Abstract
Triplex formation is a promising strategy for realizing artificially controlling of gene expression, reversible assembly of nanomaterials and DNA nanomachine and single-walled nanotubes (SWNTs) have been widely used as gene and drug delivery vector or as ‘building blocks’ in nano-/microelectronic devices. CGC+ triplex is not as stable as TAT triplex. The poor stability of CGC+ triplex limits its use in vitro and in vivo. There is no ligand that has been reported to selectively stabilize CGC+ triplets rather than TAT. Here, we report that SWNTs can cause d(CT)•d(AG) duplex disproportionation into triplex d(C+T)•d(AG)•d(CT) and single-strand d(AG) under physiological conditions. SWNTs can reduce the stringency of conditions for CGC+ triplex formation studied by UV–vis, CD, DNA melting, light scattering and atomic force microscopy. Further studies indicate that electrostatic interaction is crucial for d(CT)•d(AG) repartition into triplex d(C+T)•d(AG)•d(CT). Our findings may facilitate utilization of SWNTs–DNA complex in artificially controlling of gene expression, nanomaterials assembly and biosensing.
INTRODUCTION
Nucleic acid triplex formation has received much attention because of their potentials in biomedical and biotechnological applications, such as exploiting the third strand binding as an artificial mechanism for the selective inhibition of gene expression and as a tool for site-specific delivery of reagents to genomes (1–3). Stretches of duplex sequence d(A–G)6 d(C–T)6 are well known in eukaryotic genomes (4), particularly in promoter and gene switch regions (4,5), where they are thought prone to form triplex through protonation of dC residues in the third strand d(C-T)6 (6). Although forming the isostructural triplet, the interaction of C+ with GC base pairs is much weaker than T with AT under physiological conditions (6). The poor stability of CGC+ triplex limits its use in vitro and in vivo (7,8). To overcome this drawback, one way is to screen small molecules to increase triplex stability. A series of natural and synthetic compounds have been reported to bind specifically to triplex DNA or RNA, some even can induce triplex formation (9–13). However, there is no ligand that has been reported selectively to stabilize CGC+ triplets rather than TAT, presumably because the positive charge on the protonated cytosine prevents binding of the cationic ligand (14). The other way is to use artificial DNA analogues. Although many DNA analogues remove pH dependency, none of these have been widely used (7). Greater efforts are still needed on how to selectively stabilize CGC+ triplex as well as how to promote this triplex formation under physiological conditions.
Single-walled carbon nanotubes (SWNTs) have been considered as the leading candidate for nanodevice applications because of their one-dimensional electronic band structure, molecular size, biocompatibility, controllable property of conducting electrical current and reversible response to biochemical reagents (15–20), SWNTs are widely used as gene and drug delivery vector or as ‘building blocks’ in nano-/micro-electronic devices. Among the molecules that can non-covalently bind to SWNTs, DNA has been the research focus, which adsorbs as single-stranded (ssDNA) or double-stranded DNA (dsDNA) complex. Previous studies have shown that ssDNA can helically wrap around SWNTs by means of π-stacking interactions between nucleotide bases and SWNT sidewalls (21), while dsDNA can bind to SWNTs via groove binding and DNA end absorption (22,23). Further studies demonstrate that DNA binding to SWNTs may influence DNA structure (24–31). In a recent report, duplex DNA can absorb on SWNTs to give an ordered multilayered structure (24). We have reported that SWNTs can induce a sequence dependent DNA B–A transition and selectively induce human telomeric i-motif DNA formation (25–30). In addition, SWNTs can cause single-stranded poly(rA) to self-structure and form a duplex structure (31). However, to our knowledge, there is no report to show that SWNTs can facilitate triplex DNA formation.
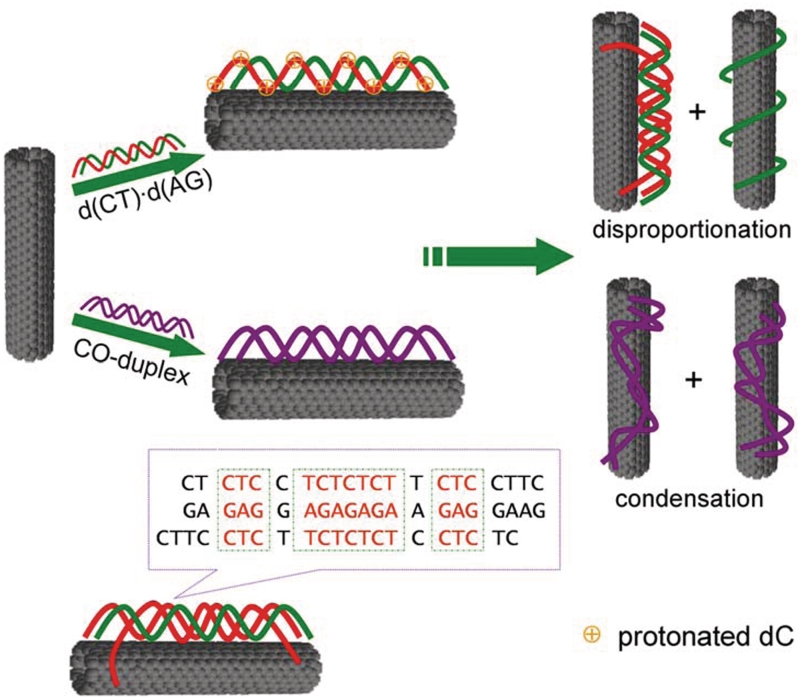
In the present work, we report that duplex d(CT)•d(AG) can be disproportioned into triplex d(C+T)•d(AG)•d(CT) and single-strand d(AG) in the presence of carboxyl-modified SWNTs (SWNTs–COOH) at pH 6.5 (Scheme 1). The disproportionation increases SWNTs stability in solution. However, at pH 8.5, duplex DNA is condensed on the surface of SWNTs, which decreases SWNTs stability. Carboxyl-/hydroxyl-modified SWNTs can induce the formation of triplex d(C+T)•d(AG)•d(CT), while positively charged amino-modified SWNTs cannot. This indicates that electrostatic interaction is crucial for d(CT)•d(AG) repartition into triplex d(C+T)•d(AG)•d(CT) except hydrophobic interaction and π–π stacking.
