Abstract
The minichromosome maintenance (MCM) helicase complex is essential for the initiation and elongation of DNA replication in both the eukaryotic and archaeal domains. The archaeal homohexameric MCM helicase from Sulfolobus solfataricus serves as a model for understanding mechanisms of DNA unwinding. In this report, the displaced 5′-tail is shown to provide stability to the MCM complex on DNA and contribute to unwinding. Mutations in a positively charged patch on the exterior surface of the MCM hexamer destabilize this interaction, alter the path of the displaced 5′-tail DNA and reduce unwinding. DNA footprinting and single-molecule fluorescence experiments support a previously unrecognized wrapping of the 5′-tail. This mode of hexameric helicase DNA unwinding is termed the steric exclusion and wrapping (SEW) model, where the 3′-tail is encircled by the helicase while the displaced 5′-tail wraps around defined paths on the exterior of the helicase. The novel wrapping mechanism stabilizes the MCM complex in a positive unwinding mode, protects the displaced single-stranded DNA tail and prevents reannealing.
INTRODUCTION
The eukaryotic minichromosome maintenance protein complex (MCM2-7) is essential for DNA replication initiation and elongation (1,2) by participating in the licensing of chromatin (3) and subsequently functioning as a DNA helicase for unwinding (4,5). The archaeal MCM helicase generally exists as a homohexameric form of a single subunit product that is homologous to each of the six sequence-distinctive subunits in eukaryotes (6,7). All MCM helicases are members of the AAA+ superfamily (ATPases associated with a variety of cellular activities) that share common ATP binding motifs (8,9). The archaeal MCM helicases from Sulfolobus solfataricus (Sso) and Methanothermobacter thermoautotrophicus (Mth) serve as simplified models for understanding mechanisms of ATP hydrolysis, DNA binding and unwinding for these classes of hexameric DNA replication helicases.
MCM hexamers share a common architecture consisting of a central channel that encircles DNA, an N-terminal tier and a C-terminal AAA+ ATPase tier (7,10,11). The unwinding polarity for the MCM helicase is 3′–5′, where critical residues at the tips of β-hairpins in the central channel contribute to DNA binding and ATP hydrolysis to drive unwinding (12,13). The N-terminal tier acts to increase the processivity of the helicase by modulating interactions with DNA throughout the hexamer (13). The C-terminal AAA+ domain contains conserved motifs involved in ATP hydrolysis, demonstrated by a mutation in a lysine residue at the tip of a β-hairpin that abolishes unwinding activity (12). The ATP binding site is positioned in trans between conserved domains from adjacent MCM subunits that couple ATP binding and hydrolysis to helicase activity (14). Communication between the N- and C-terminal tiers is facilitated by a conserved allosteric loop that senses the presence of bound nucleotide and controls conformational changes between tiers coupled with ATP hydrolysis (15,16) and presumably provides the underlying energy for unwinding.
Upon elucidation of the SsoMCM crystal structure, two limiting models of unwinding were proposed: steric exclusion and side channel extrusion (7). The extrusion model is based on homology and similarities in DNA unwinding to the SV40 large T antigen, which forms a double hexamer structure where DNA is pumped out of the interface between the two hexamers (17). In this model, double-stranded DNA (dsDNA) enters the central channel where it is separated into single-stranded DNA (ssDNA) and one strand is extruded out through a side channel. Interestingly, MthMCM has also been shown to form a double hexamer through self-association at the N-termini (10,18), but the oligomeric state is easily modulated by the salt concentration, and the active form is thought to be hexameric, similar to SsoMCM (12,19). In the more classical representation of hexameric helicases, the steric exclusion model predicts that separation of the DNA strands occurs prior to entry into the central channel: one strand proceeds through the central channel, while the other is displaced away from the exterior surface of MCM (20,21). In either model, after separation of the two strands, the excluded or extruded 5′-strand was not known to interact further with MCM or to play any other role in the unwinding mechanism. Previously, the close spatial relationship between the ends of each separated ssDNA tail was observed by single-pair fluorescence resonance energy transfer (spFRET) (22). It was hypothesized that after separation, the 5′-tail makes significant contact with the exterior of MCM, although the role of this putative interaction was unknown.
Helicase unwinding models have principally focused on defining the location and importance of the encircled DNA strand(s), while the role of the displaced strand has made relatively minor contributions to current models. In this study, we demonstrate that the 5′-tail is more than just a passive structure in the unwinding mechanism. We provide evidence that SsoMCM binds to and unwinds DNA using a modified steric exclusion mechanism that includes wrapping of the 5′-tail along specific paths on the exterior hexameric surface. Mutation of conserved SsoMCM surface residues alters the path of the 5′-tail and reduces its unwinding ability. We show that the length of the 5′-tail is important in the stabilization of the hexameric MCM structure on DNA. Furthermore, 5′-tails that are more than twice the longitudinal length of the MCM helicase are protected from nuclease degradation, suggesting that wrapping of ssDNA around the exterior of the MCM complex occurs during unwinding. We propose a steric exclusion and wrapping (SEW) model for MCM helicases, which the hexamer complex is stabilized by wrapping of the displaced 5′-strand around the exterior surface, resembling a spool of thread.
MATERIALS AND METHODS
Materials
ATP was obtained from Invitrogen. Mung bean nuclease, T4 polynucleotide kinase (PNK) and terminal transferase (TdT) were purchased from NEB. Optikinase was purchased from USB. All other materials were from commercial sources and were analytical grade or better. Helicase buffer is used in all unwinding and binding reactions and consists of 125 mM potassium acetate, 25 mM Tris acetate (pH 7.5) and 10 mM magnesium acetate.
Cloning and protein purification
K323A and R440A single and double mutants were cloned by overlap extension and insertion into pET30a (NdeI/XhoI). Mutations were confirmed using the DNA sequencing faculty at the University of Pittsburgh. Full-length wild-type (WT) and mutant SsoMCM were purified as previously described using 70°C heat treatment as well as MonoQ, heparin and gel filtration columns to isolate the hexameric species (12).
DNA substrates
Oligonucleotides (Supplementary Table S1) were purchased from IDT Corp and gel purified (23). [γ-32P]ATP and [α-32P]dATP were purchased from MP Biomedicals and used with PNK/Optikinase or TdT to 32P label the 5′- or 3′-ends of DNA, respectively. Fluorescent DNA was synthesized and HPLC purified by IDT. Complementary DNA was added in a ratio of 1.2:1. 32P-labeled DNA substrates were heated at 95°C for 5 min and then cooled to room temperature after turning off the heat block.
