Abstract
Familial Adenomatous Polyposis (FAP [OMIM 175100]) is an autosomal dominant colorectal cancer predisposition syndrome characterized by hundreds to thousands of colonic polyps and, if untreated by a combination of screening and/or surgical intervention, a ~99% lifetime risk of colorectal cancer. A subset of FAP patients develop an attenuated form of the condition characterized by lower numbers of colonic polyps (highly variable, but generally less than 100) and a lower lifetime risk of colorectal cancer, on the order of 70%. We report the diagnosis of three attenuated FAP families due to a 1.4-kb deletion within intron 14 of the APC, originally reported clinically as a variant of unknown significance (VUS). Sequence analysis suggests that this arose through an Alu-mediated recombination event with sequences from chromosome 6q22.1. This mutation tracks to members who presented with an attenuated FAP phenotype, with variable age of onset and severity. Sequence analysis of mRNA revealed an increase in the level of aberrant splicing of exon 14, resulting in the generation of an exon13-exon15 splice-form that is predicted to lead to a frameshift and protein truncation at codon 673. The relatively mild phenotypic presentation and the intra-familial variation are consistent with the leaky nature of exon 14 splicing in normal APC. The inferred founder of these three families may account for as-yet undetected affected branches of this kindred. This and similar types of intronic mutations may account for a significant proportion of FAP cases where APC clinical analysis fails because of the current limitations of testing options.
Keywords: Familial adenomatous polyposis (FAP), variant of unknown significance (VUS), RNA splicing, Colon cancer, Founder mutation
INTRODUCTION
Familial Adenomatous Polyposis (FAP; [MIM 175100]) is an autosomal dominant colorectal cancer predisposition syndrome characterized by hundreds to thousands of colonic polyps of the colonic epithelium. It has an average age of onset between the late teens and the early thirties, depending on the site of the mutation, and, in the absence of surgical intervention, a median age of colon cancer of 38 years (Cao, et al., 2006; Friedl, et al., 2001; Petersen, et al., 1991). Affected individuals also bear increased risk for extra-colonic malignancies including gastric, duodenal, pancreatic, thyroid, and CNS cancers (Elkharwily and Gottlieb, 2008; Galiatsatos and Foulkes, 2006; Knudsen, et al., 2003; Nieuwenhuis and Vasen, 2007). FAP is primarily caused by a constitutional mutation of the adenomatous polyposis coli (APC) gene. A subset of patients have a clinically attenuated presentation and course of disease progression, including lower numbers of polyps, generally less than 100 (Leppert, et al., 1990; Lynch, et al., 1993; Lynch, et al., 1990; Lynch, et al., 1992) and a lower lifetime risk of colon cancer (~70% in the absence of therapeutic intervention (Burt, et al., 2004)). In this report, a family with an attenuated FAP (AFAP) phenotype was investigated for the possible causality of a complex deletion within intron 14 of APC. The deletion was originally detected by clinical genetic testing of the proband using Southern blot analysis, and reported as a variant of unknown significance (“del portion intron 14”). Analysis of the mRNA demonstrated the association of the variant with reduced normal splicing of exon 14. Subsequently, two additional families with intron 14 deletions were referred to the study, and found to share the identical mutation, raising the possibility that this is a previously unrecognized founder mutation accounting for a large number of unidentified AFAP patients, among descendants dating back ~6 generations.
METHODS
This study was approved by the Institutional Review Board of the University of Utah. Informed consent was obtained from all research participants.
Research Subjects
The three families (K7652, K495, and K485) were referred through research studies and medical centers in Missouri, Texas, and California. The proband of the original family (K7652) under investigation presented for genetic counseling and testing. She reported that she had a clinical diagnosis of AFAP, had a right hemi-colectomy at the age of 44 and underwent a further partial colectomy, at the age of 66. Cancer history in extended family was reported by the proband and her brother, and confirmed when possible by other enrolled family members and available medical records.
Genetic testing
Genomic DNA for research was extracted from fresh blood using Puregene® DNA Purification Kit, Gentra Systems (Minneapolis, Minnesota) according to manufacturer’s protocols. PCR amplification of genomic DNA (spanning nucleotides c.1862 to c.1959-529 according to established nomenclature guidelines ((den Dunnen and Antonarakis, 2000); as recently updated at http://www.hgvs.org/mutnomen/recs-DNA.html), was performed using primers, APC exon 14 Forward, (5’-CTTACCGGAGCCAGACAAAC-3’) and APC intron 14 Reverse, (5’-GCCATCCTTCTTACGAGTGC-3’). Standard PCR conditions were followed, using PrimeStar Enzyme (Takara Bio Inc. Otsu, Shiga, Japan) with a PCR cycle of 98°C for 10 seconds, 60°C for 5 seconds and 72°C for 2 minutes. PCR products were gel-purified using Qiagen QiaQuick® PCR Purification Kit #28104 (Valencia, CA) according to the manufacturer’s protocols. Sequence reactions were performed in both directions on gel-purified PCR products using APC exon 14 Forward and APC intron 14 Reverse primers. Reaction products were run on Applied Biosystems 3730XL capillary sequencer.
Analysis of mRNA
RNA was extracted from EBV-transformed lymphoblasts using TRIZOL® reagent according to manufacturer’s protocol (Invitrogen Carlsbad, California). cDNA was generated using the random hexamer protocol of SuperScript™ III First-StrandKit # 18080-051 (Invitrogen Carlsbad, California) and PCR products were generated from the APC cDNA from coding nucleotide 1688 to 2099 using primers, APC 13 Forward, (5’-TGCGAGAAGTTGGAAGTGTG-3’) and APC 15 Reverse, (5’-TCCCATAATGCTTCCTGGTC-3’). Standard PCR conditions were followed, using 2.5 mM MgCl2 and an annealing temperature of 58°C. Sequence reactions were performed in both directions on gel-purified PCR products using APC 13 Forward and APC 15 Reverse primers. Reaction products were run on Applied Biosystems 3730XL capillary sequencer. Sequence traces were compiled and aligned using Sequencher™ DNA Sequence Assembly Software, version 4.8, Build 3767 (Gene Codes Corporation, Ann Arbor, Michigan).
Haplotype analysis
Genomic DNA was genotyped by PCR amplification at 4 short tandem repeat (STR) loci; D5S2027, D5S2501, D5S346 and D5S421, which span the APC locus (http://genome.ucsc.edu) using published primer and reaction conditions (Schwab, et al., 2008). Reaction products were run on Applied Biosystems 3730XL capillary sequencer. Allele frequencies used to calculate the probability of inheriting the specific haplotype were based on those reported by the CEPH genotype database browser V2.1 http://www.cephb.fr/en/cephdb/search.php, derived from a minimum of 27 unrelated Caucasian individuals from 8 kindreds (1992).
