Abstract
Most DNA double-strand breaks (DSBs) in S- and G2-phase cells are repaired accurately by Rad51-dependent sister chromatid recombination. However, a minority give rise to gross chromosome rearrangements (GCRs), which can result in disease/death. What determines whether a DSB is repaired accurately or inaccurately is currently unclear. We provide evidence that suggests that perturbing replication by a non-programmed protein–DNA replication fork barrier results in the persistence of replication intermediates (most likely regions of unreplicated DNA) into mitosis, which results in anaphase bridge formation and ultimately to DNA breakage. However, unlike previously characterised replication-associated DSBs, these breaks are repaired mainly by Rad51-independent processes such as single-strand annealing, and are therefore prone to generate GCRs. These data highlight how a replication-associated DSB can be predisposed to give rise to genome rearrangements in eukaryotes.
INTRODUCTION
The path to complete genome duplication is littered with obstacles that can impede the progression of the replication fork (1). These replication fork barriers (RFBs) include natural programmed RFBs, which are designed to ensure unidirectional replication of a given locus, as well as numerous accidental RFBs, including various DNA lesions, secondary structures in the template DNA, and protein–DNA complexes (2).
Cells appear to have a variety of mechanisms for coping with RFBs, and it is thought that the nature of the barrier determines which is used in any given situation (1). In many cases it appears that the response is to protect the fork and its protein components (the replisome), until such time as the block has been removed and DNA synthesis can resume, or replication is completed by the arrival of the opposing fork (3). However, when fork protection fails or the barrier proves persistent homologous recombination (HR) can be used to enable template switching to bypass the RFB or to promote replication restart if the fork has collapsed (= dissociation of the replisome from DNA) or broken (4,5). This recombination typically occurs between equivalent DNA sequences on the sister chromatids, however occasionally HR acts between allelic or ectopic sites and consequently can result in changes to the genome such as loss of heterozygosity (LOH) and gross chromosome rearrangements (GCRs) including deletions, duplications, inversions and translocations that in humans can promote diseases such as cancer (5–8). Whether this misdirected recombination is solely stochastic or can be driven by the nature of the fork impediment is unknown.
A key requirement in HR is the presence of a stretch of single-stranded (ss) DNA that is bound by recombinases, such as Rad51 and Rad52, which then promote either strand invasion of a homologous duplex DNA or annealing to a complementary DNA strand (9). Replication forks naturally contain a region of ssDNA on the lagging template strand, and it is thought that this can become accessible to recombinases following fork stalling. Alternatively, the stalled fork can be remodelled through a process of fork reversal, which causes the nascent DNA strands to anneal to each other forming a duplex DNA end that can be resected to produce a ssDNA tail by an exonuclease (10). A duplex DNA end can also be generated by structure-specific nucleases such as Mus81-Eme1 cleaving the stalled fork (11). In all of the cases above one sister chromatid is essentially left intact and therefore can act as the donor DNA molecule during HR. Indeed it has been shown that accurate sister chromatid recombination, which is catalysed by Rad51, is the predominant pathway for repairing broken replication forks (12). In the absence of Rad51 repair proceeds by alternative low-fidelity mechanisms such as single-strand annealing (SSA) or non-homologous end joining (NHEJ) (13).
Much of our current understanding of how protein–DNA RFBs can induce HR in eukaryotes has come from recent studies of a site-specific RFB in the fission yeast Schizosaccharomyces pombe called RTS1 (5,6,14,15). This programmed RFB has evolved to work closely with the replisome and components of the so-called fork protection complex to elicit a very strong polarised block to replication (16). Here replication restart is inhibited by the Rtf2 protein, which promotes replication termination at RTS1 by the opposing fork merging with the stalled fork (17). However, some forks blocked at RTS1 can undergo recombination-dependent replication restart, and in so doing occasionally elicit genome rearrangements (5). Importantly most of this recombination in wild-type cells occurs independently of DSB formation, and therefore presumably involves the unwinding of DNA strands at the stalled fork.
To determine whether replication restart is the only way in which protein–DNA RFBs give rise to recombination we have investigated what the cellular response is to a persistent non-programmed bi-directional protein–DNA barrier. We show that, unlike RTS1, a persistent non-programmed barrier frequently results in DNA breakage even in the presence of a functional fork protection complex. However, unlike previously studied replication-associated DSBs, these breaks are not repaired by sister-chromatid recombination. Instead they are repaired by SSA and consequently are associated with DNA deletion. Intriguingly replication fork stalling and DNA breakage correlate with the appearance of structures that are analogous to ultrafine anaphase bridges (UFBs), which frequently form at fragile sites during mitosis in human cells, and are thought to occur when replication is incomplete (18,19). We propose that a failure of replication restart at non-programmed bi-directional protein–DNA barriers results in a region of unreplicated DNA that becomes broken during mitosis or cytokinesis. Such breaks can only be repaired by allelic recombination, SSA or end-joining pathways due to the absence of an intact sister chromatid, demonstrating how some replication-associated DSBs can be predisposed to give rise to LOH and GCR.
MATERIALS AND METHODS
Media and genetic methods
Media and genetic methods are based on established protocols (20). The complete and minimal media were yeast extract with supplements (YES) and Edinburgh minimal medium plus 3.7 mg/ml sodium glutamate (EMMG) plus appropriate amino acids (0.2475 mg/ml), respectively. Low adenine media (YES-LA) was supplemented with 0.01 mg/ml adenine. Ade+ recombinants were selected on YES lacking adenine and supplemented with 200 mg/l guanine to prevent uptake of residual adenine (21). Where indicated 4 µM thiamine was added to media to repress expression from the nmt promoter.
Strains, plasmids and probes
The lacO array, family of repeats (FR) and MATa-containing strains were made by integration of derivatives of pFOX2 (22) (see below), which had been linearised with BlpI, at ade6-M375 in FO126 or FO1236. Southern blotting and/or colony PCR was used to determine that the linear plasmid had integrated at the correct site. The lacO array-containing derivative of pFOX2, pMW731, was constructed by subcloning a ∼4.5 kb XhoI lacO array fragment from pLAU43 (23) into the SalI site in pFOX2. Derivatives of pMW731 containing shorter lacO arrays were constructed by digestion and re-ligation to delete the appropriate number of repeats from the 115 repeat array. The digests were: NheI (to retain 2 lacO repeats in pMW892); NheI + BstEII (to retain 12 lacO repeats in pMW893); NheI + AfeI (to retain 15 lacO repeats in pMW894); and NheI + PshAI (to retain 21 lacO repeats in pMW895). The MATa cleavage site-containing derivative, pFOX16, was constructed by inserting a 117 bp EcoRV–HincII MATa fragment from pCla7 (24) into the HincII site that overlaps the SalI site in pFOX2. The derivative of pFOX2 containing 10 FR repeats (pMW897) was constructed by inserting a 838 bp XbaI–MluI fragment from AGP73 (25) into the SalI site in pFOX2. DNA ends were filled in using Klenow prior to ligation. pREP41/42/81-NLS-LacI-eCFP was constructed by PCR amplification of lacI-eCFP from pLAU53 (23) using primers oMW708 (5′-TTTGTCGACATGGGAAGTCCTAAGAAGAAACGAAAGGTGTTCACCGTGAAACCAGTAACGTTATACG-3′) and oMW710 (5′-TTTGGATCCTAATCTAGACACCATGG-3′), which were designed to introduce a NLS and a SalI site at the 5′-end of lacI-eCFP and a BamHI site at the 3′-end. The amplified gene was then cloned into pREP41, pREP42 and pREP81 (26) using the SalI and BamHI sites. pREP41-NLS-EBNA1 was constructed by PCR amplification of a portion of the EBNA1 gene encoding its DNA binding domain from plasmid 1160 (25) using primers oMW1367 (5′-TATTACATATGGGAAGTCCTAAGAAGAAACGAAAGGTGTTCACCCGGGGTCAGGGTGATGGAGGCAG-3′) and oMW1366 (5′-TTGGATCCTCACTCCTGCCCTTCCTCAC-3′). The amplified gene was then cloned into pREP41 using the NdeI and BamHI sites. pACYCREP81-HO has been described (24). All plasmids were sequenced to confirm that no mutations had been introduced during the cloning. Probe A used for two-dimensional (2D) gel analysis in Figure 1 is a ∼3.7 kb BlpI–XbaI fragment from pMW731 and probe B is ∼3.1 kb SacII–BamHI fragment from pMW731. For the CHEF gel in Figure 4B probe I is a fragment spanning mus81 and probe II is a fragment spanning brl1. The probe for the CHEF gel in Figure 4D is a ∼1 kb fragment amplified from genomic DNA using primers oMW1355 (5′-GCGCAACTCCTTTAGGGGTAG-3′) and oMW1356 (5′-GCGTATTCTAACGTCTTCGTG-3′). A full list of S. pombe strains used in this study is given in Table 1.
