Abstract
Histone modifications play an important role in epigenetic gene regulation and genome integrity. It remains largely unknown, however, how these modifications dynamically change in individual cells. By using fluorescently labeled specific antigen binding fragments (Fabs), we have developed a general method to monitor the distribution and global level of endogenous histone H3 lysine modifications in living cells without disturbing cell growth and embryo development. Fabs produce distinct nuclear patterns that are characteristic of their target modifications. H3K27 trimethylation-specific Fabs, for example, are concentrated on inactive X chromosomes. As Fabs bind their targets transiently, the ratio of bound and free molecules depends on the target concentration, allowing us to measure changes in global modification levels. High-affinity Fabs are suitable for mouse embryo imaging, so we have used them to monitor H3K9 and H3K27 acetylation levels in mouse preimplantation embryos produced by in vitro fertilization and somatic cell nuclear transfer. The data suggest that a high level of H3K27 acetylation is important for normal embryo development. As Fab-based live endogenous modification labeling (FabLEM) is broadly useful for visualizing any modification, it should be a powerful tool for studying cell signaling and diagnosis in the future.
INTRODUCTION
Post-translational protein modifications play a critical role in the regulation of many biological processes, like cell metabolism and signaling. These modifications can change rapidly and dramatically in response to internal and external stimuli, altering the properties of proteins, including their enzymatic activity, subcellular localization and intermolecular interactions (1). Whereas modification-specific antibodies and mass spectrometry can detect the dynamics of modifications in fixed and biochemically isolated samples, real-time imaging of modifications in single living cells is important for understanding their spatiotemporal regulation (2).
Histones are some of the most highly modified proteins in the cell nucleus. Together with DNA they form nucleosomes, which are subject to a wide range of modifications, including acetylation, methylation and phosphorylation. These modifications help regulate gene expression and maintain genome integrity (3). Transcriptionally active chromatin is generally associated with histone H3 lysine acetylation and H3 lysine 4 (H3K4) methylation, whereas silent chromatin typically harbors methylation on H3 lysine 9 or 27 (H3K9 or H3K27). The levels of histone modifications change both locally and globally throughout the cell cycle, as well as during development and differentiation, pathogenesis, and in response to external stimuli (3,4). For example, immunofluorescence studies using modification-specific antibodies have shown the drastic changes in the acetylation and methylation levels of histone H3 and H4 during mouse embryo development and germ cell formation (5–7).
Until recently little was known about the dynamics of histone modifications in single living cells, but new developments are beginning to change this (8). A Förster/fluorescence resonance energy transfer (FRET)-based sensor has been developed to monitor histone H4 diacetylation in response to histone deacetylase (HDAC) inhibitors (9). Although this sensor can monitor the balance of modifying and demodifying enzymes, it is unable to detect the modifications of endogenous proteins. Concurrently, we have used fluorescently labeled antigen-binding fragments (Fabs) to visualize endogenous histone H3 phosphorylation during the cell cycle (10).
In this study, we demonstrate that the localization and global level of a variety of different epigenetic modifications of endogenous histone H3 can be monitored in single living cells using specific monoclonal antibodies (mAbs) (11). Soon after being loaded into the cytoplasm of living cultured cells, fluorescently labeled Fabs can enter the nucleus and produce distinct nuclear patterns that are characteristic of their target modifications. Fabs bind to their epitopes transiently in living cells and the binding time correlates with their in vitro epitope-binding affinity. Thus, when nuclear modification levels increase, so too does the concentration of nuclear Fabs (since there are now more target binding sites). It is therefore possible to monitor rapid changes in global modification levels by simply comparing the concentration of Fabs in the nucleus and cytoplasm.
Fabs with higher affinities are also suitable for imaging modifications in mouse preimplantation embryos without disturbing normal development. Monitoring H3K9 and H3K27 acetylation levels in mouse preimplantation embryos revealed their differential behavior. While H3K9 acetylation level remains relatively constant throughout zygote and two-cell stages, H3K27 acetylation is reduced drastically in zygotes. Furthermore, the H3K27 acetylation level in somatic nuclei transferred into oocytes is much lower than pronuclei in zygotes, but is increased by histone deacetylase inhibitor treatments, which are known to improve the cloning efficiency of embryos transferred with somatic nuclei (12). These data suggest that a high level of H3K27 acetylation is important for normal embryo development, possibly by counteracting the methylation on the same site. In general, Fab-based live endogenous modification labeling (FabLEM) can be used to monitor any endogenous protein modification in single living cells. We therefore anticipate it will find many future applications.
MATERIALS AND METHODS
Antibody and dye-conjugation
To generate monoclonal antibodies directed against histone H3K9me1, H3K9me2, H3K9me3 and H3K27me3, mice were immunized with synthetic peptide ARTKQTAR(me1-K)STGGKAPRKQC, ARTKQTAR (me2-K)STGGKAPRKQC, ARTKQTAR(me3-K)STGGKAPRKQC and KQLATKAAR(me3-K)SAPATGGVKC, respectively, coupled to keyhole limpet hemocyanin. After generating hybridomas, clones were screened by ELISA using peptides listed in (11). Clones CMA316, CMA317/6D11, CMA318/2F3 (13) and CMA323 reacted specifically with H3K9me1, H3K9me2, H3K9me3 and H3K27me3, respectively (e.g. Supplementary Figure S1). CMA316 was isotyped as IgG3-κ and the other clones were isotyped as IgG1-κ using a kit (AbD Serotec). For antibody purification, hybridomas were grown in CD Hybridoma medium (Invitrogen) supplemented with 2 mM glutamine. Except CMA316, the culture supernatant (250 ml) was filtrated through a 0.45-µm pore filter, added with NaCl at a final concentration of 4 M and applied to a HiTrap Protein A HP Sepharose column (1 ml; GE Healthcare). After washing the column with Protein A IgG1 binding buffer (Thermo Fisher Scientific), IgG was eluted using Mouse IgG1 Mild Elution Buffer (Thermo Fisher Scientific) and concentrated up to 4–8 mg/ml in PBS using an Amicon Ultra filter (50 k-cut off; Millipore). As CMA316 was sensitive to low pH elution, it was purified through a HiPrep CM Sepharose (GE Healthcare) using a linear NaCl gradient (20–300 mM) in 50 mM 2-morpholinoethanesulfonic acid butter (pH 6.5), followed by gel filtration chromatography using HiLoad 16/60 Superdex 200 pg (GE Healthcare) in PBS.
The affinity of antibodies to the epitope was measured by surface plasmon resonance using BIACORE X100 (GE Healthcare Biosciences). The interaction experiments were performed in PBS, at flow rate of 30 µl/min. A 60 µl aliquot of each of the different concentrations of antibodies (1, 4, 16, 64 and 256 nM) was injected onto the chip sequentially without regeneration. Although a previous ELISA study showed CMA310 cross-reacted with acetylated H3K27 (H3K27ac) (11), the binding to H3K27 was several orders of magnitude lower according to BIACORE measurements (6.2 × 10−6 M to H3K27ac peptide versus 4.4 × 10−11 M to H3K9ac; Table 1); thus CMA310 is now regarded as H3K9ac specific mAb.
Table 1.
Epitope-binding affinity of mAbs in vitro by surface plasmon resonance measurements
| mAba | Peptideb | ka (M−1s−1)c | kd (s−1)d | KD (M)e |
|---|---|---|---|---|
| CMA303 | H3K4me2 | 7.6 × 109 | 8.2 × 101 | 1.1 × 10−8 |
| CMA309 | H3K27ac | 2.4 × 105 | 3.4 × 10−7 | 1.4 × 10−12 |
| CMA310 | H3K9ac | 1.6 × 105 | 6.8 × 10−6 | 4.4 × 10−11 |
| H3K27ac | 7.5 × 103 | 4.7 × 10−2 | 6.2 × 10−6 | |
| CMA317 | H3K9me2 | 1.6 × 109 | 2.4 × 101 | 1.5 × 10−8 |
| CMA323 | H3K27me3 | 2.6 × 105 | 3.2 × 10−3 | 1.2 × 10−8 |
amAb analyzed.
bSensor tip was coated with bovine serum albumin conjugated with indicated peptide.
cAssociation rate.
dDissociation rate.
eDissociation constant (kd/ka).