RESULTS
Clinical Presentation
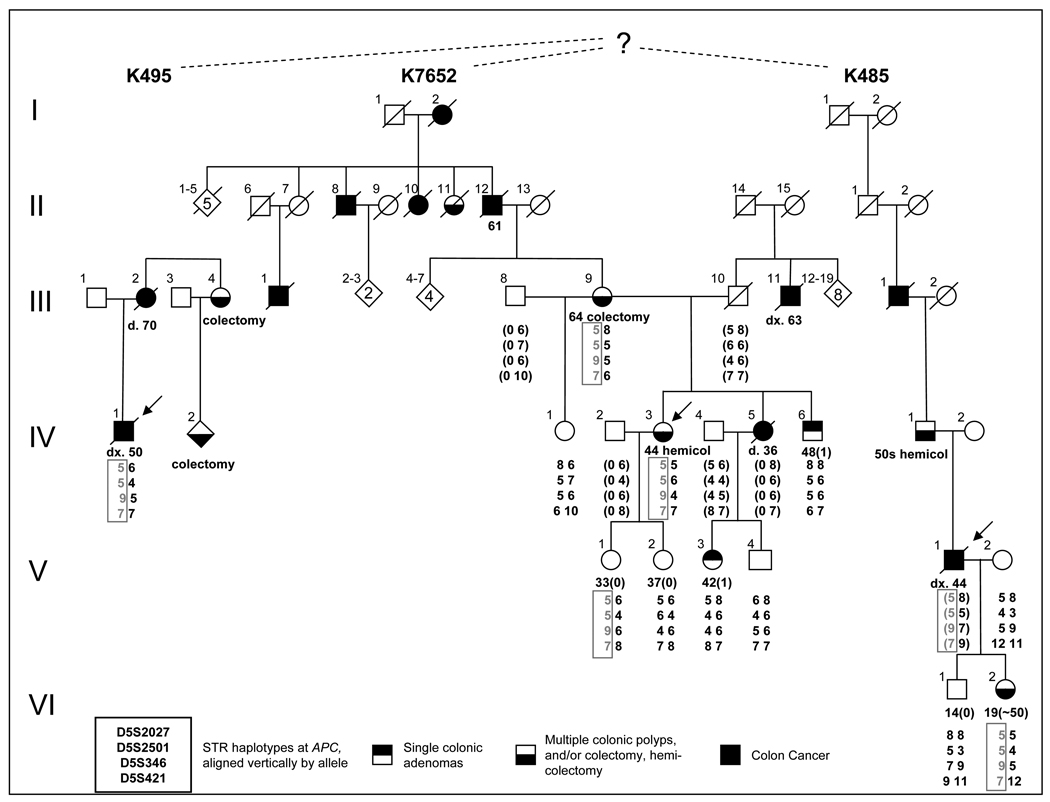
The phenotypic information collected on the available family members of each kindred is detailed in Table 1 and in the pedigree in Figure 1, including cancer, the presence or absence of adenomatous polyps, surgeries, and the age at the time of observation.
Table 1.
Summary of clinical phenotypes of affected individuals in K485, K495 and K7652, where reported. Self- and relative-reported information is listed under "Diagnosis" and "Diagnosis age", (with cancer diagnoses in UPPER case lettering) and results from verified medical records are repored in "Procedure" and "Procedure age" columns.
| Kindred | Pedigree location |
Diagnosis (self, or relative report) |
Diagnosi s age (if known) |
Procedure type (confirmed by medical records) |
Procedur e age |
Polyp number |
|---|---|---|---|---|---|---|
| K485 | III-1 | COLON, NOS | ||||
| K485 | IV-1 | Hemicolectomy | ||||
| K485 | V-1 | COLON, NOS | 44 | R Hemicolectomy | 44 | 51–100 |
| K485 | V-1 | ASCENDING COLON | 44 | R Hemicolectomy | 44 | 51–100 |
| K485 | V-1 | LIVER | 44 | |||
| K485 | VI-1 | Colonoscopy | 14 | 0 | ||
| K485 | VI-2 | EGD (Endview) | 16 | 0 | ||
| K485 | VI-2 | EGD (Endview) | 18 | 0 | ||
| K485 | VI-2 | EGD (Endview) | 20 | 0 | ||
| K485 | VI-2 | Colon Polyps | Colonoscopy | 16 | 1 | |
| K485 | VI-2 | Colon Polyps | Colonoscopy | 18 | 11–20 | |
| K485 | VI-2 | Colon Polyps | Colonoscopy | 19 | 21–50 | |
| K485 | VI-2 | Colon Polyps | Colonoscopy | 20 | 11–20 | |
| K485 | VI-2 | Capsule Endoscopy | 19 | 0 | ||
| K495 | III-2 | COLON, NOS | 70 | |||
| K495 | III-4 | Colon Polyps | ||||
| K495 | IV-1 | ASCENDING COLON | 50 | Hosp Summary sheet | ||
| K495 | IV-2 | Colon Polyps | ||||
| K7652 | I-2 | COLON, NOS | 65 | |||
| K7652 | II-8 | COLON, NOS | ||||
| K7652 | II-10 | COLON, NOS | ||||
| K7652 | II-11 | BREAST, NOS | ||||
| K7652 | II-11 | Colon Polyps | ||||
| K7652 | II-12 | COLON, NOS | 61 | |||
| K7652 | II-13 | GALLBLADDER | 71 | |||
| K7652 | II-13 | UNKNOWN | 75 | |||
| K7652 | II-14 | PROSTATE GLAND | 70 | |||
| K7652 | III-1 | COLON, NOS | ||||
| K7652 | III-9 | Colon Polyps | Subtotal Colectomy | 64 | unknown | |
| K7652 | III-9 | Flex Sigmoidoscopy | 75 | 0 | ||
| K7652 | III-9 | Flex Sigmoidoscopy | 78 | 0 | ||
| K7652 | III-9 | EGD (Endview) | 86 | 0 | ||
| K7652 | III-9 | Flex Sigmoidoscopy | 86 | 0 | ||
| K7652 | III-11 | COLON, NOS | 63 | |||
| K7652 | III-11 | LUNG, NOS | ||||
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 43 | 1 | |
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 44 | 11–20 | |
| K7652 | IV-3 | R Hemicolectomy | 44 | 0 | ||
| K7652 | IV-3 | Colonoscopy | 45 | 0 | ||
| K7652 | IV-3 | Colonoscopy | 48 | 0 | ||
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 51 | 5 | |
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 53 | 4 | |
| K7652 | IV-3 | Small Bowel Series | 53 | 0 | ||
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 54 | 1 | |
| K7652 | IV-3 | Colonoscopy | 55 | 0 | ||
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 56 | 3 | |
| K7652 | IV-3 | Colonoscopy | 58 | 0 | ||
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 61 | several | |
| K7652 | IV-3 | Colon Polyps | Colonoscopy | 63 | 1 | |
| K7652 | IV-3 | Colonoscopy | Colonoscopy | 65 | several | |
| K7652 | IV-3 | Colonoscopy | Colonoscopy | 65 | numerous | |
| K7652 | IV-3 | EGD (Endview) | 65 | 0 | ||
| K7652 | IV-3 | Colon Polyps | Partial Colectomy | 66 | numerous | |
| K7652 | IV-5 | COLON, NOS | 32 | |||
| K7652 | IV-5 | CECUM | 36 | Hosp Summary sheet | 36 | |
| K7652 | IV-6 | Single adenoma | Colonoscopy | 48 | 1 | |
| K7652 | IV-6 | Colonoscopy | 59 | 0 | ||
| K7652 | V-1 | Colonoscopy | 33 | 0 | ||
| K7652 | V-2 | Colonoscopy | 32 | 0 | ||
| K7652 | V-3 | Single adenoma | Colonoscopy | 42 | 2 | |
Figure 1. Family pedigrees.