Scheme 1.
Schematic representation of disproportionation of d(CT)·d(AG) and condensation of control duplex DNA (CO-duplex) in the presence of SWNTs under physiological conditions. The control duplex cannot form triplex DNA.
MATERIALS AND METHODS
Materials
Single-walled carbon nanotubes (φ = 1.1 nm, purity >90%) were purchased from Aldrich (St Louis, MO, USA). The details of preparation of SWNT–COOH, SWNT–OH and SWNT–NH2 were described (25,31) in Supplementary Data. The stock solution of these three kinds of modified SWNTs (0.10 mg ml−1) was obtained by sonicating the samples for 8 h in pH 7.0 aqueous solution (25–30).
DNA oligomers were synthesized by Sangon Biotechnology Co. (Shanghai, China) and used as received: d(AG), 5′-GAG AGG AGA GAG AAG AGG AAG-3′; d(CT), 5′-CTT CCT CTT CTC TCT CCT CTC-3′; d(TC), 5′-CTC TCC TCT CTC TTC TCC TTC-3′; 20A, 5′-GAA CGA AAC CAT TAT ACG AT-3′; 20B, 5′-ATC GTA TAA TGG TTT CGT TC-3′. Concentrations of DNA oligomers were determined by measuring the absorbance at 260 nm after melting using extinction coefficient: ε260nm = 242 100 M−1 cm−1 (d(AG)), ε260nm = 160400 M−1 cm−1(d(CT)), ε260nm = 160 400 M−1 cm−1(d(TC)), ε260nm = 207 900 M−1 cm−1 (20A) and ε260nm = 192 900 M−1 cm−1 (20B). The extinction coefficient was calculated from mononucleotide and dinucleotide data by using nearest-neighbor approximation (25–30). Single-strand d(CT) and single strand d(AG) were mixed at pH 6.5 in a 1:1 ratio, heating to 95°C and slowly cooling to room temperature. The formed duplex DNA is denoted as d(CT)·d(AG). Control duplex DNA (CO-duplex) refers to mixing single strand 20A and single strand 20B at pH 6.5 in a 1:1 ratio, heating to 95°C and slowly cooling to 4°C. Unlike d(CT)·d(AG) DNA, CO-duplex cannot form triplex DNA. All experiments were carried out in aqueous cacodylic buffer (1 mM cacodylic acid/sodium cacodylate/200 mM NaCl) unless stated otherwise (25–30).
In order to illustrate, SWNTs can facilitate CGC+ triplex DNA formation, we used a reversible sequence d(TC)(3′-CTCTCCTCTCTCTTCTCCTTC-5′) instead of d(CT) (5′-CTT CCT CTT CTC TCT CCT CTC-3′), which can form a perfect matched CGC+ triplex DNA with d(CT)·d(AG). In the mismatched triplex or the perfect matched triplex, the same duplex d(CT)·d(AG) was used to perform the experiment, while the other ssDNA are different.
Bioassay
UV absorbance measurements and melting experiments were carried out on a Jasco-V550 UV/Vis spectrophotometer equipped with a Peltier temperature control accessory. All UV/Vis spectra were measured in 1.0 cm path length quartz cuvettes with the same concentration of SWNTs aqueous solution as the reference. Dry purified nitrogen was passed through the cell compartment to prevent condensation on the cells at low temperature. Flow rate was set low enough so as not to create a temperature gradient between the sample and the Peltier, which was confirmed by monitoring the temperature in the sample and the Peltier during trial melting profiles (6). Absorbance changes at 260 nm versus temperature were collected at a heating rate of 1°C min−1 for DNA melting experiments. The sample solution was prepared by mixing SWNT–COOH and DNA and then stood overnight at 4°C to perform melting experiment.
CD spectra were measured on a JASCO J-810 spectropolarimeter equipped with a temperature-controlled water bath at 4°C. The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen for 45 min before use and kept the nitrogen atmosphere during experiments. Three scans were accumulated and automatically averaged.
The light scattering (LS) was measured with a JASCO FP-6500 spectrofluorometer. The sample solution was prepared by mixing 15 μg ml−1 SWNT and 1 μM d(CT)·d(AG) and then stood for 24 h at 4°C followed by centrifugation (1000 rpm, 111.5g). The supernatant was decanted and collected for LS measurements. The LS spectra were obtained by simultaneously scanning the excitation and emission monochromators from 220 nm to 700 nm with the slit width for the excitation and emission of 5 nm (32).
Atomic force microscopy (AFM) measurements (25,31) were performed using Nanoscope V multimode atomic force microscope (Veeco Instruments, Santa Barbara, CA, USA). The sample solution was stood overnight at 4°C, then deposited onto a piece of freshly cleaved mica with APTES and rinsed with water and dried before measurements. Tapping mode was used to acquire the images under ambient conditions (25,31,33). Images were collected at a 1 Hz scan rate and 512 × 512 pixel resolution. Image analysis of SWNTs-DNA was performed using Nanoscope v7.30 software (Veeco Instruments). Measurements were made for the height of over 100 peaks and are reported as the Gaussian center (calculated using Origin6.0, OriginLab Corp., Northampton, MA, USA).