Fab was prepared using a kit (Thermo Fisher Scientific; Pierce Fab Preparation Kit or Pierce Mouse IgG1 Fab and F(ab′)2 Preparation Kit) according to the manufacturer’s instruction. After the buffer was replaced to PBS using PD10 desalting column (GE Healthcare), the samples were concentrated up to ∼1 mg/ml using an Ultrafree 0.5 filter (10 k-cut off; Millipore). The purity and integrity of Fab were analyzed by SDS–PAGE using a 10–20% gradient gel (Wako). Purified Fab or IgG was conjugated with a fluorescent dye using Alexa Fluor 488 tetrafluorophenyl ester (Invitrogen), Cy3 N-hydroxysuccinimide ester (GE Healthcare), or Cy5 N-hydroxysuccinimide ester (GE Healthcare). Dried fluorescent dye esters (for labeling 1 mg protein) were dissolved into 50 (Alexa488) or 100 (Cy3 and Cy5) µl dimethyl sulfoxide (DMSO; Wako) and stored at −20°C. Fab (100 µg) was diluted into 100 mM NaHCO3 (pH 8.3) in 100 µl. After addition of a dye solution (5, 1.5 and 4 µl for Alexa488, Cy3 and Cy5, respectively), the mixture was incubated for 1 h at room temperature with gentle rotation. The sample was passed through a PD-mini G-25 desalting column (GE Healthcare), which was pre-equilibrated with PBS, to remove unconjugated dyes, and dye-conjugated Fab was concentrated up to ∼1 mg/ml using an Ultrafree 0.5 filter (10 k-cut off; Millipore). The Fab concentration and dye:protein ratio were calculated from the absorbance at 280 and 494, 552, or 650 nm, using the extinction coefficient of IgG and correction factor at 280 nm provided by the manufacturers (i.e. 0.11, 0.08, or 0.05). Fluorescent dye-labeled Fab samples that yielded dye:protein ratio ∼1–2 were used for live imaging.
Purified recombinant PCNA (14) was conjugated with Cy3 as described above (0.6 mg/ml; dye:protein ratio 0.3).
Cells and microscopy
Somatic cells were routinely grown in DME (Dulbecco’s modified Eagle’s medium; Sigma-Aldrich) supplemented with penicillin/streptomycin (10 U/ml penicillin, 50 µg/ml streptomycin; Sigma-Aldrich) and 10% FCS. Skin fibroblasts derived from normal female (WI-38; passage 14), Klinefelter Syndrome (Detroit 529; passage 14 and Dempsey; passage 25) and Turner Syndrome (Detroit 525; passage 22) were purchased from American Type Culture Collection. HeLa and U2OS cells have been described previously (10). HeLa cells stably expressing histone H2B-mRFP were established by transfecting an H2B-mRFP expression construct (15,16). Conditional G9a knockout ES cells based on Mer-Cre-Mer system (17) were cultured in DME (Nacalai Tesque) supplemented with leukemia inhibitory factor (1000 U/ml; Invitrogen), non-essential amino acids, sodium pyruvate, 2-mercaptoethanol, penicillin/streptomycin and 10% FCS. To delete the G9a gene, ES cells were cultured in 800 nM 4-hydroxytamoxifen (Sigma-Aldrich) for 4 days.
For live cell imaging, cells were plated on a glass-bottom dish with a coverslip (Mat-Tek) and the next day fluorescent Fab was loaded using a bead-loading method (10,18,19), as follows. The medium was removed from the dish, fluorescent Fab was pipetted onto the center of the coverslip (1 mg/ml in PBS; 2–4 µl), and glass beads (106 µm; Sigma-Aldrich; G-4649) were sprinkled on top. After tapping the dish four to eight times, DME was immediately added to the dish and beads were washed away. The medium was then replaced to phenol red-free DME (Nacalai Tesque) containing the supplements. The dish was placed either on an inverted microscope (Ti-E; Nikon) or on a confocal microscope (FV-1000; Olympus). Either microscope featured a culture system (Tokai Hit) at 37°C under 5% CO2. Wide-field fluorescence images were captured using a Ti-E (Nikon) under the operation of NIS Elements ver 3.0 (Nikon) with a PlanApo VC ×100 (NA = 1.4) oil-immersion objective lens equipped with an electron multiplying charge-coupled device (iXon+; Andor; normal mode; gain ×5.1) with filter sets (Semrock; LF488-A for Alexa488, LF561-A for Cy3 and Cy5-4040 A for Cy5); the exposure period was set to 100–1000 ms. A 75 W Xenon lamp was used as a light source and attenuated through neutral-density and 440 nm long-pass filters to achieve a light intensity of 6–10 μW at the specimen. Phase-contrast images were also collected (100 ms) using an external phase ring. In some cases, three different focal planes were imaged with 2 µm intervals. For Figure 3A–D and Supplementary Figure S2, optical sections were collected using an FV-1000 (Olympus) with a PlanSApo 60× (NA = 1.35) oil-immersion lens (512 × 512 pixels; 10 µs/pixel; four-line Kalman; pinhole 120 µm). Multicolor fluorescence images were collected using line sequential mode, and differential interference contrast images were collected simultaneously with Alexa488 images (Figure 3C).
Figure 3.
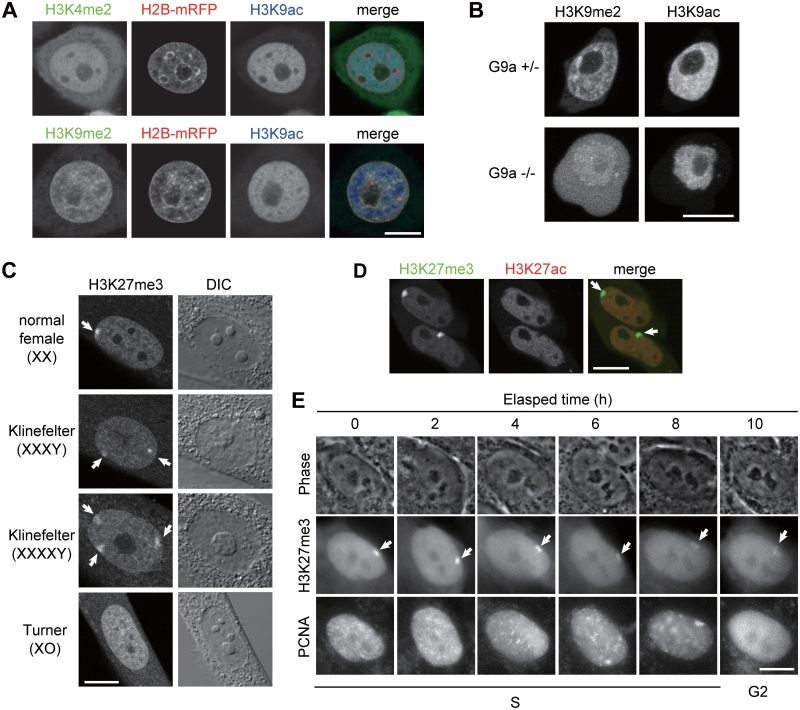
Localization of methylated histone H3 in living cells. (A–D) Confocal microscope images of living cells. (A) H2B-mRFP-expressing HeLa cells loaded with FabH3K4me2-488, or FabH3K9me2-488, together with FabH3K9ac-Cy3. (B) Nuclear concentration of FabH3K9me2 is diminished in G9a knockout ES cells. FabH3K9me2-488 and FabH3K9ac-Cy3 were loaded into conditional G9a knockout ES cells before (top) and after (bottom) the gene deletion. (C) FabH3K27me3 is concentrated in inactive X chromosomes. Arrows indicate FabH3K27me3 foci. (D) Mouse MC12 cells loaded with FabH3K27me3-488 and FabH3K27ac-Cy3. Arrows indicate FabH3K27me3 foci. (E) Behavior of inactive X chromosomes during S phase. hTERT-RPE1 cells were loaded with FabH3K27me3-488 and PCNA-Cy3, and phase contrast and wide-field fluorescent images were collected every 15 min (Movie 4 in Supplementary Data). Arrows indicate FabH3K27me3-enriched inactive X chromosomes. Bars, 10 µm.
