Abstract
Purpose of Review
It has been known for decades that in order to grow tumors need to activate quiescent endothelial cells to form a functional vascular network, a process termed 'angiogenesis'. However, the molecular determinants that reverse this endothelial quiescence to facilitate pathological angiogenesis are not yet completely understood. This review examines a critical regulatory switch at the level of Ras that activates this angiogenic switch process and the role that microRNAs play in this process.
Recent Findings
In the last few years, microRNAs, a new class of small RNA molecules, have emerged as key regulators of several cellular processes including angiogenesis. MicroRNAs such as miR-126, miR-296 and miR-92a have been shown to play important roles in angiogenesis. We recently described how miR-132, an angiogenic growth factor inducible microRNA in the endothelium facilitates pathological angiogenesis by downregulating p120RasGAP, a molecular brake for Ras. Importantly, targeting miR-132 with a complementary, synthetic anti-microRNA restored the brake and decreased angiogenesis and tumor burden in multiple tumor models. Taken together, emerging evidence suggests a central role for microRNAs downstream of multiple growth factors in regulating endothelial proliferation, migration and vascular patterning.
Summary
Further research into miR-132-p120RasGAP biology and more broadly, microRNA regulation of Ras pathways in the endothelium will not only advance our understanding of angiogenesis but also provide opportunities for therapeutic intervention.
Keywords: Pathological angiogenesis, microRNA-132, p120RasGAP, endothelial quiescence
“Cells are required to stick precisely to the point. Any ambiguity, any tendency to wander from the matter at hand, will introduce grave hazards for the cells, and even more for the host in which they live.”
-Lewis Thomas “The lives of a cell: Notes of a Biology Watcher”
Introduction
It is estimated that an adult human has about 1–10 × 1012 endothelial cells lining the insides of blood vessels. Among these cells, only about 0.01% are estimated to be in the cell cycle at any given time [1, 2]. This exquisite degree of quiescence of the vascular endothelium is maintained by a balance of pro and anti-angiogenic factors. Such quiescence is also one of the bottlenecks that constrain the growth and sustenance of tumors beyond 1–2 mm diameter [3]. However, during certain physiological events such as wound healing or pathological stimuli such as inflammation or cancer, these endothelial cells get activated and resume an active proliferation program that results in growth of new blood vessels. This process of reversing endothelial quiescence and restoring a proliferative state is governed by a complex milieu of growth factors and signaling networks to generate new blood vessels.
Pathological angiogenesis: a complex host response to diverse stimuli
The circulatory system is one of the first organs to develop in an embryo so that organogenesis can be sustained with the delivery of oxygen and nutrients. During early development, blood vessels arise de novo from endothelial precursor cells (angioblasts) that form primitive capillary networks in a process broadly termed `vasculogenesis' [4]. These early capillaries can then sprout and branch into a capillary network in a process often referred to as `angiogenesis' [5]. These vascular networks that arise as a part of normal development often go on to mature and become stable with a robust interaction with perivascular cells. Many of the same developmental pathways of attracting precursor cells and/or inducing sprouting of pre-existing vessels have been adapted during pathological angiogenesis. For instance, a developing tumor begins to secrete angiogenic factors such as vascular endothelial growth factor (VEGF) and Angiopoietin in response to hypoxia. This leads to the activation of the dormant endothelial cells that begin to proliferate, migrate and establish a robust capillary network. However, in contrast to normal vasculature, pathological neovascularization results in leaky immature vessels with poor pericyte coverage. This phenomenon of a small dormant tumor acquiring a vascular network has been historically referred to as the `angiogenic switch' [6]. While early evidence suggests that the angiogenic switch occurs at an early stage during tumorigenesis, more recent work indicates that this process can occur during later stages of tumor progression. It is clear that despite the wealth of data on pathological angiogenesis we do not fully understand the molecular mechanisms that govern the angiogenic switch in a context specific manner.
Targeting microRNAs in pathological angiogenesis
Current strategies targeting angiogenesis using VEGFR-2 antibodies has shown varying degrees of success in different diseases. Treatment of patients with age-related macular degeneration (AMD) with anti-VEGFR2 antibody has shown significant clinical benefit [7]. However, the anti-angiogenic effects of VEGFR2 blockade in cancer seem to be context dependent. It has been shown that in some glioblastoma patients, treatment with anti-VEGFR2 leads to the development of evasive resistance where the tumors have upregulated production of bFGF [8]. Several studies have suggested that inhibition of multiple angiogenic pathways might have a better, synergistic effect than targeting a single growth factor dependent pathway [9]. In this context, emerging evidence suggests that targeting microRNAs (miRs) provide unique advantages by being downstream of multiple growth factor pathways. Moreover, since the endogenous angiogenic miRs are activated only in response to growth factors, there is a degree of specificity inherent in this approach to the proliferating endothelium.