RESULTS AND DISCUSSION
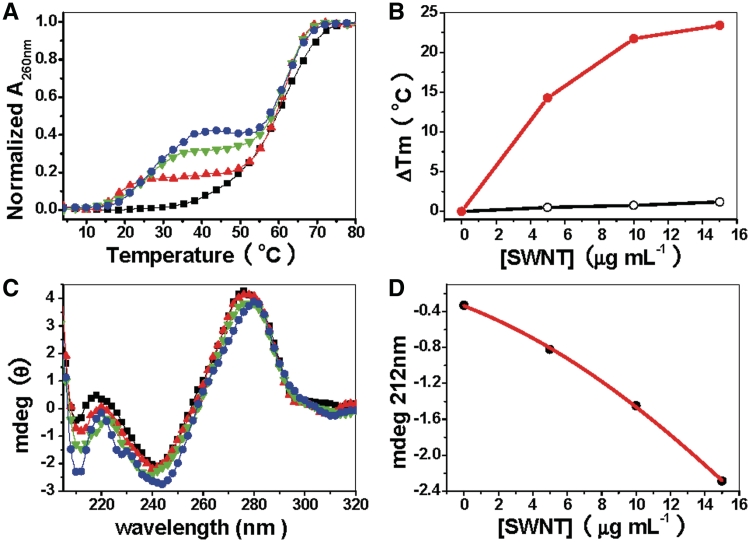
UV melting profiles of d(CT)•d(AG) at pH 6.5 in the absence or presence of SWNTs are shown in Figure 1A. In the absence of SWNTs, there is only one transition (Tm = 60.8°C). This transition corresponds to the dissociation of a Watson–Crick duplex, which reveals triplex is not formed under these conditions (1 mM cacodylic, 200 mM NaCl, pH 6.5). However, in the presence of SWNTs, there are two well resolved transitions: the first transition at low temperature (Tm1) is relative to the dissociation of the Hoogsteen base-paired strand from the target duplex, the second higher temperature transition (Tm2) is attributed to the denaturation of the Watson–Crick double helix (6,34,35). This is because, we have investigated the melting at different pH values and found that Tm1 is pH dependent. According to our melting profiles, the lower pH value gives a higher melting temperature, which is the character of Hoogsteen base pairs (6). Furthermore, the negative band at 212 in CD spectrum also suggests the formation of Hoogsteen base pairs of triplex DNA (6,34,35).
Figure 1.
UV melting profiles and CD spectra of d(CT)d(AG) at pH 6.5 illustrate triplex formation in the presence of SWNTs. (A) UV melting profiles of 1 µM d(CT)·d(AG) in the absence (black line) or presence of SWNTs: 5 μg ml−1 (red line), 10 μg ml−1 (green line), 15 μg ml−1 (blue line) in cacodylic buffer (1 mM cacodylic acid /sodium cacodylate/200 mM NaCl/pH 6.5) (B) Plot of ΔTm (the difference in the apparent Tm in the presence of SWNTs relative to d(CT)·d(AG). Filled circles are for the transition for dissociation of the third strand. ΔTm3-2 (triplex → duplex + single strand) is calculated by assuming a ΔTm3-2 of 4°C in the absence of SWNTs (no transition seen). Open circles are for the duplex melting transition. (C) CD spectra of 2 µM d(CT)·d(AG) at pH 6.5 in the absence (black line) or presence of SWNTs: 5 μg·ml−1 (red line), 10 μg ml−1 (green line), 15 μg ml−1 (blue line) in cacodylic buffer (1 mM cacodylic acid /sodium cacodylate/200 mM NaCl/pH 6.5). (D) Plot of CD intensity at 212 nm (solid circle) versus concentration of SWNTs. The data were adopted from C.
The two obvious transitions suggest that SWNTs binding drives d(CT)•d(AG) disproportionation into triplex DNA. Further studies indicate that both Tm1 and hyperchromicity are increased as SWNTs concentration increased. Upon increasing concentration of SWNTs, Tm1 for triplex transition is systematically increased from 18°C up to a maximal shift of 28°C. In marked contrast, Tm2 for duplex transition is hardly changed (Figure 1B). The melting and cooling curves indicate that the triplex cannot be re-formed after thermal denaturation (Supplementary Figure S2B) (34), that might be due to the irreversible aggregation of SWNTs occurred at high temperature.
CD spectra of d(CT)•d(AG) with or without SWNTs are shown in Figure 1C. Without SWNTs, CD spectrum shows that d(CT)•d(AG) forms canonical B-form duplex DNA: a large positive band at 276 nm, a small negative band at 242 nm and a small positive band at 220 nm (25,34). Upon addition of SWNTs, the positive band at 276 nm is reduced in magnitude and shifted to higher wavelength while the negative band at 242 nm is reduced slightly in magnitude. As the concentration of SWNTs goes up to 15 μg ml−1, the spectrum has a positive band at 280 nm and negative bands at 246 nm and 212 nm. Negative band at 212 nm has been used to indicate triple-stranded DNA formation (6,34,35).
