Abstract
Convenient and well-performing protein detection methods for a wide range of targets are in great demand for biomedical research and future diagnostics. Assays without the need for washing steps while still unaffected when analyzing complex biological samples are difficult to develop. Herein, we report a well-characterized nucleic acid proximity-based assay using antibodies, called Proximity Extension Assay (PEA), showing good performance in plasma samples. Target-specific antibody pairs are linked to DNA strands that upon simultaneous binding to the target analyte create a real-time PCR amplicon in a proximity-dependent manner enabled by the action of a DNA polymerase. 3′Exonuclease-capable polymerases were found to be clearly superior in sensitivity over non-3′exonuclease ones. A PEA was set up for IL-8 and GDNF in a user-friendly, homogenous assay displaying femtomolar detection sensitivity, good recovery in human plasma, high specificity and up to 5-log dynamic range in 1 μL samples. Furthermore, we have illustrated the use of a macro-molecular crowding matrix in combination with this homogeneous assay to drive target binding for low-affinity antibodies, thereby improving the sensitivity and increasing affinity reagent availability by lowering assay development dependency on high-affinity antibodies. Assay performance was also confirmed for a multiplex version of PEA.
INTRODUCTION
Detection and quantification of proteins in complex biological samples can pose several technical challenges. Assay performance as of recovery, specificity and linearity can especially be negatively affected. The homogenous proximity ligation assay (PLA) has been shown to provide sensitive protein detection in very small sample volumes (1,2). PLA is based on the analyte becoming bound by two so called proximity probes which are antibodies coupled with DNA strands, and when these strands come in close proximity upon by target binding they are united through the activity of a DNA ligase enzyme. The newly formed joint ligation product then serves as a template for quantitative PCR reflecting the amount of target protein present. As this assay is homogeneous, meaning no washing steps, all the components of the sample are present throughout the assay. Such components might interfere with the assay and lower the efficiency of the enzymatic processes involved, thereby impairing assay recovery. The use of DNA ligases for PLA has proven difficult in human plasma requiring data normalization (3). Therefore, we investigated the use of DNA polymerases to improve homogenous nucleic acid proximity reactions when using biological samples. A few previous reports have used DNA polymerase-based proximity assays (4–6). However, those assays were not based on antibodies, nor were their analyses performed on human plasma samples, and only modest low pM sensitivity was reported.
In endeavors to develop a wider range of sensitive protein detection assays ultimately covering the entire human proteome, suitable affinity reagents are lacking. Antibody affinities vary greatly with different target antigens and different batches. This directly affects assay performance as of sensitivity and causes prolonged assay development time in a search for better antibodies. In an attempt to enable the use of antibodies of lower affinities in proximity extension assay (PEA), we sought to use a water exclusion matrix to drive target analyte binding by increasing the effective concentration of the PEA probes by the use of sephadex bead expansion (7).
MATERIALS AND METHODS
Plasma samples, antigen standards and antibodies
EDTA blood samples were collected from healthy subjects. The samples were centrifuged at 2500g for 10 min at 4°C. After centrifugation the plasma was aspirated, aliquoted and stored at −20°C. All polyclonal antibody (Ab) and their respective recombinant human proteins were purchased from RnD Systems, besides CA19-9 that was a kind gift from Fujirebio Diagnostics AB. Recombinant proteins were all reconstituted in PBS +0.1% BSA and stored at −80°C.
Proximity probes and antigen standards
Proximity probes were prepared by covalently linking a single batch of affinity-purified polyclonal antibody or matched monoclonal antibody pairs to 3′-hydroxyl free and 5′-phosphate free 40-mer oligonucleotide sequences. These antibody-oligonucleotide conjugates were generated by Innova Biosciences (Cambridge, UK) using their Lightning-Link™ technology. Conjugation quality was analyzed by SDS–PAGE (data not shown). The 40-mer 3′ and 5′ oligonucleotide sequences used for the antibody conjugation comprised a 20-bp universal sequence used as hybridization site and target for the Molecular Beacon (8), and a unique 20-bp sequence for primer targeting in qPCR. The extension primer was hybridized to the 5′-free oligonucleotide of one of the proximity probe conjugates, at a 2:1 oligo-to-Ab ratio. All sequences used are reported in (Supplementary Table S1).