Unwinding reactions
Helicase unwinding reactions were incubated at 60°C for 5 min and initiated upon addition of either SsoMCM or ATP. Final reaction conditions included helicase buffer, 2 mM ATP, 15 nM radiolabeled DNA and varying concentrations of helicase, totaling 10 μl/reaction.
Reactions were quenched with an equal volume of glycerol quench (0.5% w/v SDS, 50% v/v glycerol, 0.1% w/v bromophenol blue, 100 mM EDTA pH 8.0 and 150 nM trap ssDNA), and then stored on ice until loading. Denaturing gels [14% acrylamide (29:1 acryl:bisacryl), 8 M urea and 1× TBE buffer] or native gels [20% acrylamide and 1× TBE buffer] were used to monitor the unwinding of fork or tailed DNA, respectively, for enhanced resolution. The gels were exposed to phosphor screens, imaged using a Storm 820 Phosphorimager (GE Healthcare), and the fraction unwound was calculated.
Biotin–streptavidin unwinding assays
Biotin–streptavidin unwinding experiments were conducted as above with the exception that near the duplex region of the forked DNA, a thymidine residue was biotinylated on either the 3′- or 5′-strand (Supplementary Table S1). Streptavidin concentrations were 100-fold higher than the biotinylated forked substrates to ensure a 1:1 streptavidin:biotin stoichiometry. Unwinding reactions were initiated by addition of SsoMCM and 20-fold excess biotin to trap the unwound DNA. Background streptavidin displacement from ssDNA was measure and corrected as described previously (24).
Nuclease footprinting
Stoichiometric concentrations (6:1) of SsoMCM (540 nM) were incubated with 90 nM DNA (spiked with 32P-labeled DNA) for 5 min at 30°C, followed by addition of mung bean nuclease for 30 min before quenching with an equal volume of formamide quench (0.1% w/v SDS, 78% v/v formamide, 0.1% w/v bromophenol blue, 100 mM EDTA pH 8.0 and 900 nM trap ssDNA). Reactions were boiled for 5 min at 95°C, followed by 2 min on ice, before resolving and imaging as above.
Fluorescence anisotropy
Anisotropy experiments were performed using a Fluoromax-3 fluorometer (HORIBA Jobin Yvon) as described previously (25). Cy3-labeled DNA was annealed to unlabeled DNA, forming forked, 3′- and 5′-tailed substrates. Anisotropy values were collected with an integration time of 0.5 s for eight consecutive readings. Final values from at least three independent experiments were averaged, normalized and fit to a single binding equation:
| (1) |
where Amax is the maximal anisotropy and Kd is the dissociation constant.
Off-rate anisotropy experiments were monitored as a function of time after addition of 100-fold excess of salmon sperm DNA to a preformed MCM/fork DNA–Cy3 complex. Anisotropy values were collected every ∼12.6 s for 2000 s with an integration time of 0.5 s. Results were fit to a double exponential decay equation:
| (2) |
where Amin is the final anisotropy value, A1 and A2 are the change in anisotropy, k1 and k2 are the observed rate constants. The final values are the average of at least three separate experiments.
Stopped flow fluorescence resonance energy transfer
Stopped flow experiments were performed on an SX.18MV (Applied Photophysics). SsoMCM was fluorescently labeled at the N-terminus with Alexa 488 or Alexa 555 succinimidyl esters (Invitrogen) as previously described (12). Preformed Alexa 488-SsoMCM bound to fork DNA with various length 5′-tails were rapidly mixed with Alexa 555-SsoMCM at different concentrations and the fluorescence sensitization was monitored over time using a 570 nm cut-off filter. Changes in fluorescence were fit to single or double exponential equations using the included software.
Single-pair fluorescence resonance energy transfer
Cy3 and Cy5 fluorophores were placed on a 30 base 3′-termini and variable length (30, 50 and 70 bases) 5′-termini, respectively, of forked DNA substrates (Supplementary Table S1). A 5′-biotin on the Cy3-labeled strand was used to immobilize the DNA onto a PEG-passivated quartz slide as described previously (26). Experiments were performed on a prism-based total internal reflection fluorescence microscope (27,28) employing a 532 nm diode laser. Donor and acceptor emission signals were separated by a 610 nm dichroic longpass mirror, a 580/40 nm bandpass filter and a 660 nm longpass filter and subsequently imaged using an EM-CCD camera (Andor Technologies). Images from >10 regions (∼50–200 molecules/region) were acquired at 10 Hz for several minutes and corrected for thermal/mechanical drift (29,30). Regions surrounding individual peaks (7×7 pixels) were identified by goodness of fit to a 2D Gaussian, and time-dependent intensity traces, corrected for local background, were extracted. Apparent E (Eapp) was measured according to:
| (3) |
where IA and ID represent acceptor and donor intensities, respectively. Histograms were produced from manually identified regions of traces displaying anti-correlated donor/acceptor signals and single-step dye photobleaching. Imaging buffer included helicase buffer and an oxygen-radical scavenging system consisting of 0.1 mg/ml glucose oxidase, 0.02 mg/ml catalase, 0.4% wt/v β-d-glucose and 2 mM Trolox. For experiments including SsoMCM, protein was infused into the flow cell containing tethered fork substrates and incubated for 5 min before imaging.
RESULTS
The 5′-tail is excluded from the central channel of MCM during unwinding
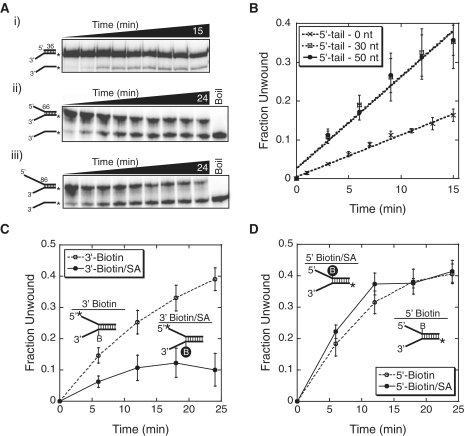
We examined the helicase activity of SsoMCM on DNA substrates with different length 5′-tails (0, 30 or 50 bases). Consistent with previous results (12,13,31), the presence of any length 5′-tail resulted in efficient unwinding, while the absence of any DNA bases on the displaced strand reduced the unwinding efficiency (Figure 1A and B; Table 1). In order to differentiate between the two limiting models of helicase unwinding (steric exclusion and side channel extrusion), we utilized biotin/streptavidin as a physical block to unwinding that can identify which strand(s) proceeds through the central channel. Similar experiments have been performed previously with other hexameric helicases, Escherichia coli DnaB and yeast MCM4/6/7, and the results were consistent with the steric exclusion model for unwinding (32).
