Abstract
Mutations in ATP13A2 (PARK9) have been linked to juvenile parkinsonism with dementia or Kufor–Rakeb syndrome (KRS). The ATP13A2 gene encodes at least three protein isoforms that arise by alternate splicing. A previous study indicated the Atp13a2Isoform-1 protein is localized to lysosomes, whereas three separate mutations involved in disease cause retention of the protein in the ER. One speculation is that the mutant Atp13a2Isoform-1 proteins are misfolded and eliminated by the ER-associated degradation pathway (ERAD), which involves the dislocation of proteins from the ER to the cytoplasm for proteasome degradation. We examined whether Atp13a2 proteins are degraded by ERAD and whether the Atp13a2Isoform-3 protein has similar localization to the Atp13a2Isoform-1 protein. Through analysis of protein turnover and by disrupting different steps in the ERAD pathway we demonstrate that mutant Atp13a2Isoform-1 proteins are indeed eliminated by ERAD. Thus, siRNA-mediated knockdown of erasin, a platform for assembly of an ERAD complex, or expression of a dominant negative form of p97/VCP, a protein essential for dislocation of ERAD substrates, or inhibition of the proteasome all slowed degradation of the mutant Atp13a2Isoform-1 proteins, but not the wild-type Atp13a2Isoform-1 protein. Immunoprecipitation assays confirmed that the Atp13a2 proteins are ubiquitinated in accord with degradation by ERAD. In contrast to Atp13a2Isoform-1, we show Atp13a2Isoform-3 is localized to the ER and rapidly degraded. Lastly, we show Atp13a2 mutants have increased cytotoxicity and predispose cells to ER-stress-induced cell death. These results provide new insight into the properties of wild-type and mutant Atp13a2 proteins involved in KRS.
INTRODUCTION
Parkinson's disease (PD) is a progressive neurodegenerative disorder characterized by tremor at rest, bradykinesia, rigidity and postural instability (1,2). The etiology of PD has been traced primarily to Mendelian inheritance of mutations in a number of different genes and to exposure to environmental toxins (1,3). The focus of this study is ATP13A2 (PARK9), in which mutations cause a Parkinson's-like disease called Kufor–Rakeb syndrome (KRS) (4–10).
The function of the ATP13A2 gene is not known (4), although based on studies of two yeast proteins with limited homology (∼22–27% sequence identity), it was suggested it might be involved in protecting cells against manganese and mutant α-synuclein toxicity (11). The gene is widely expressed in the body with highest expression in the brain. It is predicted to encode a protein with similarity to P-type ATPases (12). According to sequences deposited in the NCBI database, at least three alternatively spliced ATP13A2 transcripts are expressed in humans, each encoding a different protein (Supplementary Material, Fig. S1). Isoform-1 encodes a protein of 1180 amino acids with 10 transmembrane domains (∼130 kDa). Isoform-2 differs from isoform-1 by containing a small five amino acid in-frame deletion near the N-terminus. Isoform-3, which is 1158 amino acids long, contains an additional downstream in-frame deletion of 117 bases and an out-of-frame deletion of 170 bases removing 39 amino acids and generating a highly diverged C-terminus, respectively (Supplementary Material, Fig. S2). The consequence of these changes on the function of the proteins is not known.
The first three mutations identified in the ATP13A2 gene were a deletion of a cytosine residue at position 3057, a duplication of residues 1632–1653 in exon 16, and a guanine-to-adenine transition in the donor splice site of exon 13 leading to the skipping of exon 13, listed relative to isoform-1 of the protein, which we refer to here as ▵C, Dup22 and Ex13, respectively (4). The ▵C and Dup22 mutations cause frameshift mutations that result in the loss of the last three and six transmembrane domains of the Atp13a2 protein, respectively, whereas the exon13 splice-site mutation results in an in-frame deletion of part of the third transmembrane domain (Supplementary Material, Fig. S3). An initial characterization of the wild-type (wt) and mutant proteins expressed in COS7 cells revealed that the wild-type Atp13a2Isoform-1 has a long half-life (in excess of 12 h) and is localized to lysosomes, whereas all three mutants were unstable and localized in the endoplasmic reticulum (ER). Treatment with MG132 resulted in stabilization of the mutant proteins suggesting degradation by the proteasome. The properties of the mutants are consistent with substrates that are eliminated via the ER-associated degradation (ERAD) pathway (13), although this was not established. There was uncertainty about this issue, because the authors could not detect any ubiquitination of the mutants, which would be expected for proteins that are degraded by ERAD (4).
During ERAD, misfolded proteins are retained and selectively exported from the ER to the cytoplasm for degradation by the proteasome (reviewed in 14–16). Numerous factors are involved in this process, including proteins that are believed to form the conduit for protein export from the ER, ubiquitin ligases, the AAA-ATPase p97/VCP molecular chaperone protein, scaffold proteins, ubiquitin-shuttle factors and factors on opposite sides of the ER membrane (i.e. in the lumen and cytosol). Defects in ERAD lead to a buildup of misfolded proteins in the ER causing ER stress, chronic activation of which can induce cell death. Interestingly, ER stress has been linked to neurodegenerative diseases including PD (17–21).
Here, we report on differences in the localization and degradation of Atp13a2 isoform 1 and 3 proteins carrying PD-linked mutations. The isoform 2 protein was not studied because it contains a five amino acid deletion compared with isoform 1, which we speculated would exhibit similar properties to the isoform 1 protein in our assays. The studies revealed that the wild-type Atp13a2Isoform-3 protein is retained in the ER and rapidly degraded, unlike the wild-type Atp13a2Isoform-1 protein, which is stable and localized in the lysosome. We also show Atp13a2isoform-1 and -3 proteins carrying PD-linked mutations are ubiquitinated and rapidly degraded via ERAD. Finally, we show that overexpression of Atp13a2Isoform-1 proteins carrying PD-linked mutations are more cytotoxic than the wild-type protein, and that they hypersensitize cells to ER-stress induced cell death.