For Immunofluorescence using fixed samples, cells grown on coverslips were fixed with 1% formaldehyde (Electron Microscopy Sciences) in 250 mM HEPES–NaOH (pH 7.4) for 10 min at room temperature, washed with PBS, incubated in 2 µg/ml Cy3-conjugated IgG and 10 ng/ml Hoechst 33342 (Nacalai Tesque) in PBS containing 10% Blocking One-P (Nacalai Tesque) and 0.5% Triton X-100 for 2 h at room temperature. After washing three times with PBS, coverslips were mounted using Prolong-Gold (Invitrogen). Fluorescence images were sequentially collected using a confocal microscope (FV-1000; Olympus; operated by the built-in software ver. 1.7) as above.
FRAP was performed using a confocal microscope (FV-1000; Olympus) with a PlanSApo 60× (NA = 1.35) oil-immersion lens (10). For Figure 2, 10 images were collected (0.4% 488-nm laser transmission; 1 s/frame; 2 µs/pixel; 256 × 256 pixels; pinhole 800 µm; 6× zoom), a 2 µm diameter spot was bleached (100% 488-nm laser transmission; four iteration), and a further 90 images were collected using the original settings. For Supplementary Figure S5, 10 images were collected (0.3% 488-nm and 3% 543-nm laser transmission; 0.38 s/frame; 2 µs/pixel; 128 × 128 pixels; pinhole 800 µm; 10× zoom), a 2 µm diameter spot was bleached (25% 405-nm, 100% 488-nm and 100% 543-nm laser transmission; 53 ms), and a further 290 images were collected using the original settings. To quantify the bound/free fraction, the pure-diffusion coefficient of each Fab was required. For this purpose, measurements of diffusion in the cytoplasm were made. To monitor fast diffusion, a single line (0.8% 488-nm and 39% 543 nm laser transmission; 2 µs/pixel; 128 × 1 pixels; pinhole 800 µm; 10× zoom) passing through the center of the bleach spot (2 µm diameter, 100% 405-nm laser transmission; 77 ms) was continually scanned to quantify the bleach spot profile with time. 100 pre-bleach scans were followed by 2000 post-bleach scans.
Figure 2.
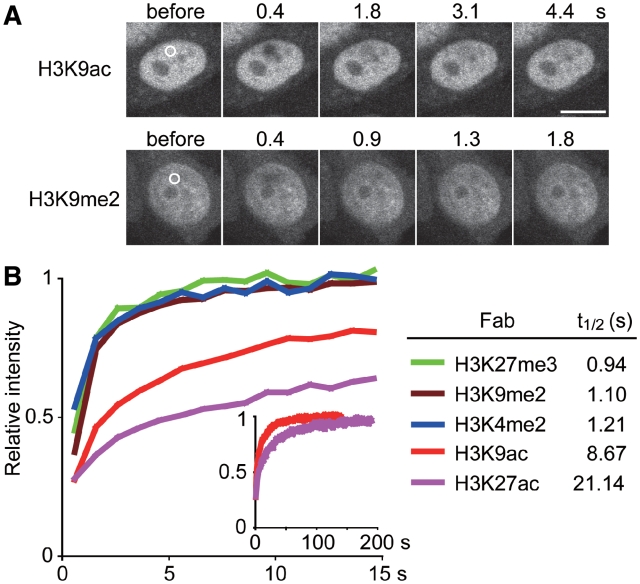
Kinetics of Fabs in living cells. (A) FRAP examples. A small area (2 µm diameter) is bleached and the fluorescence intensity in the bleached area is measured. Bar, 10 µm. (B) FRAP results. The relative intensity of bleached area for the indicated Fabs was plotted (averages of >15 cells) with the recovery t1/2.
Fluorescence intensity was measured using Metamorph ver 7.5 (Molecular Devices) and plotted using Origin ver 7 (OriginLab). Mathematica (Wolfram Research) was used for the fitting analysis in Supplementary Figure S5.
Imaging mouse preimplantation embryos
Mouse oocyte preparation, in vitro fertilization (IVF), microinjection, and imaging were described previously (10,20). Briefly, fertilized oocytes at anaphase II-telophase II stage were injected with a mixture of fluorescently-labeled Fab and 5 µg/ml of histone H2B-mRFP1 mRNA and placed on the imaging system (Figure 6A and B). For the imaging of the SCNT embryos, metaphase II stage oocytes were injected with the above mixture, enucleated and injected with a somatic nucleus derived from a cumulus cell. The reconstructed embryos were artificially activated with Ca2+-free Chatot-Ziomek-Bavister medium containing 10 mM SrCl2 and 5 µg/ml cytochalasin B in the presence or absence of 50 nM TSA for 6 h. According to the original protocol (12), the embryos were further incubated for 3 h in the presence of 50 nM TSA. As an imaging system, an inverted microscope (IX-71; Olympus) with UPlanApochromat 20× (NA = 0.8) oil-immersion lens was attached with a Nipkow-disk scanning confocal unit (CSU10; Yokogawa Electric Corp.) and an electron multiplying charge-coupled device (iXON BV-887; Andor). Three-dimensional and multipoint acquisition of images were performed using a z motor (Mac5000; Ludl Electronic Products) and an auto-xy stage (Sigma Koki). MetaMorph ver 7.5 (Molecular Devices) was used for device control and image analysis. Fifty-one images in the z-axis and two color images (excited with 488- and 561-nm laser lines) were captured at 15 min intervals. After the imaging, morula–blastocyst stage embryos produced by IVF were transferred to the uterus of the recipient pseudopregnant mothers. All animals were maintained in accordance with the Animal Experimental Handbook at the Kobe Center for Developmental Biology (RIKEN).
Figure 6.