Proximity extension assay
One microliter sample (PBS + 0.1% BSA buffer ± antigen spike or human EDTA plasma) was mixed with 1 µl plasma dilution buffer (Olink Bioscience, Sweden). Samples were incubated at 25°C for 20 min. To these samples, 2 µl of probe mix [25 mM Tris–HCl, 4 mM EDTA, 0.016 mg/ml single-stranded salmon sperm DNA (Sigma Aldrich), 0.02% sodium azide and 100 pM of each PEA conjugate] was added and incubated at 37°C for 1 h.
After the probe incubation, the samples were transferred to a thermal cycler and put on hold at 37°C. An amount of 76 µl of dilution mix containing 70.5 mM Tris–HCl, 17.7 mM ammonium sulfate, 1.05 mM dithiothreitol and 40 µM of each dNTP's were added. After a 5-min incubation at 37°C, a 20 µl extension mix containing 66.8 mM Tris–HCl, 16.8 mM ammonium sulfate, 1 mM dithiothreitol, 33 mM magnesium chloride, 62.5 U/ml T4 DNA Polymerase [or 62.5 U/ml Klenow fragment exo(−), 125 U/ml Klenow fragment, 125 U/ml DNA Polymerase I (Fermentas), 250 U/ml Exonuclease I (New England Biolabs)] was added. The extension reactions were run at 37°C for another 20 min followed by a 10-min heat-inactivation step at 80°C.
Sephadex G100 experiments were performed as above, but using incubation tubes containing 500 µg of lyophilized sephadex G100 at the bottom. Sephadex G100 (Pharmacia) was resolved at 2.5 w/v% in 20% EtOH, and kept shaking at 200 rpm at room temperature (RT) for at least 24 h. Before each experiment, 20 µl of the sephadex G100 slurry was added per well and lyophilized using a SpeedVac SPD1010 (Thermo Scientific), 60 min at 65°C and 10 torrs.
PEA was also performed in multiplex using the 23-plex panels given in Supplementary Table S1. All steps were performed as above, but using a set of 23 unique probe oligo pairs, 23 corresponding extension primers and 23 unique primer pairs for qPCR detection and a universal molecular beacon probe for detection (Supplementary Table S1). Each multiplex PEA sample was split across the individual qPCRs readouts each using a target-specific primer pair.
Detection by quantitative PCR
For the qPCR detection, 4 µl of the extension products was transferred to a qPCR plate and mixed with 6 µl qPCR mix; 25 mM Tris–HCl, 7.5 mM magnesium chloride, 50 mM potassium chloride, 8.3 mM ammonium sulfate, 8.3% Trehalose (Acros Organics), 333 µM (each) dNTP's, 1.67 mM dithiothreitol, 833 nM of each primer (forward: 5′-TCGTGAGCCCAAGTGTTAATTTGCTTCACGA-3′ and reverse: 5′-TGCAGTCTGTAGCGAAGTTCTCATACTGCA-3′; or the hairpin primers forward: 5′-TCGTGAGCCCAAGTGTTAATTTGCTTCACGA-3′ and reverse 5′-TGCAGTCTGTAGCGAAGTTCTCATACTGCA-3′), 417 nM Molecular Beacon (FAM-CCCGCTCGCTTATGCTACCGTGACCTGCGAATCCCGAGCGGG-DABSYL, Biomers), 41.7 U/ml recombinant Taq polymerase (Fermentas) and 1.33 µM ROX reference (ROX-TTTTTTT, Biomers). A two-step qPCR was run with initial denaturation at 95°C for 5 min, followed by 15 s denaturation at 95°C; and 1 min annealing/extension at 60°C for 45 cycles. Raw real-time PCR profiles are shown in Supplementary Figure S1.