RESULTS
Expression and localization of wild-type and mutant Atp13a2Isoform-1 proteins in HeLa cells
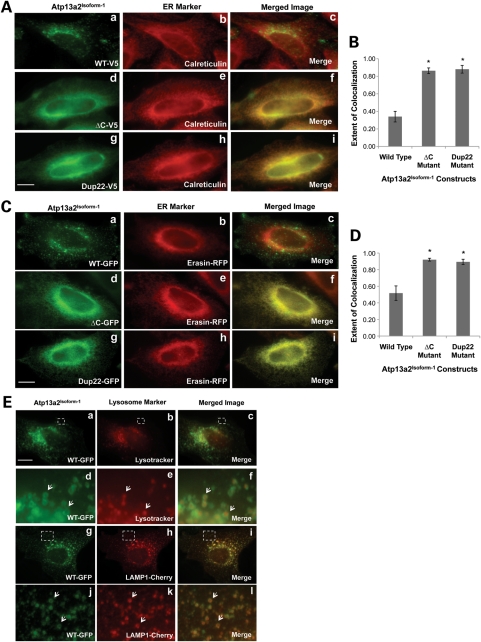
We utilized HeLa cells to determine whether mutations in Atp13a2 proteins that are linked to PD are targeted for degradation through the ERAD pathway. We chose HeLa cells to investigate this possibility because of the different reagents we possess for interfering with ERAD in the cells. We first examined whether wild-type and mutant Atp13a2Isoform-1 proteins expressed in HeLa cells have similar localization as reported in COS7 cells (4). Of the three KRS mutants studied previously, we focused on two random representatives of them, ▵C and Dup22 for the Atp13a2Isoform-1, and the ▵C and Ex 13 mutant for the Atp13a2Isoform-3 mutant (as discussed later). Accordingly, we transfected HeLa cells with V5-tagged Atp13a2-WTIsoform-1, Atp13a2-▵CIsoform-1 and Atp13a2-Dup22Isoform-1 expression constructs and visualized the proteins using immunofluorescence microscopy (Fig. 1). Double immunofluorescence staining confirmed that both mutant Atp13a2Isoform-1 proteins are localized to the ER as evidenced by the strong colocalization of their staining with calreticulin, an ER marker (Fig. 1A, d–i and B). In contrast, the Atp13a2-WTIsoform-1 protein displayed a vesicular staining pattern that did not colocalize with the calreticulin staining (Fig. 1A, a–c and B). To ensure that the staining was not an artifact of the staining protocol, we examined the localization of GFP-tagged versions of the Atp13a2Isoform-1 constructs. Fluorescence of the GFP-tagged Atp13a2Isoform-1 mutants colocalized with that of mRFP-tagged erasin, an ER localized protein (22), while the wild-type did not (Fig. 1C, D). In contrast, GFP-tagged Atp13a2-WTIsoform-1 fluorescence was localized to vesicles that stained positive for Lysotracker as well as cotransfected Lysosomal-associated membrane protein 1 (LAMP1)-Cherry (Fig. 1E). These results confirmed that wild-type Atp13a2Isoform-1 localizes to lysosomes while the mutants are retained in the ER.
Figure 1.
Expression and localization of Atp13a2Isoform-1 constructs in HeLa cells. (A) HeLa cells were transfected with V5-tagged wild-type (a–c), ▵C (d–f) or Dup22 (g–i) Atp13a2Isoform-1 and visualized using immunofluorescence microscopy. (B) Quantification of the extent of colocalization of the indicated Atp13a2Isoform-1 proteins and calreticulin calculated from three different cells (*P < 0.0005). (C) HeLa cells were cotransfected with mRFP-tagged erasin and GFP-tagged wild-type (a–c), ▵C (d–f) or Dup22 (g–i) Atp13a2Isoform-1 and detected using florescence microscopy. (D) Quantification of the extent of colocalization of the indicated Atp13a2Isoform-1 proteins and erasin-mRFP calculated from three different cells (*P < 0.005). (E) (a–f) HeLa cells were transfected with GFP-tagged wild-type Atp13a2Isoform-1 and incubated with the lysosomal dye Lysotracker before imaging. (d–f) Magnification of region outlined in panels (a–c). Arrows indicate examples of colocalization of Atp13a2Isoform-1 and Lysotracker. (g–l) HeLa cells were cotransfected with GFP-tagged Atp13a2Isoform-1 and LAMP1-Cherry and detected using florescence microscopy. (j–l) Magnification of region outlined in panels (g–i). Arrows indicate examples of colocalization of Atp13a2Isoform-1 and LAMP1. Bar, 5 µm.
Mutant Atp13a2Isoform-1 proteins are unstable and degraded by the proteasome
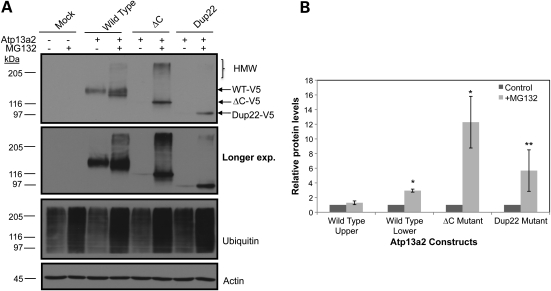
We next examined whether accumulation of the Atp13a2Isoform-1 proteins in HeLa cells were sensitive to proteasome inhibition, as this is characteristic of ERAD substrates (Fig. 2). Accordingly, we compared the accumulation of transiently expressed wild-type and mutant V5-tagged Atp13a2Isoform-1 proteins in HeLa cells treated with or without the proteasome inhibitor MG132. An anti-ubiquitin immunoblot confirmed that the MG132 treatment induces a build-up of ubiquitinated substrates, as expected. A parallel immunoblot of the cell lysates revealed that the levels of both Atp13a2-▵CIsoform-1 and Atp13a2-Dup22Isoform-1 mutant proteins were increased dramatically after MG132 treatment. Interestingly, two closely migrating forms of the wild-type Atp13a2Isoform-1 protein were seen in cells treated with MG132: an upper band which increased only modestly after MG132 treatment and a lower band that increased more dramatically. We speculate that the upper band is the mature glycoslyated form of the protein, whereas the lower band is the immature and/or incorrectly folded form of the protein. Quantification of the two bands after MG132 treatment indicated that the lower band comprises ∼25% of the total wild-type Atp13a2Isoform-1 protein, which is in accord with the estimate of the frequency in errors that is known to occur during protein synthesis (23). Evidence suggesting the upper band is indeed glycosylated was found upon treatment of cells with tunicamycin to inhibit N-glycosylation, which resulted in a time-dependent disappearance of the upper band and a concomitant increase in the lower band (Supplementary Material, Fig. S5). Interestingly, high molecular weight species of the Atp13a2Isoform-1 proteins were observed after MG132 treatment, which is characteristic of ubiquitinated proteins and further supports degradation of the proteins by the proteasome.
Figure 2.
Proteasome inhibition and Atp13a2Isoform-1 protein levels. (A) HeLa cells were either mock transfected or transfected with the indicated V5-tagged Atp13a2Isoform-1 expression constructs. Twenty hours after transfection, the cells were treated with or without MG132 (50 mm) for 6 h. The indicated Atp13a2Isoform-1 proteins were detected by immunoblotting of the lysates using an antibody to V5. The lysates were also immunoblotted for actin (to assess loading) and ubiquitin (to confirm proteasome inhibition) as shown. HMW indicates migration of high molecular weight species. (B) Quantification of the relative change in protein levels before and after MG132 treatment for each Atp13a2 construct from three independent experiments. Data are shown as the mean ± standard deviation of the mean (SDM) (*P < 0.005) (**P < 0.05).
