI fusion protein. (A) SSB–ExoI fusion protein specific activity is plotted as a function of SSB (black), SSB113 (red) or SSBΔC8 (green) [concentrations are nM (monomers)]. (B) Equilibrium SSB–ExoI binding to F-SSB-Ct or F-SSB113 peptide as monitored by fluorescence anisotropy. All data points are the average of three experiments. Error bars are 1 SD from the mean and are sufficiently small to be occluded by the symbols in most cases. Concentrations of ExoI–SSB are expressed in nM (monomers). (C) SSB-Ct peptide was incubated at indicated concentrations in SSB–ExoI nuclease assays with 200 nM SSB included (filled squares) or omitted (open squares). Specific activity values are presented as the mean of three measurements with errors bar depicting 1 SD.
Experiments were next performed to test whether the modest SSB-dependent increase in ExoI–SSB activity was due to binding of the fusion protein to SSB. Previous experiments showed that a peptide comprising the SSB-Ct sequence acts as a competitive inhibitor that selectively abrogates SSB-stimulation of ExoI activity by blocking complex formation (31). To test whether recruitment is responsible for SSB-stimulation of ExoI–SSB, the effects of the SSB-Ct peptide inhibitor on nuclease activity were measured. First, the ability of the fusion protein to bind to the SSB-Ct was confirmed using a previously established fluorescence-anisotropy assay (Figure 3B). The ExoI–SSB protein binds a fluorescently labeled SSB-Ct peptide with an apparent dissociation constant of 1.1 ± 0.09 µM. Similarly to ExoI (15), binding to the SSB-Ct peptide was specific; the ExoI–SSB fusion protein only bound an SSB113 control peptide at the highest concentrations tested (>5µM, Figure 3B). Next, the ability of the SSB-Ct to abrogate SSB stimulation of ExoI–SSB activity was tested. Similarly to its effects with ExoI (15), the SSB-Ct peptide abrogated the modest SSB stimulation of the ExoI–SSB fusion protein and had no effect on the activity of the fusion protein in the absence of SSB (Figure 3C). These data indicated that the modest SSB enhancement of activity in the fusion protein is likely due to a recruitment effect that is reduced relative to that observed with ExoI. We note that this modest effect could reflect stimulation of a small amount of ExoI that has been cleaved from the fusion protein by proteolysis rather than SSB recruitment of the ExoI–SSB fusion protein.
DISCUSSION
SSB/ssDNA structures are central substrates in cellular genome maintenance pathways. To better understand how SSB/ssDNA substrates are processed, we have measured the effects of SSB and several SSB variant proteins on the steady-state nuclease activity of ExoI. Our findings show that SSB stimulates ExoI-catalyzed ssDNA hydrolysis through effects on both kcat, app and Km, app, leading to a model in which SSB interactions with both ssDNA and ExoI are responsible for SSB stimulation of ExoI activity. Interestingly, the specific activity of a fusion protein in which ExoI is tethered to SSB is nearly equivalent to that of SSB-stimulated ExoI. This result has implications on the possible importance of SSB mobility in stimulating ExoI nuclease activity as described below.
Based on the data presented in this manuscript and elsewhere (15,29,31,32), we propose a model for SSB stimulation of ExoI nuclease activity (Figure 4). ExoI catalyzes hydrolysis of ssDNA in a 3′–5′ direction (25). When acting on homopolymeric ssDNA substrates, which lack secondary structure, ExoI processively hydrolyzes DNA at a previously measured maximal rate of 10 400–16 500 min−1 (35,36). However, with more biologically relevant mixed-sequence ssDNA substrates, secondary structures exist in the free DNA that attenuate ExoI nuclease activity (26–28). When ExoI encounters such structures, hydrolysis would likely be halted and the enzyme can dislodge from the substrate as has been described (27,28) (Figure 4A). Our model attributes the low relative kcat, app of ExoI on the heteropolymeric ssDNA substrate used in these experiments [880 ± 70 min−1 (Table 1)] to this effect. Consistent with this interpretation, inclusion of saturating concentrations of either E. coli SSB or T4 gp32, which melt out secondary structural elements in the substrate (Figure 4B), led to significant enhancements in kcat, app values [6930 ± 260 min−1 for SSB and 14 900 ± 5800 min−1 for gp32 (Table 1)]. The kcat, app values for ExoI activity on SSB/ssDNA and gp32/ssDNA substrates were very similar to those measured with homopolymeric ssDNA substrates.