Figure 1.
MCM unwinding of DNA with variable length 5′-tails and physical blocks on either the 3′- or 5′-strand. (A) DNA unwinding by WT SsoMCM (700 nM hexamer) on different length 5′-tail forked DNA substrates with (i) no, (ii) a 30 base or (iii) a 50 base 5′-tail and a constant 30 base 3′-tail. (B) Quantification of the average of three separate unwinding reactions for each DNA substrate in (A). The rates were linear for the first 12 min and equal to 0.012 ± 0.001 min−1 for 0 nt 5′-tail (times symbol), 0.027 ± 0.002 min−1 for 30 nt 5′-tail (open square) and 0.026 ± 0.002 min−1 for 50 nt 5′-tail (open triangle). The effect of biotin (open circle) or biotin/streptavidin (SA) (closed circle) blocks on the (C) 3′-tail or (D) 5′-tail strands on MCM unwinding. Cartoon DNA inset represents the 32P-labeled DNA template used for each experiment. Data were from at least three independent experiments and corrected for experimentally determined streptavidin displacement as described in ‘Materials and Methods’ section and shown in Supplementary Figure S1.
Table 1.
WT SsoMCM unwinding activity, binding affinity and hexamer stability on fork DNA with different length 5′-tails
| DNA type | Helicase |
MCM subunit exchangea |
MCM Off-rateb |
|||
|---|---|---|---|---|---|---|
| Activityc | Kd (nM)d | kobs1 (10−3 s−1) | kobs2 (10−3 s−1) | k−1 (10−3 s−1) | k−2 (10−3 s−1) | |
| No DNA | n/d | n/d | 192 ± 3 | 23.7 ± 0.2 | n/d | n/d |
| ssDNA (66 nt) | n/d | 26 ± 5 | n/d | n/d | 40 ± 6 | 1.7 ± 0.2 |
| 5′-tail–0 nte | ++ | 56 ± 6 | 196 ± 5 | 23.8 ± 0.5 | 14 ± 3 | 1.9 ± 0.5 |
| 5′-tail–30 nte | +++ | 60 ± 13 | 55.6 ± 1.1 | 3.5 ± 0.1 | 12 ± 2 | 1.3 ± 0.2 |
| 5′-tail–50 nte | +++ | 75 ± 13 | 47.4 ± 1.0 | 3.5 ± 0.1 | 7 ± 1 | 0.6 ± 0.1 |
aAveraged from at least seven individual stopped flow FRET experiments. bAveraged from at least three separate fluorescence anisotropy off-rate experiments.
c+++ represents full activity, ++ and + are 2-fold and 4-fold decreases, respectively.
dMeasured from equilibrium fluorescence anisotropy. Fits are shown in Supplementary Figure S2A.
eUsing a 30-mer 3′-tail. n/d, not determined.
Biotin was incorporated on the ssDNA tail two nucleotides downstream of the ssDNA–dsDNA junction. Binding of streptavidin to this site creates a sizeable steric block to unwinding. When a biotin/streptavidin block was included on the 3′-tail, the rate of SsoMCM unwinding was significantly reduced (Figure 1C). Conversely, an identical block on the 5′-tail did not affect the unwinding rate (Figure 1D). We also found that SsoMCM was able to background displace streptavidin from a biotin-labeled ssDNA template under our experimental conditions, artificially increasing the unwinding rate when on the 3′-strand (Supplementary Figure S1). This has already been noted for other DNA helicases and been suggested as a means to remove bound proteins from the DNA template during unwinding (24). The background rate of streptavidin displacement was used to correct the raw unwinding data to yield the results shown in Figure 1C. These results are consistent with SsoMCM sterically excluding the 5′-strand from the central channel during unwinding.
The 5′-tail stabilizes the MCM hexamer on DNA
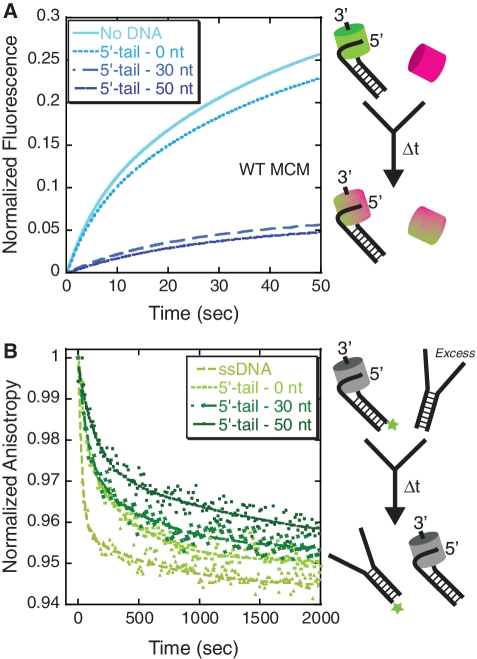
We then decided to measure specific kinetic parameters associated with DNA binding to assess the role of the 5′-tail, including a measure of SsoMCM subunit exchange and the off-rate (k−1) of the SsoMCM complex from DNA. Previously, individual MCM subunits labeled at the N-terminus with fluorescent dyes were used to examine the subunit arrangement of the SsoMCM helicase on DNA assembled from a subunit exchange mechanism (12). We took advantage of the ability of individual SsoMCM subunits to exchange in solution to kinetically monitor the stability of the SsoMCM hexamer on DNA templates with various 5′-tail lengths using stopped-flow FRET.
Rapid mixing of acceptor-labeled SsoMCM with preformed donor-labeled SsoMCM on fork DNA resulted in an exchange of subunits and an increase in FRET (Figure 2A). The first exponential rate was concentration dependent and therefore attributed to the direct exchange of SsoMCM subunits. Doubling the concentration of acceptor SsoMCM while holding donor SsoMCM and 30-mer 5′-tail fork DNA constant gave observed rates of 0.125 ± 0.005 min−1 and 0.0039 ± 0.0004 min−1, while halving the concentration of acceptor SsoMCM resulted in observed rates of 0.031 ± 0.002 min−1 and 0.0029 ± 0.0003 min−1. The second exponential was unchanged with concentration and likely results from a conformational rearrangement between subunits to form the final hexameric state. Interestingly, as the 5′-tail length increased from 0 to 30 to 50 nucleotides while holding the MCM concentration constant, the subunit exchange rate decreased (Table 1). DNA substrates lacking 5′-tails had very similar and more rapid subunit exchange kinetics to that of SsoMCM alone.
