Abstract
Vascular endothelial growth factor-2 (VEGFR-2 or KDR) is a known endothelial target also expressed in NSCLC tumor cells. We investigated the association between alterations in the KDR gene and clinical outcome in patients with resected NSCLC (n=248). KDR copy number gains (CNGs), measured by quantitative PCR and fluorescence in situ hybridization, were detected in 32% of tumors and associated with significantly higher KDR protein and higher microvessel density than tumors without CNGs. KDR CNGs were also associated with significantly increased risk of death (HR=5.16; P=0.003) in patients receiving adjuvant platinum-based chemotherapy, but no differences were observed in patients not receiving adjuvant therapy. To investigate potential mechanisms for these associations we assessed NSCLC cell lines and found that KDR CNGs were significantly associated with in vitro resistance to platinum chemotherapy as well as increased levels of nuclear HIF-1α in both NSCLC tumor specimens and cell lines. Furthermore, KDR knockdown experiments using small interfering RNA reduced platinum resistance, cell migration, and HIF-1α levels in cells bearing KDR CNGs, providing evidence for direct involvement of KDR. No KDR mutations were detected in exons 7, 11 and 21 by PCR-based sequencing; however, two variant SNP genotypes were associated with favorable overall survival in adenocarcinoma patients. Our findings suggest that tumor cell KDR CNGs may promote a more malignant phenotype including increased chemoresistance, angiogenesis, and HIF-1α levels, and that KDR CNGs may be a useful biomarker for identifying patients at high risk for recurrence after adjuvant therapy, a group that may benefit from VEGFR-2 blockade.
Introduction
Tumor growth is critically dependent on neovascularization (1). The ligand vascular endothelial growth factor (VEGF) is an endothelial cell-specific mitogen known to be a highly potent and specific mediator of angiogenesis, and has two identified tyrosine kinase receptors, VEGF receptor-1 and 2 (2–5). The VEGF receptor-2 (VEGFR-2) coded by the gene KDR (located in 4q12) is the predominant mediator of VEGF-stimulated endothelial cell functions, including cell migration, proliferation, survival, and enhancement of vascular permeability (6, 7). VEGFR-2 exhibits robust protein-tyrosine kinase activity in response to the VEGF ligand (3).
In human epithelial tumors, including lung, VEGFR-2 has shown to be expressed in malignant cells as well as in the endothelial cell of tumor vasculature (8–11). In non-small cell lung carcinoma (NSCLC), VEGFR-2 has been found to be overexpressed in malignant cells of tumor tissues, and associated with a poor outcome (8–12). The mechanism and biological impact of VEGFR-2 overexpression of NSCLC cells, however, is not known. Recent work from our group and others has demonstrated that tumor cell expression of VEGFR-1 may drive tumor cell invasiveness (13, 14) and promote hypoxia-independent upregulation of hypoxia inducible factor-1α (HIF-1αbut it is not known whether VEGFR-2 signaling directly impacts the tumor cell phenotype in NSCLC.
Recently, a relatively high frequency (9%) of mutation and amplification of KDR has been detected in lung adenocarcinoma histology (15); however, the presence of these abnormalities in squamous cell carcinomas of the lung is unknown. In addition, there is no data available on the correlation of KDR abnormalities with tumor and patients’ characteristics in lung cancer, including outcome and response to therapy.
The objective of this study was to characterize the molecular abnormalities of VEGFR-2 in epithelial malignant cells of NSCLC major histology types, adenocarcinoma and squamous cell carcinoma, and correlate with patients’ clinical characteristics. We studied KDR copy number gain (CNG), mutation, and genetic variations in malignant cells of surgically resected NSCLC tumor tissues and correlated results with pathological features in NSCLC patients’ tumors and with their platinum adjuvant treatments and outcomes. In addition, using a series of NSCLC cell lines and tissue specimens, we investigated molecular mechanisms associated with KDR CNG in resistance to platinum, particularly the potential role of HIF-1α, a key regulator of angiogenesis in malignant tumors (16).
Material and Methods
NSCLC Tumor Specimens
We obtained archived frozen and formalin-fixed and paraffin-embedded (FFPE) tissues from NSCLC patients who were surgically resected with curative intent from the Lung Cancer Specialized Program of Research Excellence (SPORE) tissue bank at The University of Texas M. D. Anderson Cancer Center (Houston, TX). The tissue banking and the study were approved by the Institutional Review Board. We randomly selected 248 NSCLC specimens (159 adenocarcinomas and 89 squamous cell carcinomas) to test KDR abnormalities. Detailed clinical and pathologic information of the cases is presented in Supplementary Table 1.
KDR Copy Number Analysis in Tumor Specimens
We utilized two methodologies to test KDR CNG in NSCLC tumor specimens: real-time quantitative PCR (qPCR) and fluorescence in situ hybridization (FISH). To enrich for malignant cell content for qPCR analysis, tumor tissues were manually microdissected from optimal cutting temperature compound-embedded frozen tissue sections for subsequent DNA extraction. Tumor DNA was extracted using Pico Pure DNA Extraction Kit (Arcturus, Mountain View, CA) according to the manufacturer’s instructions. DNA samples with proportions of microdissected tumor cell greater than 70% were qualified for qPCR analysis. KDR gene copy number was detected by qPCR using the ABI 7300 real time PCR system (Applied Biosystems, Foster City, CA). The primers used to amplify KDR were KF-GACACACCCTCAGGCTCTTG, and KR-ACTTTTCACCGCCTGTTCTC. Each PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) at 50ºC for 2 min and 95ºC for 10 min followed by 40 cycles at 95ºC for 15 s and 60ºC for 1 min. β-Actin was introduced as the endogenous reference gene and TaqMan Control Human Genomic DNA (Applied Biosystems, Foster City, CA) was amplified as a standard control for calibration. All sample and standard DNA reactions were set in triplicate to gauge reaction accuracy. The target gene copy number was quantified using the comparative Ct method. Gene copy number of greater than 4 was considered as CNG, as previously reported (17).
