Abstract
We conducted a genome-wide association study on cutaneous basal cell carcinoma (BCC) among 2045 cases and 6013 controls of European ancestry, with follow-up replication in 1426 cases and 4845 controls. A non-synonymous SNP in the MC1R gene (rs1805007 encoding Arg151Cys substitution), a previously well-documented pigmentation gene, showed the strongest association with BCC risk in the discovery set (rs1805007[T]: OR (95% CI) for combined discovery set and replication set [1.55 (1.45–1.66); P= 4.3 × 10−17]. We identified that an SNP rs12210050 at 6p25 near the EXOC2 gene was associated with an increased risk of BCC [rs12210050[T]: combined OR (95% CI), 1.24 (1.17–1.31); P= 9.9 × 10−10]. In the locus on 13q32 near the UBAC2 gene encoding ubiquitin-associated domain-containing protein 2, we also identified a variant conferring susceptibility to BCC [rs7335046 [G]; combined OR (95% CI), 1.26 (1.18–1.34); P= 2.9 × 10−8]. We further evaluated the associations of these two novel SNPs (rs12210050 and rs7335046) with squamous cell carcinoma (SCC) risk as well as melanoma risk. We found that both variants, rs12210050[T] [OR (95% CI), 1.35 (1.16–1.57); P= 7.6 × 10−5] and rs7335046 [G] [OR (95% CI), 1.21 (1.02–1.44); P= 0.03], were associated with an increased risk of SCC. These two variants were not associated with melanoma risk. We conclude that 6p25 and 13q32 are novel loci conferring susceptibility to non-melanoma skin cancer.
INTRODUCTION
Basal cell carcinoma (BCC), a basal keratinocyte tumor in the epidermis, is the most common form of non-melanoma skin cancer, followed by squamous cell carcinoma (SCC). BCC is the most commonly diagnosed cancer among populations of European ancestry, with more than 1 million new cases each year in the USA, representing ∼80% of all skin cancer cases (1). Despite this high incidence, BCC is rarely fatal and uncommonly metastasizes. However, it can cause clinically significant destruction of surrounding tissues if not treated adequately. BCC typically occurs in areas exposed to the sun, and ultraviolet (UV) exposure is the most important and common environmental risk factor. The major host susceptibility risk factor of BCC is lighter pigmentation (2). UV-induced somatic p53 mutations have frequently been found in BCC cases. In addition, somatic mutations in the patched 1 (PTCH1) gene, a receptor in the hedgehog signaling pathway, have been found in most BCC cases (3). In addition to these rare high-penetrance alleles, common low-penetrance alleles also contribute to the genetic susceptibility to BCC. For example, genetic variants in the melanocortin 1 receptor (MC1R) gene, the major known contributor to skin pigmentation, were associated with an increased risk of BCC as well as melanoma and SCC (4–10).
Recent genome-wide association studies (GWASs) identified several genetic loci (including 1p36, 1q42, 5p15, 7q32, 9p21, 12q13 and 11q14) that confer susceptibility to BCC (6,11–13). We have presented the results of these previously identified susceptibility loci (except for 11q14) in the discovery set of our study in Supplementary Material, Table S1. To identify additional genetic loci, we performed a multistage GWAS of BCC. First, to obtain a discovery set, we conducted a GWAS among 2045 cases of BCC in both men and women and 6013 controls of European ancestry in the USA (Supplementary Material, Table S2). We combined data from five case–control studies nested within the Nurses' Health Study (NHS) and the Health Professionals Follow-up Study (HPFS): a type 2 diabetes case–control study nested within the NHS (T2D_NHS, BCC cases = 665, BCC controls = 2,162); a type 2 diabetes case–control study nested within the HPFS (T2D_HPFS, BCC cases = 597, BCC controls = 1555); a coronary heart disease case–control study nested within the NHS (CHD_NHS, BCC cases = 253, BCC controls = 765); a coronary heart disease case–control study nested within the HPFS (CHD_HPFS, BCC cases = 282, BCC controls = 715) and a postmenopausal invasive breast cancer case–control study (controls only) nested within the NHS (BC_NHS, BCC cases = 248, BCC controls = 816). Second, we conducted a fast-track replication of eight promising SNPs in the replication set of 1426 BCC cases and 4845 controls (Supplementary Material, Table S2). These cases and controls in the replication set were from three studies: a study of 24 h urine composition in individuals with and without a history of kidney stones within the NHS and HPFS (KS_NHS_HPFS, BCC cases = 232, BCC controls = 703); a BCC case–control study nested within the NHS (BCC_NHS, BCC cases = 588, BCC controls = 2026) and a renal function study nested within the NHS (RF_NHS, BCC cases = 606, BCC controls = 2116). There was no sample overlap among the five studies of the discovery set and the three studies of the replication set, nor between the discovery and replication sets. The study protocol was approved by the Institutional Review Board of Brigham and Women's Hospital and the Harvard School of Public Health.