Figure 2.
The presence of a 5′-tail stabilizes the MCM hexamer. (A) Stopped flow FRET experiments were performed to detect an exchange of donor (Alexa488) and acceptor (Alexa555)-labeled MCM subunits on DNA forks with different length 5′-tails through an increase in FRET. Concentrations of SsoMCM (1.2 µM) and DNA (200 nM) were stoichiometric and were held well above the Kd value to promote the DNA-bound state. The increase in FRET was fit to two exponentials for no DNA, 5′-tail, 0 nt; 5′-tail, 30 nt; and 5′-tail, 50 nt; reported in Table 1, and attributed to the exchange of MCM subunits from solution. The cartoon shows the result of the exchange of a donor-labeled MCM bound to DNA with a free acceptor-labeled MCM complex giving rise to a mixed donor and acceptor MCM hexamer and an increase in FRET. (B) Kinetic anisotropy experiments monitoring the off-rate of the MCM complex from the fluorescently labeled DNA templates with different length 5′-tails upon addition of excess unlabeled DNA. The data was fit to Equation 2, and the individual rates are reported in Table 1. The cartoon shows the result of the dissociation of the MCM complex after trapping with unlabeled DNA leading to a decrease in the fluorescence anisotropy.
Analogous experiments were performed to monitor the dissociation (k−1) of the entire SsoMCM complex from DNA substrate with variable 5′-tail lengths after addition of a high concentration of unlabeled DNA trap using fluorescence anisotropy. Again, the presence of a longer 5′-tail reduced the off-rate of the MCM complex from DNA (Figure 2B and Table 1). As above, the change in anisotropy with time also fit better to a double exponential equation and is indicative of at least a two-step process, whereby faster dissociation of multiple subunits precedes the slower removal of any remaining MCM subunits. The second exponential rate was roughly 10-fold slower than the first rate (Table 1). Comparing the first and second observed off-rates between substrates with or without a 50 base 5′-tail shows that there is an equal 2- to 3-fold reduction for each rate. Therefore, stabilization of the SsoMCM hexamer on DNA through interaction with the 5′-tail is a concerted process that affects both steps and includes subunit dissociation as a mechanism for disassembly.
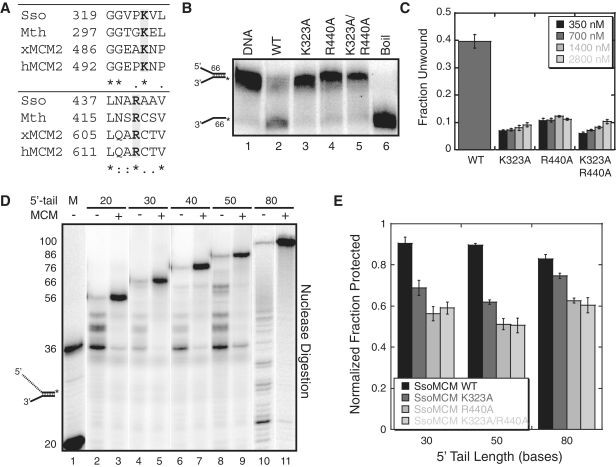
Mutations of conserved external residues on the surface of MCM abrogate unwinding
The recent availability of the SsoMCM crystal structure (7) allowed us to identify several conserved residues (Figure 3A) on the exterior surface that could interact with the displaced 5′-tail to stabilize the complex. Mutation of K323 and/or R440 to alanine resulted in a reduction of unwinding activity (Figure 3B and C; Table 2). These residues make up a positively charged patch on the exterior waist of the SsoMCM hexamer. K323 resides on the exterior of a previously identified exterior hairpin (EXT-hp) proposed to be involved in contacting DNA during unwinding (33). Consistent with previous results (33), mutation of K323A resulted in reduced unwinding over a variety of concentrations (Figure 3C). R440 is positioned on the external surface at the base of the presensor-1 hairpin (PS-1 hp). Alanine mutations of the lysine at the tip of the PS-1 hp have been shown to reduce DNA binding affinity, and eliminate unwinding (12). Both the R440A mutation and the double mutant (K323A/R440A) reduced unwinding activity similar to K323A (Figure 3B and C; Table 2).
Table 2.
SsoMCM mutant DNA unwinding and binding parameters
| SsoMCM | Helicasea | Kd (nM)b | Oligomeric state |
|---|---|---|---|
| WT | +++ | 75 ± 13 | Hexamer |
| K323A | + | 93 ± 10 | Hexamer |
| R440A | + | 92 ± 13 | Hexamer |
| K323A/R440A | + | 90 ± 9 | Hexamer |
a+++ represents full activity, ++ and + are 2-fold and 4-fold decreases, respectively.
bMeasured using fork DNA with a 30-mer 3′- and 50-mer 5′-tail. Fits are shown in Supplementary Figure S2B.
Figure 3.
External mutations on the surface of MCM disrupt unwinding and protection of the 5′-tail. (A) Alignment of proposed exterior surface residues on MCM that interact with ssDNA using CLUSTAL W2 (http://www.ebi.ac.uk/Tools/clustalw2). Aligned are MCM exterior surface residues proposed to bind ssDNA from Sulfolobus solfataricus (Sso), Methanothermobacter thermoautotrophicus (Mth), Xenopus laevis MCM2 (xMCM2) and human MCM2 (hMCM2). (B) DNA unwinding assays comparing wild-type and mutant MCM activities at 700 nM hexamer. Fork DNA with 30 base 3′- and 5′-tails were examined for unwinding at 60°C for 30 min as described in ‘Materials and Methods’ section. (C) Quantification of fraction unwound in (B) for WT at 700 nM and the three mutants at four separate concentrations (350, 700, 1400 and 2800 nM) from at least three independent experiments. (D) Nuclease assays were performed in the presence and absence of SsoMCM with different length 5′-tails as described in ‘Materials and Methods’ section. DNA was labeled at the 3′-end with [α-32P]dATP. DNA markers (M) are shown in lane 1. The length of the 5′-tail was varied from 20, 30, 40, 50 and 80 bases. The duplex region (36 bases) and 3′-tail (30 bases) were identical for lanes 2–9. The duplex region for lanes 10–11 were 20 bases and 3′-tail were 30 bases. (E) Quantification of the fraction protected from at least three independent mung bean nuclease assays comparing WT SsoMCM to mutants (K232A, R440A and K323A/R440A) with 30, 50 or 80 base 5′-tails and shown and reported in Supplementary Table S2.