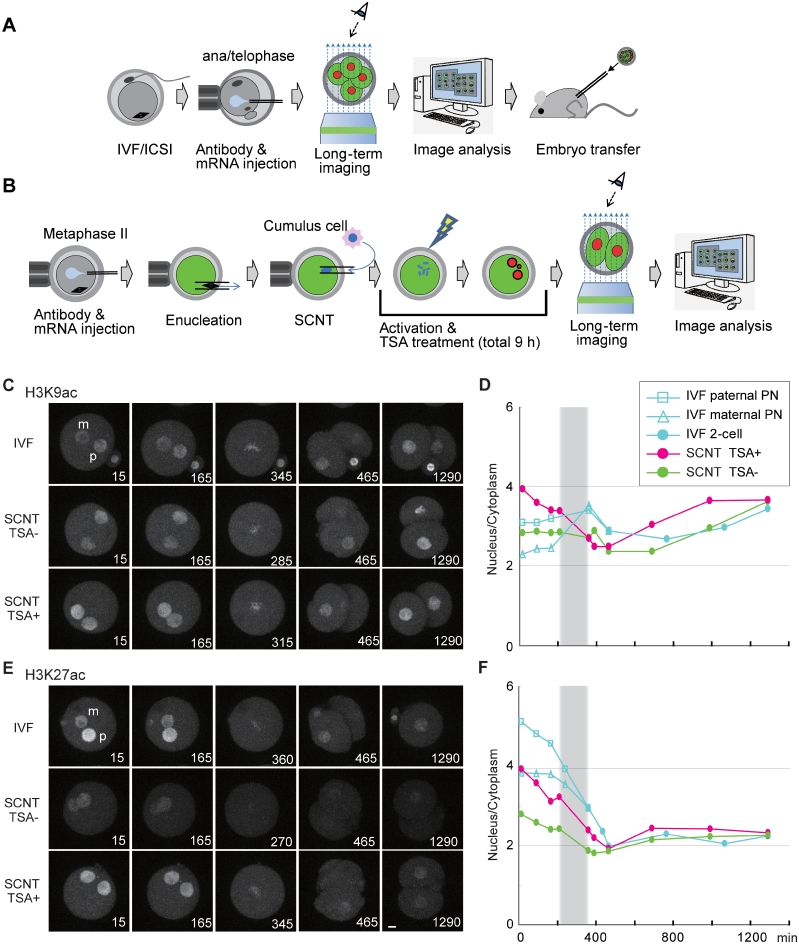
Monitoring the levels of histone H3 acetylation in living mouse preimplantation embryos. (A and B) Schematic drawing of mouse embryo imaging. (A) FabLEM perfomed on eggs with in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI). Oocytes were fertilized in vitro, injected with a mixture of fluorescently labeled Fab and histone H2B-mRFP1 mRNA at anaphase II-telophase II stage, and then placed on the imaging system. After the imaging, morula–blastocyst stage embryos were transferred to the uterus of the recipient pseudopregnant mothers. (B) FabLEM perfomed on embryos with somatic cell nuclear transfer (SCNT). Metaphase II stage oocytes were injected with a mixture of fluorescently labeled Fab and histone H2B-mRFP1 mRNA, enucleated, and injected with a somatic nucleus derived from a cumulus cell. The reconstructed embryos were activated with SrCl2 and cytochalasin B (±50 nM TSA) for 6 h. The embryos were further incubated (±50 nM TSA) for 3 h. (C–F) Monitoring the levels of histone H3 acetylation. Time-lapse confocal images were acquired for IVF and SCNT (±TSA treated) embryos injected with FabH3K9ac-488 (C and D; Movie 8 in Supplementary Data), or FabH3K27ac-488 (E and F; Movie 9 in Supplementary Data), and mRNA encoding H2B-mRFP. Typical images before and after the first division are shown in (C) and (E). The intensity ratio of nucleus to cytoplasm was measured and the averages (n > 10) are plotted (D and F). Bars, 10 µm.
RESULTS
Visualizing acetylated histone H3 in living cells using a specific monoclonal antibody
To visualize protein modifications in living cells, purified monoclonal antibody (mAb), either as a form of immunoglobulin G (IgG) or Fab, was conjugated with a fluorescent dye and loaded into cells (Figure 1A). If the antibody specifically binds to nuclear protein modifications, it should localize to regions of chromatin that are enriched in that modification. We first used acetylated histone H3K9 (H3K9ac)-specific mAb CMA310 (11) (Table 1) to compare the properties of IgG (IgGH3K9ac) and Fab (FabH3K9ac). When loaded into the cytoplasm of HeLa cells at ∼1 µM by bead-loading (10), Alexa Fluor 488-labeled IgGH3K9ac was excluded from the nucleus because it could not pass through the nuclear pore due to its large size (∼150 kD). Only during mitosis, when the nuclear membrane breaks down, could IgGH3K9ac access target chromatin and be incorporated into the nucleus in daughter cells (Figure 1B and Movie 1 in Supplementary Data). In contrast, Alexa Fluor 488-labeled H3K9ac-specific Fab (FabH3K9ac-488) immediately diffused into the nucleus due to its smaller size (∼50 kD) and exhibited nucleoplasmic concentrations in less than an hour (Figure 1C and Movie 2 in Supplementary Data). Hence, Fab appears to be more convenient than the whole IgG molecule for live-cell imaging. The monovalency of Fab may also be an advantage, because the divalent IgG could form artificial aggregations when two antigens are bridged (21).
Figure 1.
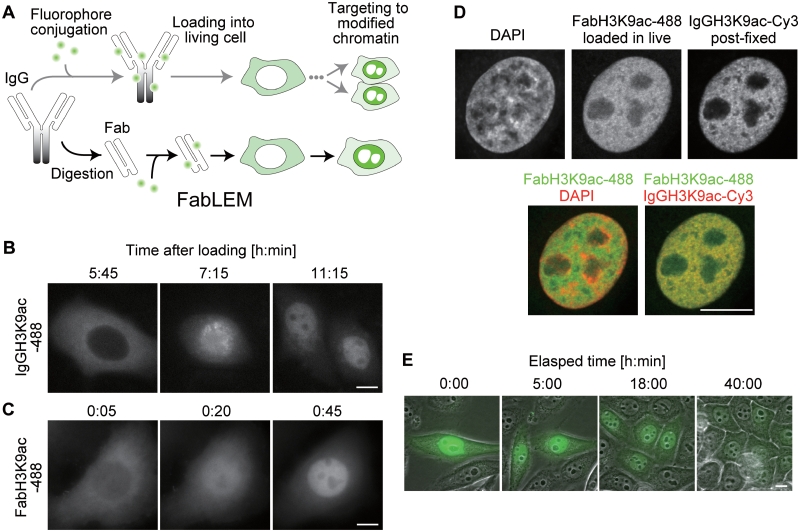
Histone H3K9 acetylation is visualized in living cells.(A) Schematic drawing showing how histone modifications are visualized in living cells. In the upper path, modification-specific mAb (IgG) is directly conjugated with a fluorescent dye, loaded into the cytoplasm and targeted to nuclear epitopes after cell division. In the lower path, illustrating FabLEM, an additional digestion step is required to make Fab, but targeting to nuclear epitopes takes less than an hour. (B and C) Distribution of Alexa488-labeled H3K9ac specific IgG and Fab. HeLa cells were loaded with IgGH3K9ac-488 (B) or FabH3K9ac-488 (C), and fluorescence images collected every 30 or 5 min (Movies 1 and 2 in Supplementary Data). (D) FabH3K9ac-488 loaded in living HeLa cells distributed similarly to IgGH3K9ac-Cy3 in fixed cells. Confocal images are shown. (E) Time lapse imaging of HeLa cells loaded with FabH3K9ac-488 through multiple cell divisions (Movie 3 in Supplementary Data). Bars, 10 µm.
To validate whether FabH3K9ac is indeed targeted to H3K9ac on chromatin in living cells, HeLa cells were loaded with FabH3K9ac-488, cultured for 4 h, and then fixed with paraformaldehyde for immunostaining with the same mAb labeled with Cy3 (IgGH3K9ac-Cy3). FabH3K9ac-488 and IgGH3K9ac-Cy3 were colocalized outside DAPI-dense heterochromatin (Figure 1D), consistent with H3K9ac being an active chromatin mark, although FabH3K9ac-488 was distributed more diffusely due to an unbound fraction. This result indicates that FabH3K9ac is concentrated in H3K9ac-rich chromatin in living cells. Since IgGH3K9ac-Cy3 could still bind to the same modifications post-fixation, the binding sites appear to be unsaturated.
To investigate whether FabH3K9ac binds to its epitope transiently or permanently, its mobility was investigated by fluorescence recovery after photobleaching (FRAP) (10). In cells loaded with FabH3K9ac-488, a small spot was bleached and the rate of fluorescence recovery was measured. As the recovery t1/2 was 8.7 s (Figure 2), FabH3K9ac did not appear to permanently block its epitope from accessing other cellular proteins. This binding time was shorter than that of phosphorylated H3 Ser10 (H3S10ph)-specific Fab (residence time ∼44 s) (10) and a CpG-methylated DNA specific probe (∼20 s) (22), both of which have proven useful for live imaging without affecting cell growth and embryo development (8). Consistently, FabH3K9ac did not appear to be toxic for cells. After Fab loading, HeLa cells went through a few cell divisions over 40 h (Figure 1E and Movie 3 in Supplementary Data) and mouse preimplantation embryos developed to birth like controls (Table 2) (20). Taken together with the above immunofluorescence data showing that not all H3K9ac is occupied by FabH3K9ac (Figure 1D), this suggests that Fab-binding is transient and does not saturate available binding sites. For these reasons, Fabs do not appear to block the association of other cellular proteins to H3K9ac, which may explain the lack of a detrimental phenotype in Fab-loaded cells.