Figure 4.
Model of SSB stimulation of ExoI nuclease activity. Model depicting SSB-free and SSB-stimulated ExoI activity are shown. In (A) ExoI activity is limited by the presence of secondary structures within the ssDNA substrate that preclude processive hydrolysis of the substrate. In (B) SSB stimulates ExoI activity by stabilizing the ssDNA structure and by recruiting ExoI to the substrate though direct protein interaction.
Experiments involving SSB proteins with altered ExoI binding sites were used to better dissect the role of ExoI/SSB interactions on ExoI activity. These showed that ExoI binding to SSB is important for stimulation. Substrates formed between ssDNA and either SSB113 or T4 gp32 are hydrolyzed by ExoI with significantly higher Km, app values than SSB/ssDNA but without significant effects on kcat, app (Table 1). The relatively poor Km, app of the gp32/ssDNA substrate was improved by changing its C-terminus to the E. coli SSB-Ct sequence so as to enable ExoI binding to the protein (Table 1). The Km, app is dependent on the binding rate constant (k1), dissociate rate constant (k-1) and hydrolysis rate constant (kcat) as is delineated in Equation (1). Since kcat, app is very similar among the various SSB/ssDNA substrates tested (Table 1), the differences in Km, app observed with SSB113 and gp32 most likely arise from changes in k1, k−1 or both. As such, these changes appear to reflect the contribution of the ExoI/SSB interaction in stimulating ExoI nuclease activity. These experiments, coupled with earlier observations showing that peptide and small-molecule inhibitors that block ExoI/SSB complex formation increase ExoI Km, app on SSB/ssDNA substrates (31,32), indicate that ExoI engages both the ssDNA and SSB in processing the nucleoprotein complex (Figure 4B).
A fusion protein in which ExoI is added to the C-terminus of SSB further tested the importance of SSB interaction with ExoI in stimulating activity. As predicted by the model in Figure 4, the specific activity of the fusion protein with free heteropolymeric ssDNA was nearly identical to that of ExoI on SSB-saturated substrates (Table 2 and Figure 3). The fusion protein's activity was minimally stimulated by adding more SSB. This latter point highlights a possible arrangement in which ExoI relies on SSB mobility to melt out secondary structures in ssDNA. Since the ExoI portion of the fusion protein must engage the 3′-end of ssDNA wrapped around its SSB domain, nuclease activity would seem to require the fusion protein to slide along the ssDNA. In this model, as the protein moves along DNA, the SSB domain of the fusion protein would melt out inhibitory structures in advance of substrate encountering the ExoI domain. This action would proceed in a 3′–5′ direction as determined by the polarity of ExoI activity. For native ExoI activity on SSB/ssDNA substrates, it would follow that ExoI could engage the SSB tetramer present on the 3′-end of the ssDNA substrate and maintain contact with the same SSB throughout hydrolysis of the ssDNA substrate, pushing more 5′-proximal SSB tetramers along the ssDNA as it proceeds. An analogous model in which RecA protein filaments push SSB tetramers along ssDNA in a polar manner has been proposed based on single-molecule experiments (5). These observations could indicate that SSB mobility on ssDNA is an important feature for utilization of SSB/ssDNA substrates by other genome maintenance enzymes as well.
We propose that the striking inhibition of ExoI activity by SSBΔC8 (Figure 2 and Table 1) could be related to the importance of SSB mobility for proper biochemical stimulation. Variants of E. coli SSB lacking the SSB-Ct bind ssDNA with higher affinity and in a more highly cooperative manner than wild-type E. coli SSB (37–39). These differences could modulate SSBΔC8 mobility, making it act as a roadblock to ExoI progression (or, potentially, as an impediment to any SSB/ssDNA-processing enzyme). Similar inhibition by E. coli SSB variants lacking the SSB-Ct has been observed with RecQ and PriA (16,18). Future studies will be required to examine the role of SSB mobility in processing of SSB/ssDNA substrates and the physical basis underlying inhibition in SSBΔC8.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM068061 to J.L.K.). Funding for open access charge: National Institutes of Health (GM068061).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank John Denu and members of the Keck lab for critical reading of the manuscript.