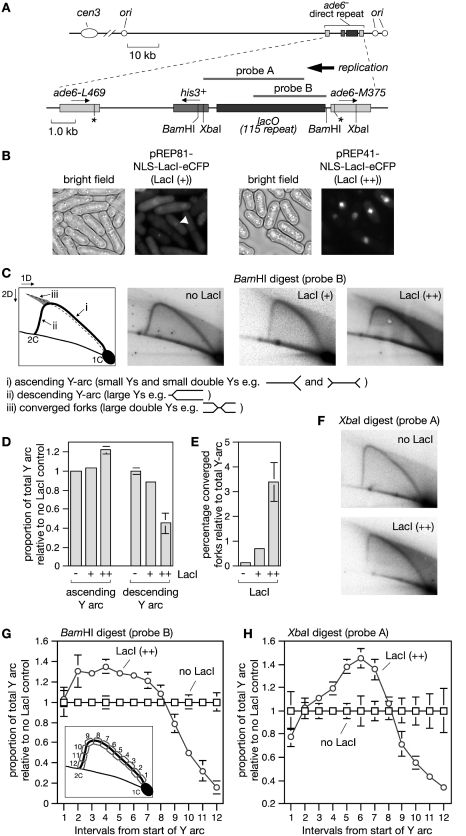
Figure 1.
A lacO-LacI array is an effective RFB in eukaryotes. (A) Schematic showing the position of the ade6- direct repeat and lacO array on chromosome 3 as well as the probes used for the 2D gel analysis in C and F. (B) Images showing (+) and (++) levels of NLS-LacI-eCFP in cells containing the lacO array. The arrowhead indicates an example of a lacO-LacI focus. Cells were analysed after 20 h of LacI induction. (C) Two-dimensional gel analysis of replication intermediates in the BamHI lacO-containing fragment from a wild-type strain with the indicated amount of LacI. Note that the shadow behind the Y-arc is due to shearing of DNA during sample preparation, and, in the case of LacI (++), also contraction of the lacO array (see also Figure 2B). (D and E) Quantification of data like in (C). The data are mean values from three independent experiments. Error bars represent standard deviations. (F) Two-dimensional gel analysis of replication intermediates in the XbaI lacO-containing fragment from a wild-type strain with the indicated amount of LacI. (G and H) Quantification of data like in (C) and (F). The diagram inset in (G) shows the regions of the Y-arc that were quantified. The data are mean values from three or four independent experiments. Error bars represent standard deviations.
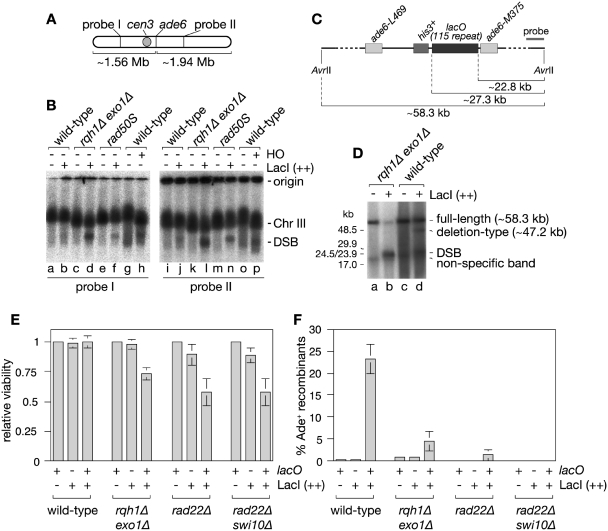
Figure 4.
Detection of DSBs caused by the lacO-LacI array. (A) Schematic of chromosome 3 showing the position of the ade6 locus and probes used for the Southern blot analysis of the CHEF gel in (B). (B) Detection of DSBs by whole chromosome CHEF analysis. The DNA in lanes a–f and i–n are from strains containing the lacO array with and without LacI (++) expression. The DNA in lanes g, h, o and p are from a strain that contains a MATa cleavage site in place of the lacO array and either pREP81 (HO −) or pACYCREP81-HO (HO +). Cultures were grown in EMMG lacking leucine, histidine and thiamine at 30°C for 20 h (lacO array strains) or 24 h (MATa strains) before isolating genomic DNA in agarose plugs. (C) Schematic showing the position of the lacO array relative to AvrII sites flanking the ade6 locus and the probe used for the Southern blot analysis of the CHEF gel in (D). (D) CHEF analysis of AvrII digested genomic DNA from strains with and without LacI (++) expression. (E) Viability of strains relative to minus LacI control after 24 h of LacI (++) induction. (F) The percentage of viable colonies in E that are Ade+. In both (E) and (F) the error bars are the standard deviations about the mean.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Mating type | Genotype | Source |
|---|---|---|---|
| FO126 | h− | ura4-D18 leu1-32 his3-D1 ade6-M375 | Lab strain |
| MCW1221 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW429 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/ade6-M375 | Lab strain |
| FO1236 | h− | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-M375 | Lab strain |
| MCW1988 | h− | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW2087 | h+ | rad51Δ::arg3+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW2088 | h+ | rad22Δ::ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW2089 | h+ | rad22Δ::ura4+ rad51Δ::arg3+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW3350 | h− | swi10Δ::kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW3409 | h+ | swi10Δ::kanMX6 rad22Δ::ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW2189 | h+ | rqh1Δ::kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW2186 | h− | exo1Δ::ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW4087 | h+ | rqh1Δ::kanMX6 exo1Δ::ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW2372 | h+ | rad50S ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| FO1751 | h− | rad22+-YFP-kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 | This studya |
| MCW2669 | h+ | rad22+-YFP-kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW4916 | h+ | leu1-32::pJK148-leu1-pfh1+ pfh1Δ::kanMX6 ura4-D18 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This studyb |
| MCW4949 | h+ | leu1-32::pJK148-leu1-pfh1-mt* pfh1Δ::kanMX6 ura4-D18 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This studyc |
| SAS67 | h+ | chk1Δ::ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
| MCW5157 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO2/ade6-M375 | This study |
| MCW5158 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO12/ade6-M375 | This study |
| MCW5159 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO15/ade6-M375 | This study |
| MCW5160 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO21/ade6-M375 | This study |
| MCW5162 | h+ | ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/FR10/ade6-M375 | This study |
| MCW5320 | h− | hht2+::GFP-ura4+ ura4-D18 leu1-32 his3-D1 arg3-D4 ade6-L469/pUC8/his3+/lacO115/ade6-M375 | This study |
Recombination assays
Spontaneous, lacO-LacI-induced and FR-EBNA1-induced Ade+ recombinant frequencies were measured essentially as described (14,21). To select for plasmids and allow for expression of LacI/EBNA1, cells were grown on EMMG lacking leucine and thiamine for 6–7 days at 30°C prior to selecting Ade+ recombinants. Recombinant frequencies represent the mean value from at least 15 colonies for each strain, and for strains containing plasmids at least three independent transformants were tested. The statistical significance of differences between recombinant frequencies was calculated with two-sample t-tests.
Viability assay
Strains with and without the 115 repeat lacO array containing either pREP41 or pREP41-NLS-LacI-eCFP were grown on EMMG plates lacking histidine and leucine and containing thiamine. Colonies from these plates were used to inoculate EMMG liquid media lacking histidine, leucine and thiamine, and these cultures were grown for 24 h before cells were harvested, washed in sterile water, counted and plated onto YES-LA containing thiamine. Plates were incubated at 30°C for 3–5 days before colonies were counted to determine the percentage of the plated cells that were viable. In addition colonies were scored for whether they were Ade+ (white/pale pink) or Ade− (dark pink/red) to provide a measure of the percentage of viable cells that were recombinants.
S. pombe genomic DNA preparation
Genomic DNA for neutral/neutral 2D gels was purified from logarithmically growing cultures as described (27), and enriched for replication intermediates on benzoylated naphthoylated DEAE (BND)-cellulose (Sigma). Genomic DNA for standard 1D agarose gels and CHEF gels was prepared in agarose plugs either as described by Win et al. (28) (Figures 2B and 4B) or as described by Hyppa and Smith (29) (Figure 4D). For the data in Figure 4D the plugs were melted at 65°C for 5 min prior to digestion of the agarose with ß-agarase I for 2.5 h at 37°C. The DNA was then digested over night with 40 units of AvrII. For the data in Figure 2B the DNA was digested in the plugs by overnight incubation with 40–100 U of restriction enzyme at 37°C.