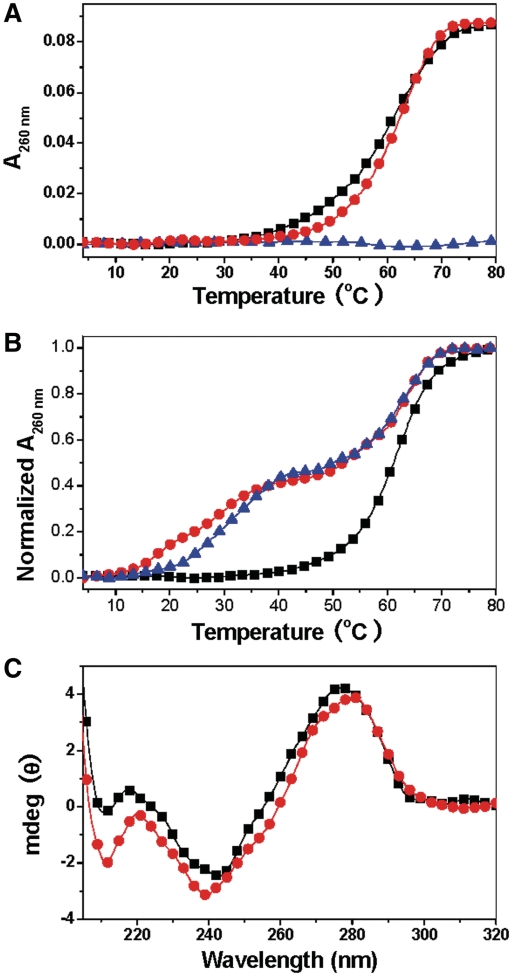
To verify that SWNTs can facilitate d(C+T)•d(AG)•d(CT) formation, we investigate d(C+T)•d(AG)•d(CT) formation under basic conditions. Without SWNTs, d(CT) can interact with d(CT)•d(AG) to form the triplex structure d(C+T)•d(AG)•d(CT) at pH 6.5 (Supplementary Figure S3). At pH 8.5, SWNTs cannot disproportion d(CT)•d(AG) into triplex DNA (Figure 2A). Only in the presence of d(CT) strand, two obvious transitions are observed (Figure 2B). As d(CT) cannot form a high order structure induced by SWNTs (Figure 2A), the two transitions suggest d(CT) interact with d(CT)•d(AG) to form a triplex structure. This assignment agrees with the CD result (Figure 2C), which reveals that, in the presence of SWNTs, a band at 212 nm with large magnitude is characteristic for the formation of CGC+ triple helices (6,34,35). As shown in Scheme 1, the single-strand d(CT) sequence has overhang around the triplex formation region. The overhang may act as the arms to immobilize the triplex structure on the surface of SWNTs and to promote the triplex structure formation. In the control experiment, we used perfect-matched triplex sequence. SWNTs can also induce a complete CGC+ triplex formation (Supplementary Figure S4A). However, the melting experiment suggests that the weak binding between perfect-matched DNA and SWNTs is not strong enough to induce CGC+ triplex formation (Supplementary Figure S4B).
Figure 2.
UV melting profiles and CD spectra of d(CT)d(AG) and d(CT) at pH 8.5 illustrate triplex formation in the presence of SWNTs. (A) UV melting profiles: d(CT)·d(AG) (black), d(CT)·d(AG) in the presence of 15 μg ml−1SWNTs (red) and d(CT) in the presence of 15 μg ml−1SWNTs (blue) in cacodylic buffer (1 mM cacodylic acid /sodium cacodylate/200 mM NaCl/pH 8.5) (B) UV melting profiles of solution containing a 1:1 stoichiometry of d(CT)·d(AG) and d(CT) in the absence (black line) or presence of SWNTs: 5 μg ml−1 (red line), 10 μg ml−1 (blue line) in cacodylic buffer (1 mM cacodylic acid /sodium cacodylate/200 mM NaCl/pH 8.5). (C) CD spectra of triplex in the absence (black line) or presence of 10 μg ml−1 SWNTs (red line) in cacodylic buffer (1 mM cacodylic acid /sodium cacodylate/200 mM NaCl/pH 8.5).
Our results indicate that the mismatched triplex DNA is much more easily formed in the presence of SWNTs. As shown in Supplementary Figure S4B, formation of the perfect matched triplex needs much more SWNTs than that of partial triplex structure. This result indicates that the weak binding between perfect-matched triplex DNA and SWNTs may be not be strong enough to induce CGC+ triplex formation. The overhang and mismatched structure help the triplex to immobilize on SWNTs by increasing the interaction between SWNTs and DNA. Therefore, the single-strand overhang and mismatch structure are important and can enhance DNA binding to SWNTs that can induce CGC+ triplex formation. Although the mismatched DNA cannot form a complete CGC+ triplex DNA, some bases in d(CT) can be compatible with d(CT)·d(AG) to form a partial triplex structure (Scheme 1). The partial triplex structure contains the repeating sequences 3′-TCTCTCT-5′/5′-AGAGAGA-3′; therefore, our results would be helpful for the application of carbon nanotubes in nanomedicine.
As SWNTs can induce the perfect matched triplex DNA formation while MWNTs (Supplementary Figure S4C) do not have the effect, which suggests the diameter of nanotubes is very important for DNA binding. MWNTs (10–20 nm-sized) are too large to bind to the major groove (26). In our previous study, SWNTs (1.1 nm-sized) can bind to the groove of TAT triplex DNA and decrease the stability of TAT triplex DNA (25). In the present study, this selectivity can be attributed to the size of the groove and negatively charged carboxyl-modified SWNTs binding to the groove can further stabilize CGC+ triplex DNA.