MicroRNA regulation of angiogenesis
MicroRNAs (miRs) represent a unique mechanism to regulate gene expression by exerting a rapid and powerful response to various stimuli. miRs are endogenous RNAs that are encoded in the genome. They are transcribed like regular messenger RNAs by transcription factors binding to upstream regions. These miR transcripts are subsequently processed by the RNAse Drosha, exported out of the nucleus, and then cleaved by a second RNAse known as Dicer [10]. The first hints of miR function during vascular development came from mice with hypomorphic expression of Dicer that show embryonic lethal vascular malformations [11]. Similarly, knockdown of Dicer in zebrafish results in pericardial edema and vascular defects [12]. Furthermore, knockdown of Dicer and/or Drosha in vitro suppresses endothelial cell proliferation, migration, capillary sprouting, and tube formation [13, 14]. Together, these early studies demonstrate that miR expression in general is essential for blood vessel development and remodeling.
Subsequent studies have identified specific miRs that play key roles in angiogenesis or which are dysregulated in human cardiovascular diseases [15–17]. miR-195 and miR-133 are involved in cardiac hypertrophy [18, 19], miR-1 and miR-133 in arrhythmias [20, 21], and miR-126 in vascular development and angiogenesis [22]. miR-92a blocks angiogenesis and impairs functional recovery of ischemic tissue [23]. In some cases, individual target genes have been identified and validated. The endothelial-specific miR-126 inhibits sprouty related protein 1 (SPRED1), a negative inhibitor of VEGF signaling [24]. Interestingly, recent work has shown that blood flow induced upregulation of miR-126 by a mechano-sensitive transcription factor Klf2 in endothelial cells activated VEGF signaling pathways and leads to sprouting and remodeling of the aortic arch in developing zebrafish [25]. miR-296 is elevated during angiogenesis and regulates growth factor receptor overexpression in angiogenic endothelial cells [26]. Hypoxia has been shown to trigger miR-424 expression which leads to degradation of an ubiquitin ligase scaffold protein cullin-2. Degradation of cullin-2 prevents HIF-1α downregulation facilitates a cascade of pro-angiogenic signaling [27]. These representative examples illustrate both the scope and diversity of extracellular cues that trigger miR expression in the endothelium enabling the activation of broad angiogenic signaling pathways.
miR-132 as a genomic `first responder' to activation
We recently characterized miR-132 as an activating switch for quiescent endothelium that functions by downregulating p120RasGAP [28]. miR-132 is encoded on human chromosome 17, and is transcribed by the transcription factor, cAMP-response element binding protein (CREB) in multiple cell types [28, 29]. We showed that angiogenic growth factors such as VEGF, bFGF and conditioned media from a variety of tumors lead to phosphorylation of CREB and rapid transcription of miR-132 that peaks about 3–6 hours after activation. Interestingly, Lagos et al also observed upregulation of miR-132 with similar kinetics in lymphatic endothelial cells during viral infections [30]. Several groups have noted the early induction of miR-132 downstream of neurotropic growth factors [29, 31–34]. These observations highlight the fact that multiple cell types have co-opted miR-132 as an integral part of an early cellular response to a diverse group of activating stimuli.
We observed that the ectopic expression of miR-132 was sufficient to increase endothelial proliferation and tube formation in vitro. Conversely, a complementary anti-miR-132 decreased endothelial proliferation, tube formation in vitro and both developmental and pathological angiogenesis in vivo. Our observations highlight that miR-132 is not only among the early response genes in endothelial activation but also a critical regulator of the downstream events that control endothelial proliferation, tube formation and angiogenesis in vivo.
