Abstract
Escherichia coli is an enteric bacterium that is capable of growing over a wide range of pH values (pH 5 - 9)1 and, incredibly, is able to survive extreme acid stresses including passage through the mammalian stomach where the pH can fall to as low as pH 1 - 22. To enable such a broad range of acidic pH survival, E. coli possesses four different inducible amino acid decarboxylases that decarboxylate their substrate amino acids in a proton-dependent manner thus raising the internal pH. The decarboxylases include the glutamic acid decarboxylases GadA and GadB3, the arginine decarboxylase AdiA4, the lysine decarboxylase LdcI5, 6 and the ornithine decarboxylase SpeF7. All of these enzymes utilize pyridoxal-5'-phospate as a co-factor8 and function together with inner-membrane substrate-product antiporters that remove decarboxylation products to the external medium in exchange for fresh substrate2. In the case of LdcI, the lysine-cadaverine antiporter is called CadB. Recently, we determined the X-ray crystal structure of LdcI to 2.0 Å, and we discovered a novel small-molecule bound to LdcI the stringent response regulator guanosine 5'-diphosphate,3'-diphosphate (ppGpp) 14. The stringent response occurs when exponentially growing cells experience nutrient deprivation or one of a number of other stresses9. As a result, cells produce ppGpp which leads to a signaling cascade culminating in the shift from exponential growth to stationary phase growth10. We have demonstrated that ppGpp is a specific inhibitor of LdcI 14. Here we describe the lysine decarboxylase assay, modified from the assay developed by Phan et al.11, that we have used to determine the activity of LdcI and the effect of pppGpp/ppGpp on that activity. The LdcI decarboxylation reaction removes the α-carboxy group of L-lysine and produces carbon dioxide and the polyamine cadaverine (1,5-diaminopentane)5. L-lysine and cadaverine can be reacted with 2,4,6-trinitrobenzensulfonic acid (TNBS) at high pH to generate N,N'-bistrinitrophenylcadaverine (TNP-cadaverine) and N,N′-bistrinitrophenyllysine (TNP-lysine), respectively11. The TNP-cadaverine can be separated from the TNP-lysine as the former is soluble in organic solvents such as toluene while the latter is not (See Figure 1). The linear range of the assay was determined empirically using purified cadaverine.
Protocol
1) Reagents and Equipment
First, prepare the following three solutions: 1 mL of Solution A which is made up of 8 mM L-lysine, 100 mM sodium 2-(N-Morpholino)ethanesulphonic acid (MES) pH 6.5, 0.2 mM nucleotide, where the nucleotide is either: guanosine diphosphate (GDP), guanosine triphosphate (GTP), guanosine 5'-diphosphate,3'-diphosphate (ppGpp), or guanosine 5'-triphosphate,3'-diphosphate (pppGpp), 0.1 mM pyridoxal 5'-phosphate (PLP), and 1 mM β-mercaptoethanol/ (β-ME). 1 mL of Solution B which consists of 100 mM sodium MES pH 6.5, 0.1 mM PLP, 1 mM β-ME, and 50 nM LdcI. LdcI was purified as described in Snider et al.6 and Kanjee et al. 14. 1 mL of Solution C which is identical to Solution B but does not contain LdcI.
Prepare 100 mL of the stop solution consisting of 1 M sodium carbonate (10.6 g/100 mL) and aliquot 50 μL into each well of a 96-well polystyrene plate using a multi-channel pipette. Add 30 μL of water to the plate and then cover. Note that the sum of the volume of water added and the volume of sample extracted during the enzyme reaction must equal 50 μL. In this case, 20 μL of enzyme reaction sample will be removed.
Prepare 5 mL of TNBS solution at 10 mM by diluting 294 μL of 5% (w/v) TNBS stock with 4706 μL water, wrap in aluminium foil and keep on ice.
Equilibrate the Eppendorf ThermoStat Plus at 37°C. The ThermoStat Plus has 24 wells in 6 columns. See Figure 2 for schematic. Label 1.5 mL Eppendorf tubes indicating the identity of the nucleotide that will be tested (one per column): GTP (column A), GDP (column B), ppGpp (column C), pppGpp (column D), no nucleotide (column E), and protein samples (column F).
Rack several boxes of 200 μL tips such that each alternate row is omitted giving a total of four rows of tips. This is necessary for the use of the multi-channel pipette with the ThermoStat Plus heatblock during the enzyme assay.