RESULTS AND DISCUSSION
Proximity extension assay
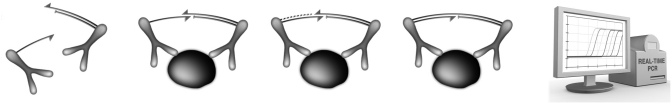
We have previously developed a highly sensitive and specific protein detection method based on antibodies coupled to DNA oligos (proximity probes) (1,2). Proximity probes joined with a DNA ligase can in some cases suffer from recovery loss in some complex biological fluids, such as blood plasma (3). Therefore, we set out to develop a new method for protein detection not based on DNA ligases but instead based on a proximity-dependent DNA polymerization event taking place between the two proximity probes (Figure 1). Antibodies [either two matched monoclonal antibody (mAb), or one batch of polyclonal antibody (pAb) split in two fractions] are covalently linked with two different 40-mer oligonucleotides, one being attached at the 3′-end, and the other at the 5′-end (for details on the sequence design, see ‘Materials and Methods’ section and Supplementary Table SI). To the 3′-linked probe, a 56-mer DNA oligo comprising 40-nt complementary to that probe, a 7-nt spacer and 9-nt complementary to the corresponding 5′-linked probe is hybridized. Next, the hybridized proximity probe pair is incubated with a sample containing the antigen of interest. This results in binding between the proximity probe pair and the antigen, and as a result, the oligonucleotides come in close proximity and are hybridized to each other. The addition of a DNA polymerase leads to an extension of the hybridizing oligo over the other probe arm. Finally, the DNA template generated can be detected and quantified by qPCR.
Figure 1.
Schematic description of the PEA. Upon sample incubation, the proximity probe pair binds its specific antigen. As a result, the probe oligos come in close proximity and hybridize to each other. The addition of a DNA polymerase leads to an extension of the hybridizing oligo. Finally, this results in a DNA template that can be detected and quantified by qPCR.
To enable an effective hot start when using native Taq DNA polymerase in the qPCR reaction, some primer pairs where designed as hairpins with limited stability, thereby not exposing free 3′-ends at room temperature during reaction set up (9). The hairpin comprised 6 nt of the 3′-end, and 6 nt of the 5′-end of each primer (see ‘Material and Methods’ section).
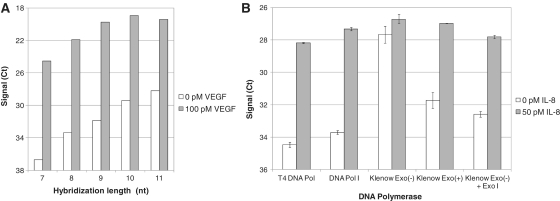
To optimize the sensitivity of the PEA reaction, different designs of the extension primer were evaluated, in this experiment using T4 DNA polymerase. This included varying the length of the sequence being complementary to the 5′-linked probe (n = 7–11) as shown in Figure 2A. The extension primer that generated high signal, enabling robust qPCR data, in combination with good sensitivity for detecting different analytes was chosen for the experiments presented below. The selected primer had a 9-nt sequence complementary to the 5′-linked probe. The PEA reaction was further optimized by varying the salt concentrations and comparing different reaction temperatures (data not shown).
Figure 2.
Exonuclease activity and hybridization length affects assay sensitivity. (A) VEGF assays were designed with different lengths of the hybridization site and compared with respect to sensitivity. A 9-nt hybridization site was found to give the best signal-to-noise levels and was selected for further studies. (B) Different DNA polymerases were tested with regards to their ability to generate good sensitivity in an IL-8-specific assay. T4 DNA polymerase I, DNA polymerase I and Klenow fragment exo+ all possess a 3′→5′ exonuclease activity and performed well in the IL-8 detection. Klenow fragment exo−, on the other hand, generated a background signal that was almost at the level of the antigen-induced signal. When exogenous Exonuclease I was added to the reaction, the signal-to-noise level was restored.