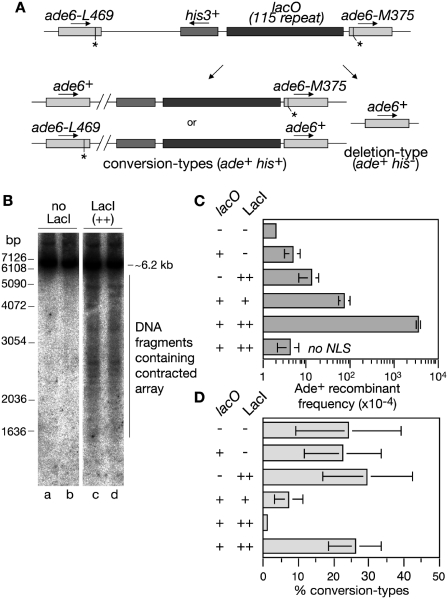
Figure 2.
Direct repeat recombination in strains containing a lacO-LacI array. (A) Schematic showing the ade6− direct repeat and lacO array on chromosome 3 and the two classes of Ade+ recombinant. Asterisks indicate the position of the point mutations in ade6-L469 and ade6-M375. (B) Neutral gel analysis of the XbaI lacO-containing fragment (see Figure 1A) from strain MCW1998 containing either pREP41 (LacI −) or pREP41-NLS-LacI-eCFP (LacI ++). Cells were grown for ∼20 h in the absence of thiamine prior to DNA extraction in agarose plugs. The Southern blot is probed with probe A (see Figure 1A). Lanes a and b, and c and d contain duplicate samples. (C) Ade+ recombinant frequencies in wild-type strains with and without the lacO array and LacI as indicated. (D) The percentage of Ade+ recombinants in C that are conversion-types. In all cases error bars are the standard deviations about the mean (see also Supplementary Table S1).
Gel electrophoresis
Two-dimensional gels were run according to Brewer and Fangman (30). Agarose gels of 0.4 and 1% were run for the first and second dimension, respectively. Neutral 1D gels were 0.8% agarose and were run in 1× TBE at 50–60 V for ∼16 h. The CHEF gels in Figure 4B were 0.7% agarose in 1× TAE, and were run for 48 h at 2 V/cm with an initial switch time of 20 min and a final switch time of 30 min using a CHEF II apparatus (Bio-Rad). The CHEF gel in Figure 4D was 1.0% agarose in 0.5× TBE and was run for 17 h at 6 V/cm with a switch time of 1–6 s using a CHEF II apparatus (Bio-Rad). All gels were Southern blotted and probed with 32P-labelled probe as indicated. Analysis of the blots was performed using a Fuji FLA3000 PhosphoImager and Image Gauge software.
Microscopy
For imaging UFBs, cells were grown in EMMG lacking leucine and thiamine for 24 h at 30°C to allow for NLS-LacI-eCFP expression. Cells were then fixed in 70% ethanol, stained with DAPI or SYBR Green and analysed using an Olympus BX50 epifluorescence microscope equipped with the appropriate filter sets to detect blue and green fluorescence. Black and white pictures of the single fluorescence channels were acquired with a cooled CCD camera (Princeton Instruments Inc., NJ, USA) controlled by Metamorph software (version 6.1r6; Molecular Devices, Sunnyvale, CA, USA). Single channel pictures were assigned pseudocolours and merged using Adobe Photoshop 7.0 (Adobe Systems Inc., San Jose, CA, USA). Live cells were imaged on an inverted Olympus IX71 microscope controlled by a DeltaVision Core using DeltaVision softWoRx 4.0.0 software (Applied Precision Inc., Issaquah, WA, USA), all subsequent image manipulations were performed with this software suite as well. Images were taken with a CCD camera (CoolSNAP HQ; Photometrics, Tucson, AZ, USA) using an oil-immersed Olympus 100× UPlanSApo objective with a NA of 1.40. For time-lapse analysis cells were mounted with soybean lectin (Sigma-Aldrich Ltd., Dorset, UK) in a glass bottom culture dish (MatTek Corp., Ashland, MA, USA) and observed in the appropriate medium at 30°C. A stack of 26 focal planes at a step-size of 0.2 µm was taken every 5 min for 4 h.
RESULTS
A lacO array bound by LacI can perturb replication fork progression in eukaryotes
We have previously shown that replication fork stalling at the polar RFB RTS1 induces Rad51-dependent recombination in S. pombe (14). As RTS1 is a programmed protein–DNA barrier we wanted to see whether a non-programmed protein–DNA barrier provokes a similar response. In Escherichia coli both lac and tet operator arrays when bound by their cognate repressor can act as site-specific protein–DNA RFBs (31). To determine whether the same is true in eukaryotes we inserted 115 lacO repeats between a direct repeat of mutant ade6- heteroalleles on chromosome 3 of S. pombe in approximately the same position that RTS1 had been placed previously (Figure 1A) (14). The Lac repressor protein (LacI), with an N-terminal nuclear localization signal (NLS) and C-terminal enhanced cyan fluorescent protein (eCFP) tag, was then expressed from the thiamine-repressible nmt promoter in either pREP81 or pREP41. The nmt promoters in these vectors are differently mutated resulting in an ∼10-fold difference in expression levels, which is evident from the amount of fluorescence in cells following induction (Figure 1B). pREP81 levels of LacI [hereafter referred to as LacI (+)] result in a discrete focus of fluorescence due to a high proportion of the induced protein binding to the lacO array, whereas pREP41 levels [hereafter referred to as LacI (++)] fill the entire nucleus (Figure 1B). Induction from the nmt promoter following the removal of thiamine is quite slow with the first mRNA detectable after 10 h and maximal protein expression achieved after 16 h (32). In our hands the maximum level of LacI is achieved after ∼20 h (data not shown). Much of our analyses therefore focus on samples taken after 20 or 24 h, with the latter time point ensuring that most cells have completed at least one cell cycle with LacI at its maximum level.
After 20 h of LacI induction, replication across the BamHI DNA fragment (∼5.2 kb) containing the lacO array was analysed by native 2D gel electrophoresis (Figure 1A and C). In both the presence and absence of LacI an arc of Y-shaped replication intermediates is detected. However, LacI (++) results in a greater accumulation of replication intermediates in the ascending portion of the Y-arc than when it is absent (Figure 1D). Replication across the ade6 locus is essentially unidirectional [moving towards the centromere (cen3)] (Figure 1A) (14), and therefore the accumulation of replication intermediates in the ascending Y-arc indicates that the progression of forks is perturbed as they enter the array. The absence of a defined point of DNA accumulation on the ascending Y-arc indicates that forks stall at multiple sites within the array. There is also a corresponding decrease in the descending Y-arc signal together with an increase in converged forks indicating that a significant percentage of stalled forks fail to restart in the presence of LacI (++) (Figure 1D and E). It is also important to note that the ascending Y-arc can contain small double Y structures, where both opposing forks are blocked and separated by at least half the total length of the DNA fragment being analysed. The same trends are seen with the lower level of LacI [LacI (+)], albeit they are far less marked than with the higher level (Figure 1C–E).
To provide further evidence that replication forks stall within the lacO array we compared the changes in signal intensity along the Y-arc in a BamH1 fragment with those in an Xba1 fragment where the start of the lacO array is shifted ∼1 kb in from the fragment end (Figure 1G and H). In the BamH1 fragment a relative increase in replication intermediates can be detected near to the start of the Y-arc (Figure 1G), whereas in the Xba1 fragment this increase is shifted along the Y-arc (Figure 1F and H), which is consistent with stalling occurring within the lacO array. Altogether these data indicate that the lacO-LacI interaction can act as a RFB in eukaryotes like in E. coli.
Replication fork stalling at the lacO array correlates with an induction of recombination
Having established that the lacO-LacI interaction hinders fork progression, we next tested whether this has any effect on recombination both between the repeats in the lacO array itself and the ade6− heteroalleles that flank it (Figure 2A). In the case of the lacO repeats, LacI expression results in an increase in contraction of the array as evident from the ladder of shorter lacO array-containing fragments seen on 1D gels (Figure 2B). In the case of the ade6− repeats, recombination was measured by determining the frequency of Ade+ recombinants (Figure 2A and C). Here recombinant formation increases by 15-fold with LacI (+) and almost 700-fold with LacI (++) (Figure 2C and Supplementary Table S1). Indeed with LacI (++) the frequency of Ade+ recombinants appears to be at or near its maximum with the remaining cells either being Ade− deletion-types or having lost the lacO array or LacI expression (data not shown). Heightened recombination is dependent on the presence of both the lacO array and the expression of LacI that is able to localize to the nucleus by the inclusion of a NLS (Figure 2C and Supplementary Table S1). The presence of a his3+ gene between the ade6 repeats enables us to distinguish two types of Ade+ recombinant—those that retain his3+ (conversion-types) and those that lose it (deletion-types). In the absence of LacI ∼78% of spontaneous recombinants are deletion-types (Figure 2D and Supplementary Table S1). Strikingly this percentage increases significantly with expression of LacI reaching 99% of recombinants with LacI (++) (Figure 2D and Supplementary Table S1). Altogether these data suggest that replication fork stalling at the lacO-LacI RFB triggers recombination, which mostly results in deletions. Intriguingly, this contrasts with recombination induced by the RTS1 RFB, which yields approximately equal amounts of deletion- and conversion-types (14), and suggests that programmed and accidental protein–DNA RFBs might trigger recombination in different ways.