Pedigrees of three kindreds, K495, K7652 and K485, presumed to be related by a common ancestor, showing AFAP clinical diagnoses. Haplotype markers are presented as 4X2 arrays, using allele numbers based on the number of repeats observed for each allele in the CEPH families for the four markers D5S2027, D5S2501, D5S346 and D5S421, which span the APC locus (http://genome.ucsc.edu). Where possible, parental haplotypes inferred from the available data are presented in parentheses. The intron 14-deletion-associated haplotype is presented in grey, and boxed, for ease of identification. Colonic phenotypes, including both single adenomas and multiple polyps detected on colonoscopy, reported colon cancers and surgeries and age at diagnosis are noted where known.
The proband of K7652 (IV-3) had more than 36 polyps detected by successive colonoscopies at regular intervals, including those noted at her hemi-colectomy at age 44. She underwent an additional partial colectomy at the age of 66 at which time numerous adenomas were noted. Her mother (III-9) was reported to have had several polyps at the time of her subtotal colectomy at the age of 64, but medical records have since been destroyed. Two maternal aunts and two maternal uncles of the proband were reported to have had colon cancer and/or polyps; however medical records were not available. The proband’s brother, IV-6, had one adenoma at the age of 48, and none at the age of 66, as verified by medical records. Oral family history included reports that the proband’s maternal grandfather, several of his siblings and his mother had colon cancer; however no medical records are available. The proband’s sister, IV-5 was reported and confirmed to have died from complications of colon cancer at the age of 36. This sister’s daughter (V-3) had a colonoscopy at the age of 42 which showed the presence of a single small rectal tubular adenoma (<5 mm). Other enrolled individuals on whom medical records were available were confirmed to be adenoma-free at the ages noted.
Limited family history was available for the proband of K495, and no expansion was possible. The proband, previously reported as MDA046 (Su, et al., 2000), was diagnosed with colon cancer at 50 and died at 52, reported a mother who died of colon cancer at 70, and a maternal family history of one aunt and one cousin who were each reported to have had colectomies. No confirming records were available.