RESULTS
Detailed descriptions of the population for each study in the discovery set and replication set are presented in Supplementary Material, Methods. Both the NHS and HPFS collected information on self-reported diagnosis of BCC. The definitions of BCC for each study of discovery set and replication set are provided in Supplementary Material, Methods.
In each GWAS of the discovery set, those imputed SNPs with minor allele frequency (MAF) >2.5% and imputation R2 > 0.3 were selected for combined meta-analysis. The detailed number of SNPs used in each study of the discovery set was presented in Materials and Methods. A total of 2 318 094 SNPs were finally available for meta-analysis. The quantile–quantile (Q–Q) plots based on the five individual GWASs and combined meta-analysis in the discovery set are presented in Supplementary Material, Figure S1. The Q–Q plots did not demonstrate a systematic deviation from the expected distribution, consistent with a minimal likelihood of systematic genotype error or bias due to underlying population substructure. The overall genomic control inflation factor was λGC= 0.996.
We selected top four regions (chromosomes 3, 6, 9 and 13) for a fast-track replication. To ensure the validity of genotyping, in each region except for the region of UBAC2 on chromosome 13, we selected two top SNPs in linkage disequilibrium (LD) as mutual surrogates (r2> 0.9 in HapMap CEU). These SNPs were at P-value <1.5 × 10−6 in the discovery set. We excluded SNPs with P-value for heterogeneity test <0.01. Near the region of UBAC2, an SNP rs7335046 was ranked number 2 for the association with BCC risk in the discovery set. Although there were other SNPs in complete LD with the SNP rs7335046, those SNPs showed the P-value for heterogeneity test <0.01 in the discovery set of this study. Hence, we selected the SNP rs12019494 in this region with P-value for heterogeneity >0.01 (Pheterogeneity= 0.28) and presenting a modest LD with the SNP rs7335046 (r2= 0.4 in HapMap CEU). Moreover, in the discovery set, we found that a non-synonymous SNP in the MC1R gene (rs1805007), a previously well-documented pigmentation gene, was ranked number 1 for the highest association with BCC risk (rs1805007, P= 5.9 × 10−9). We included the SNP rs1805007 for further replication as well. The imputation R2 and association results of these nine SNPs with BCC risk in the discovery set are presented in Supplementary Material, Tables S3 and S4.
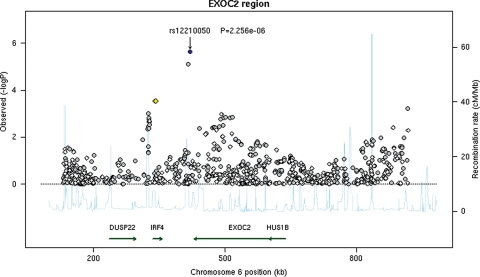
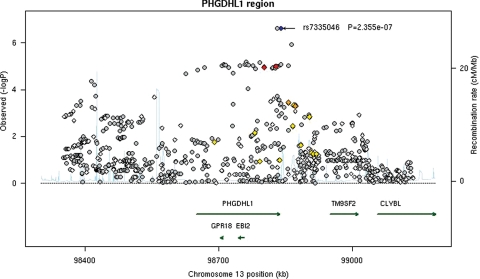
We attempted to replicate the associations of those selected nine SNPs with BCC risk in a replication set of 1426 cases and 4845 controls. Out of nine SNPs selected, in addition to the SNP rs1805007 in the MC1R gene, two SNPs near the EXOC2 gene on 6p25 (rs12210050 and rs12202284) and two SNPs near the UBAC2 gene on 13q32 (rs7335046 and rs12019494) were replicated with P-value <0.05 (Supplementary Material, Table S5). After combining the discovery set with the replication set, the SNP rs1805007 in the MC1R gene was identified as having the smallest P-value [rs1805007[T]: OR (95% CI), 1.55 (1.45–1.66); P= 4.3 × 10−17] (Table 1). Two SNPs, one near the EXOC2 gene (rs12210050, P= 9.9 × 10−10) and the other near the UBAC2 gene (rs7335046, P= 2.9 × 10−8), were also found to reach genome-wide significant association at the 5.0 × 10−8 threshold. The ORs (95% CI) for SNP rs12210050[T] and rs7335046[G] were 1.24 (1.17–1.31) and 1.26 (1.18–1.34), respectively (Table 1). No genome-wide significant results were found for the remaining six SNPs in the combined set (Supplementary Material, Table S5). The regional association plots for both the EXOC2 and UBAC2 regions in the discovery set are presented in Figures 1 and 2. For the region EXOC2, after adjusting for rs12210050 in the discovery set, none of the remaining 989 SNPs in this region was significant at P < 0.001. Similarly, in the region UBAC2, after adjusting for rs12210050 in the discovery set, none of the remaining 802 SNPs was significant at P < 0.001. It is likely that these identified markers are both in LD with the causal variants in these regions.