KDR copy number analysis in NSCLC malignant tumor cells was also performed using a dual-color FISH assay developed by one of the co-authors (M. V-G). The KDR probe was prepared from the BAC clone RP11-21A18 obtained from CHORI (Oakland, CA). The FISH assay was performed as we have previously published (18). Copy number analysis was done in approximately 50 nuclei per tumorin at least four areas. Greater than 2 gene copies per cell on average was considered as CNG.
KDR Copy Number and VEGFR-2 and HIF-1α Expression Analyses in Cell Lines
All NSCLC cell lines were authenticated by DNA-fingerprinting. Whole-genome SNP array profiling was performed in 75 NSCLC cell lines using the Illumina Human1M-Duo DNA Analysis BeadChip (Illumina, Inc., San Diego, CA). Prior to analysis, SNP data were normalized to the regional baseline copy number to account for aneuploidy. For VEGFR-2 reverse phase protein array (RPPA) analysis performed in 63 NSCLC cell lines, protein lysate was collected from sub-confluent cultures after 24 hours growth in media with 10% fetal bovine serum (FBS) and assayed by RPPA as previously described (19, 20). Cisplatin and carboplatin sensitivity was determined in triplicate by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay for each cell line and the concentration required for 50% growth inhibition (IC50) was determined. For HIF-1α expression analysis, the cells were serum-starved for 24 h and stimulated with 50 ng/ml VEGF-A (R&D Systems, MN, USA). Cells were incubated in normoxia and protein lysates were collected after 8 h. HIF-1α ELISA (R&D Systems, MN, USA) was performed according to the manufacturer’s instructions (13).
Microvascular Density (MVD), VEGFR-2 and HIF-1α Expression Analyses in Tumors
Histology sections were incubated with primary antibodies against VEGFR-2 (dilution 1:50, Abcam, Cambridge, MA) for 90 min, CD34 (dilution 1:100, Lab Vision, Fremont, CA) for 35 min, and HIF-1α (dilution 1:100, Novus Biologicals, Littleton, CO) for 65 min. Tissue sections were then incubated with the secondary antibody (EnVision Dual Link+; DAKO, Carpinteria, CA) for 30 min, after which diaminobenzidine chromogen was applied for 5 min.
Protein expression was quantified by immunohistochemistry using light microscopy with a ×200 magnification by two observers (F. Y. and I. W.). Tissue samples were analyzed for VEGFR-2 expression in the cytoplasm and membrane of malignant cells, and for HIF-1α in the nucleus. We used a 4-value intensity score (0, 1+, 2+, 3+) and the percentage (0% to 100%) of the extent of reactivity. The final score was obtained by multiplying the intensity and extent-of-reactivity values (range, 0 to 300). MVD was assessed by AriolR 2.0 Image System (AriolR, Genetix, San Jose, CA) using the criteria of Weidner et al (21).
Small Interfering RNA (siRNA)Transfection, Platinum Cytotoxicity and Cell Migration Assays in Cell Lines
We transfected NSCLC cells with three KDR gene-specific siRNA duplexes and control siRNA (OriGene Technology, MD, USA), at a final concentration of 10 nM using Lipofectamine RNAiMAX (Invitrogen, CA, USA) according to the manufacturer’s instructions. To verify the knockdown efficiency, mRNA and protein of transfected cells were collected for real-time RT-PCR and Western blot analyses. The assessment of in vitro resistance to cisplatin and carboplatin was determined by the MTS assay. NSCLC cell lines were seeded in octuplicate at a density of 2,000 per well in 96-well plates. The following day, cells were treated with cisplatin and carboplatin at various concentrations ranging from 0 to 120 μmol/L for cisplatin and 0 to 200 μmol/L for carboplatin. After 72 h of drugs exposure, 20 μl of MTS solution were added per well. Cells were Incubated for 1–4 hours at 37ºC and read at a wavelength of 490 nm. The cell migration assay using NSCLC cell lines was performed as previously reported (13).
KDR Mutation and SNPs Genotyping Analyses
For KDR mutation and SNP genotyping analysis in NSCLC cell lines we examined exons 7, 11, 21, 26, 27 and 30, using PCR-based sequencing and intron-based PCR primers as detailed in the Supplementary Table 2.
Statistical Analysis
Demographic and clinical information were compared using the Chi-square or Fisher exact tests for category variables, and Wilcoxon rank-sum or Kruskal-Wallis tests for continuous variables. The distributions of overall survival (OS) and recurrence-free survival (RFS) were estimated by the Kaplan-Meier method and compared between groups using the log-rank test. Cox proportional hazard models were used for regression analyses of survival data and conducted on OS defined as time from surgery to death or last contact, and on RFS defined as time from surgery to recurrence or last contact. Follow-up time was censored at 5 years. For the correlation analysis of KDR CNG in NSCLC cell lines using the whole-genome SNP arrays data with cisplatin sensitivity we used the Wilcoxon rank sum test. The NSCLC cell lines RPPA data was quantified using the SuperCurve method which detects changes in protein level as previously reported (22).
Results
KDR Gene CNG Analysis
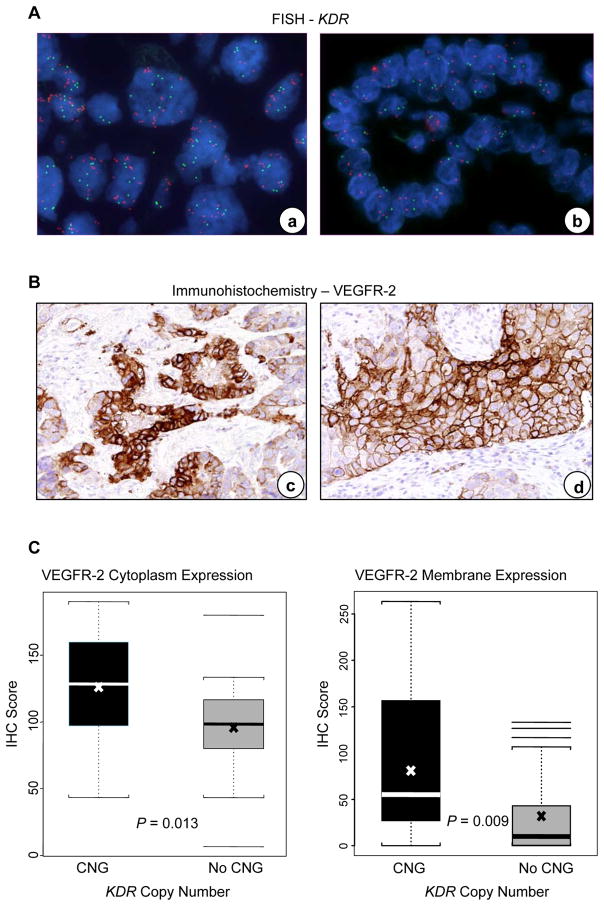
In epithelial malignant NSCLC cells microdissected from tumor tissues, KDR CNG was detected in 45 (32%) of 139 tumors examined. Similar frequency of KDR CNG was found in adenocarcinoma (26/85, 31%) and squamous cell carcinoma (19/54, 35%) histologies (P=0.572). The range of increased KDR copy numbers was from 4 to 11 gene copies. None of 15 normal tissue samples adjacent to the NSCLC tested showed KDR CNG. To confirm KDR CNG results by qPCR, 20 tumor specimens with KDR CNG by qPCR were examined by FISH. KDR copy gains in the malignant cells were confirmed by FISH in all 20 NSCLC specimens detected by qPCR (Figure 1, Panel A).