The proband of K485 was diagnosed with adenocarcinoma, confirmed by pathology reports at the age of 44, and died at 45. Family history included a father who underwent a hemi-colectomy in his 50s and a paternal grandfather who died of colon cancer and who was reported at autopsy to have had multiple polyps and metastasized abdominal cancer at the time of death at the age of 60. The proband’s daughter was confirmed by pathology records to have had multiple tubular adenomas by successive colonoscopies by age 19.
Germline genetic changes
The original indication for the research study was the variant, determined by Southern blot analysis of the APC gene from peripheral blood, initially reported as a deletion of a portion of intron 14, using the commercially available COLARIS AP® genetic test (Myriad Genetic Laboratories, Inc.). Full APC sequence analysis, as well as MUTYH testing for the two common mutations (Y179C; c.536A>G and G396D; c.1187G>A) was included in the test; no mutations were found.
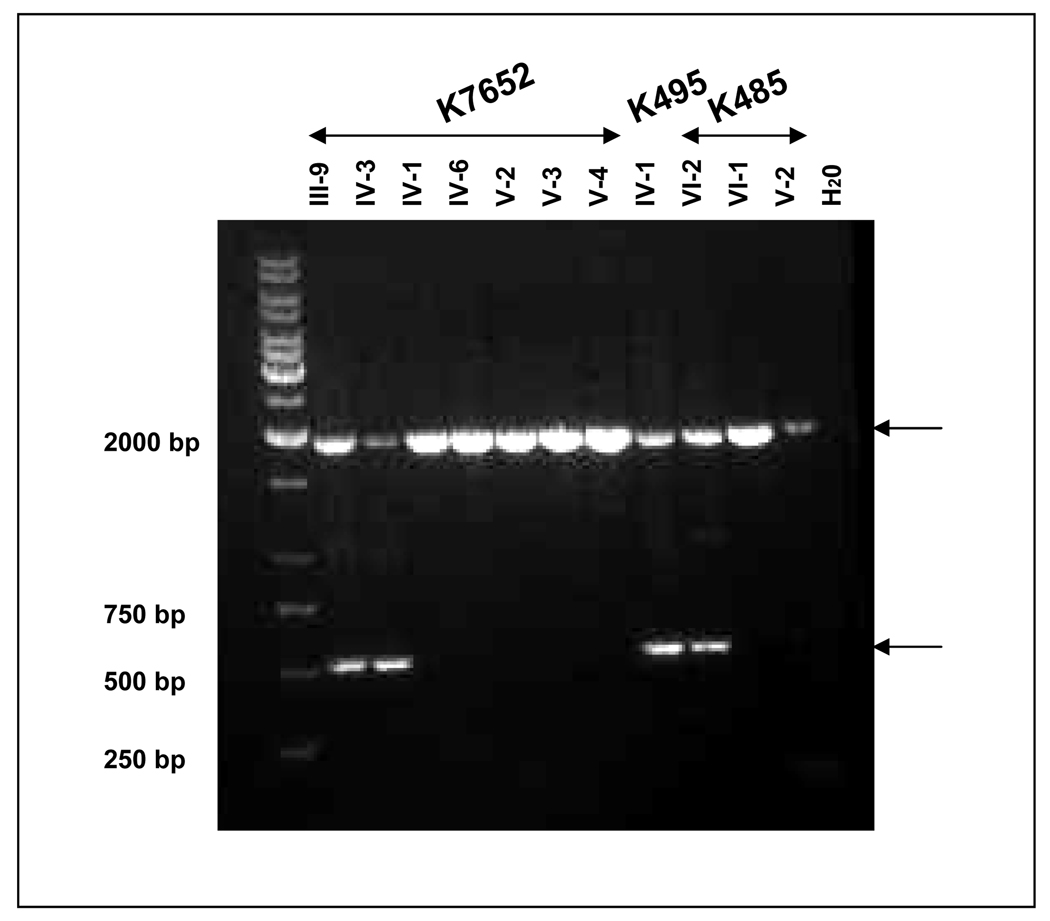
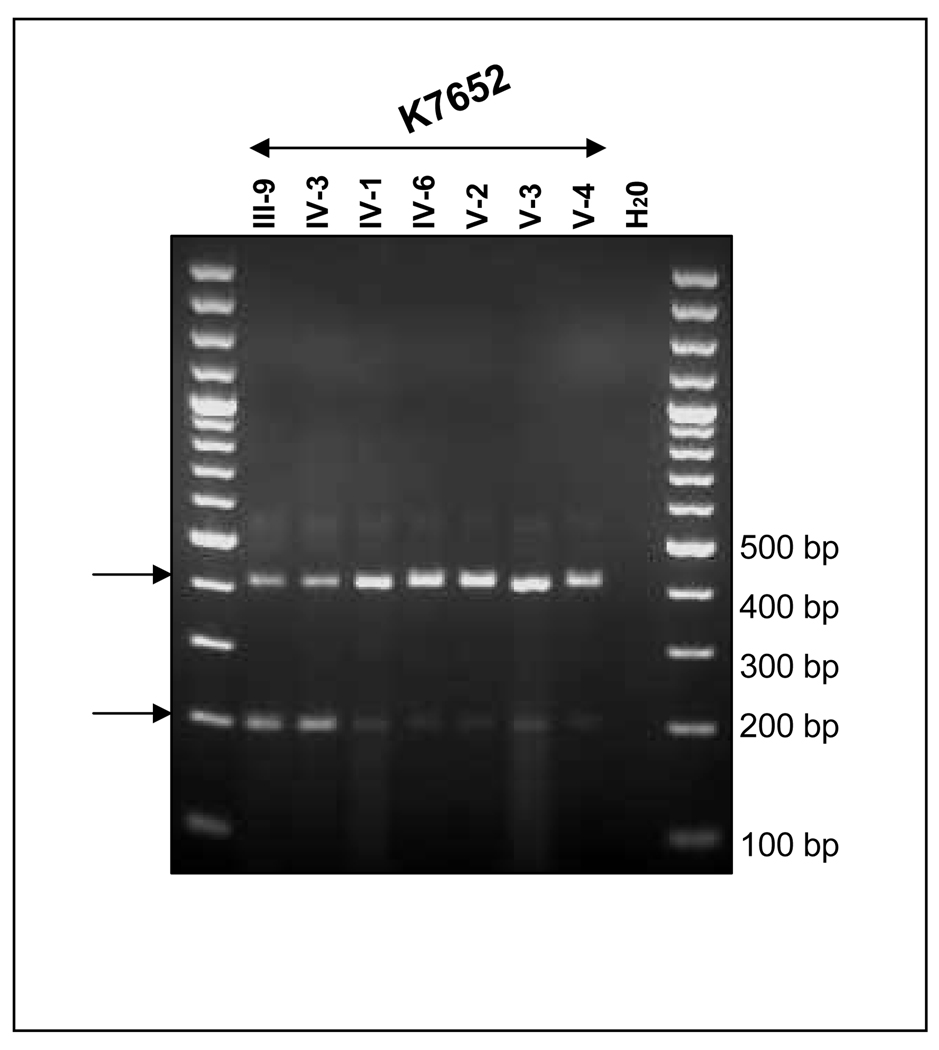
Under a research protocol, genetic testing for the deletion was performed on blood samples from the proband, IV-3, her mother, III-9, her unaffected siblings, IV-1, and IV-6, as well as her daughters V-1 and V-2, and a niece V-3, and nephew V-4, through her deceased affected sibling IV-5. PCR amplification from exon 14 to the distal portion of intron 14 resulted in the predicted fragment of 2 kb in unrelated and unaffected related controls (Figure 2A). The deletion identified in the affected proband through clinical genetic testing was verified to be approximately 1.4 kb, resulting in a 0.5-kb product. Sequence analysis of this 0.5-kb product confirmed that the deletion was entirely within intron 14; c.1958+28_1958+1660 (Figure 2B).
Figure 2. PCR and sequence amplification of genomic DNA.
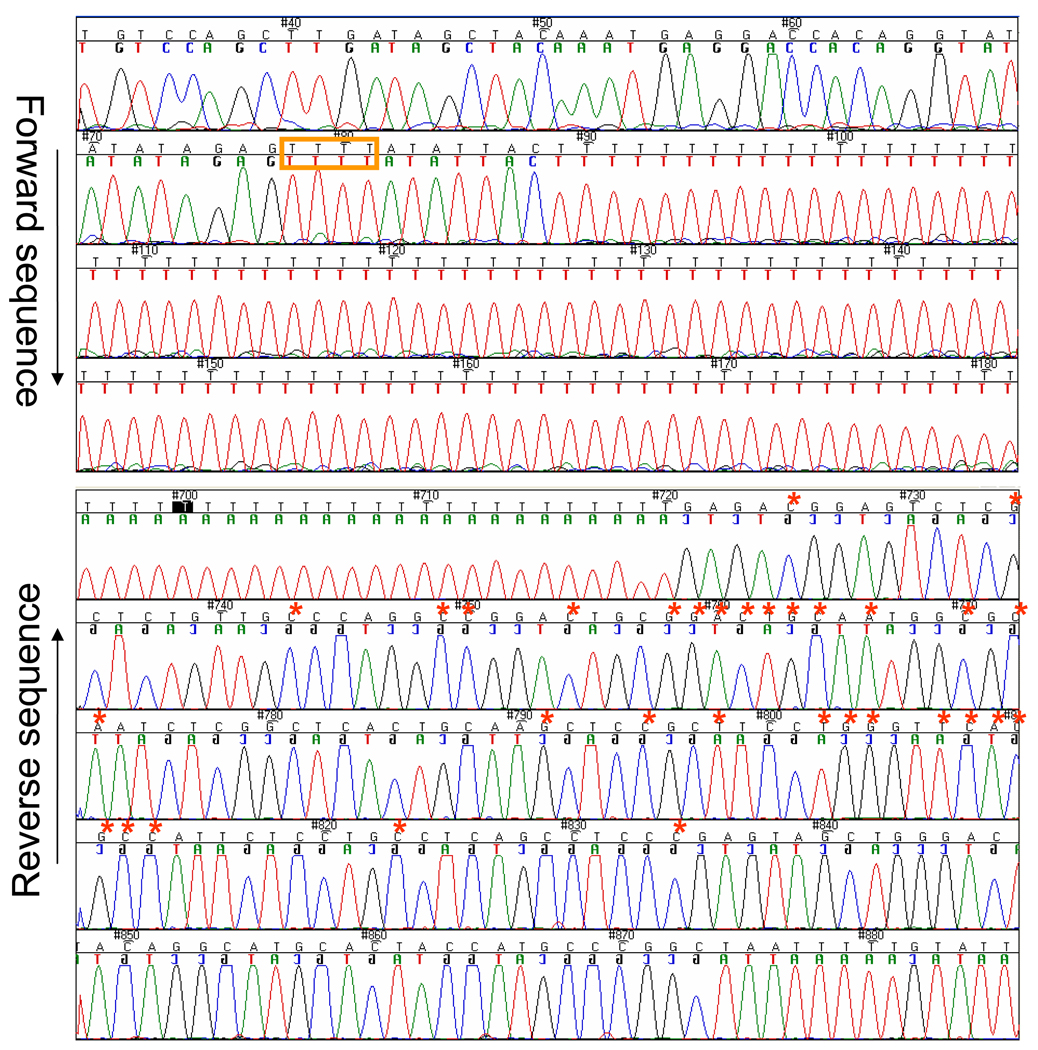
PCR analysis of genomic DNA from proband and related family members, as noted from each kindred, showing the normal 1936-bp allele, as well as the 534-bp allele detected in affected family members, carrying the shared haplotype. Samples are marked according to the position of the participant on the pedigree in Figure 1; unaffected; H2O: no template control. B. Sequence of mutant allele. The upper panel shows the proximal portion of the rearranged allele (forward strand) and the lower panel, the distal portion (reverse strand). The upstream run of Ts depicted in Figure 3A is marked with a tan-colored box. The nucleotides that diverge from APC intron 14 sequence and share identity with the repetitive element on chromosome 6q22.1 are marked by red asterisks.