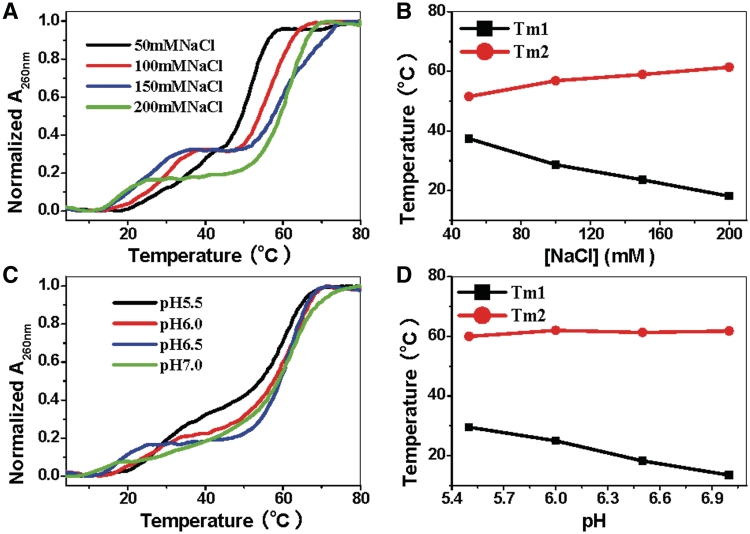
Lavelle and Fresco (6) have shown that increasing NaCl concentration from 0.1 M to 0.4 M does not influence CGC+ triplex stability. Nevertheless, for the triplex formed by d(CT)·d(AG) duplex disproportionation, increasing NaCl can decrease triplex stability. Figure 3A shows the melting profiles of d(CT)·d(AG) in the presence of SWNTs at different salt concentration. As NaCl concentration increased from 50 mM to 200 mM, Tm1 of d(C+T)·d(AG)·d(CT) decreased from 30°C to 14°C. Meanwhile, duplex dissociation temperature, Tm2, increased from 52°C to 61°C. According to polyelectrolyte theory, increasing NaCl concentration can have two effects: one is to shield the negative charges of phosphate that can increase d(CT)·d(AG) duplex stability (36); the other is to shield the negative charges of SWNTs that can decrease the favorable electrostatic interaction (36–38) between SWNTs and d(C+T)·d(AG)·d(CT). Therefore, inverse dependence of triplex stability on NaCl concentration shows that electrostatic attraction is important for d(CT)·d(AG) duplex disproportionation switched by SWNTs. Since CGC+ triplex formation requires protonation of the third strand dC residues (6), it is not surprising that increasing proton concentration favors triplex stability (6,34,35). Figure 3C shows the melting profiles of d(CT)·d(AG) in the presence of SWNTs at different pH. When increasing pH values from 5.5 to 7.0, Tm1 of triplex DNA decreased to 16°C while Tm2 of duplex hardly changed (Figure 3D). These observations provide further evidence that electrostatic attraction between SWNTs and d(C+T)·d(AG)·d(CT) is important.
Figure 3.
UV melting profiles of d(CT)·d(AG) in the presence of SWNTs at different salt concentration and pH values. (A) UV melting profiles of 1 µM d(CT)·d(AG) added 5 μg ml−1 SWNTs in 1 mM Cacodylic, 50 mM NaCl (black line), 100 mM NaCl (red line), 150 mM NaCl (blue line) and 200 mM NaCl (green line) at pH 6.5. (B) The third strand melting temperature (Tm1) and the duplex melting temperature (Tm2) as a function of NaCl concentration. (C) UV melting profiles of 1 µM d (CT)·d (AG) added 5 μg ml−1 SWNTs in 1 mM Cacodylic, 200 mM NaCl at pH 5.5 (black line), pH 6.0 (red line), pH 6.5 (blue line) and pH 7.0 (green line). (D) The third strand melting temperature (Tm1) and the duplex melting temperature (Tm2) as a function of pH.
Since ssDNA can wrap on the surface of SWNTs, this results in SWNTs surface more negatively charged and SWNTs become more stable, while dsDNA does not have the effect (39,40). So, ssDNA can prevent SWNTs from aggregation in electrolyte solution while dsDNA cannot. With this in mind, if duplex d(CT)•d(AG) disproportionate into triplex d(C+T)•d(AG)•d(CT) and single-strand d(AG), single-strand d(AG) would wrap on SWNTs and enhances their stability. In our experiment, SWNTs in the absence or presence of d(CT)•d(AG) show different pH dependent response. In the absence of dsDNA, SWNTs–COOH sample is stable at pH 8.5 over 24 h, while obvious aggregation occurs at pH 5.0 (Figure 4A). Light scattering signals of the supernatant also support the results, the signal intensity decreases with decrease of pH (Figure 4C). This result is attributed to the surface modification with the carboxylate groups that serve as pH-sensing groups through protonation and deprotonation. So at low pH value, SWNTs come into contact and aggregate without electrostatic repulsion (41). However, in the presence of d(CT)·d(AG), the dispersed SWNTs sample in low pH buffers is much more stable than at pH 8.5 (Figure 4B and D). The sample at pH 6.5 shows fewer amounts of aggregations and the supernatant has stronger scattering signals than that at pH 8.5, showing that d(CT)·d(AG) disproportionation enhanced SWNTs stability.
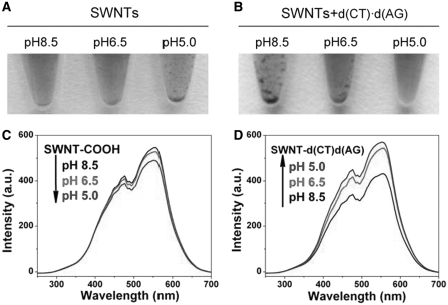
Figure 4.
Photo images of (A) SWNT–COOH and (B) SWNTs-d(CT)·d(AG) of different pH values after 24 h at 4°C. Light scattering spectra of supernatants for (C) SWNT–COOH and (D) SWNT-d(CT)·d(AG) at different pH values after 24 h at 4°C.