Table 1.
Association of rs12210050 near the EXOC2 gene, rs7335046 near the UBAC2 gene and rs1805007 in the MC1R gene with the risk of BCC
| SNP (major, minor allele) | Number of participants |
MAF |
OR (95% CI) | P-value | P-value for heterogeneity | ||
|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||
| rs12210050 (C, T) | |||||||
| Discovery set | |||||||
| BC_NHS (female) | 248 | 816 | 21.6 | 17.2 | 1.19 (0.94–1.45) | 0.18 | |
| T2D_NHS (female) | 665 | 2162 | 20.5 | 16.8 | 1.18 (1.02–1.33) | 0.04 | |
| T2D_HPFS (male) | 577 | 1504 | 17.1 | 13.5 | 1.25 (1.08–1.42) | 0.01 | |
| CHD_NHS (female) | 253 | 765 | 22.1 | 16.0 | 1.42 (1.16–1.67) | 0.01 | |
| CHD_HPFS (male) | 282 | 715 | 18.6 | 14.4 | 1.32 (1.07–1.58) | 0.03 | |
| All (meta-analysis) | 2025 | 5962 | 19.6 | 15.6 | 1.25 (1.15–1.34) | 2.3E − 06 | 0.77 |
| Replication set | |||||||
| KS_NHS_HPFS (female and male) | 232 | 703 | 19.0 | 15.1 | 1.28 (1.00–1.56) | 0.08 | |
| BCC_NHS (female) | 568 | 1998 | 20.7 | 16.4 | 1.28 (1.08–1.51) | 4.6E − 03 | |
| RF_NHS (female) | 572 | 2030 | 22.0 | 19.5 | 1.19 (1.01–1.40) | 0.04 | |
| All (meta-analysis) | 1372 | 4731 | 21.0 | 18.0 | 1.24 (1.13–1.34) | 1.1E − 04 | 0.80 |
| Combined set (meta-analysis) | 3397 | 10 693 | 20.2 | 16.6 | 1.24 (1.17–1.31) | 9.9E − 10 | 0.93 |
| rs7335046 (C, G) | |||||||
| Discovery set | |||||||
| BC_NHS (female) | 248 | 816 | 17.5 | 10.5 | 1.81 (1.51–2.10) | 9.9E − 05 | |
| T2D_NHS (female) | 665 | 2161 | 13.0 | 11.4 | 1.15 (0.97–1.34) | 0.14 | |
| T2D_HPFS (male) | 596 | 1555 | 15.9 | 10.7 | 1.56 (1.36–1.76) | 1.1E − 05 | |
| CHD_NHS (female) | 253 | 765 | 14.0 | 12.1 | 1.26 (0.96–1.56) | 0.13 | |
| CHD_HPFS (male) | 282 | 715 | 12.9 | 13.5 | 0.98 (0.68–1.27) | 0.88 | |
| All (meta-analysis) | 2044 | 6012 | 14.5 | 11.4 | 1.32 (1.22–1.43) | 2.4E − 07 | 0.01 |
| Replication set | |||||||
| KS_NHS_HPFS (female and male) | 232 | 703 | 12.1 | 10.8 | 1.20 (0.87–1.52) | 0.28 | |
| BCC_NHS (female) | 580 | 1999 | 14.3 | 12.3 | 1.14 (0.94–1.39) | 0.19 | |
| RF_NHS (female) | 601 | 2083 | 13.0 | 11.2 | 1.20 (0.98–1.47) | 0.07 | |
| All (meta-analysis) | 1413 | 4785 | 13.4 | 11.6 | 1.18 (1.05–1.30) | 0.01 | 0.93 |
| Combined set (meta-analysis) | 3457 | 10 797 | 14.1 | 11.5 | 1.26 (1.18–1.34) | 2.9E − 08 | 0.55 |
| rs1805007 (C, T) | |||||||
| Discovery set | |||||||
| BC_NHS (female) | 248 | 816 | 11.1 | 7.0 | 1.55 (1.20–1.90) | 0.02 | |
| T2D_NHS (female) | 660 | 2149 | 11.1 | 6.4 | 1.75 (1.54–1.96) | 3.5E − 07 | |
| T2D_HPFS (male) | 594 | 1553 | 7.2 | 6.0 | 1.24 (0.97–1.51) | 0.12 | |
| CHD_NHS (female) | 253 | 765 | 8.7 | 7.6 | 1.17 (0.80–1.54) | 0.41 | |
| CHD_HPFS (male) | 282 | 715 | 8.9 | 6.4 | 1.42 (1.06–1.79) | 0.06 | |
| All (meta-analysis) | 2037 | 5998 | 9.4 | 6.5 | 1.47 (1.34–1.60) | 5.9E − 09 | 0.22 |
| Replication set | |||||||
| KS_NHS_HPFS (female and male) | 232 | 703 | 11.4 | 7.3 | 1.79 (1.42–2.16) | 2.0E − 03 | |
| BCC_NHS (female)a | 290 | 840 | 11.4 | 6.9 | 2.02 (1.41–2.88) | 6.6E − 04 | |
| RF_NHS (female) | 597 | 2083 | 10.7 | 6.8 | 1.63 (1.30–2.05) | 2.5E − 05 | |
| All (meta-analysis) | 1119 | 3626 | 10.2 | 7.2 | 1.70 (1.53–1.87) | 5.4E − 10 | 0.89 |