Below, a detailed characterization and evaluation of the PEA method are described. Immunoassay parameters including sensitivity, precision, recovery, specificity, assay range and detectability in plasma are assessed for several analytes.
Exonuclease activity reduces non-specific background and improves assay sensitivity
An experiment was designed to assess whether 3′→5′ exonuclease activity possessed by some polymerases can affect the extension reaction. We investigate the signal-to-noise ratio for the detection of 50 pM interleukin-8 (IL-8). T4 DNA Polymerase, DNA Polymerase I, Klenow Fragment, Klenow Fragment exo− were used in the extension reaction, and compared with respect to the resulting signal-to-background for IL-8 detection. When using DNA polymerases that have a 3′→5′ exonuclease activity (T4 DNA Polymerase, DNA Polymerase I, Klenow Fragment), there was a clear difference in signal relative to the background (Figure 2B). In contrast, the Klenow Fragment lacking exonuclease activity (Klenow exo−) generated a much higher background than the Klenow Fragment (Klenow exo+). By adding Exonuclease I to the Klenow Fragment exo− reaction, the background was lowered and, thereby, the signal-to-noise level was restored. This showed that the reduced background observed with the Klenow exo+ compared to Klenow exo− was due to the exonuclease activity and not due to intrinsic differences between the polymerases. The increased sensitivity observed for exonuclease-able assays, can be explained by a degradation of the free non-proximal DNA ends, which prevent them from accumulating extension products over time caused by continuous random proximity events occurring during the extension reaction.
DNA polymerase I and T4 DNA polymerase were the most potent polymerases for this assay, based on the signal-to-noise levels. However, the recovery in plasma was generally somewhat lower for DNA polymerase I (Figure 2B; and data not shown). Therefore, T4 DNA polymerase was selected for the remaining experiments of this study. Two additional DNA polymerases, T7 and phi-29, were also tested in this initial experimental setup. These enzymes generated a much lower signal over all, and were therefore not considered for further studies (data not shown). However, both of these polymerases possess a strong 3′→5′ exonuclease activity, and should still be considered as potential candidates for PEA and could benefit from further optimization of their specific reaction conditions.
Precision, recovery and detection of low-abundant analytes in blood plasma
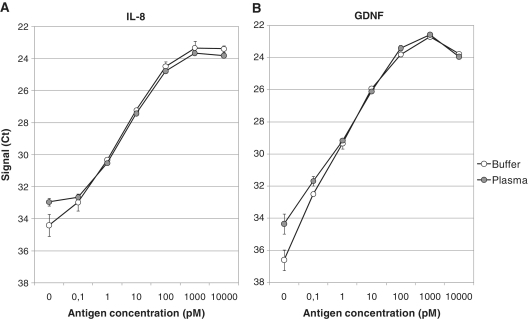
One of the major challenges in biomarker research is to develop highly sensitive methods that allow for the detection of proteins present only in minute amounts in biological samples. Therefore, we examined the ability of PEA to accurately detect low-abundant proteins in a complex matrix, human EDTA-prepared plasma, using the 3′→5′ exonuclease-efficient T4 DNA polymerase. Human IL-8 was spiked at increasing concentrations into either a non-complex matrix, PBS with 0.1% BSA, or into plasma and quantified by PEA (Figure 3A). As a second example, the human protein glial cell line-derived neurotrophic factor (GDNF) was spiked into human plasma (Figure 3B). The sensitivity of both assays was found to be very good. Both analytes were detected and could be quantified at concentrations as low as 0.1 pM. In fact, the signal-to-noise for the GDNF detection at 0.1 pM was more than eight times over background. This suggests that even lower concentrations of GDNF would be detectable. The detection dynamic range of these IL-8 and GDNF assays spanned at least four orders of magnitude (between 0.1 and 100 pM), and the lower limit of detection (2× SD above buffer background) for was determined to 48 and 9 fM, respectively. Both proteins are of very low abundance (low picomolar) in plasma and therefore difficult to detect with some standard technologies (10,11). Despite this, both analytes were readily detected in human blood plasma demonstrating the sensitivity of PEA. In addition, as PEA consumes only 1 µl of sample for analysis, it is highly suitable for biomarker research studies when clinical samples often are in shortage.