It is possible that the reduced helicase activity observed with these mutations is a consequence of reduced DNA binding ability. In fact, the K323A mutation has been shown previously to have slightly reduced binding affinity as determined from EMSA experiments (33). We instead chose to use fluorescence anisotropy assays to more accurately quantify the binding affinities of wild-type (WT) and mutant SsoMCMs under equilibrium solution conditions. The measured binding affinity for WT SsoMCM is slightly tighter than previously measured (7,12,33,34), perhaps owing to our more quantitative fluorescent experimental approach. Changing the length of the 5′-tail from 0 to 30–50 nt did not significantly change the binding affinity of WT SsoMCM (Table 1 and Supplementary Figure S2A) suggesting that the majority of binding energy results from encircling the 3′-strand. Interestingly, we detected only slight decreases in the DNA binding affinities of the mutants compared to WT SsoMCM using a forked DNA substrate with a 30 base 3′-tail and a 50 base 5′-tail (Table 2 and Supplementary Figure S2B).
Long 5′-tails are protected from nuclease digestion
The stability of the interaction of the 5′-tail on the exterior of the SsoMCM hexamer was probed using a nuclease footprinting assay. Mung bean nuclease was selected since it is a single-strand-specific DNA endonuclease. We designed forked DNA substrates with a constant 30 base 3′-tail and 5′-tails ranging from 20 to 80 nt (Supplementary Table S1). The 3′-end of the 5′-tail was labeled with [α-32P]dATP to visualize the length of the digested 5′-tail.
In Figure 3D, a mung bean nuclease mapping experiment was performed in the absence and presence of WT SsoMCM. Varying lengths of the 5′-tail (20, 30, 40, 50 and 80 nucleotides) were probed for protection from nuclease digestion. The stoichiometry of protein and DNA were carefully controlled to allow for 1:1 binding. To be certain, a titration of WT SsoMCM (270–1080 nM) with constant DNA (90 nM) was examined using these nuclease assays (Supplementary Figure S3A). In the absence of protein, there are digested bands of varying sizes down to the ssDNA-dsDNA junction (either 20 or 36 nt), whereas in the presence of protein, the DNA becomes almost fully protected (Figure 3D and Supplementary Figure S3). A limited amount of digestion of the 5′-tail occurred at the ssDNA-dsDNA junction in all cases when SsoMCM was bound.
We also measured the protection of the 5′-tail with the external ssDNA binding site mutants (K323A, R440A, K323A/R440A) in identical assays (Supplementary Figure S3B). The fraction of mung bean digested 5′-tail was quantified and compared between WT and mutant SsoMCMs for 30, 50 and 80 base 5′-tails (Figure 3E and Supplementary Table S2). There is a significant increase in 5′-tail digestion for each of the SsoMCM mutants compared to WT. The change in the digestion pattern is most significant for the 50 base 5′-tail. In other cases, there is a more distributed nuclease digestion pattern observed along the length of the 5′-tail for the mutants.
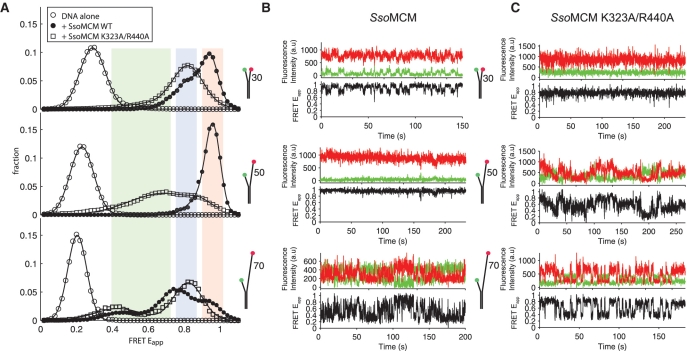
External surface mutations disrupt the path and stability of binding the 5′-tail
In order to characterize the 5′-tail binding path on the SsoMCM exterior in the absence of ensemble averaging, we utilized spFRET. Previous results from spFRET experiments indicated a dynamic interaction of the 5′-tail with the exterior of the SsoMCM hexamer (22), but the roles of 5′-tail length or specific SsoMCM residues involved in this interaction were not investigated. Based on the perturbation of unwinding by K323A/R440A shown above, we predicted that the path of the 5′-tail would be altered compared to that of WT. Here, we report results from experiments in which we monitored spFRET from the free ends of DNA with variable length 5′-tails in complex with WT or K323A/R440A SsoMCM.
Forked DNA substrates with Cy3 on the end of a 30 base 3′-tail and Cy5 on the 5′ end of different length 5′-tails (30, 50 and 70 bases) were attached to the surface of a flow chamber through a biotin–streptavidin interaction. As expected, the FRET values of the forked substrates alone decreased with increasing 5′-tail length (Figure 4A). Subsequently, WT or K323A/R440A SsoMCM was flowed into the chamber containing tethered fork substrates. In all instances, FRET values dramatically increased after SsoMCM was added, consistent with the 3′- and 5′-tails coming together in closer proximity. In complex with WT SsoMCM, different length 5′-tails (30, 50 and 70 bases) displayed a consistent high FRET state (∼0.95) (Figure 4A and Supplementary Figure S5). The complex with the 50-mer 5′-tail exists almost entirely in this high FRET state, with most traces rarely or never visiting the medium FRET state (highlighted in Figure 4B by a brief excursion to the medium state), whereas this high FRET state is transiently, but stably visited during the course of the trajectory for the case of the WT SsoMCM bound to 30- and 70-mer 5′-tail forked substrates (Figure 4B). Deconvoluted Gaussian fits for each FRET population are shown in Supplementary Figure S5.
Figure 4.
Single-pair FRET experiments monitoring the change in position of the 3′- and 5′-tails upon binding wild-type SsoMCM or external surface mutant SsoMCM (K3232A/R440A). (A) Shows the single-molecule distributions of the FRET populations of DNA alone (open circle), DNA bound to WT SsoMCM (filled circle) or DNA bound to SsoMCM (K323A/R440A) (open square) for a constant 30-mer 3′-tail and variable length 5′-tails (30, 50 or 70 bases). For all histograms, points represent binned data and solid lines represent composite Gaussian fits. Colored panes denote the high (red), medium (blue) and low (green) states as discussed in the text. Representative kinetic single-molecule fluorescence and FRET traces for each DNA substrate for (B) WT SsoMCM or (C) SsoMCM (K323A/R440A), indicated as donor (green), acceptor (red) and FRET (black).