The genetic requirements of lacO-LacI-induced recombination indicate that deletions arise by SSA
We have previously shown that the majority of recombinants that are induced by the RTS1 RFB (both conversion- and deletion-types) are dependent on Rad51 (14). To see if the same is true with the lacO-LacI RFB we assessed Ade+ recombinant formation with different levels of LacI expression in a rad51Δ background (Figure 3A and C and Supplementary Table S1). In all cases loss of Rad51 has little or no effect on the frequency of deletion-types, although the low-level of conversion-types is abolished (Figure 3B and D). This indicates that deletion-types induced by the lacO-LacI RFB occur mainly by a mechanism other than Rad51-mediated strand invasion. One possibility is SSA, which in Saccharomyces cerevisiae is known to be dependent on Rad52 and the Rad1–Rad10 structure-specific endonuclease (33). We therefore assessed lacO-LacI-induced recombinant formation in rad22Δ (Rad22 is the S. pombe orthologue of Rad52) and swi10Δ (Swi10 is the S. pombe orthologue of Rad10) single and double mutant strains (Figure 3A and C and Supplementary Table S1). Deletion-types are reduced in both single mutants and almost abolished in the double mutant. This loss of recombinant formation is not due to fluctuations in the levels of LacI expression, which, based on western blot analysis, remain fairly constant in the strains under investigation here (Supplementary Figure S1). Therefore these data indicate that lacO-LacI-induced deletion-types are likely to be formed by SSA.
Figure 3.
Genetic requirements for lacO-LacI-induced direct repeat recombination. (A) Ade+ recombinant frequencies in lacO array-containing strains expressing LacI (+). (B) The percentage of Ade+ recombinants in A that are conversion-types. (C) Ade+ recombinant frequencies in lacO array-containing strains expressing LacI (++). (D) The percentage of Ade+ recombinants in C that are conversion-types. In all cases error bars are the standard deviations about the mean (see also Supplementary Table S1 and Supplementary Figure S1).
The finding that lacO-LacI-induced deletion-types are formed by SSA suggests that replication fork stalling at the lacO-LacI RFB can result in DSBs. Indeed an HO-induced DSB between the ade6− repeats has previously been shown to give rise to deletion-types via SSA (24). For such breaks to be repaired by SSA between the ade6 repeats, several kilobases of DNA would have to be resected on either side of the DSB. Extensive 5′–3′ DSB resection requires either of two alternative pathways—one dependent on the exonuclease Exo1 and the other on a RecQ family DNA helicase (Sgs1 in S. cerevisiae or BLM in humans) (34). To determine whether lacO-LacI-induced deletion-types depend on DNA resection by Rqh1 (= S. pombe RecQ helicase) and/or Exo1 we assessed recombinant formation in rqh1Δ and exo1Δ single and double mutant strains (Figure 3A and C and Supplementary Table S1). Loss of Rqh1 reduces lacO-LacI(+)-induced deletion-type formation by ∼2-fold (P < 0.00001), whereas recombinant formation remains largely unaffected in the exo1Δ single mutant (P = 0.17) (Figure 3A). Importantly the rqh1Δ exo1Δ double mutant exhibits a synergistic reduction (∼10-fold) in deletion-types (P < 0.00001) (Figure 3A). With LacI (++) the effect of rqh1 and exo1 deletion is less pronounced with only a slight reduction in recombinants in each single mutant and an almost 2-fold reduction in the double mutant (P < 0.0001). We suspect that this less dramatic reduction in recombinants with LacI (++) is due to the greater frequency of DNA breakage, which can occur in successive cell cycles if repair is mediated by annealing between the lacO repeats without deleting the entire array. This means that there are multiple opportunities of forming an Ade+ recombinant, and therefore mutants that are not completely deficient in SSA will accumulate Ade+ recombinants to a similar level as wild-type albeit at a slower rate. Consistent with this there is a 5-fold difference in Ade+ recombinant frequency between wild-type and rqh1Δ exo1Δ strains after 24 h of LacI (++) induction in liquid culture (Figure 4F) as opposed to only a 2-fold difference after 6 days induction on solid media (Figure 3C).
Overall the data indicate that Rqh1 or Exo1 are needed for efficient SSA, presumably because they promote DNA resection. However, RecQ helicases and Exo1 are also implicated in the processing of stalled replication forks and therefore we cannot discount the possibility that they affect recombinant formation by other mechanisms (35–37).
lacO-LacI promotes DSB formation
A requirement for SSA is the presence of two DNA ends, which are resected to expose complementary nucleotide sequences. CHEF gel and Southern blot analysis of chromosome 3 revealed a band that migrates ahead of the chromosome 3 band, indicative of a DSB and amounting to 3% of the total DNA, in samples from cells with the lacO array after 20 h of LacI (++) induction (Figure 4A and B, compare lanes a and b, and i and j). Moreover, the detection of this band using probes either side of ade6 indicate that the chromosome is broken into two fragments. If the band is indicative of broken DNA, which is subject to active repair, then it should accumulate to higher levels in rqh1Δ exo1Δ and rad50S mutants that are defective in processing DSBs. In both mutants, but especially in the rqh1Δ exo1Δ double mutant, the DSB band accumulated to a higher levels (16.8 and 6.1%, respectively) than in the wild-type consistent with the expected impairment in DSB repair (Figure 4B, lanes c–f and k–n). However, it should be noted that RecQ helicases and Exo1 have roles in promoting the stability of stalled forks (35–37) and therefore the accumulation of high levels of DSBs in the rqh1Δ exo1Δ double mutant could also be due to increased levels of DNA breakage. To confirm that the detected band is indeed the result of a DSB at ade6, we analysed samples from a strain containing the HO endonuclease cleavage site MATa inserted in place of the lacO array (Figure 4B, lanes g, h, o and p). Following induction of the HO endonuclease a DSB band was detected, which migrated similarly to the band detected in the samples from the lacO-LacI(++)-containing strains (lanes h and p). Finally, we analysed DNA after digestion with AvrII, which liberates the ade6 region as a ∼58.3 kb fragment (Figure 4C). In addition to the full-length band, a ∼24 kb band was detected in samples where LacI (++) was induced, which maps to within the lacO array (Figure 4D, lanes b and d). The amount of DNA breakage is much higher than in Figure 4B with ∼20% of the DNA broken in the wild-type and 84% in the rqh1Δ exo1Δ double mutant. This is due to a longer induction time for LacI (++) (24 h instead of 20 h), which ensures that all cells have undergone at least one cell division in the presence of its maximum level (see earlier). Altogether these data suggest that replication fork stalling at the lacO-LacI RFB causes DNA breakage in a significant percentage of cells.