| Combined set (meta-analysis) | 3156 | 9624 | 9.7 | 6.8 | 1.55 (1.45–1.66) | 4.3E − 17 | 0.58 |
Results for each GWAS of the discovery set were calculated based on the unconditional logistic regression adjusted for age and top-three principal components of genetic variance. Results for the KS_NHS_HPFS of the replication set were calculated based on the unconditional logistic regression adjusted for age, gender and top-three principal components of genetic variance. Results for the BCC_NHS and RF_NHS of the replication set were calculated based on unconditional logistic regression adjusted for age.
BC_NHS, postmenopausal invasive breast cancer case–control study nested within the NHS; T2D_NHS, type 2 diabetes case–control study nested within the NHS; T2D_HPFS, type 2 diabetes case–control study nested within the HPFS; CHD_NHS, coronary heart disease case–control study nested within the NHS;
CHD_HPFS, coronary heart disease case–control study nested within the HPFS; KS_NHS_HPFS, kidney stone study nested within the NHS and HPFS; BCC_NHS, BCC case–control study nested within the NHS; RF_NHS, renal function study nested within the NHS.
aGenotyping data used in the previous publication were included for data analysis (7).
Figure 1.
Regional association plot in the 600 kb neighborhood of EXOC2. The left-hand Y-axis shows the association P-value of individual SNPs in the discovery set, which is plotted as −log10(P) against chromosomal base–pair position. The right-hand Y-axis shows the recombination rate estimated from the HapMap CEU population. Genotyped SNPs are plotted as diamonds, and imputed as circles in gray. Blue highlights the SNP rs12210050; bright red indicates high LD (r2 ≥ 0.8) with rs12210050; orange, moderate LD (r2 ≥ 0.5 but <0.8); yellow, weak LD (r2 ≥ 0.2 but <0.5) and white, no LD (r2 < 0.2). The genomic coordinate is in NCBI35/hg17.
Figure 2.
Regional association plot in the 600 kb neighborhood of PHGDHL1 (UBAC2). The left-hand Y-axis shows the association P-value of individual SNPs in the discovery set, which is plotted as −log10(P) against chromosomal base–pair position. The right-hand Y-axis shows the recombination rate estimated from the HapMap CEU population. Genotyped SNPs are plotted as diamonds, and imputed as circles in gray. Blue highlights the SNP rs7335046; bright red indicates high LD (r2 ≥ 0.8) with rs7335046; orange, moderate LD (r2 ≥ 0.5 but <0.8); yellow, weak LD (r2 ≥ 0.2 but <0.5) and white, no LD (r2 < 0.2). The genomic coordinate is in NCBI35/hg17. The PHGDHL1 is alternatively called UBAC2.
We further evaluated the associations of the three SNPs that reached genome-wide significance (rs1805007, rs12210050 and rs7335046) with the risk of SCC in 783 incident cases and 2026 controls nested within the NHS and HPFS (Table 2). Details of the study population are provided in Supplementary Material, Methods. All three SNPs were significantly associated with the risk of SCC: rs1805007 (P= 0.002), rs12210050 (P= 7.6 × 10−5) and rs7335046 (P= 0.03). The ORs (95% CI) for the SNPs rs1805007[T], rs12210050[T] and rs7335046[G] were 1.37 (1.12–1.68), 1.35 (1.16–1.57) and 1.21 (1.02–1.44), respectively (Table 2).
Table 2.