Figure 1.
KDR copy number gain (CNG) correlated with VEGFR-2 protein expression in NSCLC tumors. Panel A, representative examples of KDR copy number examined by FISH in NSCLC tissue specimens. a, copy number gain; b, no copy number gain. Red signals represent the KDR gene probe, and green signals the internal control probe (magnification ×1000). Panel B, representative example of immunohistochemical expression of VEGFR-2 in NSCLC tissue specimens. VEGFR-2 protein expression was present both in the cytoplasm and membrane of tumor cells in (c) adenocarcinoma and (d) squamous cell carcinoma (magnification ×200). Panel C, expression of VEGFR-2 in tumors with KDR CNG compared with lung cancers without CNG. The box-plots depict scores of immunohistochemical (IHC) expression of VEGFR-2 cytoplasm and VEGFR-2 membrane comparing 26 lung cancers having KDR CNG with 26 lung cancers without CNG. In the box plots, bars indicate median score, x indicates mean scores, and dashed line s.d.
Correlation between KDR CNG and VEGFR-2 Protein Expression and MVD
To assess the immunohistochemical expression of VEGFR-2 in NSCLC malignant cells and the MVD (CD34) in lung tumor tissue stroma, we selected 52 lung tumor specimens with whole histologic sections from FFPE tissues. Of these, 26 cases had KDR CNG and 26 cases did not. VEGFR-2 protein expression was present both in the cytoplasm and membrane of malignant cells as well as in vessel endothelial cells (Figure 1, Panel B).
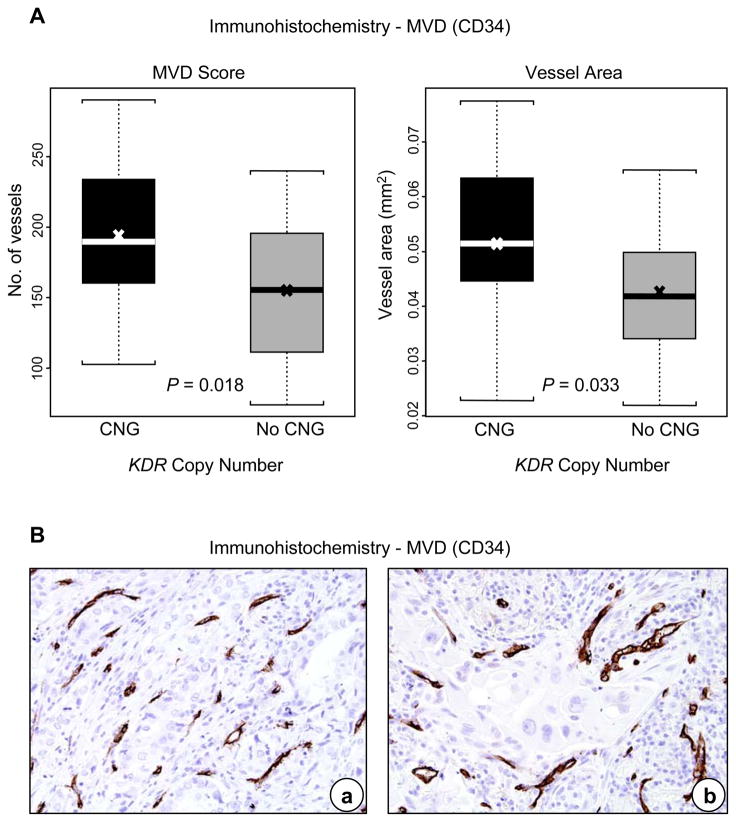
Levels of VEGFR-2 expression in cytoplasm and in membrane were associated with KDR CNG in malignant cells of NSCLC. Tumors with KDR CNG showed significantly higher cytoplasmic (P=0.013) and membrane (P=0.009) VEGFR-2 protein expression in the malignant cells (Figure 1, Panel C), and higher MVD (P=0.018) and larger vessel areas (P=0.033) in the tumor stroma than cases without KDR CNG (Figure 2, Panels A and B).
Figure 2.
KDR copy number gain (CNG) correlated with microvascular density (MVD) in NSCLC tumors. Panel A, expression of MVD in tumors with KDR CNG compared with lung cancers without CNG. The box-plots depict scores of immunohistochemical assessment of MVD and vessel area (mm2) comparing tumors with and without KDR CNG. In the box plots, bars indicate median score, x indicates mean scores, and dashed line standard deviation. Panel B, representative example of immunohistochemical expression of CD34-positive vessels (MVD) in (a) adenocarcinoma and (b) squamous cell carcinoma (magnification ×200).