The lacO-LacI array reduces the viability of mutants defective in SSA
The high levels of DSBs detected in strains containing the lacO array upon LacI (++) induction should correlate with a reduction in viability in mutants deficient in DSB repair. Indeed a rad22Δ mutant containing the lacO array grows more slowly upon LacI (++) induction than when LacI is repressed, whereas a wild-type strain grows similarly whether LacI (++) is induced or not (Supplementary Figure S2). Intriguingly the growth of a rad51Δ mutant containing the lacO array is largely unaffected by LacI (++) induction indicating that Rad51 is not essential for the repair of lacO-LacI-induced DSBs (Supplementary Figure S2), which suggests that like recombinant formation viability might depend on SSA. If true then mutants deficient in SSA that contain the lacO array should show a reduction in viability when LacI (++) is induced. To test this we compared the viability of wild-type, rqh1Δ exo1Δ, rad22Δ and rad22Δ swi10Δ strains with and without the lacO array after 24 h of LacI (++) induction (Figure 4E). Each of the mutant strains containing the lacO array exhibit a reduction in viability after LacI (++) induction compared to strains without the lacO array (Figure 4E). Moreover this reduced viability correlates with a reduction in Ade+ recombinants suggesting that SSA between the ade6− genes that flank the lacO array is a major pathway for repairing the DSBs that are induced within it (Figure 4F). Based on the data in Figure 4D, at least 80% of rqh1Δ exo1Δ mutant cells containing the lacO array experience a DSB after 24 h of LacI (++) induction. In contrast viability is reduced only by ∼25% in this strain (Figure 4E). This indicates that the majority of the DSBs formed in an rqh1Δ exo1Δ double mutant are repaired successfully, albeit their higher accumulation suggests that the kinetics of repair may be slower than in wild-type. Furthermore, the low level of Ade+ recombinants suggests that SSA between the ade6− repeats is not the main repair mechanism (Figure 4F). Instead we suspect that repair occurs mostly by annealing between the lacO repeats. The larger reduction in viability (∼40%) observed in rad22Δ and rad22Δ swi10Δ mutants compared to the rqh1Δ exo1Δ double mutant suggests that this alternative means of repair is at least partly dependent on Rad22 (Figure 4E). Altogether these data show that genes required for SSA are also needed for the efficient repair of DSBs induced in the lacO array following LacI (++) induction.
lacO-LacI induces the formation of structures analogous to UFBs
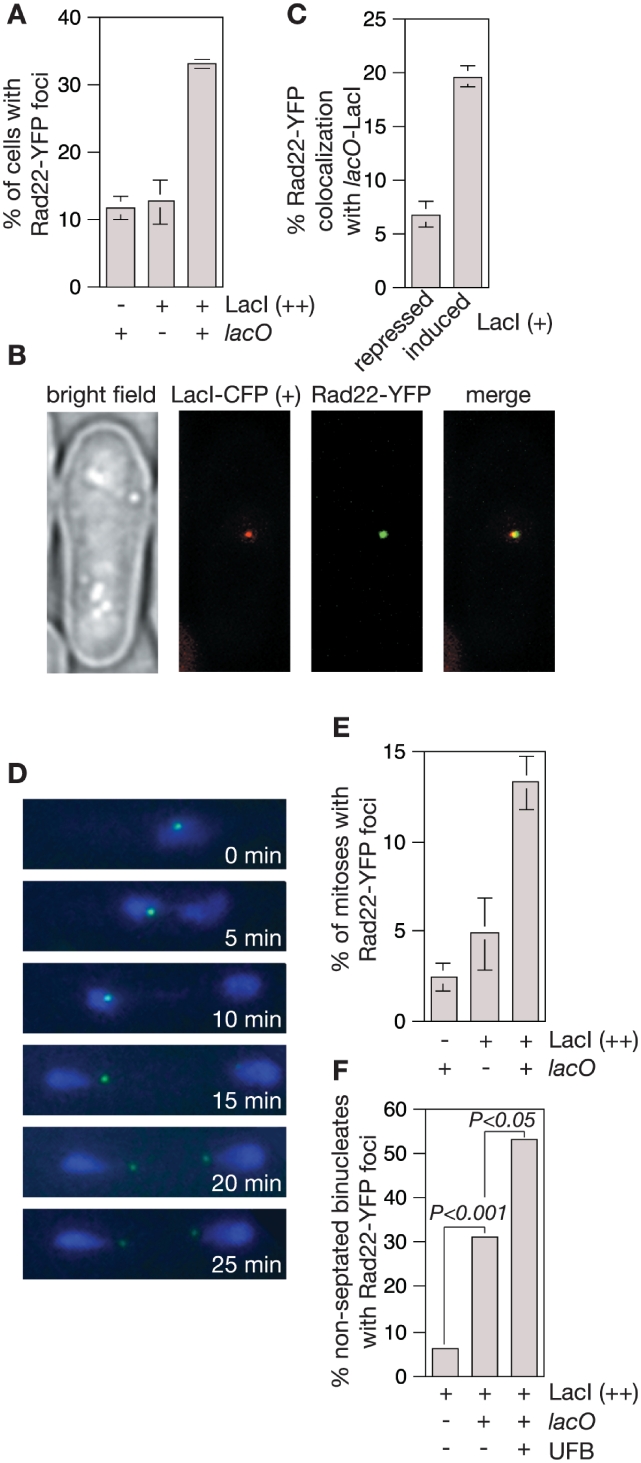
Whilst monitoring the expression of LacI-eCFP by epifluorescence microscopy we observed that ∼15% of cells undergoing mitosis display a bridge of LacI-eCFP fluorescence spanning the interval between the segregating DNA masses (Figure 5A and B). Unlike classical anaphase bridges, the bridges highlighted by LacI-eCFP fluorescence are generally undetectable by DNA specific dyes such as SYBR Green and DAPI (Figure 5A and data not shown). Like other fungi, S. pombe undergoes a closed mitosis where the nuclear membrane does not break down, and therefore the bridge of fluorescence could simply represent a tendril of nucleoplasm devoid of DNA. However, Histone H3.2 fused to GFP localizes to the bridges (Figure 5C) suggesting that they contain chromatin, and more importantly they are only prevalent in wild-type cells that contain the lacO array (Figure 5B), which indicates that the interaction between LacI and lacO is necessary for their formation. Furthermore, we observe lacO-LacI-dependent bridges that persist until the formation of the cell division septum suggesting that unresolved anaphase bridges could be broken during cytokinesis thereby providing a mechanism for the formation of DSBs in the lacO array (Figure 5D).
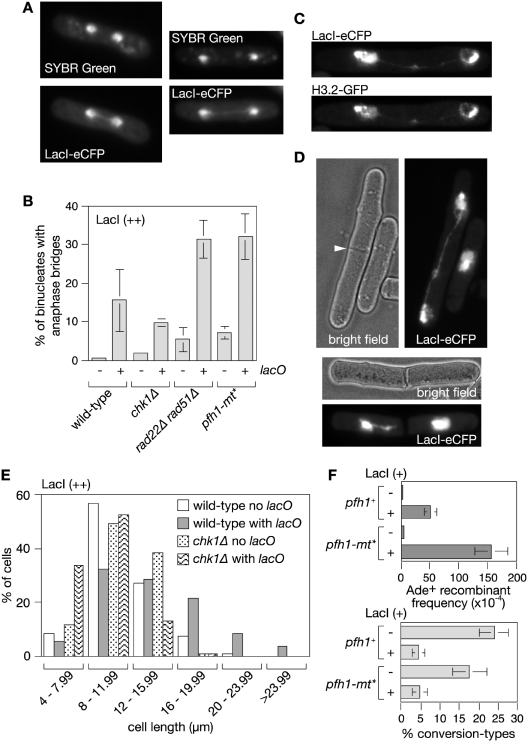
Figure 5.
lacO-LacI induces UFB formation. (A) Examples of UFBs in wild-type cells with the lacO array after 24 h of LacI (++) induction. (B) Quantification of the percentage of dividing cells with UFBs in wild-type and mutant strains with and without the lacO array after 24 h of LacI (++) induction. Mean values are from at least three independent experiments with ≥100 binucleate cells scored in each experiment. Error bars are the standard deviations. (C) Example showing localization of Histone H3.2-GFP to a UFB. (D, top panel) Example of cell with a UFB in which a division septum (indicated by the arrowhead) is in the process of forming. (D, bottom panel) Example of a cell in which a division septum has formed and appeared to have broken a UFB. (E) The distribution of cell lengths in asynchronously growing populations of wild-type and chk1Δ strains with and without the lacO array after 24 h of LacI (++) induction (n ≥ 300). (F) Ade+ recombinant frequencies (top panel) and percentage of Ade+ recombinants that are conversion-types (bottom panel) in lacO array-containing strains expressing LacI (+). Error bars are the standard deviations about the mean.