Association of rs12210050 near the EXOC2 gene, rs7335046 near the UBAC2 gene and rs1805007 in the MC1R gene with the risk of SCC
| SNP (major, minor allele) | Genotype | Cases (%) | Controls (%) | OR (95% CI) |
|---|---|---|---|---|
| rs12210050 (C, T) | ||||
| Co-dominant model | CC | 483 (62.6) | 1401 (70.1) | 1.00 |
| CT | 257 (33.3) | 539 (27.0) | 1.42 (1.18–1.70) | |
| TT | 32 (4.2) | 58 (2.9) | 1.59 (1.01–2.48) | |
| Dominant model | CC | 483 (62.6) | 1401 (70.1) | 1.00 |
| CT or TT | 289 (37.5) | 597 (29.9) | 1.43 (1.20–1.71) | |
| Additive model | T allele | 1.35 (1.16–1.57) | ||
| P-value for trend | 7.6E − 05 | |||
| rs7335046 (C, G) | ||||
| Co-dominant model | CC | 568 (73.2) | 1535 (76.8) | 1.00 |
| CG | 195 (25.1) | 437 (21.9) | 1.23 (1.01–1.50) | |
| GG | 13 (1.7) | 27 (1.4) | 1.34 (0.68–2.62) | |
| Dominant model | CC | 568 (73.2) | 1535 (76.8) | 1.00 |
| CG or GG | 208 (26.8) | 464 (23.2) | 1.24 (1.02–1.50) | |
| Additive model | G allele | 1.21 (1.02–1.44) | ||
| P-value for trend | 0.03 | |||
| rs1805007 (C, T) | ||||
| Co-dominant model | CC | 616 (79.5) | 1676 (84.7) | 1.00 |
| CT | 154 (19.9) | 289 (14.6) | 1.46 (1.17–1.82) | |
| TT | 5 (0.7) | 13 (0.7) | 0.96 (0.34–2.74) | |
| Dominant model | CC | 616 (79.5) | 1676 (84.7) | 1.00 |
| CT or TT | 159 (20.5) | 302 (15.3) | 1.44 (1.16–1.78) | |
| Additive model | T allele | 1.37 (1.12–1.68) | ||
| P-value for trend | 0.002 | |||
The ORs (95% CIs) were calculated based on the unconditional logistic regression adjusted for age and gender.
Moreover, we evaluated the association of these three SNPs with melanoma risk in 586 melanoma cases and 2026 controls nested within the NHS and HPFS (set 1). Details of the study population are described in Supplementary Material, Methods. The SNP rs1805007[T] was significantly associated with the risk of melanoma [rs1805007[T]: OR (95% CI), 1.63 (1.32–2.01); P= 6.0 × 10−6]. For rs12210050 and rs7335046, we also have data from a case–control study of 1804 melanoma cases and 1027 controls from the MD Anderson Cancer Center (set 2). Details of the study population are described in Supplementary Material, Methods. For both rs12210050 and rs7335046, a meta-analysis was used to combine the results from the two sets. As shown in Supplementary Material, Table S6, we did not identify significant associations between either rs12210050 or rs7335046 and melanoma risk. The OR (95% CI) for rs12210050 and rs7335046 was 1.07 (0.96–1.19) and 1.01 (0.88–1.15), respectively.
DISCUSSION
In this study, the SNP rs1805007 was identified with the strongest associations with both melanoma and non-melanoma skin cancers. MC1R encodes a 317-amino acid seven-pass transmembrane G-protein-coupled receptor, and the SNP rs1805007 encodes an Arg151Cys substitution. A well-known red hair color variant, the SNP rs1805007, along with other genetic variants in the MC1R gene, was shown to confer susceptibility to both melanoma and non-melanoma (BCC and SCC) skin cancers in our previous study and studies performed by other groups (4–10). This supports the validity of our GWAS data and further validates our self-reported BCC data set. Also, we identified two novel alleles, rs12210050 near the EXOC2 gene at 6p25 and rs7335046 near the UBAC2 gene at 13q32, associated with non-melanoma skin cancer. EXOC2 is a component of the exocyst complex involved in the docking of exocystic vesicles with fusion sites on the plasma membrane. Some genetic variants in the EXOC2 gene (including rs12210050) were identified as contributing to human pigmentary traits such as hair color, skin color and tanning ability, in our previous GWAS on hair color and tanning ability (14,15). Hence, we performed an additional analysis for the association between rs12210050 at 6p25 and BCC risk after further adjusting for pigmentary phenotypes, tanning tendency and hair color, and the result remained to reach genome-wide significant association in the combined discovery set and replication set (P= 1.2 × 10−9). At the same locus 6p25, Sulem et al. (16) previously identified the SNP rs1540771 conferring susceptibility to pigmentary phenotypes, including freckling and skin sensitivity to sun. However, this SNP was not associated with the risks of BCC and melanoma in the other previous study conducted by Gudbjartsson et al. (6). The SNP rs1540771 and the SNP rs12210050 are not in LD (r2= 0.05 in HapMap CEU). The SNP rs1540771 showed nominal association with BCC risk in the discovery set of this study [rs1540771[C]: OR (95% CI), 0.93 (0.86–1.00); P= 0.047]. This association was eliminated after adjusting for the SNP rs12210050 (P= 0.42). The UBAC2 gene encoding ubiquitin-associated domain-containing protein 2 is alternatively called phosphoglycerate dehydrogenase-like protein 1 (PHGDHL1). This locus has been identified as a genetic susceptibility locus for Behçet's disease, a chronic systemic inflammatory disease (17).