Association Between Tumor KDR CNG, Clinicopathologic Features and Clinical Outcome
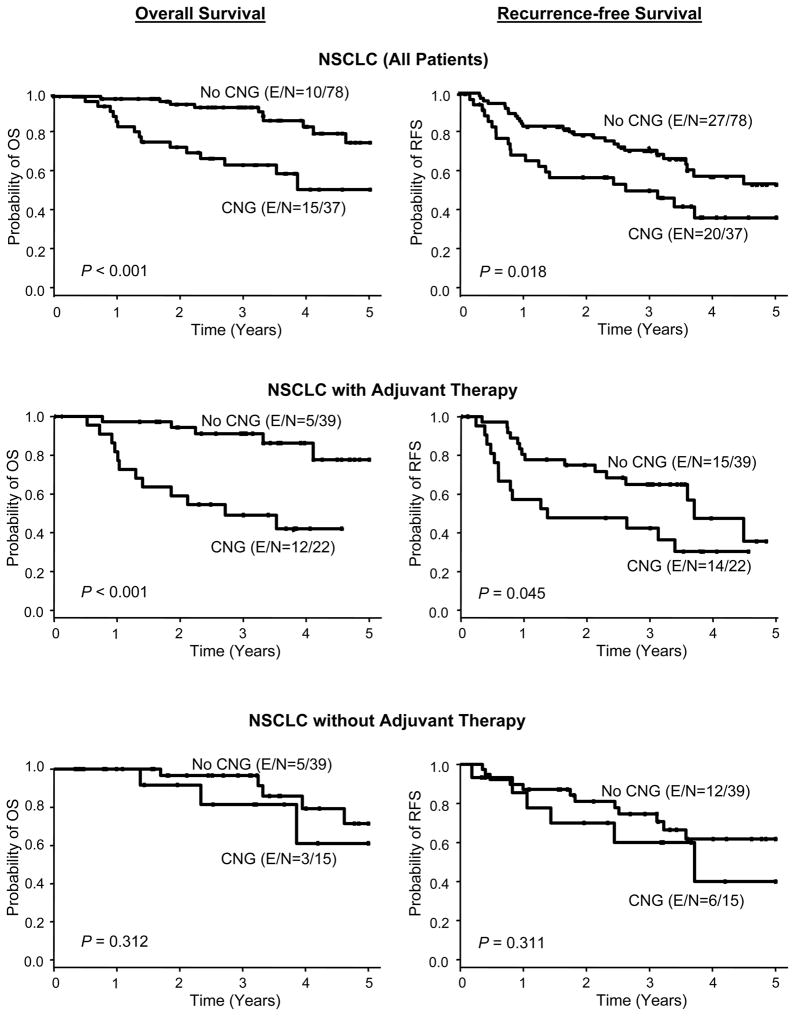
When we correlated KDR CNG with patients’ clinicopathologic features, we did not find correlation with tumor histology, smoking status, and tumor stage. In the multivariate analysis after adjusting for stage and adjuvant therapy, KDR CNG was associated with poor OS (HR=4.0; 95% CI, 1.76 to 9.07; P=0.001) and shortened RFS (HR=1.83, 95% CI, 1.02 to 3.29; P=0.044) in 115 NSCLC patients who underwent surgical resection. Strikingly, KDR CNG was associated with a significantly worse OS (HR=5.16, 95% CI, 1.75 to 15.2, P=0.003) in NSCLC patients receiving platinum adjuvant therapy, but not in patients without adjuvant therapy (P=0.349) (Figure 3 and Table 1). These data suggest that KDR CNG in malignant cells may represent a predictive marker of worse outcome in patients with surgically resected NSCLC treated with platinum-based adjuvant chemotherapy.
Figure 3.
KDR copy number gain (CNG) associated with outcome in NSCLC patients treated with adjuvant chemotherapy. Kaplan-Meier curve for overall survival (OS) and recurrence-free survival (RFS) by KDR CNG in NSCLC patients and two subgroups of platinum adjuvant therapy and without adjuvant therapy (E, event; N, total number of cases).
Table 1.
Multivariate analysis for outcome by KDR copy gain in non-small cell lung carcinoma (NSCLC) patients by adjuvant chemotherapy
| Cases | N | Comparison | Outcome | Adjusted Hazard Ratio (HR)* (95% CI) | P |
|---|---|---|---|---|---|
| All patients | 115 | Gain vs. no gain | OS± | 4.00 (1.76, 9.07) | 0.001 |
| RFS† | 1.83 (1.02, 3.29) | 0.044 | |||
| Adjuvant therapy | 61 | Gain vs. no gain | OS | 5.16 (1.75, 15.2) | 0.003 |
| RFS | 1.87 (0.9, 3.92) | 0.1 | |||
| No adjuvant therapy | 54 | Gain vs. no gain | OS | 1.99 (0.47, 8.4) | 0.349 |
| RFS | 1.83 (0.66, 5.05) | 0.243 |
Adjusting for tumor stage; follow-up is censored at 5 years.
OS, overall survival
RFS, recurrence-free survival
We also investigated examined the impact of neoadjuvant chemotherapy on KDR CNGs. The platinum neoadjuvant-treated tumors (33%, 8/24) had similar frequency of KDR CNGs than cases without neoadjuvant therapy (32%, 37/115).