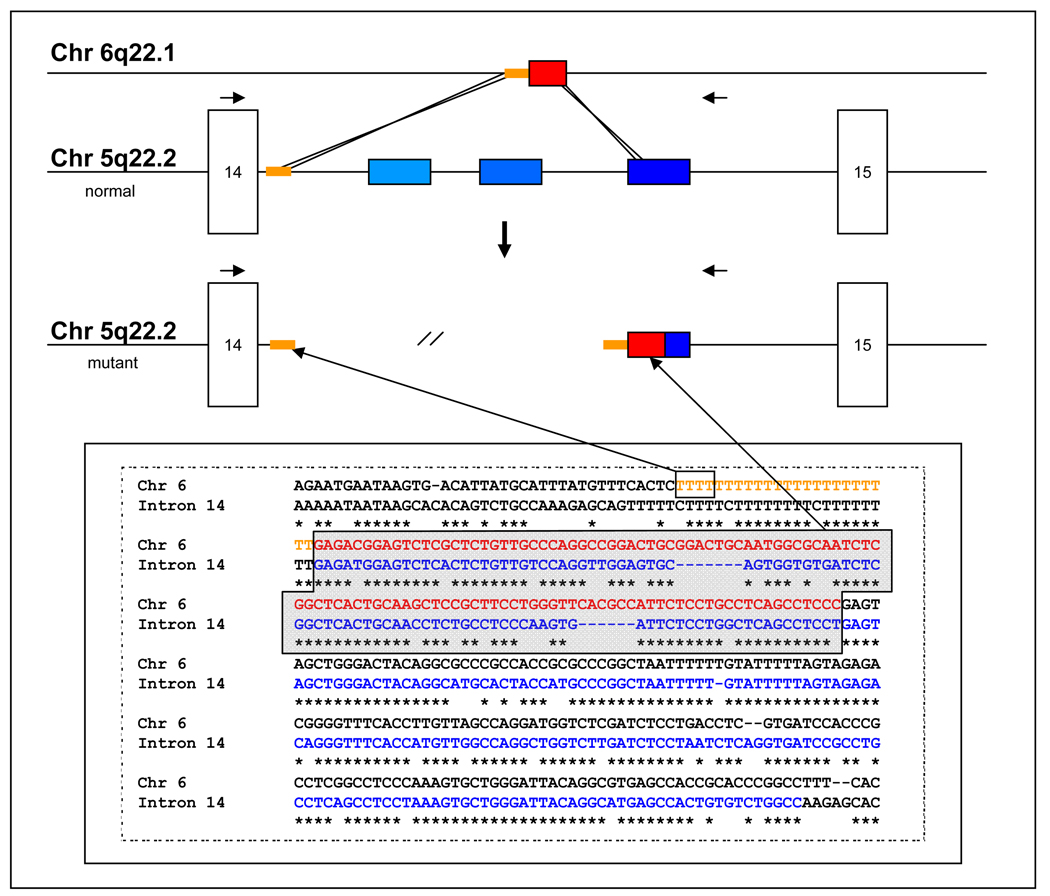
Direct sequence analysis revealed a complex insertion-deletion event, resulting from the exchange of an Alu-repetitive element-derived ~200-bp insertion originating from chromosome 6q22.1, for a 1633-bp deletion within APC (Figure 3). The insertion shared homology at the proximal end with the proximal end of the deleted sequence and at the distal end with the distal end of the deletion. This insertion contained a 114-bp portion of an Alu-containing repetitive element sequence with 100% identity to an Alu-containing repetitive element located on chromosome 6, preceded by a 87-107-bp run of Ts. The long runs of Ts that resulted from the recombination event represent a technical challenge for the polymerizing and sequencing enzymes to replicate, resulting in slippage that prevents accurate sequence reads downstream of the recombination.
Figure 3. Schematic representation of proposed recombination event leading to APC intron 14 deletion.
A. Schematic representation of recombination between Alu-containing repetitive element located on chromosome 6q22.1, which shares 100% identity with the insertion, and the APC locus on chromosome 5q22.1. The recombination appears to have been mediated on the proximal side by a run of T residues, and on the distal side by homology between the repetitive element on 6q22.1 and the third of three repetitive elements located in intron 14 of APC.
The results showed that the proband of K7652 and her clinically affected mother carried the insertion-deletion, while three clinically unaffected first-degree relatives did not. Analysis of the proband of K495 and daughter of the K485 proband also carried the identical insertion-deletion, while unaffected K485 family members did not (Figure 2A).
mRNA analysis
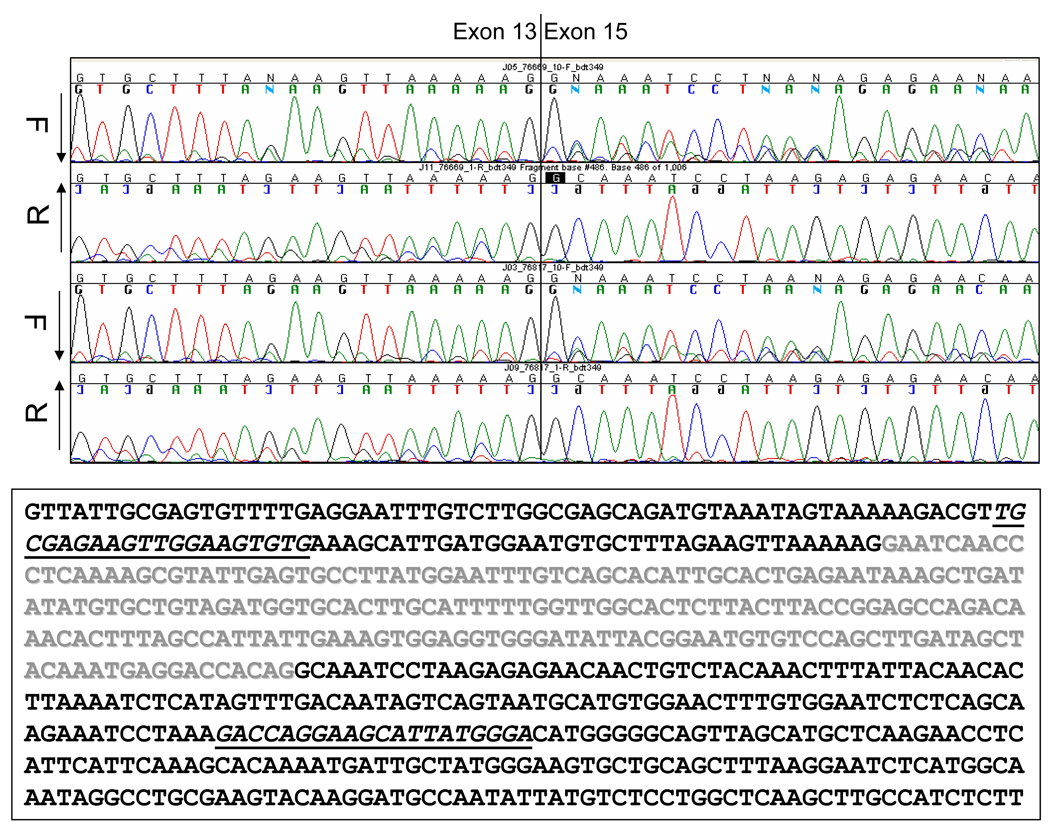
To address the possible molecular consequence of the intronic deletion on mRNA splicing, PCR was performed on cDNA derived from mRNA from available individuals in K7652 and K485. Unrelated as well as clinically unaffected family members without the deletion in question served as negative controls. PCR amplification from exon 13 to exon 15 resulted in a predicted product of 412 base-pairs in all samples (Figure 4A). The same reaction also revealed a 197-base-pair product in the K7652 proband and her affected mother, in addition to the normal-sized fragment of 412 base-pairs also found in unaffected family members. Similar analysis of mRNA from the children of the K485 proband revealed identical results (not shown). Sequence analysis of the shorter product revealed a discrete deletion of exon 14 (Figure 4B). Consistent with previous reports, the shorter fragment was also identified at low levels in the unrelated and unaffected controls representing a minor alternative splice product (Bala, et al., 1996; Bala, et al., 1997; Montera, et al., 2001). Deletion of exon 14 results in a predicted translational frameshift and protein truncation at codon 673 of the normal open reading frame, approximately 20 codons into exon 15. This results in a truncated product that lacks all of the β-catenin, microtubule, and EB-1 binding domains.