Table 2.
Full-term development of embryos injected with FabH3K9ac and subjected to long-term imaging
| Fab | Imaginga | No. of embryos transferred | No. (%) of pups |
|---|---|---|---|
| FabH3K9ac (200 µg/ml) | − | 80 | 26 (32.5)b |
| + | 40 | 16 (40.0)b |
Embryos generated by in vitro fertilization were injected with FabH3K9ac-488 together with 5 µg/ml of H2B-mRFP mRNA, imaged up to the morula/blastocyst stage (∼70 h post-insemination), and then transferred to the uterus on Day 2.5 post-coitum pseudopregnant females.
aImaging minus means the embryos were injected with antibody but not imaged.
bThe efficiency of birth rate is in a range of controls without injection (20).
Visualizing methylated histone H3K4 in living cells
Fab-based live endogenous modification labeling (FabLEM) was also applied to methylated H3K4 using Fabs specific to monomethylation (FabH3K4me1, prepared from CMA302), dimethylation (FabH3K4me2, prepared from CMA303) and trimethylation (FabH3K4me3, prepared from CMA304) (11).
When loaded into HeLa cells expressing H2B-mRFP (for visualizing chromatin), FabH3K4me2-488 exhibited a euchromatic distribution; in particular, it was excluded from H2B-mRFP-rich heterochromatin (Supplementary Figure S2). This is consistent with the notion that H3K4me2 is an active epigenetic mark (3,11). Similarly, when FabH3K4me2-488 was simultaneously loaded with another active marker FabH3K9ac-Cy5 into HeLa cells, there was considerable overlap between the two signals in euchromatic regions depleted in H2B-mRFP-rich heterochromatin (Figure 3A).
The cytoplasmic concentration of FabH3K4me2 was greater than FabH3K9ac (Figure 3A). This can be explained by its lower epitope binding affinity measured in vivo by FRAP and in vitro by surface plasmon resonance using BIACORE. Specifically, the FRAP recovery time of FabH3K4me2 was significantly faster (t1/2 1.2 s) than FabH3K9ac (t1/2 8.7 s) (Figure 2A) and the binding dissociation constant significantly higher (KD 1.1 × 10−8 M versus 4.4 × 10−11 M; Table 1).
In living HeLa cells, FabH3K4me1-488 and FabH3K4me3-488 were also concentrated in the nucleus over the cytoplasmic background; however, their distribution was less punctate in living cells compared to immunofluorescence signals by fixed cells (Supplementary Figure S2). This is probably due to the weaker epitope-binding affinity, indicated by the slightly faster FRAP recovery (Supplementary Figure S3), and/or less abundance of the target modification. Although these antibodies may not be suitable for visualizing the subnuclear localization of modifications at a high resolution, they can be used for monitoring global modification levels, as described below.
Visualizing methylated histone H3K9 in living cells
To visualize histone H3K9 methylation, we prepared Fabs from newly developed mAbs, which worked better for chromatin immunoprecipitation (13) than previously published mAbs (11). FabH3K9me1 and FabH3K9me2 were successfully prepared from CMA316 and CMA317, as were other Fabs. However, active FabH3K9me3 could not be prepared by the standard protease digestion methods using either papain or ficin because CMA318 contained protease susceptible sites in the heavy chain Fab region (Supplementary Figure S4). FabH3K9me1 distributed nearly homogenously in the nucleus, excluding nucleoli (Supplementary Figure S2), reflecting the broad distribution of H3K9me1 (11) and the presence of a large free pool (indicated by its very rapid FRAP recovery; Supplementary Figure S3). Therefore, we focused on FabH3K9me2 for localization studies.
Histone H3K9me2 is a repressive mark in euchromatin and facultative heterochromatin (3,23). Consistent with this notion, when FabH3K9me2 was loaded into living HeLa cells, it overlapped considerably with H2B-mRFP (Supplementary Figure S2), in contrast to the active marks examined earlier. Similarly, when FabH3K9me2-488 was simultaneously loaded with an active marker FabH3K9ac-Cy5, there was little overlap in signal (Figure 3A), even though binding was rapid (FRAP t1/2 1.1 s, Figure 2; KD 1.5 × 10−8 M, Table 1).
To validate whether the localization of FabH3K9me2 depended on H3K9me2, conditional G9a methyltransferase knockout cells (17) were used (Figure 3B). Mouse embryonic stem cells expressing or lacking G9a, which is responsible for H3K9 dimethylation (23), were loaded with FabH3K9me2-488 and FabH3K9ac-Cy3. Whereas FabH3K9me2 was localized to nuclear foci in control cells, it was diffuse in G9a-knockout cells. In contrast, H3K9ac was localized to the nucleus in both cells. This result directly demonstrates that FabH3K9me2 is concentrated on H3K9me2-enriched chromatin in living cells.
Visualizing trimethylated H3K27 for monitoring the dynamics of inactive X chromosomes in living cells
H3K27me3 is known to be enriched on inactive X chromosomes, which are often located near the nuclear periphery or around nucleoli in fixed female cells (24). In living cells, FabH3K27me3-488 was also concentrated on inactive X chromosomes, exhibiting single foci in normal female fibroblasts (46, XX) and two and three foci in Klinefelter Syndrome cells (X trisomy and tetrasomy, respectively) (Figure 3C, arrows). In contrast, such obvious concentrations were not observed in cells harboring only one active X, like male (46, XY) and Turner syndrome (45, XO) cells (Figure 3C and not shown). Thus, the number of FabH3K27me3 foci corresponded to that of inactive X. Simultaneous loading of FabH3K27me3-488 and FabH3K27ac-Cy3, which is enriched on active transcription start sites (11), revealed that FabH3K27ac was excluded from FabH3K27me3-enriched inactive X in living cells (Figure 3D, arrows), consistent with the transcriptional repression state of inactive X. The FRAP recovery time of FabH3K27me3 (0.94 s) was similar in range to FabH3K4me2 and FabH3K9me2, whereas FabH3K27ac exhibited more stable binding (21 s) (Figure 2). This again correlated with their epitope-binding affinity in vitro (KD 1.2 × 10−8 and 1.4 × 10−12 M for FabH3K27me3 and FabH3K27ac, respectively; Table 1).
To visualize the dynamics of inactive X chromosomes during the cell cycle, hTERT-RPE1 cells (46, XX) were loaded with FabH3K27me3-488 and PCNA-Cy3 (Cy3-labeled proliferating cell nuclear antigen), which highlighted DNA replication foci during S phase as did GFP-tagged PCNA (25). Live imaging confirmed that inactive X replicates very late in S phase (Figures 3E, 6 and 8 h and Movie 4 in Supplementary Data). Whereas previous work suggested inactive X chromosomes dynamically move from the nuclear periphery to perinucleoli during S phase (26), inactive X appeared to be replicated without changing its intranuclear positioning (Figure 3E, arrows). Such static behaviors of inactive X were also observed in other cell types like WI-38 fibroblasts and cervical squamous cell carcinoma cells (not shown), suggesting that the perinucleolar targeting is not essential for maintaining the inactive state of X chromosomes during/after replication. Similarly, autosomes are also reported to exhibit no large-scale redistribution in the nucleus in HT-1080 cells during G1 phase (27).
Quantitative measurement of changes in histone modification levels in living cells
Beyond detecting modifications at specific locations, FabLEM can also be used to monitor global modification levels throughout a single cell. For this purpose, the ratio of the nuclear to cytoplasmic Fab concentration is useful (Figure 4A). In essence, the nuclear concentration is correlated with the amount of bound Fab, whereas the cytoplasmic concentration is correlated with the amount of freely diffusing unbound Fab. Thus, when the level of modifications increase within the nucleus, so too does the nucleus:cytoplasm ratio of Fab.
Figure 4.