In humans there is a class of anaphase bridges that are detectable by immunostaining for DNA repair proteins such as BLM and FANCD2 but undetectable by DNA stains (18,19). Intriguingly these so-called UFBs are frequently associated with common fragile site loci including FRA3B, FRA16D and FRA7H, especially following inhibition of replication by aphidicolin, and are thought to be caused by incomplete replication within these regions (19). We propose that the lacO-LacI-dependent bridges are analogous to UFBs and therefore will refer to them by this name henceforth.
lacO-LacI UFBs are not caused by unrepaired DNA DSBs
In budding yeast, a single DSB induces a checkpoint-mediated arrest of the cell cycle. However, after 6–8 h cells adapt to the checkpoint and undergo mitosis even if the DSB is unrepaired, which in turn can impede proper chromosome segregation (38). In our case DSBs at the lacO array should be readily repaired by SSA during a checkpoint-mediated cell cycle arrest because the ade6− repeats are close enough to the array for sufficient DNA resection to occur prior to adaptation (38). Nevertheless, it was possible that unrepaired DSBs at the lacO array were responsible for the bridges that we had detected. If true then they should increase in frequency if the checkpoint response to the DSB is prevented as this would result in more unrepaired DSBs persisting into mitosis. Consistent with the DSB inducing a checkpoint response ∼33% of cells containing the lacO array in an asynchronously growing population are longer than 16 µm after a 24 h induction of LacI (++) compared to ∼8% without the lacO array (Figure 5E). This subpopulation of long cells is absent in the equivalent chk1Δ mutant strain indicating that lacO-LacI induces a Chk1-dependent cell cycle arrest (Figure 5E). Importantly, deletion of chk1 does not result in a greater frequency of lacO-LacI (++)-dependent bridges indicating that unrepaired DSBs are unlikely to be their cause (Figure 5B).
lacO-LacI UFBs are not caused by unresolved recombination intermediates
The UFBs detected by immunostaining for FANCD2 in human cells increase in frequency when Rad51 is depleted indicating that bridges are unlikely to be due to unresolved recombination intermediates between sister chromatids (19). Similarly lacO-LacI-dependent bridges increase in frequency in a rad22Δ rad51Δ double mutant (Figure 5B). Interestingly the frequency of bridges also increases in rad22Δ rad51Δ cells expressing LacI (++) without the lacO array compared to the equivalent wild-type strain (Figure 5B). This suggests that HR may play a role at other genomic loci to prevent the formation of UFBs caused by LacI binding to DNA non-specifically or other replication fork impediments. In the latter case, LacI-eCFP would simply highlight the presence of the UFB by occupying the nucleoplasm in the bridge.
Pfh1 prevents lacO-LacI-dependent UFB formation and hyper-recombination
In budding yeast non-nucleosomal protein–DNA complexes that could impede replication fork progression are removed by the putative ‘sweepase’ activity of the Rrm3 DNA helicase (39,40). If lacO-LacI-dependent UFBs are caused by replication fork stalling at protein–DNA barriers then they should increase in frequency in cells lacking the S. pombe orthologue of Rrm3, Pfh1 (41). Pfh1 has an essential role in the mitochondria of S. pombe and therefore we made use of a mutant pfh1 gene (pfh1-mt*) that expresses protein that localizes to the mitochondria but not the nucleus (41). A pfh1-mt* strain containing both lacO and LacI (++) shows a 2-fold increase in UFBs compared to wild-type (Figure 5B), which is consistent with the idea that bridge formation stems from an inability of replication forks to overcome the lacO-LacI barrier. Like in the rad22Δ rad51Δ double mutant LacI (++) expression in the pfh1-mt* mutant without a lacO array also results in an increase in bridge formation compared to the equivalent wild-type strain (Figure 5B). This is consistent with the idea that Pfh1 is needed at other genomic sites to aid their replication and thereby avoid bridge formation.
It is thought that unresolved UFBs in human cells may account for at least some of the DNA breakage that occurs at common fragile sites (19). If DNA breakage in the lacO array is similarly associated with a failure to properly resolve an anaphase bridge at this site then mutants such as pfh1-mt*, which manifest a greater frequency of bridges, should display more DSBs and consequently more recombination. With LacI (++) there is no difference in the frequency of lacO-LacI-induced Ade+ recombinant formation between wild-type and pfh1-mt* strains (data not shown). However, this is expected because as mentioned earlier recombinant formation is already at or near saturating levels in the wild-type. With LacI (+) a 3-fold increase in the frequency of lacO-LacI-induced Ade+ recombinants is seen in the pfh1-mt* mutant, which is consistent with Pfh1 being required to overcome the RFB and thereby avert anaphase bridge formation and associated DNA breakage (Figure 5F and Supplementary Table S1).
Evidence that the cell mounts a response to RFBs that continues through mitosis
The discovery of UFBs in human cells has revealed that replication intermediates frequently persist into mitosis, and cause the deployment of repair proteins, which act even as late as anaphase to resolve them (18,19). To see if the lacO-LacI RFB provokes a similar response we monitored sub-nuclear foci of Rad22 [fused to yellow fluorescent protein (YFP)]—a protein that plays a key role in promoting replication restart at the RTS1 RFB (5). In asynchronously growing populations of cells the presence of the lacO array together with LacI (++) results in a 3-fold increase in the number of cells with one or more foci (P < 0.01) (Figure 6A). A significant increase in the percentage of cells with foci was not seen with LacI (+) (data not shown), however increased co-localization of Rad22-YFP foci with the lacO array was (Figure 6B and C). Together these data show that the lacO-LacI RFB provokes a response that involves Rad22.
Figure 6.
Rad22-YFP foci are induced by the lacO-LacI RFB. (A) Rad22-YFP foci in strains with and without the lacO array with and without LacI (++) induction for 24 h. The data are mean values from three independent experiments with ≥100 cells assessed for foci in each experiment. Error bars represent standard deviations. (B) Example of Rad22-YFP focus co-localizing with lacO-LacI (+). (C) Co-localization of Rad22-YFP foci with the lacO array in the presence of repressed and induced levels of LacI (+). A total of between 200 and 300 Rad22-YFP foci-positive cells were assessed for co-localization in three independent experiments. The error bars are the standard deviations about the mean. (D) Images from time-lapse microscopy of Rad22-YFP foci (green) in a lacO array-containing cell undergoing anaphase after 24 h of LacI-eCFP (++) (blue) induction. (E) A comparison of the percentage of cells with and without lacO/LacI (++) that undergo mitosis with one or two Rad22-YFP foci present throughout. The data are means from three independent experiments with ∼50–100 time-lapse movies of cells undergoing mitosis assessed in each experiment. Error bars represent standard deviations. (F) A comparison of the percentage of non-septated binucleate cells with and without a lacO array and with and without a UFB (each n = 50) that contain at least one Rad22-YFP focus after 24 h of LacI (++) induction. P-values were calculated by Fisher's exact test (two-tailed).
At least some of the Rad22 foci caused by the lacO-LacI RFB are likely to be responding to the DNA DSB that is formed in the lacO array. As discussed above these DSBs should be repaired prior to mitosis. If this assumption is correct then Rad22 foci that persist into mitosis are likely to indicate the presence of unresolved replication intermediates at the lacO-LacI RFB. To investigate this we monitored foci in live cells by time-lapse microscopy (e.g. Figure 6D). Rad22-YFP foci are observed in only 2% of mitoses in the absence of LacI and 5% when LacI (++) is present in strains without the lacO array (Figure 6E). However, in strains with the lacO array LacI (++) expression results in an almost 3-fold increase in the number of mitoses with Rad22-YFP foci (P < 0.005) (Figure 6E). Typically a single Rad22-YFP focus is observed, which segregates with one DNA mass during anaphase (e.g. Figure 6D). In ∼60% of cases a second Rad22-YFP focus then appears, which is associated with the other DNA mass, and the two foci sometimes adopt a configuration where they reside at either end of an anaphase bridge (Figure 6D), which is reminiscent of the pattern of FANCD2 immunostaining observed at UFBs in human cells (19). Further analysis of fixed cells confirms that Rad22-YFP foci are more prevalent in non-septated binucleate cells containing a lacO-LacI-induced UFB than in the equivalent cells without a UFB (Figure 6F). Altogether these data suggest that Rad22 is recruited to replication forks that are blocked at the lacO-LacI array to help promote their restart, and that this response continues even during anaphase.