Figure 3.
Detection of low-abundant analytes in human blood plasma. Assays were generated for human IL-8 (A) and GDNF (B). Buffer or a blood plasma sample were spiked with either IL-8 or GDNF at concentrations between 0.1 pM and 10 nM, and measured with PEA. Signal is plotted as Ct values. Both IL-8 and GDNF, two low-abundant analytes, were detectable in plasma. Both assays showed a good sensitivity with the lower detection limit at, or below, 0.1 pM.
Another critical parameter to study in immunoassay evaluations is precision. To address the intraassay variation, the experiments in Figure 3 were run using triplicate samples. The average coefficient of variation (CV) across all concentrations was 11 and 14% for the two assays, respectively. This was similar to the results observed for PLA, for which an average CV of 11% was determined for a set of around 70 PLA assays (3).
Recovery (the difference in signal between a complex and a non-complex matrix) reflects the ability of an assay to accurately quantify an analyte in biological materials. As mentioned above, we and others have previously used PLA to detect a number of biomarkers in blood plasma (3,12–14). However, the activity of DNA ligase utilized in these assays was impaired in blood plasma, sometimes resulting in poor recovery (3). The average recovery determined for 13 PLA assays was only 33% (data not shown). By performing those PLA assays in multiplex, and using an exogenous spike-in normalizer, the recovery issue was solved. However, for measurements of single markers, PLA was still unsatisfactory. The T4 DNA polymerase used in the current PEA reaction seemed to perform well in blood plasma. At concentrations between 1 pM and 1 nM, the average recovery for IL-8 and GDNF was as high as 81 and 110%, respectively (Figure 3). It should be emphasized that these results were obtained without any optimization of the specific buffer conditions for each probe/target binding, which is standard procedure in immunoassay development. When IL-8 and GDNF were analyzed previously with PLA with the recovery was significantly lower; 18 and 24%, respectively (data not shown). This is a demonstrated a significant improvement compared to the PLA, allowing for accurate analyte quantification without the need for internal normalizations.
Enhancement of PEA performance through increased target binding upon water exclusion
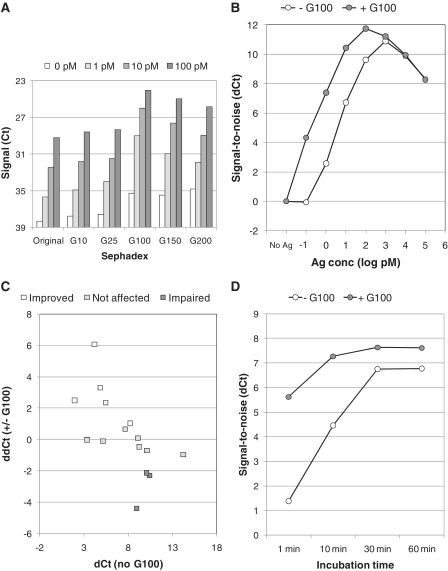
Target binding in this homogeneous immunoassay is dependent on the probe concentration and their target affinity (2). In an ideal homogeneous proximity assay, one would like to have a high probe concentration during sample incubation to drive probe/target binding. This incubation is subsequently diluted prior to the extension reaction in order to lower the probability of random proximity events. The ratio of probe concentration during incubation versus extension should be as high as possible but still within practical limits and provide sufficient amounts of template for the qPCR to maintain its robustness. Thus, simply raising the probe concentration during incubation would increase the background signal due to increased chances for random events of proximity. To get around this, the effective probe concentration was artificially raised by macro-molecular crowding, and the PEA performance was re-evaluated for a number of assays. Dry sephadex beads can upon rehydration in water expand and thereby also take up water into the beads while larger molecules, such as proteins and proximity probes, would remain outside the beads and thereby effectively enhance their concentrations and promote target binding (7). To test this, 500 µg of sephadex G10, G25, G100, G150 or G200 was lyophilized at the bottom of the PEA incubation tubes onto which the probe mix and a sample containing three different concentrations of ICAM were added, followed by a regular PEA protocol. This experiment demonstrated that while G10 to G50 did not affect the assay performance, the inclusion of G100, G150 or G200 led to a dramatic increase in signal (Figure 4A). G100 was found to give the best signal-to-noise levels at all antigen concentrations, most likely due to its high degree of swelling and a smaller pore size compared to G150 and G200. Next, a complete standard curve was generated for ICAM and analyzed with or without sephadex G100 present during the incubation step. This revealed dramatic improvements in specific target binding seen as enhanced sensitivity (Figure 4B). Most striking was the dramatic increase in signal-to-noise level obtained for the lowest antigen concentration.