A possible issue raised in this GWAS is the effect heterogeneity. Although five studies were used in the discovery set of this study, they came from only two demographically similar cohorts (NHS and HPFS). It is plausible that differences in the sampling scheme across the five case–control sub-studies could in principle introduce some effect heterogeneity, although this effect is likely to be small (18). To flag markers that show evidence of effect heterogeneity, we have calculated Cochran's Q statistic (19) and reported the corresponding P-values in the tables. Also, given the large number of SNPs (more than 2 million SNPs) analyzed in this study, nominally significant P-values for heterogeneity are difficult to interpret, and may represent false positives due to sampling variation. For example, although there is some evidence of heterogeneity for the SNP rs7335046 in the discovery set (P = 0.01), the P-value for heterogeneity of this SNP in either replication set or combined set was not significant. In addition, as mentioned above, considering the number of SNPs analyzed in this study, the P-value of 0.01 for heterogeneity in the discovery set is more likely attributable to chance. Still, we have taken a conservative approach and excluded the SNPs with P-values for heterogeneity test <0.01 from further consideration for replication.
In this study, BCC cases used for data analysis were self-reported. The validity of self-report of BCC in these medically sophisticated populations has been assessed in previous studies (20,21). Colditz et al. (20) evaluated the validity of self-reported illnesses including skin cancer in the NHS. Among 33 random samples of women who had reported non-melanoma skin cancer, medical records indicated that 30 (91%) had correctly reported the skin cancer. The three incorrect self-reports were actinic keratosis, a premalignant skin lesion. Also, Hunter et al. (21) previously examined the risk factors of BCC in the NHS using the self-reported cases. As expected, they found that lighter pigmentation (blonde or red hair color), less childhood and adolescent tanning tendency and higher tendency to sunburn were associated with an increased risk of BCC. Also, they found that women residing in California and Florida were more likely to develop BCC compared with women living in the Northeast. In addition, using the self-reported BCC cases, we identified the previously well-documented genetic variant in the MC1R gene (rs1805007) as the strongest locus in this study. These data support the validity of self-report of BCC in our study.
It is possible that the similar biases are present in both the discovery set and replication set because they were from two large cohort studies, the NHS and the HPFS. In the discovery set of this study, 43% of BCC cases were men, whereas 10% of BCC cases were men in the replication set. Also, we note that there are some differences between the two cohorts, such as gender (the NHS is female cohort, and the HPFS is male cohort), geographical background and social economic status.
In summary, in the current GWAS of individuals of European ancestry, we identified two novel loci, the EXOC2 gene on 6p25 and the UBAC2 gene on 13q32, as associated with the risks of non-melanoma skin cancer, BCC and SCC. In addition, we verified the skin cancer susceptibility locus at the MC1R gene on 16q24. Future studies are warranted to evaluate the effect of interactions between these promising SNPs and skin cancer risk factors on the risk of skin cancer. Understanding the role of these novel loci in the development of non-melanoma skin cancer could provide important insight into non-melanoma skin cancer pathogenesis and effectively improve the prevention of non-melanoma skin cancer.
MATERIALS AND METHODS
Description of study populations
Nurses' Health Study
The NHS was established in 1976, when 121 700 female US registered nurses between the ages of 30 and 55, residing in 11 larger US states, completed and returned an initial self-administered questionnaire on their medical histories and baseline health-related exposures, forming the basis for the NHS cohort. Biennial questionnaires with the collection of exposure information on risk factors have been collected prospectively. Overall, follow-up has been very high; after >20 years, ∼90% of participants continue to complete questionnaires. From May 1989 through September 1990, we collected blood samples from 32 826 participants in the NHS cohort.
Health Professionals Follow-up Study
In 1986, 51 529 men from all 50 US states in health professions (dentists, pharmacists, optometrists, osteopath physicians, podiatrists and veterinarians) aged 40–75 answered a detailed mailed questionnaire, forming the basis of the study. The average follow-up rate for this cohort over 10 years is >90%. Between 1993 and 1994, 18 159 study participants provided blood samples by overnight courier.