KDR CNG and VEGFR-2 Protein Levels and Correlation with Platinum Resistance in Cell Lines
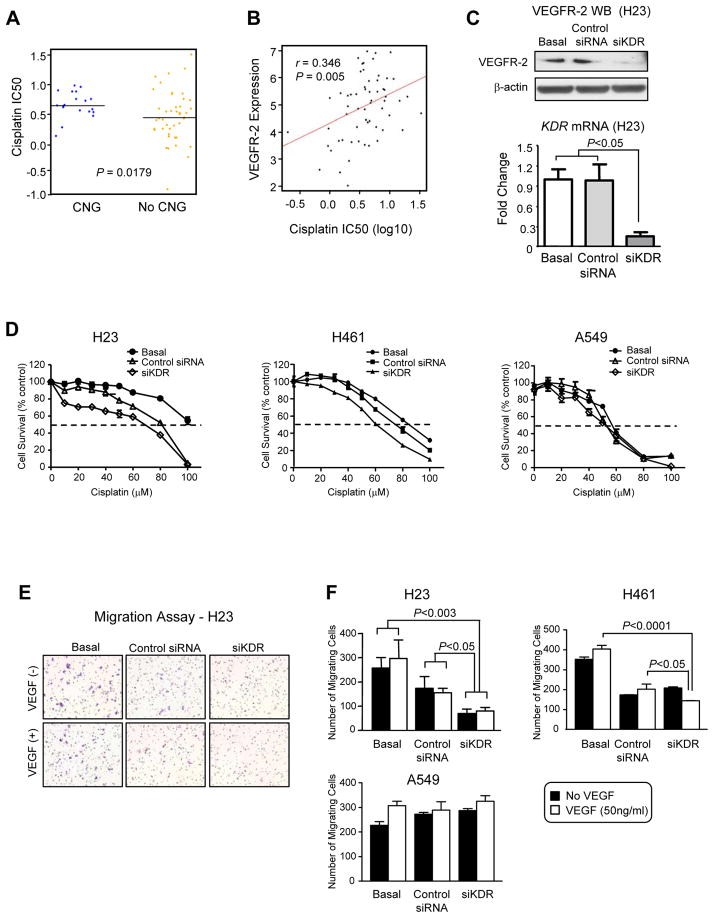
The association detected between KDR CNG and worse outcome in patients treated with platinum adjuvant therapy prompted us to examine the correlation between KDR gain and VEGFR-2 protein levels in NSCLC cell lines with in vitro resistance to platinum drugs. KDR CNG was assessed by SNP array analysis in 75 NSCLC cell lines. Cell lines with KDR copy gains of 6–9 copies or ≥10 copies above the regional baseline copy number were identified. Nineteen (25%) cell lines showed KDR CNG defined as ≥6 copies. Of these, three (4%) cell lines contained high-level gains (≥10 copies), and 16 (21%) had CNG between 6–9. Of interest, cisplatin sensitivity in cell lines with ≥6 KDR copies demonstrated significantly more resistance to cisplatin (P=0.0179) (Figure 4, Panel A).
Figure 4.
KDR copy number gain (CNG) and VEGFR-2 expression associated with resistance to cisplatin. Panel A, correlation of KDR copy number gain (CNG) with in vitro resistance to cisplatin. NSCLC cell lines demonstrating CNG (≥ 6 copies) showed significantly higher IC50 compared with cell lines without CNG. Panel B, correlation between the concentrations of cisplatin required to inhibit NSCLC cell growth (IC50) and VEGFR-2 protein expression levels by reverse phase protein array (RPPA). Panel C, siKDR in NSCLC cell line H23 inhibited significantly the expression of VEGFR-2 by Western-blot (WB) and KDR mRNA by reverse transcriptase quantitative PCR compared with basal and scrambled control siRNA (Bars: s.d.). Panel D, knocking down KDR using siRNA decreased the viability of NSCLC cell lines H23 and H461 (6–9 copies) exposed to cisplatin by MTS assay (data are graphed as mean percent increase ± percent s.d.). Knockdown of KDR in H23 cells caused 1.9-fold decrease in the cisplatin IC50 (P<0.05) and 3.5-fold decrease in the carboplatin IC50 (P<0.05). Knockdown of KDR in H461 cells caused 1.3-fold decrease in the cisplatin IC50 (P<0.05). Knockdown of KDR in A549 cells did not decrease cisplatin or carboplatin IC50. Panel E, migration of NSCLC cell line H23 by Boyden chamber assay was inhibited by siKDR in cells with and without stimulation with VEGF. Panel F, quantification of the migration assay of NSCLC cell lines before and after knocking down KDR using siKDR in cells with and without stimulation with VEGF showed decreased migration in H23 and H461 cells (6–9 KDR copies), but not in A549 cells (no KDR CNG) (Bars: s.d.).
Then, we correlated the expression of VEGFR-2 protein in a panel of 63 untreated NSCLC cell lines by RPPA with each cell line’s sensitivity to cisplatin or carboplatin. We found that higher VEGFR-2 expression levels were significantly associated with resistance to both cisplatin (Figure 4, Panel B) and carboplatin (data not shown) by Pearson correlation. The correlation coefficient (r) between VEGFR-2 expression and the concentration of cisplatin and carboplatin required to inhibit cell growth by 50% (IC50) were 0.346 (P=0.005) and 0.319 (P=0.011), respectively.
Effect of KDR Knockdown on Platinum Sensitivity and Cell Migration in Cell Lines
To investigate the role of KDR CNG and VEGFR-2 overexpression in resistance to both cisplatin and carboplatin, we utilized siRNA to knockdown KDR expression in H23 and H461 NSCLC cell lines, which contain 6–9 KDR gene copies, and as control A549 NSCLC cell line with normal KDR copy number. In both cell lines, siRNA targeting KDR significantly decreased KDR mRNA expression by real-time RT-PCR, and VEGFR-2 expression by Western blot, compared with control cells transfected with scrambled siRNA and non-transfected cells (P<0.05; Figure 4, Panel C). The in vitro sensitivity of H23 and H461 cells to cisplatin (Figure 4, Panel D) or carboplatin (data not shown) treatment was increased in siKDR transfected cells compared with control siRNA-transfected or untransfected cells, suggesting that VEGFR-2 is contributing to chemoresistance in this model. This phenomenon was not observed in cell A549 with normal KDR copy number.
In addition, we found that knockdown of reduction of VEGFR-2 expression induced by siKDR transfection significantly inhibited the migration of H23 and H461 cells compared with siRNA control-transfected or untransfected cells (Figure 4, Panels E and F).