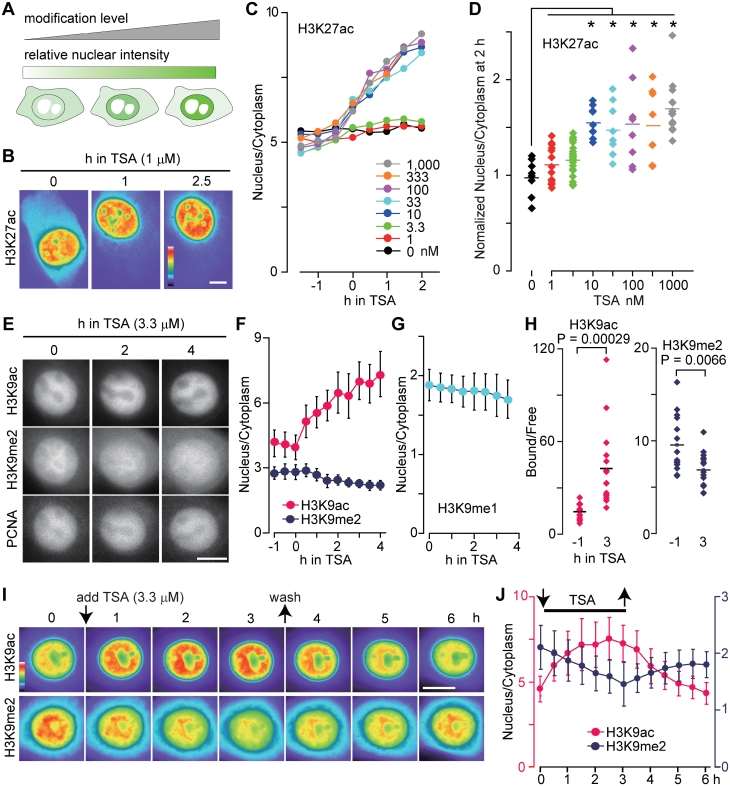
Monitoring the levels of histone H3 modifications in living cells. (A) Schematic drawing showing how modification levels are monitored in living cells using Fab. The nuclear concentration of Fab, or the intensity ratio of nucleus:cytoplasm, reflects the level of the target modification. (B–D) Monitoring H3K27ac levels in living U2OS cells. Cells loaded with FabH3K27ac-Cy3 were imaged every 15 min, and TSA was added (Movie 5 in Supplementary Data). Shown are pseudo-color images (B) and the fluorescent intensity ratio of nucleus:cytoplasm (C and D); (C) the average (n > 8); (D) the value of nucleus:cytoplasm at 2 h normalized to its value at 0 h in individual cells. Asterisks indicate P < 0.005 by two-tailed Student’s t-test; actual P-values are: 1 nM, 0.178; 3.3 nM, 0.166; 10 nM, 0.00016; 33 nM, 0.0023; 100 nM, 0.00078; 333 nM, 0.0022; and 1000 nM, 0.0000037. (E–H) Effects of TSA on H3K9 acetylation and methylation levels in HeLa cells. Cells were loaded with FabH3K9ac-Cy5, PCNA-Cy3 and FabH3K9me2-488 (E, F, H) or FabH3K9me1-488 (G), imaged every 15 min, and TSA (3.3 µM) was added. (E) Fluorescent images (Movie 6 in Supplementary Data). (F and G) The intensity ratio of nucleus:cytoplasm (averages with SD; n = 10). (H) The ratio of chromatin-bound:free FabH3K9me2 in individual cells. P-values by two-tailed Student’s t-test are indicated. (I and J) Monitoring H3K9ac and H3K9me2 levels after the removal of TSA. HeLa cells loaded with FabH3K9me2-488 and FabH3K9ac-Cy3 were imaged every 30 min, TSA (3.3 µM) was added at time point 0 and removed 3 h later. (I) Pseudo-color images of fluorescence intensity (see Movie 7 in Supplementary Data for gray scale and merged images). (J) The intensity ratio of nucleus:cytoplasm (averages with SD; n = 10). Bars, 10 µm.
To demonstrate, FabH3K27ac-Cy3 was loaded into living cells to investigate the effect of an HDAC inhibitor, trichostatin A (TSA). When TSA was added at a final concentration of 1 µM, the nucleus:cytoplasm ratio of FabH3K27ac intensity increased within 30 min and this trend continued for at least 2 h (Figure 4B, C and Movie 5 in Supplementary Data). This observation is consistent with the hyperacetylation of H3K27 induced by TSA treatment (11). The effect of TSA was detected at concentrations as low as 10 nM (Figure 4C and D). This sensitivity appears to be lower than that of a recently developed FRET-based probe for histone H4 acetylation (9), but a direct comparison is difficult as the target acetylation sites are different (H3K27 versus H4K5/8).
Live imaging also revealed that a higher concentration of TSA (3.3 µM) affected H3K9 dimethylation levels. HeLa cells were loaded with FabH3K9me2-488, FabH3K9ac-Cy5 and PCNA-Cy3, and imaged under a fluorescence microscope. After the addition of TSA, the perinucleolar concentration of FabH3K9me2 gradually decreased, as did the overall nuclear concentration (Figure 4E, F and Movie 6 in Supplementary Data). In contrast, the nuclear concentration of FabH3K9ac increased. FabH3K9me1 behaved similarly, spreading more to the cytoplasm in the presence of TSA (Figure 4G). TSA-induced H3K9 demethylation was observed in most cells, including ones outside S phase without PCNA foci (Figure 4E). This indicates that the inhibition of deacetylation induces H3K9 demethylation independently of DNA replication. These TSA-induced changes in H3K9 modification levels were also monitored by measuring the chromatin-bound fraction of Fab using FRAP (28) (Supplementary Figure S5). Consistent with the intensity ratio measurements (Figure 4F), the chromatin-bound fraction of FabH3K9ac increased in TSA for 3 h, whereas that of FabH3K9me2 decreased (Figure 4H). This demonstrates that the relatively easy-to-measure nucleus:cytoplasm ratio is appropriate for monitoring bound Fab.
We then investigated whether H3K9ac and H3K9me2 levels are restored after the removal of TSA. HeLa cells were loaded with FabH3K9ac-Cy3 and FabH3K9me2-488, incubated in 3.3 µM TSA for 3 h, washed with fresh medium, and further incubated in the absence of TSA. As seen before, the nuclear concentrations of FabH3K9ac and FabH3K9me2 increased and decreased, respectively, in the presence of TSA (Figure 4I, J and Movie 7 in Supplementary Data). Within 2 h after TSA removal, the nuclear intensity of FabH3K9ac dropped to the original level; in contrast, the nuclear intensity of FabH3K9me2 gradually increased but did not reach its original level in 3 h (Figure 4I, J and Movie 7 in Supplementary Data). Thus, deacetylation results in an increase in dimethylation, suggesting that H3K9 acetylation antagonizes dimethylation. Although it is interesting to monitor when the original H3K9me2 level is restored in long-term monitoring, many cells went into mitosis during the observation period, making quantitative analysis difficult. Perhaps the use of cells that grow more slowly will help address this question in a future study.
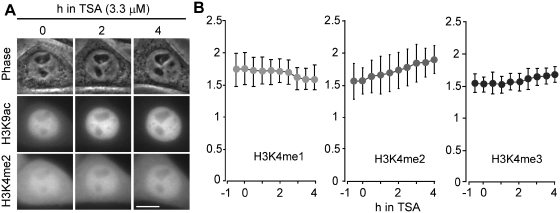
Effects of TSA on histone H3K4 methylation
It has been shown that HDAC inhibitor treatments result in an increase in H3K4 methylation, in addition to H3 and H4 acetylation (29,30). We thus used FabLEM to investigate the effects of TSA on histone H3K4 methylation. Soon after the addition of TSA, both FabH3K4me2 and FabH3K4me3 accumulated in the nucleus (Figure 5A and B). This is consistent with previous studies (29,30). In contrast, the nuclear intensity of FabH3K4me1 slightly decreased (Figure 5B). Although a long-term study suggested HDAC inhibitors could indirectly affect H3K4 methylation levels by altering H3K4 demethylase expression (30), here real-time monitoring by FabLEM demonstrated a rapid increase in H3K4me2, supporting the idea that cross-talk between H3 acetylation and H3K4 methylation is more direct and occurs through multienzyme complexes (29).