Skin cancer ascertainment in NHS and HPFS
Disease follow-up procedures are identical for both the NHS and HPFS. Along with exposures every 2 years, outcome data with appropriate follow-up of reported disease events including melanoma and non-melanoma skin cancers are collected. For melanoma and SCC, eligible cases are incident pathologically confirmed invasive cases among subjects who gave a blood specimen in the NHS and HPFS with a diagnosis anytime after blood collection. All medical records of melanoma and SCC are reviewed by dermatologists blinded to exposure information according to established criteria. Cases of BCC are not pathologically confirmed in the NHS and HPFS.
Laboratory assays
Genotyping in each GWAS of the discovery set
We performed genotyping in BC_NHS, using the Illumina HumanHap550 array, as part of the National Cancer Institute's Cancer Genetic Markers of Susceptibility (CGEMS) Project (22). For the other four GWASs of the discovery set, we performed genotyping using the Affymetrix 6.0 array.
Genotyping in the replication set
Nine promising SNPs from the discovery set were selected for further replication in the replication set. (i) The genotyping for the KS_NHS_HPFS was performed using the Illumina HumanHap610 Quad, and the imputation was performed in the same fashion as in the discovery set. The genotype data we extracted for these nine SNPs and their imputation quality data are presented in Supplementary Material, Table S3. (ii) The genotyping for the BCC_NHS and RF_NHS was performed using OpenArray assays at the Dana Farber/Harvard Cancer Center Polymorphism Detection Core.
Imputation and statistical methods
In each study of the discovery set, we used MACH v1.0.16 to impute more than 2.5 million SNPs with HapMap CEU phase II data (release 22) as the reference panel (23). Imputation results were expressed as ‘allele dosages’ (fractional values between 0 and 2). Those MACH dosage files were used for analysis of imputed data. Imputation R2 is an estimate of correlation between observed and predicted genotype. It is the ratio of observed variance to the theoretical variance (23). The number of genotyped SNPs passed quality control procedures and the imputed SNPs with MAF >2.5% and imputation R2 > 0.3 in each study of the discovery set are presented as follows:
| Study | Genotyped | Imputed |
|---|---|---|
| BC_NHS | 546 646 | 2 352 569 |
| T2D_NHS | 704 409 | 2 351 699 |
| T2D_HPFS | 706 040 | 2 356 842 |
| CHD_NHS | 721 316 | 2 350 863 |
| CHD_HPFS | 724 881 | 2 356 504 |
We fitted an unconditional logistic regression model for each SNP that passed quality control filters, using an additive model, controlling for age and the three largest principal components of genetic variation of each GWAS of the discovery set and the KS_NHS_HPFS of the replication set. These principal components were calculated for all individuals on the basis of approximately 10 000 unlinked markers, using the EIGENSTRAT software (24). In the other two replication sets of BCC (BCC_NHS and RF_NHS) as well as SCC and melanoma sets, each SNP was tested for an association with skin cancer risk by unconditional logistic regression model adjusting for age and gender.
In each study of the discovery set, those SNPs with MAF >2.5% and imputation R2 > 0.3 in each study of the discovery set were included in further meta-analysis. Estimated log odds ratios from each study of the discovery set were combined using meta-analysis, with weights proportional to the inverse variance of the estimate in each study. The same meta-analysis method was used to combine the results from the discovery set and replication set.