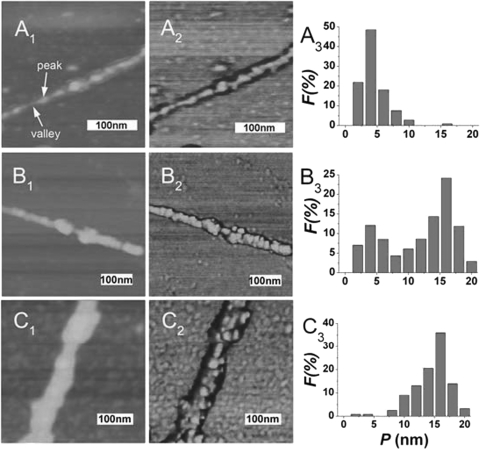
To better understand that dsDNA disproportionation enhances SWNTs stability in solution, we used AFM to study the morphology of SWNTs-dsDNA complexes at different pH values (25,31). Samples were deposited on an 3-(aminopropyl)trimethoxysilane (APTES) treated mica surface and analyzed by tapping-mode AFM (25,31,33). AFM imaging reveals a typical pattern on the SWNT–DNA surface consisting of peaks and valleys in height along the length of the tube, as well as corresponding shifts in the phase of the cantilever oscillation. Figure 5A shows that the height of SWNT–AG was 3.8 ± 0.5 nm at the peaks at pH 6.5. At pH 8.5, the peak-height distribution of SWNTs-d(CT)•d(AG) exhibits only one peak which corresponds to height of 15.0 ± 0.5 nm. However, at pH 6.5, the peak-height distribution exhibits two peaks, corresponding to heights of 3.9 ± 0.5 nm and 15.4 ± 0.3 nm, which suggests SWNT and single-strand DNA complexes were formed in the solution (Figure 5B). At pH 6.5, SWNTs are mostly dispersed on the mica surface (Supplementary Figure S5A and B). However, large SWNTs-aggregates coated by condensed DNA were observed at pH 8.5 (Supplementary Figure S5C and D). In control experiments, the CO-duplex DNA which cannot form CGC+ triplex DNA exhibits only one peak for the peak-height distribution and shows similar aggregates at different pH values (Supplementary Figure S6–8), further indicating that the enhanced SWNTs stability is due to d(CT)·d(AG) duplex disproportionation (Figure 4B and D). These results are consistent with CD and DNA melting results.
Figure 5.
AFM height (left), phase (middle) images and distribution of the peaks (right) of SWNT-d(AG) (A1,A2,A3), SWNT-d(CT)·d(AG) at pH 6.5 (B1, B2, B3) and pH 8.5 (C1, C2, C3) (n > 100 peaks). All the experiments were carried out in cacodylic buffer (1 mM cacodylic acid/sodium cacodylate/200 mM NaCl). The images are 3 µm × 3 µm.
As for DNA binding to functionalized SWNTs, various interactions of DNA bases and backbone with SWNTs, such as hydrophobic, van der Waals and electrostatic interactions, can take place (42). Simulation studies indicate that duplex DNA can bind to SWNTs via DNA groove binding or DNA end adsorption (22). Experimental results indicate that SWNTs groove binding can cause DNA condensation (25), while the interaction between DNA end base pair and SWNT wall surface involves spontaneous unzipping of partially bound strands of ds-DNA (24). Intriguingly, Cathcart et al. (24) recently reported that SWNTs can act like DNA helicase and demonstrate that long, natural dsDNA can bind to SWNTs and the dsDNA is separated into ssDNA to form an ordered helical wrapping on carbon nanotubes through long time interaction (25). In their dsDNA-SWNTs system, SWNTs can be considered as helicase to catalyze the unwinding of dsDNA. However, due to the length of the DNA sequence, this reassembly process needs long time (25). In our experiment, there are two aspects that can influence d(CT)·d(AG) disproportionation. The first is SWNTs unwinding d(CT)·d(AG) duplex, in which SWNTs act as helicase destabilizes d(CT)·d(AG) at junction. Since d(CT)·d(AG) has a short length (21-mer), the unwinding will easily take place. The second is protonation of dC residues. Previous studies have shown that SWNTs can promote protonation of conjugated polymers by changing their pKa values (43). With addition of SWNTs, the pKa of dCMP was shifted from 4.65 (44) to 5.19 (Supplementary Figure S9). In the control experiment, triplex d(C+T)•d(AG)•d(CT) is formed at low pH in the absence of SWNTs, showing that protonation can promote triplex formation (Supplementary Figures S2A and S3). Thus, when d(CT)·d(AG) binding to SWNTs, the interaction between SWNTs and d(CT)·d(AG) can promote protonation of dC residues by changing pKa values of dC residues. The protonated d(C+T) and d(CT)·d(AG) would form triplex DNA under these conditions, that will accelerate the unwinding of d(C+T)·d(AG). It should be pointed out that SWNTs-induced disproportionation is related to DNA sequence. For control duplex DNA (CO-duplex), SWNTs cannot induce disproportionation because CO-duplex cannot form CGC+ triplex DNA. In addition, the electrostatic repulsion between negatively charged SWNT–COOH and CO-duplex would decrease SWNTs helicase activity, that can cause CO-duplex condensed on SWNTs (Supplementary Figures S6–S8). These results demonstrate that electrostatic interaction is crucial for the disproportionation. To clarify the charge effect, we prepare another two functionalized SWNTs, positively charged amino group-modified SWNTs and hydroxyl–SWNTs (26,31). Our DNA melting data clearly indicate that positively charged amino group-modified SWNTs cannot induce triplex DNA formation, while carboxyl-/hydroxyl-SWNTs can induce d(CT)·d(AG) disproportionation and promote CGC+ triplex DNA formation (Figure 6) although hydroxyl-SWNTs has weaker effect than carboxyl-SWNTs.
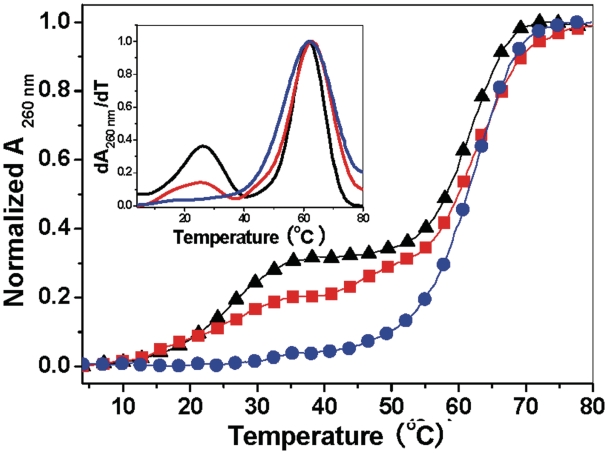
Figure 6.