Shorter arrays of lacO induce UFBs and recombination
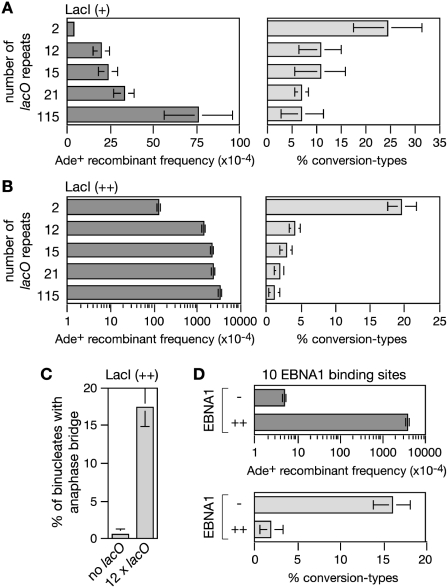
The 115 repeat lacO array was chosen because we wanted to impose a very potent barrier to replication. However, natural protein–DNA barriers are unlikely to be composed of so many adjacent protein–DNA ‘units’, and therefore to investigate whether shorter lacO arrays could generate similar problems we made a series of strains containing between 2 and 21 repeats in place of the 115 repeat array. With LacI (+) expression, no significant increase in direct repeat recombination was seen in the strain with two lacO repeats (Figure 7A and Supplementary Table S2). However, with 12, 15 and 21 lacO repeats a 5- to 8-fold increase in direct repeat recombinantion was observed with 90% or more of the recombinants being deletion-types (Figure 7A and Supplementary Table S2). With LacI (++) expression, two lacO repeats are sufficient to cause a 10-fold increase in recombinants compared to a strain with no repeats, and 12–21 repeats induce near saturating levels of recombinants similar to the 115 repeat array (Figure 7B and Supplementary Table S2). Again the majority of the induced recombinants are deletion-types, from ∼80% with two repeats to >95% with 12–115 repeats (Figure 7B and Supplementary Table S2). Similar to the 115 repeat array, the high frequency of recombinants seen with the shorter repeat arrays correlates with increased levels of UFB formation (Figure 7C and data not shown). Altogether these data indicate that the pathologies associated with problems in replicating the 115 repeat lacO-LacI array are also manifested by shorter arrays, albeit with reduced frequency especially when the array consists of only two repeats or LacI is expressed at the lower level.
Figure 7.
The effect of array length and type on recombinant frequency and UFB formation. (A) Ade+ recombinant frequencies and percentage conversion-types in strains with different length lacO arrays and LacI (+) induction. (B) The same as in (A) but with LacI (++) induction. (C) The percentage of dividing cells with UFBs in a strain with a 12xlacO array after 24 h of LacI (++) induction. (D) Ade+ recombinant frequencies and percentage conversion-types in strains with a 10xFR array with and without EBNA1 induction. In all cases error bars are the standard deviations about the mean.
EBNA1 binding to the FRs induces recombination like lacO-LacI
The interaction between LacI and lacO is similar to that of many transcription factors with their cognate DNA binding sites. However, to confirm that there was nothing unusual about the interaction between lacO and LacI that made it an especially potent RFB, we tested whether DNA binding by an unrelated protein would cause the same telltale increase in direct repeat recombination. The protein that we chose was EBNA1, which is one of only a few proteins that are expressed by the Epstein-Barr virus (EBV) during its latent infection of B lymphocytes (42). EBNA1 binds to a series of 20 repeated DNA elements known as the FRs within the viral origin of replication, and is required for the correct partitioning of the viral episomes in proliferating cells (43). An ∼800 bp DNA fragment containing 10 FR elements was inserted in place of the lacO array between the direct repeat of ade6− genes. Upon expression of the EBNA1 DNA binding domain from the nmt promoter in pREP41 a >700-fold increase in the frequency of Ade+ recombinants was observed (Figure 7D and Supplementary Table S3). Importantly >98% of these recombinants were deletion-types, which suggests that they stem from a similar set of problems as caused by the lacO-LacI repeat arrays.
DISCUSSION
It is now well established that the pausing of replication forks at RFBs can induce recombination, which can result in genome rearrangement. Recombination can be induced either through the remodelling of the stalled fork or by its cleavage to generate a DSB (11,14,44). In both cases the initiation of recombination is within replicated DNA (i.e. behind the replication fork), which makes accurate sister chromatid recombination possible and with promoting factors such as cohesin this becomes the predominant mechanism. It is thought that genome rearrangement only occurs when HR acts inadvertently between ectopic sites or low fidelity repair mechanisms such as NHEJ or SSA are used in its stead (13). However, as discussed below, our study shows how a RFB can give rise to a DSB that is predisposed to cause genome rearrangement most probably by being formed in unreplicated DNA and therefore only repairable by low fidelity mechanisms.
DSB formation at a non-programmed protein–DNA RFB
To date studies that have addressed the effect of protein–DNA RFBs on recombination in eukaryotes have focused on the programmed barriers RTS1 and the Fob1-dependent RFB from the rDNA array of S. cerevisiae (45). These have shown that such barriers can act as recombination hotspots, and in some cases this may be due to DNA breakage, although recent data shows that most recombination at the RTS1 RFB stems from template exchange events during replication restart rather than from DSB repair (5,6,14,46,47). Where DNA breakage has been observed it has generally been attributed to the action of specific nucleases, such as Slx1–Slx4 or Mus81–Eme1, cleaving the stalled fork. For example, replication fork breakage and Rad52/Rad22-dependent recombination in the rDNA repeats of both S. cerevisiae and S. pombe appear to be due to cleavage of converging forks by the Slx1–Slx4 structure-specific endonuclease (48,49). This is proposed to generate one nicked and one broken chromatid, the latter being repaired by HR (48). However, the DNA breaks that we observe in the lacO-LacI array are independent of Slx1 and Mus81 (data not shown). Moreover, cleavage by any of the known fork endonucleases would occur ‘behind the fork’ meaning that repair could occur by Rad51-dependent sister chromatid recombination. Loss of this type of repair should result in an increase in SSA and therefore in the formation of deletion-types (12). However, deleting rad51 has little or no effect on the overall frequency of recombinants suggesting that the DSBs formed at the lacO-LacI array are generally not repairable by sister chromatid recombination. This implies that DNA breakage either occurs in unreplicated DNA or at essentially the same site in both sister chromatids.
The UFBs and DSBs that are induced in strains containing a lacO-LacI array occur with similar frequency. As DSBs do not beget anaphase bridges in our experimental system, the correlation in their frequency strongly suggests that DNA breakage stems from the problems in segregating the sister chromatids containing the array. In human cells it is thought that UFBs are caused by DNA catenations between sister chromatids or regions where replication is incomplete (18,19). As the lacO-LacI array acts as a bi-directional RFB we believe that UFBs are most likely caused by a failure to complete replication in this case. This problem is far less likely with programmed barriers such as RTS1, which are unidirectional thereby allowing the completion of replication by the opposing fork if restart fails.
In human cells it is thought that entangled sister chromatids can be broken during anaphase by the pulling forces exerted by the spindle as there are generally enough microtubules (>10) attached to each kinetochore to do this (50). However, in S. pombe each kinetochore is contacted by only 2–4 microtubules and therefore the forces applied to each chromosome are insufficient to rupture them during anaphase (51). Indeed in S. cerevisiae the breakage of sister chromatids that remain catenated due to topoisomerase II inactivation depends on cytokinesis (52). In our study we observe UFBs that persist up to the point that the cell division septum is formed suggesting that DNA breaks occur during cytokinesis.
Protein–DNA arrays magnify a problem that is normally efficiently dealt with by the cell
Replication forks have to negotiate a multitude of protein–DNA interactions whilst replicating the genome. The machinery that enables them to do this includes fork protection factors such as Swi1 and Swi3 (= Tof1 and Csm3 in S. cerevisiae), the putative sweepase activity of Pfh1 (= Rrm3 in S. cerevisiae), and HR proteins that promote replication restart when replisome disassembly has occurred (5,39,40,53,54). Disabling any of this machinery can result in altered pausing at protein–DNA barriers and elevated levels of genome instability (5,15,40,55). A prime example of this is the pausing of replication forks at tRNA genes, which only elicits a strong recombinational response in the absence of fork protection or barrier removal activities (15,56). Presumably even when it is fully functional it occasionally fails to cope with the barriers that it is presented with. By using protein–DNA arrays we believe that we have increased the frequency of these normally rare, but nonetheless significant, events by creating a particularly potent barrier. Interestingly array size does not appear to be the most important factor in determining the potency of the barrier, as similar levels of anaphase bridge formation and recombination are obtained with a 12 repeat lacO array as with a 115 repeat array, and even two repeats produce measurable effects. More important than the number of repeats appears to be the amount of protein that is available to bind to them. Wild-type cells appear to cope well with a low-level of LacI-eCFP [non-induced LacI (+) level] that is sufficient to produce a detectable focus of fluroscence at the 115 repeat lacO array, however increasing the level of LacI-eCFP by ∼10-fold [LacI (+) level], which still produces just a single focus of fluorescence in cells with the array, produces significant levels of genome instability that are further increased when factors such as Pfh1 are missing. A further 10-fold increase in LacI-eCFP [LacI (++) level] is sufficient to fill the nucleus such that a discrete focus of fluorescence is no longer detectable, and at this level ∼15–20% of cells exhibit a UFB and DSB in each cell cycle. Presumably high concentrations of LacI make passage through the barrier difficult because displaced protein is more rapidly replaced. Nevertheless, even with LacI (++) the majority of cells appear to successfully replicate the lacO array in each cell cycle indicating that fork protection and replication restart are reasonably efficient even when faced with a particularly challenging barrier.
The cell's last ditch effort to restart replication?