Figure 4. Discrete deletion of Exon 14 in mRNA from mutation carriers.
(A) Results of RT-PCR analysis of mRNA from transformed lymphocytes; Amplification of transcripts encompassing exons 13–15. Unaffected individuals reveal a predominant fragment of 412 bp, and a trace of a fragment of 197 bp, while affected individuals reveal each fragment in approximately equal proportions. Samples are marked according to the position of the participant on the pedigree in Figure 1. (B) Sequence analysis, demonstrating absence of exon 14 in predominant splice-from, as indicated by vertical bar drawn through sequence trace. Forward and Reverse traces are shown for the K7652 proband and her mother. Printed sequence shows normal exon 13–15 sequence, with exon 14 colored in grey with experimental primers underlined.
Haplotype analysis
Haplotyping was used to determine the likelihood that the K495 proband, originally identified by Su et al. and K485 proband shared identity by descent at the APC locus with K7652. The disease-associated haplotype is shown in Figure 1 as alleles 5 (D5S2027), 5 (D5S2501), 9 (D5S346), and 7 (D5S421). By using the published CEPH allele frequencies of STR alleles at the four loci used, the population frequency of the deletion-carrying haplotype in the family was calculated to be 1 in 283 (giving an effective p-value of 0.0035). Both the K495 and K485 affected family members were found to share the mutation-associated haplotype of the K7652 proband, suggesting that all three kindreds descend from a common founder (Figure 1). The precise relationship was not identified but oral family histories and genealogic data from familysearch.org (www.familysearch.org) revealed that founders of K7652 and K485 were born and raised in Randolph County, IL in the 1830s supporting a common ancestral link.
DISCUSSION
We describe a family where an attenuated FAP (AFAP) phenotype is transmitted through several generations in which an APC allele, with a large intronic insertion-deletion, tracks with the phenotype (Figure 1). No other disease-causing mutations were found within APC of the proband, either by southern blot or sequence analysis. RT-PCR analysis reveals significantly enhanced prevalence of a transcript corresponding by sequence analysis to a discrete deletion of exon 14 in individuals inheriting the intronic deletion. We conclude that this deletion is the causative genetic determinant of AFAP in this family. A similar isolated case was noted previously in which an apparently identical deletion within intron 14, (described as 27–1627 base-pairs downstream of exon 14) was shown to be associated with colonic polyposis (Su, et al., 2000). This same sequence was confirmed in our kindred. We determined the breakpoint to be ≥ 21 bp downstream of this reported case, based on two further nucleotide differences between the inserted sequence. The remaining discrepancy was reconciled with the original normal intronic sequence by differences in the numbers of T residues detected in two runs of Ts, as reported by RefSeq RNA alignments against the human genome using blat, to generate the best consensus sequence (Kent, 2002; Pruitt, et al., 2007) and sequence alignment analysis run by ClustalW2 software (Larkin, et al., 2007). Accordingly, we use the nomenclature APC c.1958+28_1958+1660conNT_025741.14:c.19005463-19005598 to denote the gene conversion replacing nucleotides c.1958+28 to c.1958+1660 from intron 14 of APC with nucleotides 19005463-19005598 from chromosome 6q22.1 (den Dunnen and Antonarakis, 2000), as updated at http://www.hgvs.org/mutnomen/recs-DNA.html#indel. A third family, also sharing identical deletion and haplotype inheritance was also identified. Genealogical data revealed founders of K7652 and K485 share a common ancestor residing in Illinois around the 1830’s. We hypothesize that the deletion was originally mediated by imperfect internal Alu-mediated recombination within intron 14, similar to that reported by Cao et al (Cao, et al., 2001), in their analysis of a ~6-kb deletion that was shown to flank exon 14 of APC. There are three such repeats located within intron 14, and the proximal deletion boundary coincides with the proximal boundary of the proximal Tn-containing Alu-containing repeated element from chromosome 6 (Figure 3). The distal repeat, which also includes an Alu element, was replaced in the recombined allele with the highly related, but not identical chromosome 6 repeat. The long runs of Ts that resulted from the original recombination not only represent a technical challenge for the polymerizing and sequencing enzymes to replicate, resulting in slippage that prevents accurate sequence reads downstream of the recombination; but may also contribute to an expansion repeat element within the newly generated allele that is subject to expansion with successive generations and may contribute to the phenotype by influencing the spatial relationships of local splicing signals.
Splice signals
The mutation identified is unusual in that it is entirely intronic and relatively distant from known acceptor or donor splice signals. The intron 14 deletion ablates consensus signals for two splicing proteins, SRp40 and SC35 (Cartegni, et al., 2003; Smith, et al., 2006). Although alternative signals can be found in the sequence that remains, they are further from exon 14, weaker in consensus strength and oriented differently with respect to each other, possibly altering the efficiency with which they individually or jointly contribute to local splicing pathways.
Phenotype
Several cases of deleterious mutations in exon 14 have been reported, and include both FAP and attenuated FAP patients and families (De Rosa, et al., 2003; Enomoto, et al., 2000; Ficari, et al., 2000; Montera, et al., 2001; Scarano, et al., 1997; Won, et al., 1999). The classic FAP phenotypic cases appear to preclude normal translation of full-length APC, by the generation of a frameshift or stop codon, or by ablating normal splicing (Goncalves, et al., 2009) while the attenuated cases tend to be splice variants that would be predicted to change the balance of translated splice forms. The mutation described here results in a predominant exon13-exon15 splice form that is predicted to lead to a translational frameshift and protein truncation product at what would normally be codon 673 in the APC transcript. It is clear from our work and that of others’, that alternative splicing of exon 14 is a normal low-level event in colonic epithelium as well as in peripheral blood lymphocytes (Aretz, et al., 2004; Bala, et al., 1997; Venesio, et al., 2007). Some reports have suggested a possible functional role for such alternate splice forms in normal regulation of gene expression, perhaps by modulating total transcript or full length protein levels (Sulekova, et al., 1995).
This low-level alternate transcript generates an out-of-frame exon 13-exon 15 fusion. Other examples of attenuated FAP kindreds have been reported in which the penetrance is variable, including one with a 2-bp deletion in exon 4 (Burt, et al., 2004; Neklason, et al., 2008), in which 15% of genetically confirmed cases have ≤ 2 polyps at the time of baseline colonoscopy (ages 19–67 years; unpublished results). This may explain the normal phenotype of V-1 in K7652 at the age of 33. At the other end of the spectrum, a number of demonstrated or inferred deletion carriers died at young ages of colon cancer; V-1, in K485 at the age of 44, IV-1, in K495 at the age of 50 and IV-5 in K7652, who most likely inherited her mother’s deletion, at the age of 36. However, this is also consistent with the reported lifetime risk of colon cancer in AFAP kindreds of 70% – 80% (Burt, 2007; Burt, et al., 2004; Hernegger, et al., 2002; Knudsen, et al., 2003). Independently of the familial predisposition, the background population risk confounds the risk assessment of this allele; three individuals who do not share the deletion or the haplotype, III-11, IV-6 and V-3, were reported to have colon cancer at 63 years, a single adenoma of the ascending colon at 48 years and a single adenoma of the rectum at 42 years, respectively.