Equilibrate Digital Heatblock (VWR) at 42°C.
Place a 96-well 2.0 mL polypropylene plate on ice to cool.
2) LdcI Assay
Aliquot 50 μL of Solution A with the appropriate nucleotide to 1.5 mL Eppendorf tubes in each of the five columns of the Eppendorf ThermoStat.
Add 330 μL of Solution B to three tubes in column F and add 330 μL of Solution C (the no-protein control) to the final tube in column F.
Equilibrate the solutions at 37°C for five minutes. Tip: after 5 minutes, cut off the caps of the tubes in the center of the heating-block to prevent them from interfering with the multichannel pipette to be used next.
Using a 5-50 μL VWR multi-channel pipette, transfer 50 μL from the tubes in column F into each of the tubes in columns A-E. Start the timer as soon as the first tube is mixed.
At 2, 4, and 6 minutes remove 20 μL of sample using the multi-channel pipette and add to the stop solution.
Note: samples in stop solution can be covered in plastic wrap and frozen at -20°C for subsequent processing. The plates should be thawed at room temperature before reaction with TNBS.
3) TNBS Reaction and Color Development
Add 50 μL of 10 mM TNBS solution to the stop solution using the multi-channel pipette and then incubate at 42°C for 6 minutes. The solution will turn a dark yellow/orange color as the TNBS reacts with the lysine and the cadaverine.
After 6 minutes, cool the plate on ice to slow down the reaction.
Remove 100 μL of sample using a VWR 20-200 μL multi-channel pipette and place in the 2 mL deep well plate that was cooled on ice. Transfer ice-bucket to the fumehood.
Add 500 μL toluene to each well using a HandyStep (Brand) repeat pipettor and 12.5 mL pipette tip (Plastibrand).
Wipe away any excess toluene using a kimwipe and cover the plate with strips of packaging tape. Make sure to press down firmly to obtain a good seal. Cover the 96-well plate with a flat lid and then shake vigorously for 1 minute and 30 seconds.
Leave the solutions to settle for 5 minutes. The TNP-cadaverine is now in the upper toluene phase, while the TNP-lysine remains water soluble (see Figure 1).
Remove 200 μL of toluene using the 20-200 μL multi-channel pipette into a 96-well plate for reading. Note: make sure to check that each sample being removed is clear and does not have any of the bottom aqueous phase. Note: use barrier tips to prevent damage to multi-channel pipette by the toluene.
Read absorbance at 340 nm in a SpectraMax 340 plate reader.
To clean the quartz plate, rinse out the toluene with water and place the quartz plate in a large glass tray in the fume hood. Pour over 100 mL of a 7:3 mixture of 70% (v/v) nitric acid:95% (v/v) ethanol and cover the quartz plate with a second glass tray. Warning: the reaction is highly exothermic and sometimes explosive wear appropriate protection and ensure that the fume hood sash is set to the lower 6" level. After cooling, wash out nitric acid with water and then 95% ethanol.
4) Representative Results
1)Cadaverine Standard Curve (Figure 3) The assay was performed as described but without any L-lysine or LdcI and instead various concentrations of cadaverine were used to empirically determine the linear range of the assay. The assay was linear to an OD340 of 0.25, corresponding to ~ 22 nmoles of cadaverine (Figure 3).
2)Activity of LdcI (Figure 4) The activity of LdcI alone was determined to be 153.5 (± 18.1) nmoles cadaverine min-1 μg LdcI-1 at pH 6.5. The activity of LdcI is unaffected in the presence of 100 μM GTP or GDP but is strongly inhibited (>10-fold) in the presence of pppGpp and ppGpp (Figure 4).