Correlation Between KDR CNG and HIF-1α Expression in Cell Lines and Tumors
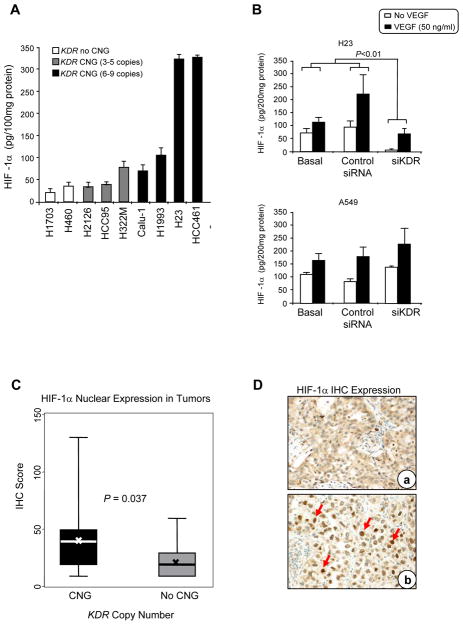
The observations that KDR CNGs were associated with increased angiogenesis, chemoresistance, and migration suggested that VEGFR-2 may be impacting the HIF-1α pathway, which is known to impact each of these cellular properties (13, 14). To investigate this further, we evaluated HIF-1α levels by ELISA in a panel of NSCLC cell lines with a range of KDR copy numbers and expression of VEGFR-2. HIF-1α levels were higher in cell lines with KDR CNG, and significantly (P=0.02) higher in cells with 6–9 gene copies, compared to cells with no CNG (Figure 5, Panel A). In H23 cells which have KDR CNG, stimulation with 50ng/ml VEGF-A for 8 h induced a rise in HIF-1α expression. Furthermore, knockdown of KDR with siRNA significantly (P=0.01) reduced HIF-1α levels (Figure 5, Panel B). This phenomenon was not detected in cell lines A549 with normal KDR copy number. These data indicated that VEGFR-2 can regulate HIF-1α in a ligand-dependent, but hypoxia-independent, manner in NSCLC cells.
Figure 5.
KDR copy number gain (CNG) correlated with HIF-1α expression in NSCLC cell lines and tumor tissue specimens. Panel A, HIF-1α protein expression determined by ELISA correlated with KDR CNG in a series of NSCLC cell lines (Bars: s.d.; cell lines with CNG 6–9 copies versus 3–5 copies and no CNG, P<0.02). Panel B, HIF-1α expression by ELISA was markedly inhibited by knocking down using siKDR in NSCLC H23 cell line, but not in A549 cell line, with and without stimulation with VEGF (Bars: s.d.). Panel C, expression of nuclear HIF-1αin tumors with KDR CNG compared with lung cancers without CNG. The box-plots depict scores of immunohistochemical (IHC) expression of nuclear HIF-1α comparing 22 lung cancers having KDR CNG with 25 lung cancers without CNG. In the box plots, bars indicate median score, x indicates mean scores, and dashed line standard deviation. Panel D, representative example of low (a, adenocarcinoma) and high (b, squamous cell carcinoma) IHC expression of HIF-1αin NSCLC tissue specimens (magnification ×200). Red arrows, positive nuclear HIF-1α immunostaining.
We next investigated the potential association between KDR CNG and HIF-1α in NSCLC clinical specimens. Similarly to the results in the NSCLC cell lines, tumor tissue specimens with KDR CNG (n=25) demonstrated a significantly (P=0.037) higher expression of nuclear HIF-1α expression by immunohistochemistry than tumors without CNG (n=22) (Figure 5, Panels C and D).
KDR Mutation and SNP Analyses
To investigate whether alterations in the KDR gene other than CNGs may impact NSCLC tumors, we assessed the KDR gene for mutations and SNPs. For KDR mutation analysis in NSCLC cell lines, we examined 6 KDR exons (7, 11, 21, 26, 27 and 30) shown to be mutant in adenocarcinoma tumors in a study published by Ding et al (15). In 37 tested NSCLC cell lines we found only two mutations in the KDR gene, an intronic T+2A exon 11 mutation in HCC2450, and a CGT946CAT point mutation in exon 21 in HCC2279. No mutation affecting exons 11 and 21 was detected in 200 NSCLC tissues specimens examined.
In addition, three KDR SNPs (889G/A, 1416A/T, and −37A/G) were genotyped in DNA extracted from 200 NSCLC tumors (Supplementary Table 3), and correlated with patients clinicopathologic features, including outcome. We did not find correlation between the SNP genotypes distribution and OS or RFS of all NSCLC patients examined. In adenocarcinoma patients both KDR 1416 AT/TT (HR=0.45; 95% CI, 0.2 to 0.99; P=0.048) and −37 AG/GG (HR=0.43; 95% CI, 0.2 to 0.92; P=0.031) variant genotypes were associated with a favorable OS in the multivariate analysis after adjusting for tumor stage and neoadjuvant therapy (Supplementary Figure 1, and Supplementary Table 4).
Furthermore, among NSCLC patients with the KDR 889 GA/AA variant genotypes, those who received platinum neoadjuvant and/or adjuvant chemotherapy showed a significantly better OS (HR=0.22; 95% CI, 0.05 to 0.94; P=0.041) than patients who did not receive chemotherapy in the multivariate analysis after adjusting for histology and tumor stage. However, no survival benefit was found in NSCLC patients with KDR 889 GG wild genotype (HR=1.23; 95% CI, 0.64 to 2.35; P=0.538).
Discussion
Our study represents the first report in lung cancer showing a high frequency of KDR CNG (32%) in both major histology types of NSCLC, adenocarcinoma and squamous cell carcinoma, by qPCR and confirmed in a subset of cases by FISH. Notably, KDR CNG predicted worse overall survival in patients who received platinum adjuvant therapy but not in untreated patients In NSCLC cell lines we found that KDR CNGs were significantly associated with in vitro resistance to platinum chemotherapy, as well as increased levels of nuclear HIF-1α. Furthermore, KDR knockdown experiments using small interfering RNA reduced platinum resistance, cell migration, and HIF-1α levels in cells bearing KDR CNGs, providing evidence for direct involvement of KDR. Our findings suggest that tumor cell KDR CNGs may promote a more malignant phenotype including increased chemoresistance, angiogenesis, and HIF-1α levels.