VUS reclassification
Reclassification of variants of unknown significance (VUS) as either deleterious or benign is an important process for good medical management. VUS present challenges at several levels. For patients and their families, they are particularly frustrating and often difficult to understand (Frost, et al., 2004). For genetic counselors and physicians, they can represent a failure; they neither provide an answer for future management choices, nor rule one option out, justifying proceeding in a different direction for answers. For basic researchers, they challenge the status quo; the paradigm within which mutations can be rationally classified into “deleterious” vs. “benign”; variants along the continuum of human genetic variation. The proportions of genetic changes classified as VUS varies from one gene to another, depending on the mutational spectrum and function of the encoded protein. Nevertheless, although it is becoming more clear that some alleles may defy the binary classification, depending on other co-factors to co-determine their penetrance, it remains a core feature of genetic services that when the bar of technological challenges is lowered and these VUS can be properly classified, satisfying answers may be provided to the families and care providers. Previous reports have clarified the contributions of APC exonic and splice-junction variants to disease (Aretz, et al., 2004; Goncalves, et al., 2009; Kaufmann, et al., 2009).
The mechanism of mutation in the kindreds examined in this study appears to fall into a small but growing class of rearrangement mutations in colon cancer predisposition genes (Halling, et al., 1999; McCart, et al., 2006; Nystrom-Lahti, et al., 1995; Yanaba, et al., 2008). Nevertheless, and despite its apparent nature as a founder mutation, several CLIA-certified genetic testing laboratories that offer genetic testing for APC mutations reported not having found this mutation in their records (Baylor College of Medicine Medical Genetics Laboratories, Mayo Clinic Molecular Genetics Laboratory; University of Pennsylvania School of Medicine Genetic Diagnostic Laboratory); however, with the exception of Myriad Genetic Laboratories, Inc., their testing methodologies, including the multiplex ligation-dependent probe amplification (MLPA) deletion assays, would not detect this mutation. The haplotyping and genealogical evidence suggests that the three kindreds may share a common ancestor dating back to the mid-1800s and may account for unidentified cases of inherited rather than sporadic polyposis and/or colon cancer.
This study is particularly important from both a clinical and research perspective, and illustrates the beneficial synergy between these two components of translational medicine. As for most genetic conditions, mutation detection in FAP and AFAP patients is incomplete using the technologies applied by clinical testing laboratories. Intronic mutations are largely ignored, undetected, and/or uncharacterized beyond the splice junction, but constitute an important clinical subset that would benefit from complete and accurate evaluation offered from a research platform. Detection and characterization of this specific intron 14 mutation is important in that it 1) links three previously independent cases to a common founder dating back to the 1800s and suggests the presence of many more affected descendents, 2) describes an Alu-mediated recombination that may underlie other APC intron deletions, and 3) assigns the attenuated FAP phenotype to mutations that cause exon 14 to be spliced out. In the future it will be important to evaluate the subset of FAP and AFAP patients that are mutation-negative for APC splice defects and determine what proportion may be attributed to this mechanism.
ACKNOWLEDGEMENTS
We acknowledge Tom Day for medial record data evaluation, Dholani Zarook, Ryan Turner, Brian Richardson and Debra Ma for family history data curation and pedigree construction.
Grant support: This research was supported by NCI grants P01-CA073992 (RWB), R01-CA040641 (RWB) and by the Huntsman Cancer Foundation.
Abbreviations
- FAP
Familial adenomatous polyposis
- APC
adenomatous polyposis coli
- VUS
variant of unknown significance
REFERENCES
- A comprehensive genetic linkage map of the human genome. NIH/CEPH Collaborative Mapping Group. Science. 1992;258(5079):67–86. [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Sun Y, Pagenstecher C, Mangold E, Caspari R, Moslein G, Schulmann K, Propping P, Friedl W. Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene. Hum Mutat. 2004;24(5):370–380. doi: 10.1002/humu.20087. [DOI] [PubMed] [Google Scholar]
- Bala S, Kraus C, Wijnen J, Meera Khan P, Ballhausen WG. Multiple products in the protein truncation test due to alternative splicing in the adenomatous polyposis coli (APC) gene. Hum Genet. 1996;98(5):528–533. doi: 10.1007/s004390050254. [DOI] [PubMed] [Google Scholar]
- Bala S, Sulekova Z, Ballhausen WG. Constitutive APC exon 14 skipping in early-onset familial adenomatous polyposis reveals a dramatic quantitative distortion of APC-gene specific isoforms. Hum Mutat. 1997;10(3):201–206. doi: 10.1002/(SICI)1098-1004(1997)10:3<201::AID-HUMU4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Burt R. Inheritance of Colorectal Cancer. Drug Discov Today Dis Mech. 2007;4(4):293–300. doi: 10.1016/j.ddmec.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt RW, Leppert MF, Slattery ML, Samowitz WS, Spirio LN, Kerber RA, Kuwada SK, Neklason DW, Disario JA, Lyon E, et al. Genetic testing and phenotype in a large kindred with attenuated familial adenomatous polyposis. Gastroenterology. 2004;127(2):444–451. doi: 10.1053/j.gastro.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Cao X, Eu KW, Seow-Choen F, Zhao Y, Cheah PY. Topoisomerase-I- and Alu-mediated genomic deletions of the APC gene in familial adenomatous polyposis. Hum Genet. 2001;108(5):436–442. doi: 10.1007/s004390100492. [DOI] [PubMed] [Google Scholar]
- Cao X, Hong Y, Eu KW, Loi C, Cheah PY. Singapore familial adenomatous polyposis (FAP) patients with classical adenomatous polyposis but undetectable APC mutations have accelerated cancer progression. Am J Gastroenterol. 2006;101(12):2810–2817. doi: 10.1111/j.1572-0241.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M, Scarano MI, Panariello L, Morelli G, Riegler G, Rossi GB, Tempesta A, Romano G, Renda A, Pettinato G, et al. The mutation spectrum of the APC gene in FAP patients from southern Italy: detection of known and four novel mutations. Hum Mutat. 2003;21(6):655–656. doi: 10.1002/humu.9151. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Elkharwily A, Gottlieb K. The pancreas in familial adenomatous polyposis. Jop. 2008;9(1):9–18. [PubMed] [Google Scholar]
- Enomoto M, Konishi M, Iwama T, Utsunomiya J, Sugihara KI, Miyaki M. The relationship between frequencies of extracolonic manifestations and the position of APC germline mutation in patients with familial adenomatous polyposis. Jpn J Clin Oncol. 2000;30(2):82–88. doi: 10.1093/jjco/hyd017. [DOI] [PubMed] [Google Scholar]