UV melting profiles of 1 µM d(CT)·d(AG) in the presence of 5 μg ml−1 SWNT–COOH (black line), 5 μg ml−1 SWNT–CH2–OH (red line) and 5 μg ml−1 SWNT–CH2–NH2 (blue line) in cacodylic buffer (1 mM cacodylic acid/sodium cacodylate/200 mM NaCl/pH 6.5). (Inset) The plot of their individual derivative of dA260nm/dT versus temperature.
CONCLUSIONS
We have shown for the first time that SWNTs can induce CGC+ triplex formation under physiological conditions. Targeting triplex structure is important and useful for developing new molecular biology tools as well as therapeutic agents, however, CGC+ triplex poor stability limits its use in vitro and in vivo. Our results indicate that SWNTs can reduce the stringency of conditions that CGC+ triplex formation requires. Therefore, this finding would facilitate utilization of SWNTs–DNA complex in artificially controlling of gene expression, nanomaterials assembly and biosensing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The 973 Project (2011CB936004); Natural Science Foundation of China (20831003, 90813001, 20833006, 90913007); the Chinese Academy of Sciences. Funding for open access charge: Natural Science Foundation of China and 973 Project.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 2.Wells RD, Collier DA, Hanvey JC, Shimizu M, Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988;2:2939–2949. [PubMed] [Google Scholar]
- 3.Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- 4.Mirkin SM, Frank-Kamenetskii MD. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 5.Beasty AM, Behe MJ. An oligopurine sequence bias occurs in eukaryotic viruses. Nucleic Acids Res. 1988;16:1517–28. doi: 10.1093/nar/16.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavelle L, Fresco JR. UV spectroscopic identification and thermodynamic analysis of protonated third strand deoxycytidine residues at neutrality in the triplex d(C+-T)6:[d(A-G)6·d(C-T)6]; evidence for a proton Switch. Nucleic Acids Res. 1995;23:2692–2705. doi: 10.1093/nar/23.14.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 8.Chan PP, Glazer PM. Triplex DNA: Fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- 9.Pilch DS, Breslauer KJ. Ligand-induced formation of nucleic acid triple helices. Proc. Natl Acad. Sci. USA. 1994;91:9332–9336. doi: 10.1073/pnas.91.20.9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren JS, Chaires JB. Preferential binding of 3,3′-diethyloxadicarbocyanine to triplex DNA. J. Am. Chem. Soc. 2000;122:424–425. [Google Scholar]
- 11.Ren JS, Bailly C, Chaires JB. NB-506, an indolocarbazole topoisomerase I inhibitor, binds preferentially to triplex DNA. Febs. Lett. 2000;470:355–359. doi: 10.1016/s0014-5793(00)01335-1. [DOI] [PubMed] [Google Scholar]
- 12.Song GT, Xing FF, Qu XG, Chaires JB, Ren JS. Oxazine 170 induces DNA : RNA : DNA triplex formation. J. Med. Chem. 2005;48:3471–3473. doi: 10.1021/jm050131g. [DOI] [PubMed] [Google Scholar]
- 13.Feng LY, Li X, Peng YH, Geng J, Ren JS, Qu XG. Spectral and electrochemical detection of protonated triplex formation by a small-molecule anticancer agent. Chem. Phys. Lett. 2009;480:309–312. [Google Scholar]
- 14.MoraruAllen AA, Cassidy S, Alvarez JLA, Fox KR, Brown T, Lane AN. Coralyne has a preference for intercalation between TA center dot T triples in intramolecular DNA triple helices. Nucleic Acids Res. 1997;25:1890–1896. doi: 10.1093/nar/25.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iijima S. Helical microtubules of graphitic carbon. Nature. 1991;354:56–58. [Google Scholar]
- 16.Liu J, Rinzler AG, Dai HJ, Hafner JH, Bradley RK, Boul PJ, Lu A, Iverson T, Shelimov K, Huffman CB, et al. Fullerene pipes. Science. 1998;280:1253–1256. doi: 10.1126/science.280.5367.1253. [DOI] [PubMed] [Google Scholar]
- 17.Keren K, Berman RS, Buchstab E, Sivan U, Braun E. DNA-templated carbon nanotube field-effect transistor. Science. 2003;302:1380–1382. doi: 10.1126/science.1091022. [DOI] [PubMed] [Google Scholar]
- 18.Wong SS, Joselevich E, Woolley AT, Cheung CL, Lieber CM. Covalently functionalized nanotubes as nanometre-sized probes in chemistry and biology. Nature. 1998;394:52–55. doi: 10.1038/27873. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Taylor S, Li HP, Fernando KAS, Qu LW, Wang W, Gu LR, Zhou B, Sun YP. Advances toward bioapplications of carbon nanotubes. J. Mater. Chem. 2004;14:527–541. [Google Scholar]
- 20.Kam NWS, O'Connell M, Wisdom JA, Dai HJ. Carbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destruction. Proc. Natl Acad. Sci. USA. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng M, Jagota A, Semke ED, Diner BA, Mclean RS, Lustig SR, Richardson RE, Tassi NG. DNA-assisted dispersion and separation of carbon nanotubes. Nat. Mater. 2003;2:338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- 22.Lu G, Maragakis P, Kaxiras E. Carbon nanotube interaction with DNA. Nano Lett. 2005;5:897–900. doi: 10.1021/nl050354u. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Johnson JK. Simulation of adsorption of DNA on carbon nanotubes. J. Am. Chem. Soc. 2007;129:10438–10445. doi: 10.1021/ja071844m. [DOI] [PubMed] [Google Scholar]