SUPPLEMENTARY MATERIAL
FUNDING
We are grateful to Merck Research Laboratories for funding of the GWAS of coronary heart disease. This work is supported by NIH grants CA122838, CA87969, CA055075, CA49449, CA100264 and CA093459.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Wei V. Chen for assistance in performing analyses in the MD Anderson Cancer Center melanoma case–control study. We thank Pati Soule and Dr Hardeep Ranu of the Dana Farber/Harvard Cancer Center High-Throughput Polymorphism Detection Core for sample handling and genotyping of the NHS and HPFS samples. We are also indebted to the participants in all of these studies. We thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Miller D.L., Weinstock M.A. Nonmelanoma skin cancer in the United States: incidence. J. Am. Acad. Dermatol. 1994;30:774–778. doi: 10.1016/s0190-9622(08)81509-5. doi:10.1016/S0190-9622(08)81509-5. [DOI] [PubMed] [Google Scholar]
- 2.Han J., Colditz G.A., Hunter D.J. Risk factors for skin cancers: a nested case–control study within the Nurses' Health Study. Int. J. Epidemiol. 2006;35:1514–1521. doi: 10.1093/ije/dyl197. doi:10.1093/ije/dyl197. [DOI] [PubMed] [Google Scholar]
- 3.Epstein E.H. Basal cell carcinomas: attack of the hedgehog. Nat. Rev. Cancer. 2008;8:743–754. doi: 10.1038/nrc2503. doi:10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastiaens M.T., ter Huurne J.A., Kielich C., Gruis N.A., Westendorp R.G., Vermeer B.J., Bavinck J.N. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am. J. Hum. Genet. 2001;68:884–894. doi: 10.1086/319500. doi:10.1086/319500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Box N.F., Duffy D.L., Irving R.E., Russell A., Chen W., Griffyths L.R., Parsons P.G., Green A.C., Sturm R.A. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J. Invest. Dermatol. 2001;116:224–229. doi: 10.1046/j.1523-1747.2001.01224.x. doi:10.1046/j.1523-1747.2001.01224.x. [DOI] [PubMed] [Google Scholar]
- 6.Gudbjartsson D.F., Sulem P., Stacey S.N., Goldstein A.M., Rafnar T., Sigurgeirsson B., Benediktsdottir K.R., Thorisdottir K., Ragnarsson R., Sveinsdottir S.G., et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 2008;40:886–891. doi: 10.1038/ng.161. doi:10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 7.Han J., Kraft P., Colditz G.A., Wong J., Hunter D.J. Melanocortin 1 receptor variants and skin cancer risk. Int. J. Cancer. 2006;119:1976–1984. doi: 10.1002/ijc.22074. doi:10.1002/ijc.22074. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy C., ter Huurne J., Berkhout M., Gruis N., Bastiaens M., Bergman W., Willemze R., Bavinck J.N. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J. Invest. Dermatol. 2001;117:294–300. doi: 10.1046/j.0022-202x.2001.01421.x. doi:10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 9.Palmer J.S., Duffy D.L., Box N.F., Aitken J.F., O'Gorman L.E., Green A.C., Hayward N.K., Martin N.G., Sturm R.A. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am. J. Hum. Genet. 2000;66:176–186. doi: 10.1086/302711. doi:10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valverde P., Healy E., Sikkink S., Haldane F., Thody A.J., Carothers A., Jackson I.J., Rees J.L. The Asp84Glu variant of the melanocortin 1 receptor (MC1R) is associated with melanoma. Hum. Mol. Genet. 1996;5:1663–1666. doi: 10.1093/hmg/5.10.1663. doi:10.1093/hmg/5.10.1663. [DOI] [PubMed] [Google Scholar]
- 11.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. doi:10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey S.N., Gudbjartsson D.F., Sulem P., Bergthorsson J.T., Kumar R., Thorleifsson G., Sigurdsson A., Jakobsdottir M., Sigurgeirsson B., Benediktsdottir K.R., et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat. Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. doi:10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 13.Stacey S.N., Sulem P., Masson G., Gudjonsson S.A., Thorleifsson G., Jakobsdottir M., Sigurdsson A., Gudbjartsson D.F., Sigurgeirsson B., Benediktsdottir K.R., et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. doi:10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J., Kraft P., Nan H., Guo Q., Chen C., Qureshi A., Hankinson S.E., Hu F.B., Duffy D.L., Zhao Z.Z., et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. doi:10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nan H., Kraft P., Qureshi A.A., Guo Q., Chen C., Hankinson S.E., Hu F.B., Thomas G., Hoover R.N., Chanock S., et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. doi:10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G., et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. doi:10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 17.Fei Y., Webb R., Cobb B.L., Direskeneli H., Saruhan-Direskeneli G., Sawalha A.H. Identification of novel genetic susceptibility loci for Behcet's disease using a genome-wide association study. Arthritis Res. Ther. 2009;11:R66. doi: 10.1186/ar2695. doi:10.1186/ar2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monsees G.M., Tamimi R.M., Kraft P. Genome-wide association scans for secondary traits using case–control samples. Genet. Epidemiol. 2009;33:717–728. doi: 10.1002/gepi.20424. doi:10.1002/gepi.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. doi:10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 20.Colditz G.A., Martin P., Stampfer M.J., Willett W.C., Sampson L., Rosner B., Hennekens C.H., Speizer F.E. Validation of questionnaire information on risk factors and disease outcomes in a prospective cohort study of women. Am. J. Epidemiol. 1986;123:894–900. doi: 10.1093/oxfordjournals.aje.a114319. [DOI] [PubMed] [Google Scholar]
- 21.Hunter D.J., Colditz G.A., Stampfer M.J., Rosner B., Willett W.C., Speizer F.E. Risk factors for basal cell carcinoma in a prospective cohort of women. Ann. Epidemiol. 1990;1:13–23. doi: 10.1016/1047-2797(90)90015-k. doi:10.1016/1047-2797(90)90015-K. [DOI] [PubMed] [Google Scholar]
- 22.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. doi:10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.