- Ficari F, Cama A, Valanzano R, Curia MC, Palmirotta R, Aceto G, Esposito DL, Crognale S, Lombardi A, Messerini L, et al. APC gene mutations and colorectal adenomatosis in familial adenomatous polyposis. Br J Cancer. 2000;82(2):348–353. doi: 10.1054/bjoc.1999.0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl W, Caspari R, Sengteller M, Uhlhaas S, Lamberti C, Jungck M, Kadmon M, Wolf M, Fahnenstich J, Gebert J, et al. Can APC mutation analysis contribute to therapeutic decisions in familial adenomatous polyposis? Experience from 680 FAP families. Gut. 2001;48(4):515–521. doi: 10.1136/gut.48.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost CJ, Venne V, Cunningham D, Gerritsen-McKane R. Decision making with uncertain information: learning from women in a high risk breast cancer clinic. J Genet Couns. 2004;13(3):221–236. doi: 10.1023/B:JOGC.0000027958.02383.a9. [DOI] [PubMed] [Google Scholar]
- Galiatsatos P, Foulkes WD. Familial adenomatous polyposis. Am J Gastroenterol. 2006;101(2):385–398. doi: 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Goncalves V, Theisen P, Antunes O, Medeira A, Ramos JS, Jordan P, Isidro G. A missense mutation in the APC tumor suppressor gene disrupts an ASF/SF2 splicing enhancer motif and causes pathogenic skipping of exon 14. Mutat Res. 2009;662(1–2):33–36. doi: 10.1016/j.mrfmmm.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Halling KC, Lazzaro CR, Honchel R, Bufill JA, Powell SM, Arndt CA, Lindor NM. Hereditary desmoid disease in a family with a germline Alu I repeat mutation of the APC gene. Hum Hered. 1999;49(2):97–102. doi: 10.1159/000022852. [DOI] [PubMed] [Google Scholar]
- Hernegger GS, Moore HG, Guillem JG. Attenuated familial adenomatous polyposis: an evolving and poorly understood entity. Dis Colon Rectum. 2002;45(1):127–134. doi: 10.1007/s10350-004-6127-y. discussion 134-6. [DOI] [PubMed] [Google Scholar]
- Kaufmann A, Vogt S, Uhlhaas S, Stienen D, Kurth I, Hameister H, Mangold E, Kotting J, Kaminsky E, Propping P, et al. Analysis of rare APC variants at the mRNA level: six pathogenic mutations and literature review. J Mol Diagn. 2009;11(2):131–139. doi: 10.2353/jmoldx.2009.080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen AL, Bisgaard ML, Bulow S. Attenuated familial adenomatous polyposis (AFAP). A review of the literature. Fam Cancer. 2003;2(1):43–55. doi: 10.1023/a:1023286520725. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leppert M, Burt R, Hughes JP, Samowitz W, Nakamura Y, Woodward S, Gardner E, Lalouel JM, White R. Genetic analysis of an inherited predisposition to colon cancer in a family with a variable number of adenomatous polyps. N Engl J Med. 1990;322(13):904–908. doi: 10.1056/NEJM199003293221306. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Smyrk TC, Lanspa SJ, Jenkins JX, Lynch PM, Cavalieri J, Lynch JF. Upper gastrointestinal manifestations in families with hereditary flat adenoma syndrome. Cancer. 1993;71(9):2709–2714. doi: 10.1002/1097-0142(19930501)71:9<2709::aid-cncr2820710904>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Smyrk TC, Lanspa SJ, Lynch PM, Watson P, Strayhorn PC, Bronson EK, Lynch JF, Priluck IA, Appelman HD. Phenotypic variation in colorectal adenoma/cancer expression in two families. Hereditary flat adenoma syndrome. Cancer. 1990;66(5):909–915. doi: 10.1002/1097-0142(19900901)66:5<909::aid-cncr2820660516>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Smyrk TC, Watson P, Lanspa SJ, Lynch PM, Jenkins JX, Rouse J, Cavalieri J, Howard L, Lynch J. Hereditary flat adenoma syndrome: a variant of familial adenomatous polyposis? Dis Colon Rectum. 1992;35(5):411–421. doi: 10.1007/BF02049396. [DOI] [PubMed] [Google Scholar]
- McCart A, Latchford A, Volikos E, Rowan A, Tomlinson I, Silver A. A novel exon duplication event leading to a truncating germ-line mutation of the APC gene in a familial adenomatous polyposis family. Fam Cancer. 2006;5(2):205–208. doi: 10.1007/s10689-006-7471-y. [DOI] [PubMed] [Google Scholar]
- Montera M, Piaggio F, Marchese C, Gismondi V, Stella A, Resta N, Varesco L, Guanti G, Mareni C. A silent mutation in exon 14 of the APC gene is associated with exon skipping in a FAP family. J Med Genet. 2001;38(12):863–867. doi: 10.1136/jmg.38.12.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklason DW, Stevens J, Boucher KM, Kerber RA, Matsunami N, Barlow J, Mineau G, Leppert MF, Burt RW. American founder mutation for attenuated familial adenomatous polyposis. Clin Gastroenterol Hepatol. 2008;6(1):46–52. doi: 10.1016/j.cgh.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis MH, Vasen HF. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol. 2007;61(2):153–161. doi: 10.1016/j.critrevonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Jarvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B, et al. Founding mutations and Alu mediated recombination in hereditary colon cancer. Nat Med. 1995;1(11):1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- Petersen GM, Slack J, Nakamura Y. Screening guidelines and premorbid diagnosis of familial adenomatous polyposis using linkage. Gastroenterology. 1991;100(6):1658–1664. doi: 10.1016/0016-5085(91)90666-9. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarano MI, De Rosa M, Gentile M, Bucci L, Ferulano GP, Carlomagno N, Renda A, Guanti G, Salvatore F, Izzo P. Three novel germline mutations in the adenomatous polyposis coli gene. Hum Mutat. 1997;9(2):191–193. doi: 10.1002/(SICI)1098-1004(1997)9:2<191::AID-HUMU16>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Schwab AL, Tuohy TM, Condie M, Neklason DW, Burt RW. Gonadal mosaicism and familial adenomatous polyposis. Fam Cancer. 2008;7(2):173–177. doi: 10.1007/s10689-007-9169-1. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15(16):2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Su LK, Steinbach G, Sawyer JC, Hindi M, Ward PA, Lynch PM. Genomic rearrangements of the APC tumor-suppressor gene in familial adenomatous polyposis. Hum Genet. 2000;106(1):101–107. doi: 10.1007/s004399900195. [DOI] [PubMed] [Google Scholar]
- Sulekova Z, Reina-Sanchez J, Ballhausen WG. Multiple APC messenger RNA isoforms encoding exon 15 short open reading frames are expressed in the context of a novel exon 10A-derived sequence. Int J Cancer. 1995;63(3):435–441. doi: 10.1002/ijc.2910630323. [DOI] [PubMed] [Google Scholar]
- Venesio T, Balsamo A, Sfiligoi C, Fuso L, Molatore S, Ranzani GN, Risio M. Constitutional high expression of an APC mRNA isoform in a subset of attenuated familial adenomatous polyposis patients. J Mol Med. 2007;85(3):305–312. doi: 10.1007/s00109-006-0127-4. [DOI] [PubMed] [Google Scholar]
- Won YJ, Park KJ, Kwon HJ, Lee JH, Kim JH, Kim YJ, Chun SH, Han HJ, Park JG. Germline mutations of the APC gene in Korean familial adenomatous polyposis patients. J Hum Genet. 1999;44(2):103–108. doi: 10.1007/s100380050118. [DOI] [PubMed] [Google Scholar]
- Yanaba K, Nakagawa H, Takeda Y, Koyama N, Sugano K. Muir-Torre syndrome caused by partial duplication of MSH2 gene by Alu-mediated nonhomologous recombination. Br J Dermatol. 2008;158(1):150–156. doi: 10.1111/j.1365-2133.2007.08233.x. [DOI] [PubMed] [Google Scholar]