Figure 5.
Effects of TSA on H3K4 methylation levels. HeLa cells were loaded with FabH3K9ac-Cy5 and FabH3K4me1-488, FabH3K4me2-488, or FabH3K4me3-488, imaged every 15 min, and TSA (3.3 µM) was added. (A) Images of phase-contrast, FabH3K4me2-488 and FabH3K9ac-Cy3. Bar, 10 µm. (B) The intensity ratio of nucleus:cytoplasm (averages with SD; n = 10).
Monitoring histone H3 acetylation in mouse preimplantation embryos
FabLEM was next used to monitor histone H3 modifications in mouse preimplantation embryos, where dynamic epigenetic changes are known to take place (5). Fluorescently labeled Fab together with mRNA encoding histone H2B-mRFP were injected into eggs produced by in vitro fertilization (IVF) or intracytoplasmic sperm injection (20) (Figure 6A). Among the Fabs investigated, only H3 acetylation-specific Fabs were concentrated in pronuclei, whereas methylation specific Fabs distributed throughout the egg cytoplasm even though some concentration of FabH3K27me3 was observed on condensed chromosomes during mitosis (Supplementary Figure S6). This suggests that higher epitope-binding affinities may be needed for monitoring interphase nuclei in mouse embryos.
When injected into IVF embryos, FabH3K9ac was concentrated in both maternal and paternal pronuclei, with more on paternal pronuclei (Figure 6C and Movie 8 in Supplementary Data). The higher level of H3K9ac in paternal pronuclei may be consistent with the absence of H3K9 methylation on paternal chromatin in zygotes (5). The H3K9ac level was relatively constant throughout zygotic and two-cell stages in IVF embryos (Figure 6D, blue symbols). FabH3K27ac was also concentrated more in paternal pronuclei, but its level was gradually and substantially decreased in zygotes before the first mitosis (Figure 6E and F, blue symbols; Movie 9 in Supplementary Data). This active deacetylation may be complementary, or prerequisite, to de novo H3K27 methylation on paternal pronuclei in zygotes (5). The distinct behaviors of H3K9ac and H3K27ac were directly compared by injecting two Fabs conjugated with different dyes (i.e. FabH3K9ac-488 and FabH3K27ac-Cy3) (Supplementary Figure S7). As seen above, H3K27ac levels decreased in zygotes. Inhibition of HDACs by TSA caused different effects on H3K9ac and H3K27ac. In the presence of TSA, the level of H3K9ac continuously increased during zygotic and two-cell stages more than in untreated embryos; in contrast, TSA prevented the decrease of H3K27ac levels (Supplementary Figure S7). Thus, TSA treatments induced hyperacetylation of H3K9 and prevented deacetylation of H3K27, suggesting that the acetylation of these sites is differentially controlled in mouse preimplantation embryos.
HDAC inhibitors like TSA are known to improve the efficiency of embryo development after somatic cell nuclear transfer (SCNT), but the mechanism of HDAC inhibitor’s effect remains unknown (12). Using FabLEM, H3 acetylation levels in SCNT embryos and the effect of TSA treatments were measured. Enucleated oocytes were injected with somatic cell nuclei, treated with or without TSA for 9 h during the activation, and then allowed to develop in TSA-free medium under a confocal microscope (Figure 6B). The levels of H3K9ac in SCNT embryo nuclei were similar to that in IVF pronuclei, and became higher upon TSA treatment (Figure 6C and D). In contrast, the levels of H3K27ac in SCNT embryo nuclei were much lower, but increased to the maternal pronuclear level upon TSA treatment (Figure 6E and F). These results suggest that different levels of H3K27ac between IVF pronuclei and SCNT embryo nuclei may at least partially account for their different development efficiencies.
DISCUSSION
This study demonstrates that FabLEM can be used to monitor the concentration of epigenetic histone modifications such as histone H3 acetylation and methylation in living somatic cells and preimplantation embryos. Under appropriate conditions, FabLEM did not affect cell growth and mouse embryo development. This implies the method does not disturb normal cell metabolism, presumably because monovalent Fabs only bind their targets transiently, so other cellular proteins maintain access to their binding sites. Fab-based live cell imaging was also used to visualize endogenous proteins in Drosophila embryos, apparently without disturbing the nuclear divisions (31,32).
In this study, we used fully characterized and highly specific mAbs (11,13) for FabLEM. In general, high-specific antibodies must be used for FabLEM because there is no blocking reagent that could suppress non-specific binding, unlike immunostaining for fixed cells. Although Fab prepared from antigen affinity-purified polyclonal antibody may suffice for FabLEM, the preparation is likely to contain various fragments with different affinities due to its polyclonal nature. Considering the importance of Fab affinity as described below, monoclonal antibodies are recommended for better control of FabLEM experiments.
Fab affinity is an important factor for the quality, versatility and interpretation of FabLEM
Whereas all methyl-specific Fabs we tested were adequate for live-imaging in cultured cells, only acetyl- and phospho-specific Fabs were successful for imaging in preimplantation embryos [this study and (10)]. This could be explained by the different affinities of Fabs, because all methyl-specific Fabs exhibit much shorter FRAP recoveries (∼1 s) than acetyl- and phospho-specific Fabs (>8 s); the residence time of Fabs does not appear to represent the turnover of the modification as H3K9 methylation is more stable than acetylation (33). Since the cytoplasmic volume of preimplantation embryos (∼270 pl; calculated from its diameter 80 µm) is more than 50-fold larger than that of HeLa cells [∼5 pl; (10)], imaging with sufficient contrast may require high affinity Fabs. As a rule of thumb, mAbs with KD ∼10−8 M can be adequate for FabLEM, but high affinity ones with 10−11−10−12 M will yield higher contrast images and generally be more versatile (contingent on the abundance of the endogenous target).
Some limitations of the FabLEM technique should be considered. First, the time resolution of FabLEM depends on the binding time of Fab. For example, changes in less than a few seconds cannot be followed using a high affinity Fab, like FabH3K9ac with toff ∼7.3 s (Supplementary Figure S5). Second, it is also possible that the turnover of modification is influenced by Fab binding and so the balance of modification–demodification may be disturbed, even though such a change may not significantly affect biological function of cells and embryos. This issue can be addressed by using two Fabs with the same specificity but with different affinities, since lower affinity Fabs should have better temporal resolution and should minimize blocking effects (at the cost of enhanced signal-to-noise).
Fabs repeatedly bind and unbind their target modifications in nuclei. As unbound Fabs can diffuse into the cytoplasm, the ratio of nuclear and cytoplasmic Fab intensity is correlated with that of bound and unbound fractions. Since the nucleus:cytoplasm ratio depends on both Fab affinity and the abundance of target modification, the degree of changes among different Fabs cannot be directly compared. The intensity measurements, for example, revealed that after the addition of TSA the level of H3K9me2 was reduced much more clearly than that of H3K9me1. This does not necessarily mean, however, that H3K9me2 is changed more significantly, because the dynamic range of FabH3K9me1 is lowered by the presence of a larger unbound fraction. Similarly, it is also difficult to conclude which state of H3K4 methylation (di or tri) is more increased in TSA. These types of more quantitiative comparisons will require knowledge of the absolute amount of each modification as well as further biophysical characterization of Fabs in living cells.
FabLEM for visualizing modification sites
As discussed above, Fabs with a wide range of epitope binding affinities can be used to monitor changes in target modification levels. Visualization of the modification sites, however, depends not only on the Fab affinity but also on the distribution and concentration of the modification. When a specific modification is massively enriched in large foci, Fabs can be concentrated locally, even if their affinity is not very high. In this case, the foci can be detected under a conventional wide-field microscope, like FabH3K27me3 on inactive X chromosomes. If the modification is scattered at numerous small foci throughout the nucleus, however, a high affinity Fab combined with a high resolution system (like confocal or deconvolution microscopy) is required. This is exemplified by H3K4me3 and H3K27ac, both of which exhibit a number of small foci in fixed and immunolabeled nuclei (Supplementary Figure S2). In FabLEM using a confocal microscope, FabH3K4me3 distributed diffusely, whereas FabH3K27ac is concentrated in foci, reflecting the differences in their affinity.