It has recently been shown that Rad22 and, to a lesser extent, Rad51 are needed for promoting replication restart at the RTS1 RFB (5). In our study we observe increased levels of UFB formation when Rad22 and Rad51 are absent consistent with them promoting replication restart at the lacO-LacI RFB. We also observe a lacO-LacI-dependent induction of Rad51-dependent conversion-type recombinants that might be generated during attempts to restart replication. Moreover, in those cells where lacO-LacI induces problems in chromosome segregation we observe Rad22-YFP foci that persist throughout mitosis suggesting that attempts to restart replication continue right up to the point of cytokinesis. As mentioned earlier this is reminiscent of the situation in human cells where DNA repair proteins such as FANCD2 and BLM are present at replication intermediates that persist into anaphase (18,19).
Even though there is a cellular response mounted to the replication problem this does not appear to include an obvious delay in cell cycle progression, since a significant percentage of cells displaying UFBs divide at a normal division cell length (data not shown). We suspect that cells dividing with UFBs at greater than normal division cell length do so as a consequence of DNA breakage caused by replication fork stalling in the previous cell cycle. As DSB repair can occur by annealing between lacO repeats, cycles of fork stalling and DNA breakage can occur in successive cell cycles until the array is completely deleted. So if cell elongation is solely due to DNA breakage then replication fork blockage at the lacO array and presumed under replication does not appear to strongly induce a checkpoint response. An absence of checkpoint activation by forks arrested at lacO-LacI would be similar to what has been observed at both the RTS1 and Fob1-mediated RFBs (6,54). There are also mutants that fail to complete replication and progress to anaphase without inducing a checkpoint response even though the checkpoint machinery is not obviously disabled (57,58). It is therefore reasonable to suppose that unreplicated DNA at the lacO array could go unnoticed by the checkpoint machinery.
EBNA1 and genome instability
EBV is associated with a variety of human malignancies (42), and this association appears to depend on the expression of EBNA1, which is known to induce chromosomal aberrations and DNA DSBs in malignant B cells (59). Such genome instability is thought to result from increased levels of DNA damage caused by transcriptional activation of NOX2 (60). However, our data suggest that EBNA1 could also promote genome instability by forming RFBs at FR-like sequences in the human genome (61,62).
A final cautionary note
Arrays of lac or tet operator sequences bound by their cognate repressor fused to a fluorescent protein are widely used as a molecular tool to tag genomic loci for live cell imaging studies. Our data show that in wild-type cells such arrays can be tolerated well as long as the level of repressor protein is low. However, in mutants where the machinery of replication fork progression/protection/restart is perturbed, even low levels of repressor protein may be sufficient to induce instability at the array. This should be borne in mind when using operator sequences for live cell imaging experiments, especially when processes involved in genome stability are under investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants 057586/Z/99/B and 090767/Z/09/Z from the Wellcome Trust. S.S. was supported by a Skaggs-Oxford scholarship. S.C. was supported by a Dorothy Hodgkin Fellowship. J.L. was supported by grant MHIRT 2T37MD001368 from the National Center on Minority Health and Health Disparities, National Institutes of Health. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ashok Aiyar (Louisiana State University), David Sherratt (University of Oxford) and Virginia Zakian (Princeton University) for strains and plasmids.
REFERENCES
- 1.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 3.Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair. 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell. Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 5.Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell. 2010;39:346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 7.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Semin. Cancer Biol. 2010;20:222–233. doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 9.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Cortes-Ledesma F, Tous C, Aguilera A. Different genetic requirements for repair of replication-born double-strand breaks by sister-chromatid recombination and break-induced replication. Nucleic Acids Res. 2007;35:6560–6570. doi: 10.1093/nar/gkm488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 14.Ahn JS, Osman F, Whitby MC. Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J. 2005;24:2011–2023. doi: 10.1038/sj.emboj.7600670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryce DW, Ramayah S, Jaendling A, McFarlane RJ. Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swi1. Proc. Natl Acad. Sci. USA. 2009;106:4770–4775. doi: 10.1073/pnas.0807739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codlin S, Dalgaard JZ. Complex mechanism of site-specific DNA replication termination in fission yeast. EMBO J. 2003;22:3431–3440. doi: 10.1093/emboj/cdg330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagawa T, Yamada-Inagawa T, Eydmann T, Mian IS, Wang TS, Dalgaard JZ. Schizosaccharomyces pombe Rtf2 mediates site-specific replication termination by inhibiting replication restart. Proc. Natl Acad. Sci. USA. 2009;106:7927–7932. doi: 10.1073/pnas.0812323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 20.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 21.Osman F, Whitby MC. Monitoring homologous recombination following replication fork perturbation in the fission yeast Schizosaccharomyces pombe. Methods Mol. Biol. 2009;521:535–552. doi: 10.1007/978-1-60327-815-7_31. [DOI] [PubMed] [Google Scholar]
- 22.Osman F, Adriance M, McCready S. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 2000;38:113–125. doi: 10.1007/s002940000145. [DOI] [PubMed] [Google Scholar]
- 23.Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 24.Osman F, Fortunato EA, Subramani S. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics. 1996;142:341–357. doi: 10.1093/genetics/142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aiyar A, Aras S, Washington A, Singh G, Luftig RB. Epstein-Barr Nuclear Antigen 1 modulates replication of oriP-plasmids by impeding replication and transcription fork migration through the family of repeats. Virol. J. 2009;6:29. doi: 10.1186/1743-422X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 27.Huberman JA, Spotila LD, Nawotka KA, el-Assouli SM, Davis LR. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 28.Win TZ, Goodwin A, Hickson ID, Norbury CJ, Wang SW. Requirement for Schizosaccharomyces pombe Top3 in the maintenance of chromosome integrity. J. Cell Sci. 2004;117:4769–4778. doi: 10.1242/jcs.01351. [DOI] [PubMed] [Google Scholar]
- 29.Hyppa RW, Smith GR. Using Schizosaccharomyces pombe meiosis to analyze DNA recombination intermediates. Methods Mol. Biol. 2009;557:235–252. doi: 10.1007/978-1-59745-527-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 31.Possoz C, Filipe SR, Grainge I, Sherratt DJ. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25:2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 33.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mimitou EP, Symington LS. DNA end resection-unraveling the tail. DNA Repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 37.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 39.Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 41.Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol. Cell Biol. 2008;28:6594–6608. doi: 10.1128/MCB.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 43.Lindner SE, Sugden B. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid. 2007;58:1–12. doi: 10.1016/j.plasmid.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The fission yeast FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labib K, Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burkhalter MD, Sogo JM. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol. Cell. 2004;15:409–421. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coulon S, Noguchi E, Noguchi C, Du LL, Nakamura TM, Russell P. Rad22/Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol. Biol. Cell. 2006;17:2081–2090. doi: 10.1091/mbc.E05-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jannink G, Duplantier B, Sikorav JL. Forces on chromosomal DNA during anaphase. Biophys. J. 1996;71:451–465. doi: 10.1016/S0006-3495(96)79247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell. Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 53.Noguchi E, Noguchi C, McDonald WH, Yates JR, 3rd, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Loza MC, Wellinger RE, Aguilera A. Stimulation of direct-repeat recombination by RNA polymerase III transcription. DNA Repair. 2009;8:620–626. doi: 10.1016/j.dnarep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- 58.Dulev S, de Renty C, Mehta R, Minkov I, Schwob E, Strunnikov A. Essential global role of CDC14 in DNA synthesis revealed by chromosome underreplication unrecognized by checkpoints in cdc14 mutants. Proc. Natl Acad. Sci. USA. 2009;106:14466–14471. doi: 10.1073/pnas.0900190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamranvar SA, Gruhne B, Szeles A, Masucci MG. Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene. 2007;26:5115–5123. doi: 10.1038/sj.onc.1210324. [DOI] [PubMed] [Google Scholar]
- 60.Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl Acad. Sci. USA. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.d'Herouel AF, Birgersdotter A, Werner M. FR-like EBNA1 binding repeats in the human genome. Virology. 2010;405:524–529. doi: 10.1016/j.virol.2010.06.040. [DOI] [PubMed] [Google Scholar]
- 62.Canaan A, Haviv I, Urban AE, Schulz VP, Hartman S, Zhang Z, Palejev D, Deisseroth AB, Lacy J, Snyder M, et al. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc. Natl Acad. Sci. USA. 2009;106:22421–22426. doi: 10.1073/pnas.0911676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meister P, Poidevin M, Francesconi S, Tratner I, Zarzov P, Baldacci G. Nuclear factories for signalling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 2003;31:5064–5073. doi: 10.1093/nar/gkg719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.