REFERENCES
- 1.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Micro. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 3.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 5.Roy R, Kozlov AG, Lohman TM, Ha T. SSB diffusion on single stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberts BM, Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 7.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 8.Chase JW, L'Italien JJ, Murphy JB, Spicer EK, Williams KR. Characterization of the Escherichia coli SSB-113 mutant single-stranded DNA-binding protein. Cloning of the gene, DNA and protein sequence analysis, high pressure liquid chromatography peptide mapping, and DNA-binding studies. J. Biol. Chem. 1984;259:805–814. [PubMed] [Google Scholar]
- 9.Meyer RR, Glassberg J, Scott JV, Kornberg A. A temperature-sensitive single-stranded DNA-binding protein from Escherichia coli. J. Biol. Chem. 1980;255:2897–2901. [PubMed] [Google Scholar]
- 10.Meyer RR, Rein DC, Glassberg J. The product of the lexC gene of Escherichia coli is single-stranded DNA-binding protein. J. Bacteriol. 1982;150:433–435. doi: 10.1128/jb.150.1.433-435.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BF. Two-dimensional electrophoretic analysis of the regulation of SOS proteins in three ssb mutants. Arch. Microbiol. 1984;138:106–112. doi: 10.1007/BF00413009. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg J, Berends LJ, Donch J, Green MH. exrB: a malB-linked gene in Escherichia coli B involved in sensitivity to radiation and filament formation. Genet. Res. 1974;23:175–184. doi: 10.1017/s0016672300014798. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg J, Donch J. Sensitivity to elevated temperatures in exrB strains of Escherichia coli. Mutat. Res. 1974;25:403–405. doi: 10.1016/0027-5107(74)90070-0. [DOI] [PubMed] [Google Scholar]
- 14.Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 15.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl Acad. Sci. USA. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 17.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J. Mol. Biol. 2009;386:612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res. 2008;36:7029–7042. doi: 10.1093/nar/gkn795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arad G, Hendel A, Urbanke C, Curth U, Livneh Z. Single-stranded DNA-binding Protein Recruits DNA Polymerase V to Primer Termini on RecA-coated DNA. J. Biol. Chem. 2008;283:8274–8282. doi: 10.1074/jbc.M710290200. [DOI] [PubMed] [Google Scholar]
- 22.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glover BP, McHenry CS. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 24.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 25.Lehman IR. The deoxyribonucleases of Escherichia coli. I. Purification and properties of a phosphodiesterase. J. Biol. Chem. 1960;235:1479–1487. [PubMed] [Google Scholar]
- 26.Lehman IR, Nussbaum AL. The Deoxyribonucleases of Escherichia Coli. V. on the Specificity of Exonuclease I (Phosphodiesterase) J. Biol. Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 27.Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- 28.Thomas KR, Olivera BM. Processivity of DNA exonucleases. J. Biol. Chem. 1978;253:424–429. [PubMed] [Google Scholar]
- 29.Molineux IJ, Gefter ML. Properties of the Escherichia coli DNA-binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J. Mol. Biol. 1975;98:811–825. doi: 10.1016/s0022-2836(75)80012-x. [DOI] [PubMed] [Google Scholar]
- 30.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat. Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 31.Lu D, Windsor MA, Gellman SH, Keck JL. Peptide inhibitors identify roles for SSB C-terminal residues in SSB/Exonuclease I complex formation. Biochemistry. 2009;48:6764–6771. doi: 10.1021/bi900361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu D, Bernstein DA, Satyshur KA, Keck JL. Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance. Proc. Natl Acad. Sci. USA. 2010;107:633–638. doi: 10.1073/pnas.0909191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor: CSHL Press; 2001. [Google Scholar]
- 34.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 35.Brody RS, Doherty KG, Zimmerman PD. Processivity and kinetics of the reaction of exonuclease I from Escherichia coli with polydeoxyribonucleotides. J. Biol. Chem. 1986;261:7136–7143. [PubMed] [Google Scholar]
- 36.Werner JH, Cai H, Keller RA, Goodwin PM. Exonuclease I hydrolyzes DNA with a distribution of rates. Biophys. J. 2005;88:1403–1412. doi: 10.1529/biophysj.104.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Biol. Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 38.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between dna binding modes of E. coli SSB protein. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozlov AG, Cox MM, Lohman TM. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 2010;285:17246–17252. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams KR, LoPresti MB, Setoguchi M, Konigsberg WH. Amino acid sequence of the T4 DNA helix-destabilizing protein. Proc. Natl Acad. Sci. USA. 1980;77:4614–4617. doi: 10.1073/pnas.77.8.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.