miR-132 targets p120RasGAP in the endothelium
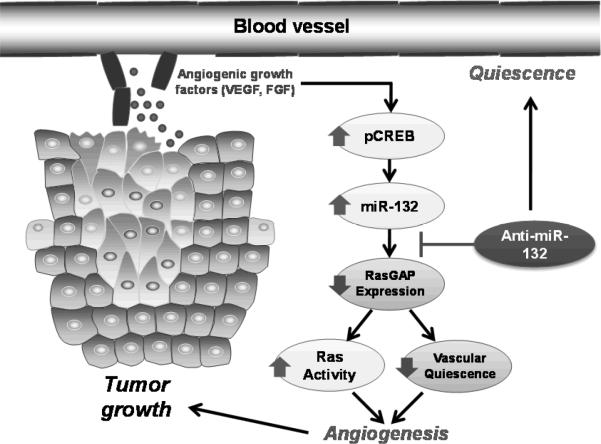
Based on target prediction algorithms, we identified at least one primary target of miR-132 in angiogenic endothelial cells to be p120RasGAP, a GTPase activating protein that attenuates p21 Ras activity. RasGAP enhances the weak intrinsic GTPase activity of Ras, resulting in the accumulation of the inactive GDP-bound form of Ras to limit activation of downstream targets PI3K and Raf-1. Consistent with the prediction databases, we found miR-132 expression was able to downregulate a luciferase reporter upstream of the 3' untranslated (UTR) region of the human p120RasGAP gene while it had no effect on a mutant 3'UTR. Importantly, we also found that expression of miR-132 directly downregulates p120RasGAP protein expression in endothelial cells. Transient knockdown of p120RasGAP phenocopied the effects of miR-132 and conversely expression of a miR-resistant p120RasGAP was able to rescue the activation of endothelial cells. Taken together, our data indicates a strong role for p120RasGAP in mediating the functions of miR-132 in the vascular endothelium as depicted in Figure 1.
Figure 1. miR-132 regulation of p120RasGAP as an angiogenic switch for tumorigenesis.
Tumors secrete growth factors that trigger phosphorylation of CREB and subsequent transcription of miR-132. miR-132 downregulates p120RasGAP thereby removing the endogenous brake on Ras activity and activates quiescent endothelium. Inhibition of miR-132 with an anti-miR restores p120RasGAP expression and dampens endothelial activation to inhibit angiogenesis and tumor growth.
Vascular role of p120RasGAP
Early studies exploring the functional role of p120RasGAP ortholog in Drosophila showed that ectopic expression of this protein decreased aberrant wing vein formation downstream of a constitutively active fibroblast growth factor receptor FGFR [35]. Deletion of p120RasGAP alone or in combination with its homolog neurofibromatosis-1 (NF-1) in mice causes a more deleterious phenotype with abnormal vascular morphogenesis in the embryo and the yolk sac leading to embryonic lethality [36]. In fact, RNA mediated silencing of p120RasGAP in mouse embryonic stem cells also recapitulated this phenotype [37]. Endothelial specific deletion of NF-1 has also been shown to be embryonic lethal with cardiovascular defects [38].
We found that angiogenic endothelium from several pathological human and mouse tissues including tumors and hemangiomas often had a significant decrease in p120RasGAP expression and a corresponding increase in miR-132 expression. We observed that endothelial specific deletion of p120RasGAP caused hypervascularization in response to bFGF in subcutaneous Matrigel plugs in mice. Conversely, ectopic expression of a miR-resistant p120RasGAP in a mouse endothelioma cell line significantly diminishes their growth in vivo (S.A and D.A.C, unpublished observations). Consistent with the mouse data, inactivating p120RasGAP mutations in humans lead to hypervascularization that manifests as capillary malformations associated with arteriovenous malformation, arteriovenous fistula, or Parkes Weber syndrome [39, 40]. Based on these observations, it is clear that p120RasGAP acts as a negative regulator of vascular patterning/morphogenesis.
Aside from binding Ras via the GAP domain, p120RasGAP also associates with a number of key signaling proteins to influence many cellular processes [41]. The SH2-SH3-SH2 region interacts with growth factor receptors, p190RhoGAP, focal adhesion kinase (FAK), and G3BP. This region is flanked by two Caspase-3 cleavage sites that result in the formation of two RasGAP fragments that have been shown to play a role in apoptosis. The pleckstrin homology (PH) domain contains interaction sites for PI3K and PKC, and is thought to influence the confirmation and accessibility of the GAP domain. The C2 domain is a calcium-responsive region that binds the FAK-related Pyk2 and AnnexinA6. p120RasGAP truncations and point mutations identified throughout the length of this sequence have been associated with vascular malformations in patients [40] suggesting a complex role for p120RasGAP and its interactants in endothelial biology.