Figure 4.
Increased target binding and enhancement of the PEA performance by upon macro-molecular crowding. (A) Different sephadex matrices were used during the probe/target incubation of an ICAM assay to test if exclusion of water from the reaction could improve assay performance. Signal is plotted as Ct values. (B) Complete standard curve for ICAM (10 nM–0.1 pM) is shown for assays performed with (gray) or without (white) sephadex G100. (C) Multiplex PEA was performed for 15 assays, and the signal-to-noise was determined at 200 pM antigen concentration. Signal-to-noise (dCt) is plotted as a function of the increase in signal-to-noise derived from sephadex G100 inclusion (ddCt). Five assays were improved (white), eight remained unchanged (gray) and three were impaired (black). (D) Buffer was spiked with 100 pM VEGF, and incubated with (gray) or without (white) sephadex G100. PEA was performed after different time points after incubation. Signal-to-noise (dCt) is plotted as a function of incubation time.
To study whether other assays would benefit from the sephadex G100 incubation, a panel of 15 different analytes were tested in multiplex. This data are displayed in Figure 4C in which the change in signal-to-noise upon sephadex G100 inclusion (ddCt) is plotted as a function of the signal-to-noise for each assay (dCt). Five assays were improved in the presence of G100, three assays were impaired and seven remained unchanged. The reduced sensitivity observed for some assays was due to a substantial increase of their background signals. Such background increase could occur if the two proximity probes have a slight affinity to each other. Overall, there was a trend that less sensitive assays (lower antibody affinities) seemed to benefit the most from water exclusion. Therefore, for improved assay performance for a certain assay, enclosure of G100 in the probe/target incubation step could be worthwhile considering.
In clinical diagnostics, rapid protocols are highly desirable. Therefore, we assessed whether sephadex G100 inclusion could increase the rapidity of probe/target binding. VEGF was spiked into buffer at 100 pM and PEA was performed at different time points after incubation (1, 10, 30 and 60 min). This demonstrated that when G100 was present during the incubation, the signal was significant already after 1 min of incubation, when compared to reactions lacking G100 (Figure 4D). Furthermore, already after 10 min of incubation as much as 80% of the maximum signal-to-noise was detected for the G100 reaction, compared to 20% for reactions lacking G100. All-in-all, these analyses bring forward macro-molecular crowding as a way to improve both assay sensitivity and velocity.
Specificity and detection in plasma
One of the hallmarks of both PEA and PLA is the requirement for dual and proximal binding of the PEA probes. In theory, this should reduce any signal derived from non-specific antibody binding. The standard curves of IL-8 and GDNF (Figure 3) indicate very low limit of detection even in plasma, indicating that the proximity probes are binding specifically even in very complex samples that contain a broad repertoire of proteins.