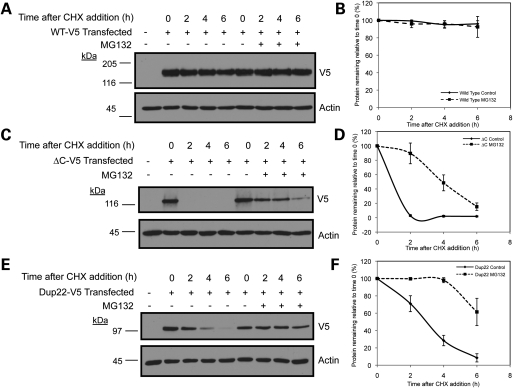
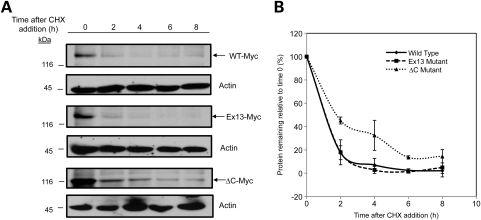
We next measured the turnover rates of the V5-tagged Atp13a2Isoform-1 expressed proteins in HeLa cells that were treated with or without MG132 using cycloheximide-chase analysis (Fig. 3). In accord with the previous findings, in the absence of MG132 treatment both the Atp13a2-▵CIsoform-1 and Atp13a2-Dup22Isoform-1 mutants turned over rapidly compared with the Atp13a2-WTIsoform-1 protein, which was remarkably stable. The Atp13a2-▵CIsoform-1 mutant turned over more rapidly than the Atp13a2-Dup22Isoform-1 mutant, having an estimated half-life of <1 h compared with 3 h, respectively (Fig. 3). In contrast, the wild-type protein was estimated to have a half-life in excess of 24 h (Supplementary Material, Fig. S7). As expected, treatment with MG132 slowed the turnover of the two mutant proteins dramatically, but had little effect on the wild-type protein (Fig. 3). We were unable to measure differences in the turnover of the immature and mature forms of the wild-type protein due to close migration of the two bands on our immunoblots. However, due to the extreme stability of the wild-type protein, we estimate that the contribution of the lower band on the turnover of the protein is minimal. These results confirmed that the ▵C and Dup22 Atp13a2Isoform-1 mutants are degraded rapidly in a proteasome-dependent manner. To confirm the results of our cycloheximide-chase experiments, we also measured the rates of turnover of the wild-type and ▵C mutant Atp13a2Isoform-1 using classical pulse-chase analysis (Supplementary Material, Fig. S6). Phosphoimage analysis of the 35S-labeled proteins yielded similar turnover rates indicating the reliability of cycloheximide-chase experiments for measuring Atp13a2 protein turnover.
Figure 3.
Turnover of Atp13a2Isoform-1 proteins. (A–F) HeLa cells were either mock transfected or transfected with V5-tagged Atp13a2-WTIsoform-1 (A), Atp13a2-▵CIsoform-1 (C), Atp13a2-Dup22Isoform-1 (E). Twenty hours after transfection, cells were treated with cycloheximide (CHX) and MG132 or DMSO vehicle and lysates collected at the indicated time points. Equal amounts of lysates were immunoblotted for V5 and actin. Quantification of Atp13a2Isoform-1 protein turnover with or without MG132 treatment relative to time point zero is shown for wild-type (B), ΔC (D) and Dup22 (F) from three independent experiments. Data are shown as mean ± SDM. Treatment with MG132 significantly slowed the turnover of the ΔC (P < 0.0005) and Dup22 (P < 0.005) mutants.
Evidence that mutant Atp13a2Isoform-1 proteins are ubiquitinated
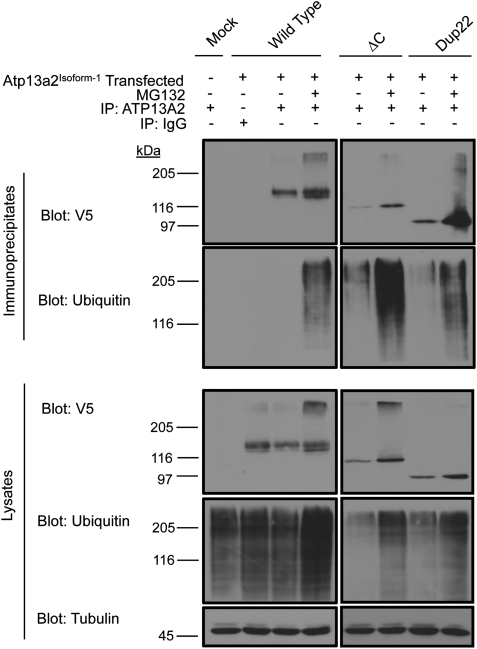
The ER localization and rapid degradation of the mutant Atp13a2Isoform-1 proteins is consistent with the degradation of the proteins by ERAD. Because ERAD substrates are typically ubiquitinated, we examined if the Atp13a2Isoform-1 proteins are similarly modified. This issue was unresolved because the previous report did not find any evidence for ubiquitin modification of Atp13a2Isoform-1 proteins (4). To determine if Atp13a2Isoform-1 proteins are ubiquitinated, we immunoprecipitated the V5-tagged wild-type and mutant Atp13a2Isoform-1 proteins from HeLa cells using an antibody to Atp13a2 that we had generated and examined the immunoprecipitates for the presence of Atp13a2Isoform-1 proteins and for ubiquitin by immunoblotting (Fig. 4). To ensure that our immunoprecipitates contained only Atp13a2 proteins and were devoid of its associated proteins, the immunoprecipitations were performed in the presence of SDS to break up any potential protein complexes. As shown in Figure 4, our Atp13a2 antibody immunoprecipitated the Atp13a2-WTIsoform-1, -▵CIsoform-1 and -Dup22Isoform-1 proteins successfully. The specificity of the immunoprecipitations is evident from the failure of the pre-immune serum to immunoprecipitate the Atp13a2-WTIsoform-1 protein. As expected, considerably more ▵C and Dup22 mutant proteins were immunoprecipitated from cells treated with MG132 than from untreated cells, confirming proteasome inhibition leads to a stabilization of the proteins. MG132 treatment also increased the amount of the wild-type protein that was immunoprecipitated, which was especially noticeable for the lower of the two bands of the protein, again suggesting it corresponds to the unstable and incorrectly folded form of the protein. The anti-ubiquitin blots produced a strong reaction with products migrating as a smear on the upper portions of the gels, especially after MG312 treatment, indicating that the Atp13a2Isoform-1 proteins are indeed modified by ubiquitination.
Figure 4.
Atp13a2Isoform-1 ubiquitination. HeLa cells were either mock transfected or transfected with V5-tagged Atp13a2-WTIsoform-1, Atp13a2-Dup22Isoform-1 or Atp13a2-▵CIsoform-1 and treated with or without MG132. Atp13a2Isoform-1 proteins were immunoprecipitated from the cell lysates using an antibody to Atp13a2 and the indicated proteins detected using antibodies to V5 and ubiquitin. Atp13a2Isoform-1 was not detected in mock transfected cells (lane 1) or in transfected cells immunoprecipitated with pre-immune serum (lane 2). Bottom panels show whole cell lysates used for the immunoprecipitations blotted for V5, ubiquitin and tubulin.