In our study, tumors with KDR CNG in the malignant cells showed significantly higher VEGFR-2 protein expression in the cytoplasm and membrane of those cells, as well as higher MVD and larger vessel areas in the tumor stroma, compared with tumors lacking the KDR CNG. One possible explanation for this association is that that tumor cell VEGFR-2 binds circulating VEGF, increasing local concentrations of the ligand which turn increases angiogenesis through effects on tumor endothelium. Another possible explanation is that VEGFR-2-overexpressing lung cancer cells may express increased levels of VEGF and other pro-angiogenic factors via upregulation of HIF-1α, which in turn could promote autocrine or paracrine signaling that further increases expression. These mechanisms are not mutually exclusive and merit further investigation. Our finding of correlations between KDR CNG and higher expression of HIF-1α in NSCLC cell lines and tumor specimens support the latter hypothesis. It has been demonstrated that activation of several receptor tyrosine kinases (RET, VEGFR-1, EGFR and PDGFR) increases HIF-1α levels in a cell-specific manner in tumors (13, 23, 24); therefore, our data represent the first evidence suggesting that VEGFR-2 may be another receptor tyrosine kinase that plays a role in increasing the levels of HIF-1α expression in cancer.
A provocative finding of this study is this first report that KDR CNG in malignant cells predicted a worse outcome of NSCLC patients receiving platinum adjuvant chemotherapy after surgical resection with curative intent, but was not predictive in patients without adjuvant therapy. These findings suggest that KDR CNG may represent a potential biomarker for predicting resistance to adjuvant platinum-based chemotherapy in NSCLC patients. It is also noteworthy that VEGFR-2 knockdown reduced chemoresistance and cell migration, and lowered HIF-1α levels, using in vitro NSCLC models. One potential implication of these findings is that VEGFR-2 blockade may sensitize tumors bearing KDR CNGs to chemotherapy through directly through effects of the tumor cells themselves, in addition to its effect on tumor endothelial cells. KDR CNGs may therefore identify a group of NSCLC patients that would receive greater relative benefit from combinations of VEGF pathway inhibitors with chemotherapy than patients lacking KDR CNGs. Further prospective studies with larger patient cohorts are needed to assess the role of KDR CNG in NSCLC tumors and outcome of NSCLC patients treated with platinum-based chemotherapy in both surgically resected and advanced metastatic tumor settings, and to determine whether KDR CNGs are predictive of either chemoresistance or benefit for VEGF inhibitor benefit/chemotherapy combinations compared to chemotherapy alone.
Our finding that KDR CNG by SNP array and higher levels of VEGFR-2 expression by RPPA in a large series of NSCLC cell lines correlated significantly with in vitro resistance to platinum dugs (cisplatin for KDR CNG, and cisplatin and carboplatin for VEGFR-2 expression) provides support to our clinical observation. The increased sensitivity of the NSCLC cell lines having KDR CNG to in vitro treatment with cisplatin or carboplatin after inhibition of KDR mRNA and protein expressions further supports the concept that KDR CNG may promote platinum resistance in NSCLC. Although the exact mechanism needs to be elucidated, we postulate that the increased expression of HIF- 1α induced by KDR CNG, and subsequent VEGFR-2 expression, in malignant NSCLC cells may explain increased platinum resistance in NSCLC. Interestingly, HIF-1α has been previously associated to chemoresistance in NSCLC (25, 26) and other solid tumor types (27, 28).
The finding that inhibition of KDR and VEGFR-2 expression resulted in decreased NSCLC cell migration points out another new interesting role of VEGFR-2 in NSCLC malignant cells. It has been established that, among other functions, VEGFR-2 is an important mediator of VEGF-stimulated endothelial cell migration (29, 30). We have also observed that that HIF-1α mediates migration driven by another receptor tyrosine kinase, EGFR, in NSCLC, independent of hypoxia (31).
In our study, the variant genotypes of KDR SNPs 1416 (AT/TT) and −37 (AG/GG) associated with a favorable OS in the multivariate analysis. Ours is the first report showing association between KDR SNP genotypes and prognosis in lung cancer. In breast cancer patients the KDR SNP 1416 A/T genotypic variant was associated with the expression of progesterone receptors, and its presence suggested a better prognosis for carriers of the T allele (32). Questions remain about the functional roles of the KDR SNPs responsible for the associations with outcome of NSCLC patients, particularly in adenocarcinoma patients, found in our study.
In summary, our findings indicate that KDR CNG was frequently detected in NSCLC tumors and associated with platinum resistance in vivo and in vitro, and may be a useful biomarker for identifying patients at high risk for recurrence after adjuvant therapy, a group that may benefit from VEGFR-2 blockade.
Supplementary Material
Acknowledgments
This study was supported by grants from the Department of Defense (W81XWH-07-1-0306 J.D.M., J.V.H., I.I.W.), the Specialized Program of Research Excellence in Lung Cancer (P50CA70907 to J.D.M., J.V.H., and I.I.W.; P50CA58187 to MVG), and the National Cancer Institute (Cancer Center Support Grant CA-16672).