Ras in endothelial quiescence
Members of the Ras superfamily of GTPases play important roles in several cellular processes including signal transduction, vesicle transport, cytoskeletal dynamics etc. In their GTP bound state, Ras proteins interact with a wide-variety of effector proteins to mediate multiple functions. Some of the early evidence for the role of Ras in regulating endothelial activation came from studies that showed that activating Ras mutations lead to the formation of endothelial-derived tumors [42, 43]. Interestingly, spontaneous development of hemangiomas was observed in a line of mice with an inducible oncogenic KrasG12D allele [44]. Moreover, mice deficient in B-Raf, a well-known Ras effector, have cardiovascular defects and die in utero [45]. Immortalized endothelial cells with an oncogenic H-Ras have been shown to gain an angiogenic phenotype and form angiosarcomas in mice [46]. Similarly, active Ras has been shown to be both necessary and sufficient to mediate an active angiogenic response including proliferation, migration and branching morphogenesis in primary endothelial cells [47]. Previous work done in our lab has shown that retroviral delivery of Ras or its effector C-Raf promotes angiogenesis in vivo [48]. Similarly, we also reported that Raf-1 plays a pivotal role in endothelial cell survival during angiogenesis [49]. Consistent with our identification of p120RasGAP as a target for miR-132, we also observed an increase in Ras activity with expression of miR-132 and a loss of Ras activity with inhibition of miR-132 suggesting that miR-132 mediates its biological effects by modulating Ras function.
Other RasGAPs and their putative microRNA regulators
In recent years, several new homologs of RasGAP have been identified as context specific regulators of Ras signaling. About 12 conserved RasGAP homologs have been identified in humans several of which contain the GAP domain. There is some evidence that multiple RasGAP family members may regulate diverse aspects of endothelial biology. For instance, endothelial specific deletion of NF-1 appears to result in elevated Ras signaling and increased nuclear localization of the transcription factor NFATc1, a known regulator of cardiovascular remodeling and lymphangiogenesis [38]. Recently, another RasGAP, AIP1, has also been shown to function as a signaling scaffold that links Ras pathway to NF-kB activation in prostate cancer [50]. Using target prediction algorithms, we analyzed the miRs that were upregulated during human ES cell vasculogenesis or angiogenic growth factor stimulation of primary endothelial cells for their ability to target any known RasGAPs. Strikingly, several highly upregulated angiogenic miRs are predicted to target RasGAPs and often the same miR family is predicted to target more than one member of the RasGAP family. It is not difficult to envision that a complex network of miRs downstream of angiogenic growth factors rapidly modulate multiple GAPs and GEFs to facilitate endothelial activation.
Evolutionary context for miR regulation of RasGAPs
In their GTP bound state, Ras proteins interact with a wide-variety of effector proteins to mediate multiple functions. It is well established that the Ras superfamily has undergone a specific and diverse expansion in eukaryotes [51]. It can be argued that evolution has adapted a small GTPase switch to exert spatial and temporal control over several interacting protein modules to mediate diverse physiological responses ranging from signal transduction to cytoskeletal dynamics [51]. Interestingly, recent work has identified microRNAs as a viable causal factor in the evolution of vertebrate complexity. For instance, miR-132 has been discovered in sea lampreys and sharks [52]. The earliest p120RasGAP ortholog has been identified in C.elegans [53]. Given these observations, it is tempting to speculate whether the evolution of specific miR families in vertebrates followed the expansion of Ras superfamily members to facilitate the control of Ras signaling pathways by miRs. Not surprisingly, several of these microRNAs and their target binding sites on the 3' untranslated regions of mRNAs are highly conserved across a broad range of species highlighting the fact that tumors have once again co-opted some evolutionarily well-conserved developmental pathways with miR-Ras switches to facilitate their survival and growth.
Conclusion
In summary, it is becoming increasingly clear that tumors are able to induce an angiogenic switch in quiescent endothelium by regulating critical miRs such as miR-132 that target Ras signaling pathways. Further identification and characterization of these miRs and their targets would not only help understand the biology of endothelial activation, but also pave way for the development of therapies to treat pathological neovascularization by targeting Ras signaling pathways.