Mutant Atp13a2Isoform-1 proteins are degraded by ERAD
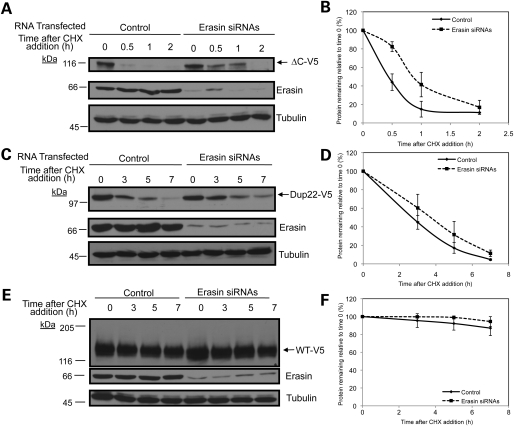
Because Atp13a2Isoform-1 mutants were retained in the ER and are degraded by the proteasome, we predicted that the proteins are eliminated from cells by the ERAD pathway. To examine this possibility, we disrupted two components involved in ERAD to see if it affected the turnover of the Atp13a2 proteins. The first protein we targeted was erasin. Previous studies had found that siRNA-mediated knockdown of erasin slows the degradation of CD3δ and alpha 1-antitrypsin substrates (22,24). We questioned whether inhibition of erasin would also slow the turnover of the Atp13a2Isoform-1 mutants. Accordingly, we transfected HeLa cells with V5-tagged Atp13a2-▵CIsoform-1 or Atp13a2-Dup22Isoform-1 after siRNA-mediated knockdown of erasin and assessed protein turnover by cycloheximide-chase analysis (Fig. 5). As shown in Figure 5A and B, siRNA knockdown of erasin in HeLa cells slowed the turnover of Atp13a2-▵CIsoform-1. Similarly, knockdown of erasin slowed the turnover of Atp13a2-Dup22Isoform-1, although the effect was marginal compared with the ▵C mutant (Fig. 5C and D). In contrast, siRNA knockdown of erasin had no affect on the turnover of wild-type Atp13a2Isoform-1 protein (Fig. 5E and F).
Figure 5.
Effects of knockdown of erasin protein on and Atp13a2Isoform-1 turnover. (A–F) HeLa cells were transfected with V5-tagged ▵C (A) Dup22 (C) or wild-type (E) Atp13a2Isoform-1 expression constructs 48 h after transfection with erasin siRNAs. At 72 h post-knockdown, cells were treated with cycloheximide for the indicated time points and the proteins detected by immunoblotting. Quantification of Atp13a2Isoform-1 protein turnover with and without erasin siRNAs relative to time point zero from three independent experiments is shown for ▵C (B), Dup22 (D) and wild type (F) proteins. Data is shown as mean ± SDM. (ΔC P < 0.05, wild-type P < 0.05.)
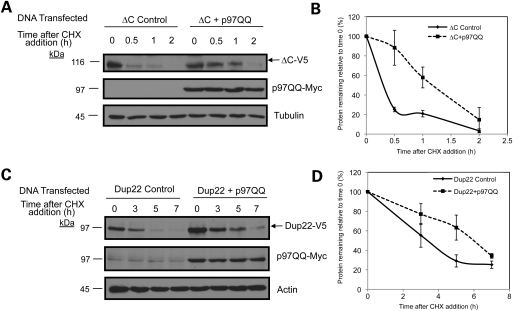
We next examined whether interference of p97/VCP function, which is essential for dislocation of proteins from the ER during ERAD, also affects turnover of the Atp13a2Isoform-1 mutant proteins. Initially, we attempted to do this by siRNA knock down of p97/VCP expression, but were hampered by encountering massive cell death (data not shown). Instead, we transfected cells with a dominant-negative ATPase deficient p97/VCP-QQ mutant, which has been shown to slow degradation of ERAD substrates (25,26). Accordingly, we cotransfected HeLa cells with myc-tagged p97/VCP-QQ and V5-tagged Atp13a2-▵CIsoform-1 or Atp13a2-Dup22Isoform-1 and assessed protein turnover by cycloheximide-chase analysis. As shown in Figure 6, coexpression of p97/VCP-QQ stabilized the turnover of both the Atp13a2-▵CIsoform-1 and Atp13a2-Dup22Isoform-1. The above results strongly indicate that the Atp13a2Isoform-1 mutant proteins are degraded by ERAD.
Figure 6.
Effects of p97/VCP-QQ expression on Atp13a2Isoform-1 turnover. (A–D) HeLa cells were cotransfected with myc-tagged p97/VCP-QQ and either ▵C (A) or Dup22 (C) V5-tagged Atp13a2Isoform-1. Approximately 20 h after cotransfection, cells were treated with cycloheximide as described in previous figures. Quantification of Atp13a2Isoform-1 protein turnover with and without p97/VCP-QQ relative to time point zero from three independent experiments for ▵C (B) and Dup22 (D). Data are shown as mean ± SDM (ΔC P < 0.05, Dup22 P < 0.05).
Characterization of Atp13a2Isoform-3 protein
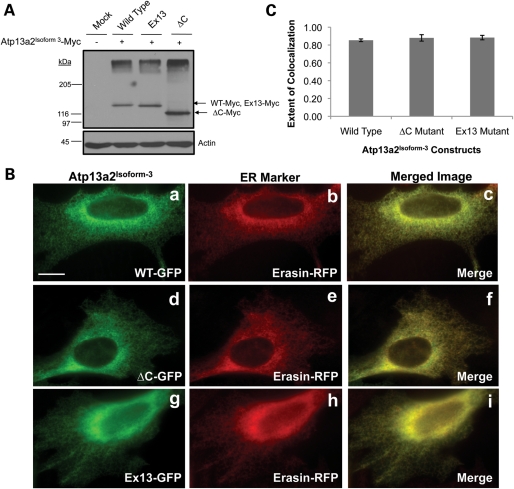
We next focused on isoform-3 form of the Atp13a2 protein to see if it behaved similar to isoform-1. In order to characterize the Atp13a2Isoform-3 protein, we generated myc-tagged expression constructs encoding the wild-type Atp13a2Isoform-3 protein and versions containing the KRS-linked ▵C and Ex13 mutations. Immunoblot analysis of transfected HeLa lysates indicated that all three constructs were expressed according to their predicted molecular weights (Fig. 7A). However, the pattern of expression of the wild-type Atp13a2Isoform-3 protein differed from that seen with the wild-type Atp13a2Isoform-1 protein. Wild-type Atp13a2Isoform-3 was expressed poorly compared with the Atp13a2Isoform-1 protein, which is evident by its lower expression compared with the mutants, particularly the ▵C mutant, suggesting that the isoform-3 protein may be less stable than its isoform-1 counterpart. Additionally, immunofluorescence staining of HeLa cells transfected with the myc-tagged constructs indicated that the wild-type Atp13a2Isoform-3 protein had a reticular staining pattern similar to that of the Ex13 and ▵C mutants (data not shown). To establish the localization of Atp13a2Isoform-3 proteins, we generated corresponding GFP-tagged Atp13a2Isoform-3 fusion constructs and cotransfected them with mRFP-tagged erasin in HeLa cells. Double fluorescence microscopy revealed that the wild-type and the two Ex13 and ▵C mutant Atp13a2Isoform-3-GFP fusion proteins colocalized with mRFP-erasin, indicating that all of the Atp13a2Isoform-3 fusion protein are localized in the ER (Fig. 7B and C). Further examination revealed that the GFP-tagged wild-type Atp13a2Isoform-3 did not colocalize with Lysotracker or cotransfected LAMP1-Cherry, unlike the wild-type Atp13a2Isoform-1 protein (Supplementary Material, Fig. S4). These results suggest that the wild-type Atp13a2Isoform-1 and Isoform-3 proteins have different localizations in the lysosome and ER, respectively, suggesting they could have unique functions in these organelles.
Figure 7.