Abbreviations
- VEGFR-2
vascular endothelial growth factor receptor-2
- KDR
kinase insert domain receptor
- SNP
single nucleotide polymorphism
- VEGF
vascular endothelial growth factor
- HIF-1α
hypoxia inducible factor-1α
- NSCLC
non-small cell lung cancer
- CNG
copy number gain
- RPPA
reverse-phase protein array
- and
RTK, receptor tyrosine kinase
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–6. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Fidler IJ, Ellis LM. The implications of angiogenesis for the biology and therapy of cancer metastasis. Cell. 1994;79:185–8. doi: 10.1016/0092-8674(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 3.Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, Heldin CH. Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem. 1994;269:26988–95. [PubMed] [Google Scholar]
- 4.Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr Rev. 1997;18:4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Terman BI, Carrion ME, Kovacs E, Rasmussen BA, Eddy RL, Shows TB. Identification of a new endothelial cell growth factor receptor tyrosine kinase. Oncogene. 1991;6:1677–83. [PubMed] [Google Scholar]
- 7.Bernatchez PN, Soker S, Sirois MG. Vascular endothelial growth factor effect on endothelial cell proliferation, migration, and platelet-activating factor synthesis is Flk-1-dependent. J Biol Chem. 1999;274:31047–54. doi: 10.1074/jbc.274.43.31047. [DOI] [PubMed] [Google Scholar]
- 8.Ishii H, Yazawa T, Sato H, Suzuki T, Ikeda M, Hayashi Y, et al. Enhancement of pleural dissemination and lymph node metastasis of intrathoracic lung cancer cells by vascular endothelial growth factors (VEGFs) Lung Cancer. 2004;45:325–37. doi: 10.1016/j.lungcan.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Ludovini V, Gregorc V, Pistola L, Mihaylova Z, Floriani I, Darwish S, et al. Vascular endothelial growth factor, p53, Rb, Bcl-2 expression and response to chemotherapy in advanced non-small cell lung cancer. Lung Cancer. 2004;46:77–85. doi: 10.1016/j.lungcan.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Seto T, Higashiyama M, Funai H, Imamura F, Uematsu K, Seki N, et al. Prognostic value of expression of vascular endothelial growth factor and its flt-1 and KDR receptors in stage I non-small-cell lung cancer. Lung Cancer. 2006;53:91–6. doi: 10.1016/j.lungcan.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Carrillo de Santa Pau E, Arias FC, Caso Pelaez E, Munoz Molina GM, Sanchez Hernandez I, Muguruza Trueba I, et al. Prognostic significance of the expression of vascular endothelial growth factors A, B, C, and D and their receptors R1, R2, and R3 in patients with nonsmall cell lung cancer. Cancer. 2009;115:1701–12. doi: 10.1002/cncr.24193. [DOI] [PubMed] [Google Scholar]
- 12.Donnem T, Al-Saad S, Al-Shibli K, Delghandi MP, Persson M, Nilsen MN, et al. Inverse prognostic impact of angiogenic marker expression in tumor cells versus stromal cells in non small cell lung cancer. Clin Cancer Res. 2007;13:6649–57. doi: 10.1158/1078-0432.CCR-07-0414. [DOI] [PubMed] [Google Scholar]
- 13.Nilsson MB, Zage PE, Zeng L, Xu L, Cascone T, Wu HK, et al. Multiple receptor tyrosine kinases regulate HIF-1alpha and HIF-2alpha in normoxia and hypoxia in neuroblastoma: implications for antiangiogenic mechanisms of multikinase inhibitors. Oncogene. 2010;29:2938–49. doi: 10.1038/onc.2010.60. [DOI] [PubMed] [Google Scholar]
- 14.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, et al. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2010;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegue E, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–20. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadara H, Behrens C, Yuan P, Solis L, Liu D, Gu X, et al. A five-gene and corresponding protein signature for stage-I lung adenocarcinoma prognosis. Clin Cancer Res. 2011;17:1490–501. doi: 10.1158/1078-0432.CCR-10-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng KW, Lu Y, Mills GB. Assay of Rab25 function in ovarian and breast cancers. Methods Enzymol. 2005;403:202–15. doi: 10.1016/S0076-6879(05)03017-X. [DOI] [PubMed] [Google Scholar]
- 20.Byers LA, Sen B, Saigal B, Diao L, Wang J, Nanjundan M, et al. Reciprocal regulation of c-Src and STAT3 in non-small cell lung cancer. Clin Cancer Res. 2009;15:6852–61. doi: 10.1158/1078-0432.CCR-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis--correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 22.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–94. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 23.Hirami Y, Aoe M, Tsukuda K, Hara F, Otani Y, Koshimune R, et al. Relation of epidermal growth factor receptor, phosphorylated-Akt, and hypoxia-inducible factor-1alpha in non-small cell lung cancers. Cancer Lett. 2004;214:157–64. doi: 10.1016/j.canlet.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 24.Phillips RJ, Mestas J, Gharaee-Kermani M, Burdick MD, Sica A, Belperio JA, et al. Epidermal growth factor and hypoxia-induced expression of CXC chemokine receptor 4 on non-small cell lung cancer cells is regulated by the phosphatidylinositol 3-kinase/PTEN/AKT/mammalian target of rapamycin signaling pathway and activation of hypoxia inducible factor-1alpha. J Biol Chem. 2005;280:22473–81. doi: 10.1074/jbc.M500963200. [DOI] [PubMed] [Google Scholar]
- 25.Mi J, Zhang X, Rabbani ZN, Liu Y, Reddy SK, Su Z, et al. RNA aptamer-targeted inhibition of NF-kappa B suppresses non-small cell lung cancer resistance to doxorubicin. Mol Ther. 2008;16:66–73. doi: 10.1038/sj.mt.6300320. [DOI] [PubMed] [Google Scholar]
- 26.Wen W, Ding J, Sun W, Wu K, Ning B, Gong W, et al. Suppression of cyclin D1 by hypoxia-inducible factor-1 via direct mechanism inhibits the proliferation and 5-fluorouracil-induced apoptosis of A549 cells. Cancer Res. 2010;70:2010–9. doi: 10.1158/0008-5472.CAN-08-4910. [DOI] [PubMed] [Google Scholar]
- 27.Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos C, Turley H, Talks K, et al. Hypoxia-inducible factor (HIF1A and HIF2A), angiogenesis, and chemoradiotherapy outcome of squamous cell head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2002;53:1192–202. doi: 10.1016/s0360-3016(02)02848-1. [DOI] [PubMed] [Google Scholar]
- 28.Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer. 2009;100:405–11. doi: 10.1038/sj.bjc.6604844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau S, Houle F, Landry J, Huot J. p38 MAP kinase activation by vascular endothelial growth factor mediates actin reorganization and cell migration in human endothelial cells. Oncogene. 1997;15:2169–77. doi: 10.1038/sj.onc.1201380. [DOI] [PubMed] [Google Scholar]
- 30.Qi JH, Claesson-Welsh L. VEGF-induced activation of phosphoinositide 3-kinase is dependent on focal adhesion kinase. Exp Cell Res. 2001;263:173–82. doi: 10.1006/excr.2000.5102. [DOI] [PubMed] [Google Scholar]
- 31.Xu L, Nilsson MB, Saintigny P, Cascone T, Herynk MH, Du Z, et al. Epidermal growth factor receptor regulates MET levels and invasiveness through hypoxia-inducible factor-1alpha in non-small cell lung cancer cells. Oncogene. 2010;29:2616–27. doi: 10.1038/onc.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forsti A, Jin Q, Altieri A, Johansson R, Wagner K, Enquist K, et al. Polymorphisms in the KDR and POSTN genes: association with breast cancer susceptibility and prognosis. Breast Cancer Res Treat. 2007;101:83–93. doi: 10.1007/s10549-006-9265-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.