- 24.Cathcart H, Nicolosi V, Hughes JM, Blau WJ, Kelly JM, Quinn SJ, Coleman JN. Ordered DNA wrapping switches on luminescence in single-walled nanotube dispersions. J. Am. Chem. Soc. 2008;130:12734–12744. doi: 10.1021/ja803273s. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Peng YH, Qu X. Carbon nanotubes selective destabilization of duplex and triplex DNA and inducing B-A transition in solution. Nucleic Acids Res. 2006;34:3670–3676. doi: 10.1093/nar/gkl513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Peng YH, Ren J, Qu X. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl Acad. Sci. USA. 2006;103:19658–19663. doi: 10.1073/pnas.0607245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng YH, Li X, Ren J, Qu X. Single-walled carbon nanotubes binding to human telomeric i-motif DNA: significant acceleration of S1 nuclease cleavage rate. Chem. Comm. 2007:5176–5178. doi: 10.1039/b710950d. [DOI] [PubMed] [Google Scholar]
- 28.Zhao C, Song YJ, Ren J, Qu X. A DNA nanomachine induced by single-walled carbon nanotubes on gold surface. Biomaterials. 2009;30:1739–1745. doi: 10.1016/j.biomaterials.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C, Ren J, Qu X. Single-walled carbon nanotubes binding to human telomeric i-Motif DNA under molecular-crowding conditions: more water molecules released. Chem. Eur. J. 2008;14:5435–5439. doi: 10.1002/chem.200800280. [DOI] [PubMed] [Google Scholar]
- 30.Peng YH, Wang XH, Xiao Y, Feng LY, Zhao C, Ren J, Qu X. i-Motif quadruplex DNA-based biosensor for distinguishing single- and multiwalled carbon nanotubes. J. Am. Chem. Soc. 2009;131:13813–13818. doi: 10.1021/ja9051763. [DOI] [PubMed] [Google Scholar]
- 31.Zhao C, Peng YH, Song YJ, Ren J, Qu X. Self-assembly of single-stranded RNA on carbon nanotube: Polyadenylic acid to form a duplex structure. Small. 2008;4:656–661. doi: 10.1002/smll.200701054. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Huang CZ, Li YF, Xiao SJ, Xie JP. Label-free detection of sequence-specific DNA with multiwalled carbon nanotubes and their light scattering signals. J. Phy. Chem. B. 2008;112:7120–7122. doi: 10.1021/jp800092r. [DOI] [PubMed] [Google Scholar]
- 33.Campbell JF, Tessmer I, Thorp HH, Erie DA. Atomic force microscopy studies of DNA-wrapped carbon nanotube structure and binding to quantum dots. J. Am. Chem. Soc. 2008;130:10648–10655. doi: 10.1021/ja801720c. [DOI] [PubMed] [Google Scholar]
- 34.Plum GE, Breslauer KJ. Thermodynamics of an intramolecular DNA triple-helix: a calorimetric and spectroscopic study of the pH and salt dependence of thermally-induced structural transitions. J. Mol. Biol. 1995;248:679–695. doi: 10.1006/jmbi.1995.0251. [DOI] [PubMed] [Google Scholar]
- 35.Soto AM, Loo J, Marky LA. Energetic contributions for the formation of TAT/TAT, TAT/CGC+, and CGC+/CGC+ base triplet stacks. J. Am. Chem. Soc. 2002;124:14355–14363. doi: 10.1021/ja026952h. [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto N, Wu P, Hara H, Kawamoto Y. pH and cation effects on the properties of parallel pyrimidine motif DNA triplexes. Biochemistry. 2001;40:9396–9405. doi: 10.1021/bi010666l. [DOI] [PubMed] [Google Scholar]
- 37.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Peng Y, Ren J, Qu X. Effect of DNA flanking sequence on charge transport in short DNA duplexes. Biochemistry. 2006;45:13543–13550. doi: 10.1021/bi061103i. [DOI] [PubMed] [Google Scholar]
- 39.Gigliotti B, Sakizzie B, Bethune DS, Shelby RM, Cha JN. Sequence-independent helical wrapping of singles-walled carbon nanotubes by long genomic DNA. Nano Lett. 2006;6:159–164. doi: 10.1021/nl0518775. [DOI] [PubMed] [Google Scholar]
- 40.Song Y, Wang X, Zhao C, Qu K, Ren J, Qu X. Label-free colorimetric detection of single nucleotide polymorphism by using single-walled carbon nanotube intrinsic peroxidase-like activity. Chem. Eur. J. 2010;16:3617–3621. doi: 10.1002/chem.200902643. [DOI] [PubMed] [Google Scholar]
- 41.Zhao W, Song CH, Pehrsson PE. Water-soluble and optically pH-sensitive single-walled carbon nanotubes from surface modification. J. Am. Chem. Soc. 2002;124:12418–12419. doi: 10.1021/ja027861n. [DOI] [PubMed] [Google Scholar]
- 42.Clarke DR, Ruhle M. Simulation of DNA-nanotube interactions. Ann. Rev. Mater. Res. 2004;34:123–150. [Google Scholar]
- 43.Steuerman DW, Star A, Narizzano R, Choi H, Ries RS, Nicolini C, Stoddart JF, Heath JR. Interactions between conjugated polymers and single-walled carbon nanotubes. J. Phy. Chem. B. 2002;106:3124–3130. [Google Scholar]
- 44.Cavalieri LF, Stone AL. Studies on the structure of nucleic acids. VIII. Apparent dissociation constants of deoxypentose nucleic acid. J. Am. Chem. Soc. 1955;77:6499–6501. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.