Expression and localization of Atp13a2Isoform-3 constructs in HeLa cells. (A) HeLa cells were mock transfected or transfected with myc-tagged Atp13a2-WTIsoform-3, Atp13a2-▵CIsoform-3 or Atp13a2-Ex13Isoform-3 and the indicated proteins detected by immunoblotting using an antibody to myc. (B) HeLa cells were cotransfected with GFP-tagged wild-type (a–c), ▵C (d–f) or Ex13 (g–i) Atp13a2Isoform-3 and Erasin-mRFP and visualized using fluorescence microscopy. Both wild-type and mutant Atp13a2Isoform-3 colocalize with the ER protein erasin. (C) Quantification of the extent of colocalization of GFP-tagged Atp13a2Isoform-3 and erasin calculated from three different cells. Bar, 5 µm.
Wild-type and mutant Atp13a2Isoform-3 proteins are ubiquitinated and rapidly degraded by the proteasome
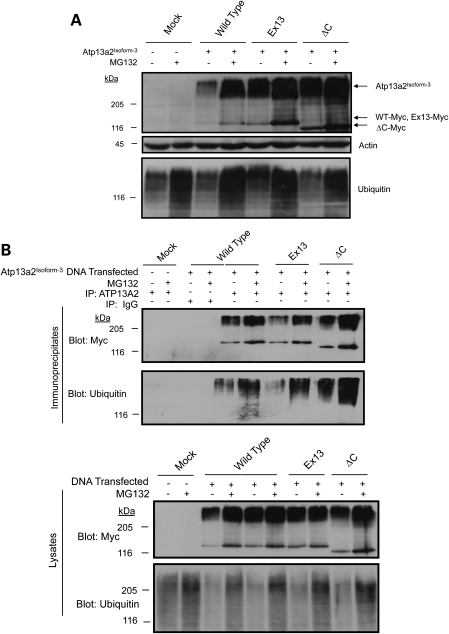
Because Atp13a2Isoform-3 proteins were localized to the ER, we next examined whether the proteins were degraded by the ubiquitin–proteasome system. Immunoblotting of the transfected cell lysates using a myc antibody revealed an increase in wild-type and mutant Atp13a2Isoform-3 protein levels after MG132 treatment, consistent with degradation of the proteins by the proteasome. Interestingly, we also observed the same increase in the high molecular weight forms of the proteins after MG132 treatment that we observed for isoform-1 proteins which suggests modification of these proteins by ubiquitin (Figs 8A and 2). Furthermore, examination of the expressed proteins following their immunoprecipitation from cell lysates in the presence of SDS confirmed that the proteins are ubiquitinated and their levels increase following inhibition of the proteasome with MG132 (Fig. 8B).
Figure 8.
Degradation of Atp13a2Isoform-3. (A and B) HeLa cells were either mock transfected or transfected with the indicated myc-tagged Atp13a2Isoform-3 expression constructs. At 20 h post-transfection, the cells were treated with MG132 (50 mm) or DMSO. The indicated proteins were detected by immunoblotting. (A) Immunoblot showing Atp13a2Isoform-3 proteins increase after MG132 treatment. (B) Immunoprecipitation of transfected myc-tagged Atp13a2Isoform-3 from HeLa cells after MG132 treatment. The immunoprecipitates were immunoblotted for myc and ubiquitin. Bottom panels show whole cell lysates used for the immunoprecipitations blotted for myc and ubiquitin.
We next measured the turnover of the Atp13a2Isoform-3 proteins using cycloheximide-chase analysis. Immunoblotting for the indicated proteins confirmed the rapid turnover of wild-type Atp13a2Isoform-3 with little protein remaining after 2 h (Fig. 9). The Atp13a2-▵CIsoform-3 and Atp13a2-Ex13Isoform-3 mutant proteins also turned over rapidly although the turnover of the ▵C mutant was slightly slower than the Ex13 mutant (Fig. 9A and B). Taken together, these results suggest that Atp13a2Isoform-3 proteins, particularly the wild-type and Ex13 containing mutant, are retained in the ER and rapidly degraded by the proteasome. The very small fraction of the ▵C mutant that has an extended half-life suggests a small amount of this mutant protein might escape from the ER or resist degradation. Analysis of cell death further revealed that overexpression of the wild-type Atp13a2Isoform-3 protein induces an increase in cell death similar to the ▵C and Ex13 mutants (Supplementary Material, Fig. S8), which is different to the properties of the wild-type Atp13a2Isoform-1 protein (see in what follows).
Figure 9.
Turnover of Atp13a2Isoform-3. (A) HeLa cells were transfected with either wild-type (top panel), Ex13 mutant (middle panel) or ▵C mutant (bottom panel) V5-tagged Atp13a2Isoform-3. Twenty hours after transfection, cells were treated with cycloheximide (CHX) and lysates collected at the indicated time points. Equal amounts of lysates were immunoblotted for Atp13a2 and actin. (B) Quantification of Atp13a2Isoform-3 protein remaining shown in (A) relative to time point zero. The graph represents an average of three experiments. Data are shown as mean ± SDM. Atp13a2Isoform-3 ΔC turns over slower than wild-type or Ex13 Atp13a2Isoform-3 (P < 0.0005).
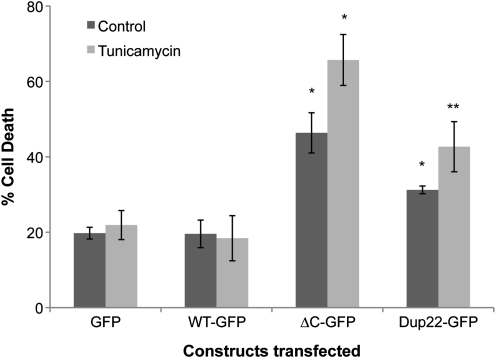
Atp13a2Isoform-1 mutant proteins are toxic and sensitize cells to increased ER stress-induced cell death
Because prolonged activation of ER-stress caused by accumulation of misfolded proteins in the ER can trigger cell death, we examined whether overexpression of wild-type and mutant misfolded Atp13a2Isoform-1 proteins enhance cell death (Fig. 10). To evaluate this possibility, we transfected HeLa cells with GFP-tagged Atp13a2Isoform-1 versions of the constructs and quantified cell death in the absence or presence of 2 μg/ml tunicamycin, which was added to induce ER stress. The quantification revealed that both ▵C and Dup22 mutations that cause KRS increased basal cell death, albeit to different levels, compared with cells transfected with constructs encoding wild-type Atp13a2Isoform-1-GFP protein or GFP protein alone (Fig. 10). Furthermore, both mutants, but not the wild-type or GFP control, were hypersensitive to ER stress-induced cell death caused by tunicamycin treatment. Immunoblots revealed that the cells transfected with the Atp13a2Isoform-1 mutants induced slightly higher BiP levels than the cells transfected with GFP alone or wild-type GFP-Atp13a2Isoform-1 (Supplementary Material, Fig. S9). These results suggest that the ▵C and Dup22 mutations increase toxicity and vulnerability of cells to ER-stress induced cell death.
Figure 10.
Expression of Atp13a2Isoform-1 and quantification of cell death. HeLa cells were transfected with either GFP alone or with the indicated GFP-tagged Atp13a2Isoform-1 constructs. Twenty hours after transfection, the cells were treated with tunicamycin or DMSO vehicle control for 8 h at a final concentration of 2 µg/ml. After 8 h, cell death was quantified by counting GFP-positive cells that had highly condensed/fragmented DNA after staining of the cells with the nuclear dye Hoechst 33342. Graph is an average of three independent experiments and the data are shown as the mean ± standard deviation of the mean. Cells expressing mutant Atp13a2Isoform-1 show a significant increase in cell death compared with GFP and WT-GFP-expressing cells, with or without tunicamycin treatment (*P < 0.0005) (**P < 0.005), respectively.