In contrast, in the presence of K323A/R440A SsoMCM, the FRET states were shifted to either medium or lower, more broadly distributed FRET states, suggesting conformations different from those seen for WT (Figure 4A). In all cases, the extremely stable high FRET conformation observed with WT SsoMCM is completely absent when the external binding site is mutated. The FRET states of the surface mutations for each of the forked substrates showed a dramatically different dynamic behavior between WT- and K323A/R440A-bound complexes (Figure 4B and C; and Supplementary Figure S5). Lifetimes of the FRET populations for each experiment also show that mutation of an external binding site in SsoMCM alters the kinetics and dynamics of 5′-tail binding (Supplementary Table S3). The bimodal switching behavior between high and medium states observed with 30-mer 5′-tail in complex with the WT SsoMCM collapses to a stable, unimodal medium FRET state when complexed with K323A/R440A. Furthermore, the stable high FRET state observed with the 50-mer 5′-tail and WT SsoMCM is shifted to a broadly distributed state upon addition of the double mutant. Addition of K323A/R440A MCM to the 70-mer 5′-tail forked substrate resulted in a bimodal distribution between a low and medium FRET state, in contrast to the highly dynamic FRET state observed with WT SsoMCM.
DISCUSSION
SsoMCM sterically excludes the 5′-strand during unwinding
Homohexameric MCMs from archaea serve as models for more complex eukaryotic MCMs for determining enzymatic mechanisms of DNA binding, ATP hydrolysis and DNA unwinding. Several models for DNA unwinding by hexameric helicases have been proposed (21,35,36). Although steric exclusion models for unwinding by hexameric helicases have been supported by results from E. coli DnaB and Yeast MCM4,6,7 (37), SV40 Large T antigen is proposed to operate as a dsDNA pump displaying ‘rabbit ear’ like protrusions of DNA out of a double hexamer complex (17,38,39). Support for the DNA extrusion model of unwinding for MCM helicases is based on homology to the SV40 large T, visualization of side channels that can accommodate ssDNA (7,40) and the functional characterization of β-hairpins that are important for DNA binding and unwinding (7,12,13,33).
Our biochemical experiments show a deceleration of DNA unwinding only when a biotin–streptavidin block is included on the encircled 3′-strand and not on the displaced 5′-strand. SsoMCM was able to displace some background amounts of streptavidin from the template similar to other non-hexameric DNA helicases (41,42), perhaps owing in part to our elevated reaction temperatures. Nevertheless, there is clear reduction in the unwinding by SsoMCM when a biotin-streptavidin block is included on the 3′-strand suggesting that only the 3′-tail enters the central channel and the 5′-tail is excluded.
SsoMCM has been shown to be more efficient at unwinding fork substrates than 3′-tail only substrates (12). The presence of any 5′-tail, regardless of length, increases the unwinding ability of SsoMCM. Tails as short as 9 nt have been shown to be just as efficient as longer 30 nt 5′-tails (31). Longer 5′-tails (50 versus 30 nt) used did not elevate the unwinding efficiency. On the contrary, the side channel extrusion mechanism should be almost as efficient at unwinding substrates without 5′-tails as efficiently those with tails if separation occurs within the central channel. Clearly, the unwinding efficiency is primarily determined by the ability to preseparate DNA before entry into the central channel. DNA templates without a 5′-tail will consequently end up encircled in the central channel of the MCM complex in a futile unwinding attempt.
External surface residues on MCM direct the binding of the displaced 5′-tail
We have identified two universally conserved residues (K323 and R440) on the exterior of SsoMCM that significantly contribute to its unwinding efficiency. These residues reside in close proximity on the external MCM surface and contribute to the structure at the base of separate β-hairpins. They are not directly adjacent to the side channels identified in the crystal structure (7) but are roughly 20 Å away. Collectively, they create a positively charged patch that likely directs binding of the displaced 5′-strand and contributes to unwinding. We measured an 6-fold decrease in unwinding activity for the K323A mutation but did not observe a decrease in binding affinity as detected previously (33). Instead, we utilized a fluorescent anisotropy DNA binding assay and found that the Kd is similar for WT and mutant MCMs (Table 1). Thus, the decrease in unwinding ability of the mutants is most likely due to a reduced stabilization of the separated 5′-tail on the exterior surface. This external stabilization does not significantly add to the binding affinity in an equilibrium state, but instead contributes to strand stabilization to promote unwinding. Conserved DNA hairpins in the central channel involved in coupling ATP hydrolysis to unwinding (12) would then act in concert with the exterior interactions to direct the helicase forward.
Single-pair FRET experiments performed with the K323A/R440A mutant showed an increase in the distribution spread of the FRET signal between the 3′- and 5′-tails, reflecting a change in 5′-tail binding dynamics. This observation was most prevalent when a 50-mer 5′-tail was used, where the surface mutations caused the distribution to change from a fairly discrete high FRET state (0.95) that was stable on the order of 100 s to a more diffuse population that visited a broad range of FRET states. We consider medium (∼0.8) and lower (∼0.4–0.7) FRET states to represent alternate surface-bound and free conformations dependent on the 5′-tail length. Collapse of the bimodal peak observed for the WT complex with the 30-mer 5′-tail to a medium peak with the double mutant suggests that the 5′-tail is in an unbound, displaced conformation. Upon mutation, the stronger surface interactions observed with the 50-mer 5′-tail, afforded by an ideal binding path are weakened, causing a metastable interaction of the 5′-tail with the surface.
Although the 70-mer 5′-tail complexed with WT MCM occasionally visited the stable, high FRET state, it also displayed a wide range of FRET states that were unstable and displayed both rapid and slower transitions, possibly representing free Brownian motions of the unbound 5′-tail and dynamic 5′-tail interactions with the surface, respectively. The decreased stability observed with the 70-mer 5′-tail may indicate a competition between the free energy of solvation and that of surface binding. For K323A/R440A, the surface binding energy is diminished, resulting in a free tail that rapidly switches between two extended conformations.
The 5′-tail stabilizes the hexameric MCM complex on DNA
Single-stranded DNA wrapping around the exterior also stabilizes the MCM hexamer complex. The off-rate (k−1) of the hexamer from DNA is reduced in the presence of a 5′-tail. Shorter (30-mer) 5′-tails still allow for dissociation to occur by sliding off the end of the DNA. This off-rate reduction is seemingly in contradiction to the similar binding affinities measured for different length tails, however, a slower on-rate (k1) of binding due to increased complexity of organization for 5′-tailed substrates would compensate for the overall dissociation constant (Kd = k−1/k1). As an aside, we measured the fastest off-rate (k−1) for ssDNA, which also possessed the tightest binding. The ease with which ssDNA can bind in multiple conformations, either on the exterior surface or in the interior channel of SsoMCM, can be explained by a fast on-rate (k1).