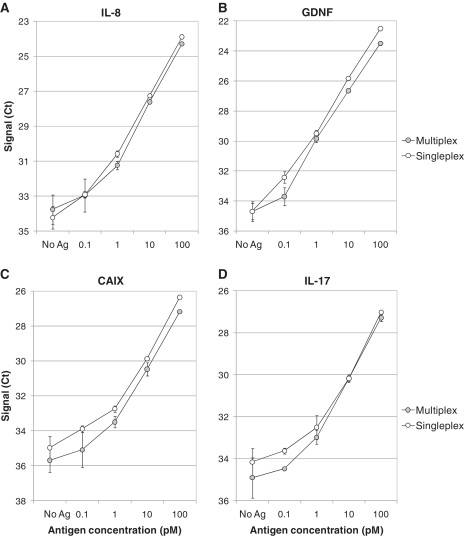
We generated 4 panels of 23 different assays using analyte-specific sequences and ran the PEA in multiplex (Supplementary Table S1). The performance of singleplex PEA was compared to that of multiplex analysis for IL-8, GDNF, CAIX and IL-17. A key feature of these multiplexed assays is that antibody cross reactivity will not result in a non-specific signal since each probe carries a unique DNA sequence and the generated DNA reporters operate independently. This is evidenced by the similarity of the standard curves generated with singleplex or multiplex PEA analysis (Figure 5).
Figure 5.
Multiplex PEA protein detection. Buffer was spiked with 23 analytes from panel 1 or 2 (see ‘Materials and Methods’ section and Supplementary Table SI) at concentrations between 0.1 and 100 pM, and measured either with singleplex (white) or multiplex PEA (gray). Results for (A) IL-8, (B) GDNF, (C) CAIX and (D) IL-17 are shown. Signal is plotted as Ct values.
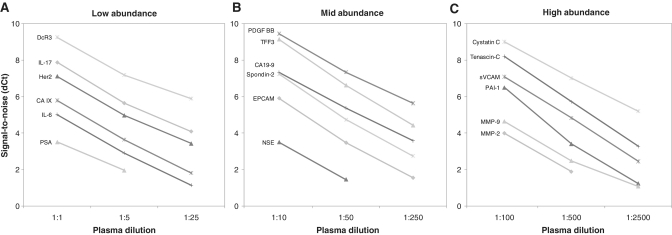
Assay linearity was also assessed using multiplex PEA. Four panels were generated each comprising 23 PEA assays: one with high-abundant, two with low-abundant and one with mid-abundant analytes. Each analyte was measured in blood plasma at three different dilutions, and a buffer background control sample. Dilutions were 5-fold, corresponding to a 2.3 theoretical Ct decrease for each dilution. Linearity was found to be good for both high and low PEA signals, for both low-abundant (Figure 6A; DcR3, IL-17, Her2, CA IX, IL-6, PSA), mid-abundant (Figure 6B; PDGF BB, TFF3, CA19-9, Spondin-2, EPCAM, NSE) and high-abundant markers (Figure 6C; Cystatin C, Tenascin C, sVCAM, PAI-1, MMP-9, MMP-2). This demonstrates that PEA is well-suited for the analyses of a broad range of analytes present in blood plasma and highlights the multiplexing capacity of PEA.
Figure 6.
Linearity of dilution in plasma. Linearity of dilution was assessed by measuring six low-abundant (A), six mid-abundant (B) and six high-abundant (C) markers at different concentrations of plasma in multiplex. Signal-to-noise values (dCt) are plotted at three different concentrations of plasma for each set of analytes. Linearity was observed for assays with either high or low signals in plasma, and also for undiluted and highly diluted plasma samples.
CONCLUSION
In an attempt to improve the assay performance in complex biological samples, two main enhancements on nucleic acid proximity-based protein detection assays were developed and evaluated. First, a DNA polymerase performing a proximity-dependent DNA polymerization forming the qPCR amplicon is used instead of using a DNA ligase. DNA polymerases proved to be less prone to be enzymatic inhibition in the presence of plasma or serum in comparison to DNA ligases, thereby improving assay recovery. Second, the choice of polymerase was found crucial for the assay sensitivity. We found that 3′→5′ exonuclease activity exhibited by certain DNA polymerases is important for this type of assay as they reduce the background by degrading remaining non-proximal DNA strands. We also discovered that for some assays, in particular those with lower sensitivity, the inclusion of sephadex G100 in the probe/target incubation significantly increased the assay sensitivity and rapidity.