DISCUSSION
Proper protein folding and processing is necessary to maintain cellular homeostasis. Protein misfolding could potentially not only affect a protein's specific function but it could also lead to aggregation and induce toxicity. Not surprising, cells have developed elaborate and complex systems to eliminate unwanted and potentially toxic proteins (27,28). One of the first quality control checkpoints is in the ER, where during synthesis, misfolded proteins are recognized and eliminated by ERAD. Although many mutations involved in human disease are thought to cause proteins to misfold, relatively few of them have been have been shown to be eliminated by ERAD (13,15). Here, we have presented evidence that strongly indicates mutations in Atp13a2 proteins linked to KRS are degraded by ERAD, consistent with the idea that the mutations cause the proteins to misfold. In fact, to our knowledge, this is the first example of a protein involved in neurodegeneration that has been shown to be eliminated by ERAD.
Four different observations support our contention that Atp13a2 proteins carrying the ▵C and Dup22 mutations are degraded by ERAD. First, we confirmed the previous report showing that the wild-type Atp13a2Isoform-1 protein localizes to lysosomes, whereas the ▵C and Dup22 mutant proteins are retained in the ER, which is a characteristic of proteins degraded by ERAD (13,14). Second, through proteasome inhibition and analysis of protein turnover, we also confirmed that the mutant Atp13a2 proteins have vastly accelerated rates of degradation compared with the wild-type Atp13a2Isoform-1 protein, and that the proteasome is responsible for the increased turnover of the proteins. Third, as expected for proteins that are degraded by the proteasome, we found Atp13a2 proteins are ubiquitinated and that the ubiquitinated forms of the proteins increase following proteasome inhibition. Fourth, interference of ERAD by overexpression of either a dominant negative p97/VCP-QQ protein or knockdown of erasin expression both slowed degradation of the mutant proteins.
The dominant-negative p97/VCP-QQ impeded the turnover of the mutant Atp13a2 proteins more potently than erasin knockdown. We speculate this is because p97/VCP is a universal and essential factor required for dislocation of all ERAD substrates (25), whereas erasin could have a more restricted role in ERAD. For example, the ERAD complex containing erasin is distinct from other ERAD complexes found in mammalian cells, suggesting it is part of the larger compendium of ERAD complexes that exist in cells (24). Furthermore, there is accumulating evidence that different ERAD complexes are dedicated to disposing different types of substrates, particularly with regard to whether the lesions that cause the proteins to misfold reside in the lumen, membrane or cytosolic sides of the ER membrane (14,29). Interestingly, we noted that erasin knockdown slowed turnover of the ▵CIsoform-1 more than the Dup22Isoform-1 mutant. The ▵C mutation occurs in the seventh transmembrane domain while the Dup22 mutation occurs in the cytoplasmic domain of the Atp13a2 protein (Supplementary Material, Fig. S3), causing a difference in the utilization of the erasin complex in disposing the two substrates. Another possibility is that the substrates might be degraded by more than one ERAD complex.
In contrast to the mutants, proteasome inhibition had little effect on the turnover of the wild-type Atp13a2Isoform-1 protein. However, we did observe a small portion of wild-type protein (∼25%) that was stabilized by proteasome inhibition, which corresponded to the lower of two closely migrating bands. Based on the disappearance of the upper band and a concomitant increase in the lower band following tunicamycin treatment, we speculate that the mature Atp13a2Isoform-1 protein is glycoslyated and that the lower band corresponds to immature and/or incorrectly folded form of the protein. Our estimate that ∼25% of Atp13a2Isoform-1 protein misfolds during synthesis is close to the 35% rate of errors that is estimated to occur during protein synthesis (23).
What we found particularly interesting was the difference in properties of the Atp13a2Isoform-3 protein compared with that of Atp13a2Isoform-1 protein. First, the Atp13a2Isoform-3 was localized exclusively to the ER instead of the lysosome as with Atp13a2Isoform-1. Second, Atp13a2Isoform-3 turned over exceedingly rapidly compared with Atp13a2Isoform-1. These results suggest Atp13a2Isoform-3 is either non-functional, misfolded or a short-lived ER-resident protein that is rapidly degraded by the proteasome. Although it is theoretically possible that the Atp13a2Isoform-3 is not translated, we believe that this is unlikely because of the abundance of the transcript (in expression databases) and the identity of most of its 5′ sequences including an intact translation start codon and sequences upstream and downstream of it compared with isoform-1. The possibility that the Atp13a2Isoform-3 generates a misfolded protein could arise because of the frame-shift event that occurs after the eight transmembrane domain of the protein creating a unique C-terminus compared with the Atp13a2Isoform-1 protein. This frame shift could generate a truncated non-functional or misfolded protein. Another possibility is that the Atp13a2Isoform-3 might have a unique function in the ER compared to the Atp13a2Isoform-1 based on differences in the localization of the two isoforms. Further studies are needed to clarify these issues.
The mechanism by which Atp13a2 mutant proteins cause disease is unknown. Our data suggest that the ▵C and the Dup22 Atp13a2 mutants are cleared by ERAD and never reach the lysosome where the normal wild-type Atp13a2Isoform-1 protein resides. The premature disposal of the mutant proteins is likely to lead to a loss of the normal Atp13a2 function in lysosomes. The lysosome is home to a variety of proteolytic enzymes that are involved in degrading unwanted cytosolic proteins, toxic aggregates and damaged organelles. Mutations in lysosomal membrane proteins have been linked to disease (30). Also, dysfunctions in autophagy, the process in which unwanted proteins in the cytosol are sequestered and delivered to the lysosomes for degradation, has been implicated in neurodegenerative diseases including PD and AD (31,32). Thus it is not surprising that loss of Atp13a2 function in lysosomes might cause disease. At present, it remains unclear whether the etiology of KRS originates from the complete or partial loss of Atp13a2 function, and/or whether it might result from toxicity intrinsic to the mutant proteins, as we have shown here. For example, although the majority of KRS cases studied so far are linked to recessive inheritance of mutations in the ATP13A2 gene, there are several reports that suggest disease symptoms can arise in patients who possess only one mutant ATP13A2 allele, suggesting that KRS does not necessarily manifest from complete loss of Atp13a2 function (5,7,9,33).
We speculate that an alternative and not mutually exclusive mode through which the mutants might induce disease is through ER stress. Accumulation of misfolded proteins in the ER has been shown to induce ER stress, which has also been implicated in human disease, including neurodegenerative diseases (17). Our results suggest ER stress may play a role in the development of KRS. First, expression Atp13a2 mutants that are mislocalized to the ER leads to an increase in cell death compared with the wild-type protein. Second, the presence of these mutants in the ER leads to increased cell death after exposure to tunicamycin, a known ER stressor. This suggests misfolded Atp13a2 proteins may negatively impact ER homeostasis causing an increase in susceptibility to ER stress-induced cell death. Therefore, it will be important to determine whether KRS is caused by complete or partial loss of Atp13a2 function, from toxicity associated with the mutations, or some combination of them.