Key Points
Endothelial cells in the adult are quiescent except during specific physiological requirements such as tissue repair and pathological growth such as tumors
This balance of quiescence and proliferation is maintained by pro and anti-angiogenic signaling pathways
microRNAs are downstream regulators of signaling across multiple angiogenic growth factors
Ras superfamily and their regulators RasGAPs and RasGEFs play important roles in angiogenesis
microRNA-132 mediated downregulation of p120RasGAP facilitates endothelial activation
Acknowledgements
We would like to thank members of the Cheresh Laboratory for useful discussions. S.A. is supported in part by a Postdoctoral fellowship from American Heart Association. Work in the Cheresh lab is supported by funding from the National Institutes of Health, USA.
Abbreviations
- VEGF
Vascular Endothelial Growth Factor
- bFGF
basic Fibroblast Growth Factor
- CREB
cyclic AMP Response Element Binding protein
- GAP
GTPase Activating Protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended reading
Papers of particular interest, published within the annual period of review have been highlighted as:
• of special interest
•• of outstanding interest
- [1].Engerman RL, Pfaffenbach D, Davis MD. Cell turnover of capillaries. Lab Invest. 1967;17(6):738–43. [PubMed] [Google Scholar]
- [2].Hobson B, Denekamp J. Endothelial proliferation in tumours and normal tissues: continuous labelling studies. Br J Cancer. 1984;49(4):405–13. doi: 10.1038/bjc.1984.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3(2):65–71. [PubMed] [Google Scholar]
- [4].Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- [5].Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6(4):389–95. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- [6].Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3(6):401–10. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- [7].Yancopoulos GD. Clinical application of therapies targeting VEGF. Cell. 2010;143(1):13–6. doi: 10.1016/j.cell.2010.09.028. [DOI] [PubMed] [Google Scholar]
- [8].Batchelor TT, Sorensen AG, di Tomaso E, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11(1):83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8(8):592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [11].Yang WJ, Yang DD, Na S, et al. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280(10):9330–5. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- [12].Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- [13].Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101(1):59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- [14].Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–73. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- [15].Latronico MV, Condorelli G. On the road to the definition of the cardiac miRNome in human disease states. J Mol Cell Cardiol. 2008;45(2):162–4. doi: 10.1016/j.yjmcc.2008.05.018. [DOI] [PubMed] [Google Scholar]
- [16].Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104(4):442–54. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bonauer A, Boon RA, Dimmeler S. Vascular microRNAs. Curr Drug Targets. 2010;11(8):943–9. doi: 10.2174/138945010791591313. [DOI] [PubMed] [Google Scholar]
- [18].Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- [19].van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48):18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103(9):919–28. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13(4):486–91. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- [22].Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a Controls Angiogenesis and Functional Recovery of Ischemic Tissues in Mice. Science. 2009 doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]; • An insightful study into an anti-angiogenic miR in the microRNA 17–92 cluster. Provides the first description of a miR that can be targeted to induce angiogenesis.
- [24].Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Together with #22 Fish et al, these papers provide a comprehensive insight into miR-126 function in angiogenesis using mouse and zebrafish studies.
- [25].Nicoli S, Standley C, Walker P, et al. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464(7292):1196–200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Uncovers a very interesting mechanism into the upregulation of miR-126 by blood flow and how that shapes development of the aortic arch in Zebrafish. While the role of growth factor receptors in regulating microRNAs is well appreciated, this paper describes a unique “mechano-transduction” signaling initiated by blood flow can modulate miR expression.
- [26].Wurdinger T, Tannous BA, Saydam O, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14(5):382–93. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ghosh G, Subramanian IV, Adhikari N, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120(11):4141–54. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper describes how hypoxia induced miR-424 is able to stablize HIF-α and contributes to both sustaining hypoxic signaling pathways and angiogenesis
- [28].Anand S, Majeti BK, Acevedo LM, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16(8):909–14. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First report of miR-132 function in regulating angiogenesis and demonstrates vascular targeted delivery of anti-miR can inhibit angiogenesis and tumor burden.
- [29].Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102(45):16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lagos D, Pollara G, Henderson S, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12(5):513–9. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]; •• This study is the first description of miR-132 function in lymphatic endothelium. The authors show how KSHV infection rapidly upregulates CREB mediated transcription of miR-132. This paper serves to highlight how diverse stimuli often upregulate the same miRs with similar kinetics but have may target different subset of proteins in different cell types in a context dependent manner.