All-in-all, PEA was found to perform well in plasma with regards to sensitivity, specificity, precision and dynamic range. Furthermore, PEA possess several advantages when compared to other immunoassays, including a fast and simple experimental protocol, multiplexing capacity, low sample consumption (1 μl) and the ability to use lower affinity antibodies without the need for optimizations of specific reaction conditions. We anticipate this method to contribute to biomarker research, especially for analysis of low-abundant proteins in precious and limited biobanked human samples, laboratory animals and other situations when only very small sample volumes are available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Seventh framework programme theme (FP7-Health-2007-B), European Commission, under the PROACTIVE project (grant number 222950).
Conflict of interest statement. M.L., A.E., B.T., E.A. and S.F. are employees of Olink AB commercializing the Proximity Extension Assay.
Supplementary Material
REFERENCES
- 1.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 2.Gullberg M, Gustafsdottir SM, Schallmeiner E, Jarvius J, Bjarnegard M, Betsholtz C, Landegren U, Fredriksson S. Cytokine detection by antibody-based proximity ligation. Proc. Natl Acad. Sci. USA. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lundberg M, Thorsen SB, Assarsson E, Villablanca A, Tran B, Gee N, Knowles M, Nielsen BS, Couto EG, Martin R, et al. Multiplexed homogeneous proximity ligation assays for high throughput protein biomarker research in serological material. Mol. Cell. Proteomics. 2011;10:M110.004978. doi: 10.1074/mcp.M110.004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Giusto DA, Wlassoff WA, Gooding JJ, Messerle BA, King GC. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005;33:e64. doi: 10.1093/nar/gni063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGregor LM, Gorin DJ, Dumelin CE, Liu DR. Interaction-dependent PCR: identification of ligand-target pairs from libraries of ligands and libraries of targets in a single solution-phase experiment. J. Am. Chem. Soc. 2010;132:15522–15524. doi: 10.1021/ja107677q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Wang L, Wang K, Tan W, Tang H, Meng X, Guo Q. Novel protein detection method based on proximitydependent polymerase reaction and aptamers. Chinese Sci. Bull. 2008;53:204–208. [Google Scholar]
- 7.Flodin P, Gelotte B, Porath J. A method for concentrating solutes of high molecular weight. Nature. 1960;188:493–494. doi: 10.1038/188493a0. [DOI] [PubMed] [Google Scholar]
- 8.Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 9.Kaboev OK, Luchkina LA, Tret'iakov AN, Bahrmand AR. PCR hot start using primers with the structure of molecular beacons (hairpin-like structure) Nucleic Acids Res. 2000;28:E94. doi: 10.1093/nar/28.21.e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinsberg J, Dembinski J, Dorn C, Behrendt D, Bartmann P, van Der Ven H. Determination of total interleukin-8 in whole blood after cell lysis. Clin. Chem. 2000;46:1387–1394. [PubMed] [Google Scholar]
- 11.Chiaretti A, Rendeli C, Antonelli A, Barone G, Focarelli B, Tabacco F, Massimi L, Ausili E. GDNF plasma levels in spina bifida: correlation with severity of spinal damage and motor function. J. Neurotrauma. 2008;25:1477–1481. doi: 10.1089/neu.2008.0638. [DOI] [PubMed] [Google Scholar]
- 12.Fredriksson S, Horecka J, Brustugun OT, Schlingemann J, Koong AC, Tibshirani R, Davis RW. Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin. Chem. 2008;54:582–589. doi: 10.1373/clinchem.2007.093195. [DOI] [PubMed] [Google Scholar]
- 13.Chang ST, Zahn JM, Horecka J, Kunz PL, Ford JM, Fisher GA, Le QT, Chang DT, Ji H, Koong AC. Identification of a biomarker panel using a multiplex proximity ligation assay improves accuracy of pancreatic cancer diagnosis. J. Transl. Med. 2009;7:105. doi: 10.1186/1479-5876-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.