Subunit exchange of MCM subunits from solution also provides a possible assembly mechanism, especially on pre-separated DNA substrates with single-stranded regions. Once loaded into a competent complex with DNA, further assembly or exchange of MCM subunits would be reduced through interactions with the 5′-tail. We suspect that binding of the 5′-tail propagates conformational changes throughout the hexamer creating a tighter, more efficient DNA unwinding complex that restricts subunit exchange. The increased stability of the MCM–DNA complex would result in a more competent helicase, able to more efficiently unwind DNA. We speculate that initial binding to fork DNA is directed by a subunit exchange mechanism primarily centered around the 3′-tail, while more subtle exterior interactions with the 5′-tail stabilize the hexamer complex to promote MCM towards an active unwinding state.
Novel SEW model of DNA unwinding for SsoMCM
Previously, it was shown that the displaced 5′-strand dynamically interacts with the exterior surface of SsoMCM (22). This result helped distinguish between the steric exclusion and side channel extrusion models of unwinding, confirming that a 5′-tail can interact in a way only possible with an external binding event. For this study, we used different 5′-tail lengths to more specifically map the exterior binding site using spFRET. Discrete high FRET populations (∼0.85 and ∼0.95) exist that are present with all 5′-tail lengths. If the 3′-tail is fixed within the central channel of MCM, then the distance to the 5′-tail must be identical for 30, 50 and 70 base 5′-tails for those FRET states. Though a stable high FRET state could also be consistent with side channel extrusion, we consider it unlikely, since the 5′-tail dynamics for the 30-mer substrate would be more restricted due to the displaced strand first passing through the central channel and then out a side channel.
The longitudinal length of the SsoMCM hexamer is roughly 100 Å (43), approximately equal to 24 bases of linear ssDNA (4.1 Å/base). If the encircled 3′-tail follows a helical conformation similar to that detected for the hexameric papillomavirus E1 helicase binding to ssDNA (44), this would place the 3′-end towards the N-terminal end of SsoMCM, but not exposed (12). Similar vertical displacement of the 30-mer 5′-strand on the exterior surface would position the donor and acceptor fluorophores in a similar location. However, if the longer 50-mer 5′-tail were to take the same longitudinal path, then the 5′-Cy3 would project past the N-terminal tier of the MCM complex. The 70-mer 5′-tail would project even further and would display an even smaller FRET value. Clearly, the linear length of the 50- and 70-mer 5′-tails precludes the appearance of such identical high FRET signals if the 5′-strand was traversing along a linear path on the MCM hexameric exterior.
Only a model that includes some wrapping of the 5′-tail around the exterior of the MCM complex could explain similar FRET states for different length 5′-tails. Complete wrapping of the 5′-tail around the exterior surface would produce an identical, unimodal high FRET state irrespective of the length. In addition to this state, we also observed similar discrete FRET states for each 5′-tail. The symmetry of the hexameric SsoMCM helicase dictates that identical repeating structural features make up the exterior surface. Therefore, wrapping of the ssDNA could interact with one, two, or more subunits before exiting past the N-terminal tier. In support of a wrapping model, electron microscopy studies have identified an external MCM hexamer binding site for dsDNA that is also proposed to wrap DNA around the exterior surface of the hexameric MCM complex (45). Although this dsDNA binding path resides along a helix–turn–helix motif in the N-terminal tier, it is possible that the unwound ssDNA 5′-tail may transverse the waist and also interact along this dsDNA binding path before being released from the helicase.
Wrapping of the displaced 5′-tail would also provide a structural mechanism to prevent reannealing. The wrapped strand would include a larger number of DNA bases interacting with SsoMCM than the encircled strand. The resulting displacement in complementary sequence between the two strands would act to prevent reannealing behind the helicase. A significant fraction of even the longest 5′-tail (80-mer) is protected from nuclease digestion when WT SsoMCM is bound. Some specific cleavage can be detected at the 5′-single-strand/double-strand junction, liberating ssDNA with a length equal to the tail. Other low-level, non-specific cleavage sites can be detected along the length of the 5′-tail. Therefore, the majority of the 5′-tail resides are in a locally protected environment on the external surface of the MCM hexamer. Presumably, rapid fluctuations between high FRET (bound) and low FRET (unbound) states would not provide a stable enough substrate for nuclease digestion.
Mutations in the positive binding patch on the surface of the MCM hexamer result in a more disperse nuclease digestion pattern suggesting a more loosely bound 5′-tail. This is also consistent with the spFRET experiments that displayed a much broader FRET distribution with the mutant MCM. It is therefore likely that the K323A and R440A mutations at each MCM subunit reduce the binding affinity of the 5′-tail, indicating these residues probably serve as positively charged guides for wrapping negatively charged DNA on the MCM surface. Other external binding patches are most likely present, as suggested from incomplete total 5′-tail digestion and the presence of distinct FRET states for the K323A/R440A mutant, which can loosely bind the 5′-tail.
Based on these results, we have developed a new model for DNA unwinding by MCM termed the SEW model, resembling a spool of thread for wrapping the 5′-tail around the exterior surface (Figure 5). This model includes steric exclusion and subsequent wrapping of the 5′-tail around the exterior of the MCM hexamer, which promotes DNA unwinding, MCM–DNA stabilization, protection of ssDNA, and strand separation. There are distinct channels on the external surface of the MCM hexamer dotted with positive charge that could direct the displaced 5′-strand around the hexamer. After significant wrapping of the 5′-tail has occurred, further DNA bases will proceed past the N-terminal tier and be released from the hexamer.
Figure 5.