A role for H3K27 acetylation in mouse embryo development
We applied FabLEM to monitor the changes of H3K9 and H3K27 acetylation in mouse preimplantation embryos. During embryo development, DNA methylation and histone modifications change drastically (5,7). After fertilization, sperm chromatin is decondensed by replacing protamines with maternal histones in egg cytoplasm. The newly assembled histones in paternal pronuclei are generally associated with acetylation without methylation, in contrast to histones in maternal pronuclei in which repressive methylation marks are inherited from oocytes. Indeed, FabLEM revealed both H3K9 and H3K27 acetylation levels are higher in paternal pronuclei. While the H3K9ac level remains relatively constant throughout zygote and two-cell stages, H3K27ac in paternal pronuclei is substantially reduced in zygotes. Furthermore, the H3K27ac level in somatic nuclei transferred into oocytes is much lower than pronuclei in zygotes, but is increased by histone deacetylase inhibitor treatments, which are known to increase the cloning efficiency of SCNT embryos (12). These data suggest that a high level of H3K27ac is important for normal embryo development, by antagonizing the methylation on the same lysine residue (34–36). Lower acetylation levels observed in transferred somatic cell nuclei might establish an abnormal epigenetic landscape, which in turn might cause aberrant gene expression profiles. Increased levels of H3K27ac by TSA may thus be a factor that contributes to the normal development of SCNT embryos. Indeed, the global level of transcription in two-cell SCNT embryos was shown to increase upon TSA treatment (37). In contrast, H3K9ac does not appear to have much antagonistic effect on methylation in zygotes, since H3K9me1 and H3K9me2 are observed in paternal pronuclei before the first division (5,34) despite the constant levels of H3K9ac.
Concluding remarks and future prospects
Besides FabLEM, real-time measurements of protein modification levels in living cells can also be achieved using FRET-based sensors (2,8,9). Although a direct comparison of these two methods has yet to be made, FabLEM generally has the following advantages: (i) endogenous modifications can be monitored; (ii) standard single-color filter sets can be used for fluorescence imaging; (iii) many antibodies are available for simultaneous detection of different modifications in the same cells; and (iv) no genetic or recombinant material is introduced, so imaged cells and embryos can act as donors for reproduction studies, although long-term experiments might be hampered by dilution of Fabs upon cell division. The requirement of protein loading into living cells might be considered a drawback of FabLEM as microinjection is tedious and other methods are not as standard as DNA-mediated expression systems; however, many different techniques and reagents have been developed for protein delivery into cells and tissues (38), possibly making it much easier to use FabLEM in the future. Alternatively, the expression of GFP-fused antigen-binding fragments, like single-chain Fabs, single-chain variable fragments, or chromobodies, might complement the limitations of protein-based methods (39).
In conclusion, we anticipate FabLEM will be a powerful tool for monitoring the dynamic changes of epigenetic modifications during the cell and developmental cycles and in response to stimuli and small chemicals. The method can be used to visualize any endogenous protein modification, providing a novel strategy to understand the dynamic nature of cell regulation and test the response of patient cells to potentially therapeutic drugs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by New Energy and Industrial Technology Development Organization of Japan (to H.K.); Japan Society for the Promotion of Science (to Y.H.-T. and T.J.S); Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan for the Osaka University Program for the Support of Networking among Present and Future Women Researchers (to Y.H-T.); and the Global Center of Excellence program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.K.). Funding for open access charge: A grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. N. Nozaki is a founder of MAB Institute Inc.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Hatanaka for help, and T. Kanda (Aichi Cancer Institute), R. Tsien (University of California, San Diego) and M.C. Cardoso (Technische Universität Darmstadt) for H2B-GFP, mRFP and PCNA-GFP constructs.
REFERENCES
- 1.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell. Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 2.Aye-Han N-N, Ni Q, Zhang J. Fluorescent biosensors for real-time tracking of post-translational modification dynamics. Curr. Opin. Chem. Biol. 2009;13:392–397. doi: 10.1016/j.cbpa.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 4.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14 doi: 10.1093/hmg/ddi114. (Spec No 1), R47–R58. [DOI] [PubMed] [Google Scholar]
- 6.Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 7.Burton A, Torres-Padilla M-E. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genomics. 2010;9:444–454. doi: 10.1093/bfgp/elq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura H, Hayashi-Takanaka Y, Yamagata K. Visualization of DNA methylation and histone modifications in living cells. Curr. Opin. Cell. Biol. 2010;22:412–418. doi: 10.1016/j.ceb.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc. Natl Acad. Sci. USA. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi-Takanaka Y, Yamagata K, Nozaki N, Kimura H. Visualizing histone modifications in living cells: spatiotemporal dynamics of H3 phosphorylation during interphase. J. Cell. Biol. 2009;187:781–790. doi: 10.1083/jcb.200904137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 12.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui H-T, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 13.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 14.Ohta S, Shiomi Y, Sugimoto K, Obuse C, Tsurimoto T. A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2-5 complex as a novel PCNA-binding protein. J. Biol. Chem. 2002;277:40362–40367. doi: 10.1074/jbc.M206194200. [DOI] [PubMed] [Google Scholar]
- 15.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 16.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc. Natl Acad. Sci. USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeil PL, Warder E. Glass beads load macromolecules into living cells. J. Cell. Sci. 1987;88(Pt 5):669–678. doi: 10.1242/jcs.88.5.669. [DOI] [PubMed] [Google Scholar]
- 19.Manders EM, Kimura H, Cook PR. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 1999;144:813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamagata K, Suetsugu R, Wakayama T. Long-term, six-dimensional live-cell imaging for the mouse preimplantation embryo that does not affect full-term development. J. Reprod. Dev. 2009;55:343–350. doi: 10.1262/jrd.20166. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka KAK, Suzuki KGN, Shirai YM, Shibutani ST, Miyahara MSH, Tsuboi H, Yahara M, Yoshimura A, Mayor S, Fujiwara TK, et al. Membrane molecules mobile even after chemical fixation. Nat. Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- 22.Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 23.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum. Mol. Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- 25.Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L-F, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 27.Müller I, Boyle S, Singer RH, Bickmore WA, Chubb JR. Stable morphology, but dynamic internal reorganisation, of interphase human chromosomes in living cells. PLoS ONE. 2010;5:e11560. doi: 10.1371/journal.pone.0011560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stasevich TJ, Mueller F, Brown DT, McNally JG. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 2010;29:1225–1234. doi: 10.1038/emboj.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 30.Huang P-H, Chen C-H, Chou C-C, Sargeant AM, Kulp SK, Teng C-M, Byrd JC, Chen C-S. Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol. Pharmacol. 2011;79:197–206. doi: 10.1124/mol.110.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swedlow JR, Sedat JW, Agard DA. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three-dimensional wide-field microscopy. Cell. 1993;73:97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- 32.Paddy MR, Saumweber H, Agard DA, Sedat JW. Time-resolved, in vivo studies of mitotic spindle formation and nuclear lamina breakdown in Drosophila early embryos. J. Cell. Sci. 1996;109(Pt 3):591–607. doi: 10.1242/jcs.109.3.591. [DOI] [PubMed] [Google Scholar]
- 33.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 35.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bui H-T, Wakayama S, Kishigami S, Park K-K, Kim J-H, Thuan NV, Wakayama T. Effect of trichostatin A on chromatin remodeling, histone modifications, DNA replication, and transcriptional activity in cloned mouse embryos. Biol. Reprod. 2010;83:454–463. doi: 10.1095/biolreprod.109.083337. [DOI] [PubMed] [Google Scholar]
- 38.Fonseca SB, Pereira MP, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv. Drug Deliv. Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.