FIGURE 1.
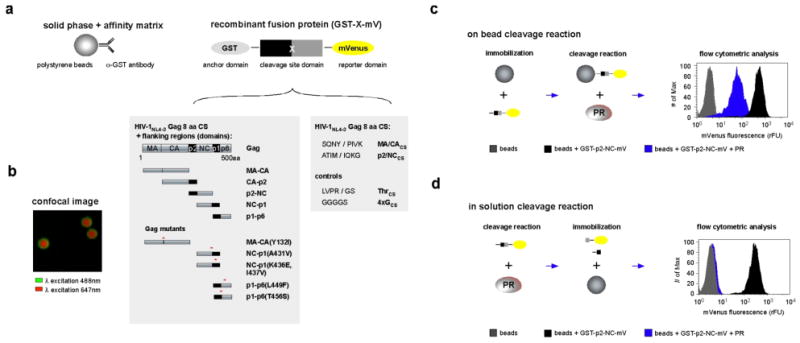
Overview of Cleavage Enzyme – Cytometric Bead Array. (a) Schematic description of the Cleavage Enzyme – Cytometric Bead Array (CE-CBA) platform containing the bead-based anti-GST antibody affinity matrix to which the recombinant fusion protein, GST-X-mV, is coupled. X refers to Gag substrate domains, with endogenous cleavage sites, or Gag cleavage site peptides all encoded from the appropriate HIV-1NL4-3 sequence. The two shaded boxes below the GST-X-mV depict Gag domains (left) or the cleavage site for various Gag domains (right) that have been inserted at the site X in GST-X-mV. Gag cleavage site peptides are referred to by the name of the Gag domain followed by a subscript CS, for the cleavage site. (b) Confocal fluorescence image of fluorescent beads (red) coupled to the GST-p2/NCCS-mVs (green). Note the homogeneous distribution of GST-p2/NCCS-mVs and the absence of discrete spots, which would be an indication of protein aggregation or clumping. (c) On bead CE-CBA of GST-p2-NC-mV by HIV-1 PR. The protease recognizes and cleaves the GST-p2-NC-mV at the specific cleavage site liberating the -NC-mV remnant containing mVenus from the bead. Loss of mVenus decreases the fluorescence signal on the bead, which can be quantified utilizing flow cytometry (histogram: right panel). (d) In solution CE-CBA of GST-p2-NC-mV by HIV-1 PR. In this assay GST-p2-NC-mV processing by protease occurs in solution and cleavage products and remaining uncleaved GST-p2-NC-mV is bound to the beads after the enzymatic reaction. Beads with a protease processed GST-p2-NC-mV protease shows the intrinsic fluorescence of the beads only after cleavage and liberation of mVenus (histogram: right panel). All experimental conditions for on-bead and in solution cleavage are fully discussed in Materials and Methods.