MATERIALS AND METHODS
Expression constructs
V5-tagged Atp13a2Isoform-1 expression plasmids were described previously (4). GFP-tagged Atp13a2 expression constructs were made by fusing GFP to the C-terminus of the Atp13a2 proteins. The LAMP1-mCherry construct was obtained from Dr. Jennifer Lippincott-Schwartz (34). The p97/VCP QQ-myc construct was described previously (35). mRFP–Erasin was generated by fusing mRFP to the C-terminus of full-length erasin. Atp13a2Isoform-3 cDNA (Origene) was cloned into a CMV expression vector upstream of a myc epitope tag. Atp13a2Isoform-3 mutants were generated by PCR mutagenesis, and verified by DNA sequencing, which was performed by the Biopolymer-Genomic Core Facility at the University of Maryland, Baltimore.
Cell culture, DNA transfection, immunofluorescence microscopy and cell death assays
HeLa cells were maintained at 37°C in DMEM supplemented with 10% FBS. Cells were transiently transfected with plasmid DNA by either the calcium phosphate coprecipitation method or Lipofectamine 2000 (Invitrogen). For proteasome inhibition experiments, cells were treated with MG132 (Calbiochem) for 6 h at a final concentration of 50 µm. For fluorescence microscopy, HeLa cells were grown on glass coverslips, fixed with 1% paraformaldehyde, treated with 70% ethanol and stained with the indicated primary antibodies and then with the appropriate florescent-conjugated secondary antibodies. The following primary antibodies were used: anti-calreticulin (Stressgen) anti-V5 (Sigma-Aldrich). The following secondary antibodies were used: Alexa Fluor goat anti-rabbit 594 and goat anti-mouse 488 (Invitrogen). For GFP microscopy work, cells were fixed with paraformaldehyde and directly mounted on slides. For colocalization with Lysotracker, cells were incubated in DMEM for 2 h at a final concentration of 50 nm prior to viewing (Molecular Probes, Invitrogen). Images were captured using a Leica DM IRB microscope and a Leica PL APO 100x/1.4 oil-immersion lens. Quantification of colocalization depicted is an average of three independent cells. Colocalization and quantification were performed using iVision-Mac software (BioVision Technologies). We used the iVision software to measure the Pearson's and M1 and M2 correlation coefficient of colocalization of red and green fluorescent signals captured of the same microscopy field. Cell death assays were performed as described previously (36).
SDS–PAGE and immunoblotting
Cells were collected in protein lysis buffer (0.5% SDS, 0.5% NP40, 0.5% sodium N-lauroylsarcosine, 50 mm Tris, pH 6.8, 150 mm NaCl, 20 mm EDTA, 1 mm EGTA, 25 mm sodium fluoride, 1 mm sodium orthovanadate, 1 mm Pefabloc (AEBSF, Boehringer Mannheim) 1 mm leupeptin and 1 mm aprotonin) (37) and further lysed by passing the lysates 10 times through a 25 G needle. Protein concentrations were determined by the bicinchonic acid assay (Thermo Fisher Scientific). Lysates were prepared with sample buffer (8 M urea, 15 mm DTT and 10% β-mercaptoethanol) and heated for 15 min at 37°C prior to loading. Equal amounts of protein were separated on a 7.5% SDS–PAGE gel and transferred onto a 0.45 mm PVDF membrane (Millipore) followed by incubation with the appropriate primary and HRP-conjugated secondary antibodies. Proteins were transferred for 3 h at 200 mA using the Mini-Trans Blot cell system (BioRad). Proteins were detected using the SuperSignal West Pico system (Thermo Fisher Scientific). The following primary antibodies were used: anti-Grp78 (BiP) (BD Biosciences), anti-tubulin and anti-V5 (both from Sigma-Aldrich), anti-V5, (Invitrogen), anti-ubiquitin and anti-actin (both from Santa Cruz Biotechnology, Inc.) and anti-myc (monoclonal 9E10), anti-Atp13a2, anti-p97/VCP and anti-erasin (generated by our laboratory). The anti-Atp13a2 antibody was generated (by Covance, Denver, PA, USA for us) against a purified GST-fusion protein composed of the Atp13a2 sequence from amino acid 81–154, which is common to all three isoforms, fused to the C-terminus of GST (Pharmacia). The antibody was affinity-purified from rabbit antiserum using the GST-fusion antigen coupled to Affi-Gel 10 (BioRad). The antibody was validated by its reaction with V5- and GFP-tagged Atp13a2 proteins expressed in HeLa cells as well as with the endogenous proteins expressed in different human cells. The anti-p97/VCP antibody was made against bacterially expressed and purified full-length His-tagged murine p97/VCP protein, and the anti-erasin antibody was made against a synthetic peptide corresponding to amino acids 476–500 of human erasin (22) and affinity-purified using the same peptide. The following HRP-conjugated secondary antibodies were used: goat anti-mouse (Thermo Fisher Scientific), bovine anti-goat (Santa Cruz Biotechnology) and donkey anti-rabbit (GE Healthcare).
Immunoprecipitations
HeLa cells were transfected with Atp13a2 expression constructs using Lipofectamine 2000 (Invitrogen). Approximately 20 h after transfection, cells were treated with MG132 or with the DMSO vehicle for 6 h. Cells were collected with SDS protein lysis buffer containing the deubiquitinating enzymes inhibitor, 10 mm N-ethylmaleimide (NEM), and sheared by passing the lysates through a 25 G needle. The lysates were spun at 14 000g for 10 min and the resulting supernatant diluted 10-fold with immunoprecipitation (IP) buffer (50 mm Tris–HCl pH7.5, 150 mm NaCl, 2 mm EDTA, 1% NP40, 10 mm NEM). The lysates were pre-cleared by incubating with rabbit serum for 30 min followed by protein A-Sepharose Cl-4B beads (GE Healthcare) for 1 h. The lysates were recovered by centrifugation at 14 000g for 10 min after which the supernatant was moved to a fresh tube. The supernatant was incubated with 7 µL of polyclonal anti-Atp13a2 antibody for 1.5 h followed by protein A-Sepharose Cl-4B beads for another 1.5 h. All incubations were carried out at 4°C with gentle rotation. The beads were recovered by centrifugation and washed four times with IP buffer. The proteins were eluted from the beads by incubation with sample loading buffer at 37°C for 15 min. Equal volumes of supernatant were separated on SDS–PAGE and the precipitated proteins detected via immunoblotting.
Analysis of protein turnover
For analysis of protein turnover, HeLa cells were transfected with the appropriate Atp13a2 expression construct by electroporation following the procedure described previously (38). Approximately 20 h after transfection, cycloheximide (Sigma-Aldrich) was added to the cultures to a final concentration of 100 µM to inhibit new protein synthesis and protein lysates were collected at appropriate time points thereafter, as shown in the figure. To measure protein turnover after proteasome inhibition, the cultures were treated in an identical manner except that MG132 was added to the cultures at the same time as cycloheximide. For analysis of protein turnover after knockdown of ERAD proteins, HeLa cells were transfected with erasin SMARTpool siRNAs (Dharmacon) using Dharmafect 1 reagent according to the manufacturer's instructions (Thermo Fisher Scientific). At 48 h post-knockdown, cells were transfected with V5-tagged Atp13a2 using calcium phosphate. At 72 h post-knockdown, cells were treated with cycloheximide and collected at the indicated time points. For turnover experiments with p97/VCP QQ-myc, the DNA construct was cotransfected with Atp13a2 plasmid DNA followed by inhibition of protein synthesis with cycloheximide. Equal amounts of protein in lysates from the different time points were separated by SDS–PAGE and immunoblotted for the proteins as shown in the figures. The rate of protein turnover was calculated from at least three independent experiments and the Microsoft Excel program was used to draw a line connecting the average values through each time point.