- [31].Kawashima H, Numakawa T, Kumamaru E, et al. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165(4):1301–11. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- [32].Magill ST, Cambronne XA, Luikart BW, et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107(47):20382–7. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nudelman AS, DiRocco DP, Lambert TJ, et al. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20(4):492–8. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Remenyi J, Hunter CJ, Cole C, et al. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J. 2010;428(2):281–91. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- [35].Feldmann P, Eicher EN, Leevers SJ, et al. Control of growth and differentiation by Drosophila RasGAP, a homolog of p120 Ras-GTPase-activating protein. Mol Cell Biol. 1999;19(3):1928–37. doi: 10.1128/mcb.19.3.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Henkemeyer M, Rossi DJ, Holmyard DP, et al. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377(6551):695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- [37].Kunath T, Gish G, Lickert H, et al. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat Biotechnol. 2003;21(5):559–61. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- [38].Gitler AD, Zhu Y, Ismat FA, et al. Nf1 has an essential role in endothelial cells. Nat Genet. 2003;33(1):75–9. doi: 10.1038/ng1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Boon LM, Mulliken JB, Vikkula M. RASA1: variable phenotype with capillary and arteriovenous malformations. Curr Opin Genet Dev. 2005;15(3):265–9. doi: 10.1016/j.gde.2005.03.004. [DOI] [PubMed] [Google Scholar]
- [40].Eerola I, Boon LM, Mulliken JB, et al. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet. 2003;73(6):1240–9. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pamonsinlapatham P, Hadj-Slimane R, Lepelletier Y, et al. P120-Ras GTPase activating protein (RasGAP): A multi-interacting protein in downstream signaling. Biochimie. 2009;91(3):320–8. doi: 10.1016/j.biochi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- [42].Hong HH, Devereux TR, Melnick RL, et al. Mutations of ras protooncogenes and p53 tumor suppressor gene in cardiac hemangiosarcomas from B6C3F1 mice exposed to 1,3-butadiene for 2 years. Toxicol Pathol. 2000;28(4):529–34. doi: 10.1177/019262330002800404. [DOI] [PubMed] [Google Scholar]
- [43].Marion MJ, Froment O, Trepo C. Activation of Ki-ras gene by point mutation in human liver angiosarcoma associated with vinyl chloride exposure. Mol Carcinog. 1991;4(6):450–4. doi: 10.1002/mc.2940040607. [DOI] [PubMed] [Google Scholar]
- [44].Fisher GH, Wellen SL, Klimstra D, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15(24):3249–62. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wojnowski L, Zimmer AM, Beck TW, et al. Endothelial apoptosis in Braf-deficient mice. Nat Genet. 1997;16(3):293–7. doi: 10.1038/ng0797-293. [DOI] [PubMed] [Google Scholar]
- [46].Arbiser JL, Moses MA, Fernandez CA, et al. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proc Natl Acad Sci U S A. 1997;94(3):861–6. doi: 10.1073/pnas.94.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Meadows KN, Bryant P, Vincent PA, Pumiglia KM. Activated Ras induces a proangiogenic phenotype in primary endothelial cells. Oncogene. 2004;23(1):192–200. doi: 10.1038/sj.onc.1206921. [DOI] [PubMed] [Google Scholar]
- [48].Hood JD, Frausto R, Kiosses WB, et al. Differential alphav integrin-mediated Ras-ERK signaling during two pathways of angiogenesis. J Cell Biol. 2003;162(5):933–43. doi: 10.1083/jcb.200304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Alavi A, Hood JD, Frausto R, et al. Role of Raf in vascular protection from distinct apoptotic stimuli. Science. 2003;301(5629):94–6. doi: 10.1126/science.1082015. [DOI] [PubMed] [Google Scholar]
- [50].Min J, Zaslavsky A, Fedele G, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16(3):286–94. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Delineates how a RasGAP AIP1 functions as a scaffold to mediate cross-talk between two well-characterized signaling pathways in cancer.
- [51].Jekely G. Small GTPases and the evolution of the eukaryotic cell. Bioessays. 2003;25(11):1129–38. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- [52].Heimberg AM, Sempere LF, Moy VN, et al. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105(8):2946–50. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stetak A, Gutierrez P, Hajnal A. Tissue-specific functions of the Caenorhabditis elegans p120 Ras GTPase activating protein GAP-3. Dev Biol. 2008;323(2):166–76. doi: 10.1016/j.ydbio.2008.08.026. [DOI] [PubMed] [Google Scholar]