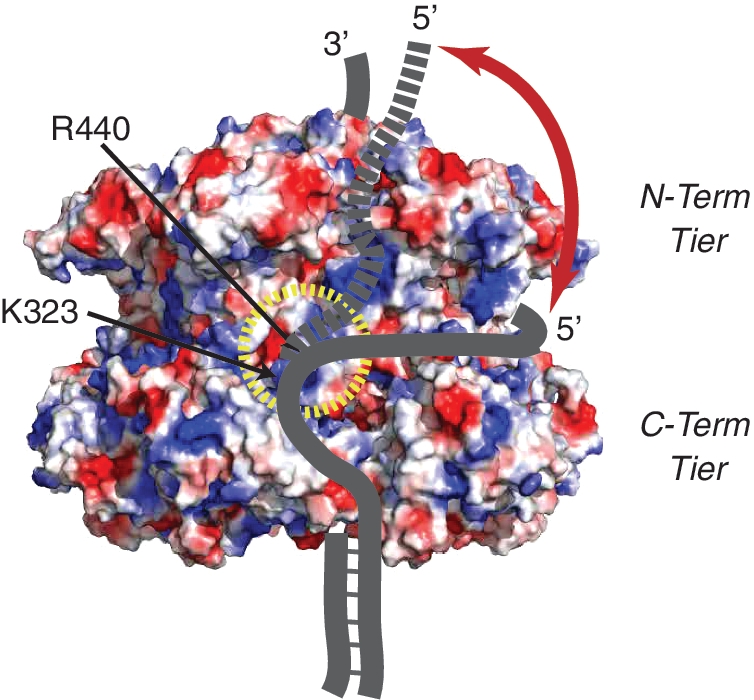
Proposed Steric Exclusion and Wrapping (SEW) model for DNA unwinding. At least two paths for wrapping ssDNA on the exterior surface of MCM are highlighted. Shown is the surface electrostatic potential representation of SsoMCM (PDBID:3F9V), where blue and red patches, respectively, represent positive and negatively charged residues, highlighting ssDNA binding residues (K323 and R440) that direct the path of the 5′-tail around a ssDNA protection channels in the waist (solid line) before exiting past the N-terminal tier (dashed line).
The displaced 5′-tail cannot wrap the MCM complex too tightly; it must be allowed to slide over the external surface of the hexamer for efficient unwinding. The spFRET dynamics for binding the 5′-tail detected here provides the basis for this dynamic binding mechanism. Therefore, binding of the 5′-tail is a compromise between adequate protection of the ssDNA, efficient stabilization of unwound DNA that contributes to the unwinding mechanism, and stabilization of the entire hexameric MCM complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Chemistry, University of Pittsburgh (M.A.T.); National Institutes of Health (GM077872 to S.H.L.). Funding for open access charge: Department of Chemistry, University of Pittsburgh.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Xiaojiang Chen for providing the coordinates for the SsoMCM symmetrical hexamer model.
REFERENCES
- 1.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell SP, Dutta A. DNA replication in eukaryotic cells. Ann. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 3.Madine MA, Khoo C-Y, Mills AD, Musahl C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- 4.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 5.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl Acad. Sci. USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 9.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 10.Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006;34:5829–5838. doi: 10.1093/nar/gkl708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Mol. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 12.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implication for the helicase mechanism. Nat. Struct. Mol. Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 13.Barry ER, McGeoch AT, Kelman Z, Bell SD. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 2007;35:988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Barry ER, Lovett JE, Costa A, Lea SM, Bell SD. Intersubunit allosteric communication mediated by a conserved loop in the MCM helicase. Proc. Natl Acad. Sci. 2009;106:1051–1056. doi: 10.1073/pnas.0809192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakakibara N, Kasiviswanathan R, Melamud E, Han M, Schwarz FP, Kelman Z. Coupling of DNA binding and helicase activity is mediated by a conserved loop in the MCM protein. Nucleic Acids Res. 2008;36:1309–1320. doi: 10.1093/nar/gkm1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, Martin CS. Polymorphism and double hexamer structure in the Archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 2005;280:40909–40915. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- 19.Shin J-H, Heo G-Y, Kelman Z. The Methanothermobacter thermautotrophicus MCM helicase is active as a hexameric ring. J. Biol. Chem. 2009;284:540–546. doi: 10.1074/jbc.M806803200. [DOI] [PubMed] [Google Scholar]
- 20.Patel SS, Picha KM. Structure and function of hexameric helicases. Ann. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem. 2007;282:34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2001. [Google Scholar]
- 24.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 25.Mikheikin AL, Lin H-K, Mehta P, Jen-Jacobson L, Trakselis MA. A trimeric DNA polymerase complex increases the native replication processivity. Nucleic Acids Res. 2009;37:7194–7205. doi: 10.1093/nar/gkp767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Axelrod D. In: Methods in Cell Biology. John JC, William Detrich H III, editors. Vol. 89. Burlington, MA: Academic Press; 2008. pp. 169–221. [Google Scholar]
- 28.Fagerburg MV, Leuba SH. Optimal Practices for Surface-tethered Single Molecule Total Internal Reflection Fluorescence Resonance Energy Transfer Analysis. 1st edn. New York, NY: Humana Press; 2011. [DOI] [PubMed] [Google Scholar]
- 29.Li K. The image stabilizer plugin for ImageJ. 2008 http://www.cs.cmu.edu/∼kangli/code/Image_Stabilizer.html (1 August 2010, date last accessed), February, 2008. [Google Scholar]
- 30. Rasband, W.S. ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/ (1 August 2010, date last accessed), 1997-2011.
- 31.Carpentieri F, De Felice M, De Falco M, Rossi M, Pisani FM. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the Crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 2002;277:12118–12127. doi: 10.1074/jbc.M200091200. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan DL, Davey MJ, O'Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 33.Brewster A, Slaymaker I, Afif S, Chen X. Mutational analysis of an archaeal minichromosome maintenance protein exterior hairpin reveals critical residues for helicase activity and DNA binding. BMC Mol. Biol. 2010;11:62. doi: 10.1186/1471-2199-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry ER, Bell SD. DNA replication in the Archaea. Microbiol. Mol. Biol. Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 36.Laskey RA, Madine MA. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4:26–30. doi: 10.1038/sj.embor.embor706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan DL, O'Donnell M. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol. Cell. 2004;15:453–465. doi: 10.1016/j.molcel.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 38.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 39.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brewster AS, Chen XS. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit. Rev. Biochem. Mol. Biol. 2010;45:243–256. doi: 10.3109/10409238.2010.484836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matlock DL, Yeruva L, Byrd AK, Mackintosh SG, Langston C, Brown C, Cameron CE, Fischer CJ, Raney KD. Investigation of translocation, DNA unwinding, and protein displacement by NS3h, the helicase domain from the hepatitis C virus helicase. Biochemistry. 2010;49:2097–2109. doi: 10.1021/bi901977k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pape T, Meka H, Chen S, Vicentini G, Heel MV, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 45.Costa A, van Duinen G, Medagli B, Chong J, Sakakibara N, Kelman Z, Nair SK, Patwardhan A, Onesti S. Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J. 2008;27:2250–2258. doi: 10.1038/emboj.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.