Statistical analysis
Student's t-tests were used for all statistical analysis. For all data, error bars represent ± the standard deviation of the mean (SDM). P < 0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a NIH grant GM066287 to M.J.M.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jennifer Lippincott-Schwartz (NICHD, NIH) for kindly providing the LAMP1-Cherry construct.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Thomas B., Beal M.F. Parkinson's disease. Hum. Mol. Genet. 2007;16(Spec no. 2)):R183–R194. doi: 10.1093/hmg/ddm159. doi:10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 2.Lees A.J., Hardy J., Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. doi:10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 3.Belin A.C., Westerlund M. Parkinson's disease: a genetic perspective. FEBS J. 2008;275:1377–1383. doi: 10.1111/j.1742-4658.2008.06301.x. doi:10.1111/j.1742-4658.2008.06301.x. [DOI] [PubMed] [Google Scholar]
- 4.Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F., Wriekat A.L., Roeper J., et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. doi:10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 5.Di Fonzo A., Chien H.F., Socal M., Giraudo S., Tassorelli C., Iliceto G., Fabbrini G., Marconi R., Fincati E., Abbruzzese G., et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology. 2007;68:1557–1562. doi: 10.1212/01.wnl.0000260963.08711.08. doi:10.1212/01.wnl.0000260963.08711.08. [DOI] [PubMed] [Google Scholar]
- 6.Ning Y.P., Kanai K., Tomiyama H., Li Y., Funayama M., Yoshino H., Sato S., Asahina M., Kuwabara S., Takeda A., et al. PARK9-linked parkinsonism in eastern Asia: mutation detection in ATP13A2 and clinical phenotype. Neurology. 2008;70:1491–1493. doi: 10.1212/01.wnl.0000310427.72236.68. doi:10.1212/01.wnl.0000310427.72236.68. [DOI] [PubMed] [Google Scholar]
- 7.Lin C.H., Tan E.K., Chen M.L., Tan L.C., Lim H.Q., Chen G.S., Wu R.M. Novel ATP13A2 variant associated with Parkinson disease in Taiwan and Singapore. Neurology. 2008;71:1727–1732. doi: 10.1212/01.wnl.0000335167.72412.68. doi:10.1212/01.wnl.0000335167.72412.68. [DOI] [PubMed] [Google Scholar]
- 8.Paisan-Ruiz C., Guevara R., Federoff M., Hanagasi H., Sina F., Elahi E., Schneider S.A., Schwingenschuh P., Bajaj N., Emre M., et al. Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov. Disord. 2010;25:1791–1800. doi: 10.1002/mds.23221. doi:10.1002/mds.23221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong C.Y., Rolfs A., Schwarzbraun T., Klein C., O'Callaghan F.J. Juvenile parkinsonism associated with heterozygous frameshift ATP13A2 gene mutation. Eur. J. Paediatr. Neurol. 2011;15:271–275. doi: 10.1016/j.ejpn.2011.01.001. doi:10.1016/j.ejpn.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Santoro L., Breedveld G.J., Manganelli F., Iodice R., Pisciotta C., Nolano M., Punzo F., Quarantelli M., Pappata S., Di Fonzo A., et al. Novel ATP13A2 (PARK9) homozygous mutation in a family with marked phenotype variability. Neurogenetics. 2011;12:33–39. doi: 10.1007/s10048-010-0259-0. doi:10.1007/s10048-010-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gitler A.D., Chesi A., Geddie M.L., Strathearn K.E., Hamamichi S., Hill K.J., Caldwell K.A., Caldwell G.A., Cooper A.A., Rochet J.C., et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 2009;41:308–315. doi: 10.1038/ng.300. doi:10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. doi:10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 13.Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. doi:10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 14.Vembar S.S., Brodsky J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. doi:10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. doi:10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 16.Bagola K., Mehnert M., Jarosch E., Sommer T. Protein dislocation from the ER. Biochim. Biophys. Acta. 2011;1808:925–936. doi: 10.1016/j.bbamem.2010.06.025. doi:10.1016/j.bbamem.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L., Ackerman S.L. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. doi:10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Lindholm D., Wootz H., Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. doi:10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- 19.Hoozemans J.J., van Haastert E.S., Eikelenboom P., de Vos R.A., Rozemuller J.M., Scheper W. Activation of the unfolded protein response in Parkinson's disease. Biochem. Biophys. Res. Commun. 2007;354:707–711. doi: 10.1016/j.bbrc.2007.01.043. doi:10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Nishitoh H., Kadowaki H., Nagai A., Maruyama T., Yokota T., Fukutomi H., Noguchi T., Matsuzawa A., Takeda K., Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. doi:10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duennwald M.L., Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. doi:10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang J., Yin C., Doong H., Fang S., Peterhoff C., Nixon R.A., Monteiro M.J. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J. Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. doi:10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- 23.Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. doi:10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 24.Lim P.J., Danner R., Liang J., Doong H., Harman C., Srinivasan D., Rothenberg C., Wang H., Ye Y., Fang S., et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J. Cell Biol. 2009;187:201–217. doi: 10.1083/jcb.200903024. doi:10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y., Meyer H.H., Rapoport T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. doi:10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 26.Ye Y., Meyer H.H., Rapoport T.A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. doi:10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. doi:10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 28.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. doi:10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho P., Goder V., Rapoport T.A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. doi:10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 30.Ruivo R., Anne C., Sagne C., Gasnier B. Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Biochim. Biophys. Acta. 2009;1793:636–649. doi: 10.1016/j.bbamcr.2008.12.008. doi:10.1016/j.bbamcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. doi:10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong E., Cuervo A.M. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. doi:10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behrens M.I., Bruggemann N., Chana P., Venegas P., Kagi M., Parrao T., Orellana P., Garrido C., Rojas C.V., Hauke J., et al. Clinical spectrum of Kufor-Rakeb syndrome in the Chilean kindred with ATP13A2 mutations. Mov. Disord. 2010;25:1929–1937. doi: 10.1002/mds.22996. doi:10.1002/mds.22996. [DOI] [PubMed] [Google Scholar]
- 34.Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl Acad. Sci. USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. doi:10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J. Biol. Chem. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. doi:10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Monteiro M.J. Ubiquilin overexpression reduces GFP-polyalanine-induced protein aggregates and toxicity. Exp. Cell Res. 2007;313:2810–2820. doi: 10.1016/j.yexcr.2007.04.006. doi:10.1016/j.yexcr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monteiro M.J., Mical T.I. Resolution of kinase activities during the HeLa cell cycle: identification of kinases with cyclic activities. Exp. Cell Res. 1996;223:443–451. doi: 10.1006/excr.1996.0100. doi:10.1006/excr.1996.0100. [DOI] [PubMed] [Google Scholar]
- 38.Mah A.L., Perry G., Smith M.A., Monteiro M.J. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J. Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. doi:10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.