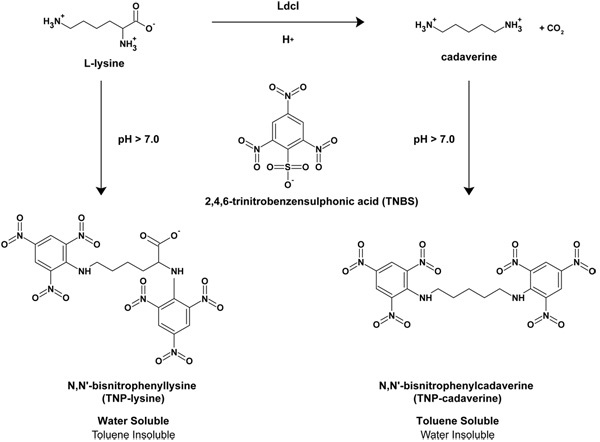 Figure 1. Schematic diagram of the LdcI reaction. The decarboxylation reaction of L-lysine to generate CO2 and cadaverine is shown as well as the subsequent reaction with TNBS at high pH to generate N,N'-bistrinitrophenylcadaverine (TNP-cadaverine) and N,N'-bistrinitrophenyllysine (TNP-lysine). Based on Phan et al.11
Figure 1. Schematic diagram of the LdcI reaction. The decarboxylation reaction of L-lysine to generate CO2 and cadaverine is shown as well as the subsequent reaction with TNBS at high pH to generate N,N'-bistrinitrophenylcadaverine (TNP-cadaverine) and N,N'-bistrinitrophenyllysine (TNP-lysine). Based on Phan et al.11
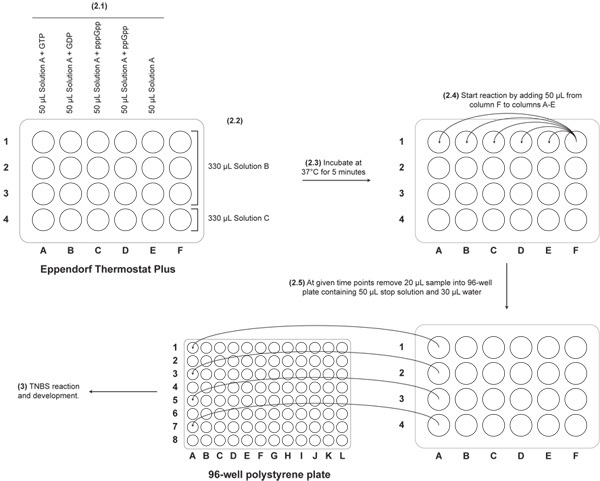 Figure 2. Setup of Eppendorf Thermostat. The layout of the samples in the 24-well Eppendorf ThermoStat is shown along with description of the steps of the assay (indicated in bold). Arrows indicate the transfer of reaction solutions according to the protocol. Removal of the reaction solution to the stop solution in the 96-well plate is also indicated.
Figure 2. Setup of Eppendorf Thermostat. The layout of the samples in the 24-well Eppendorf ThermoStat is shown along with description of the steps of the assay (indicated in bold). Arrows indicate the transfer of reaction solutions according to the protocol. Removal of the reaction solution to the stop solution in the 96-well plate is also indicated.
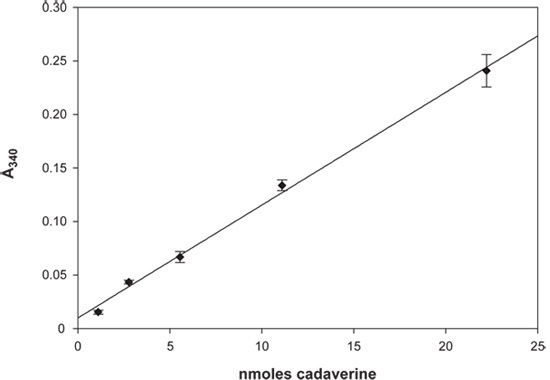 Figure 3. Cadaverine Standard Curve. A plot of absorbance at 340 nm versus nmoles of cadaverine is shown. Error bars represent the standard deviation of at least three independent measurements. The line of best fit is also shown and has an R2 of 0.996.
Figure 3. Cadaverine Standard Curve. A plot of absorbance at 340 nm versus nmoles of cadaverine is shown. Error bars represent the standard deviation of at least three independent measurements. The line of best fit is also shown and has an R2 of 0.996.
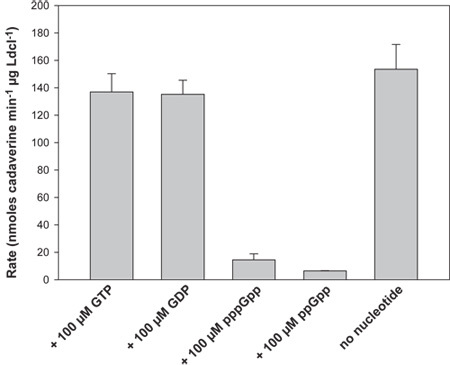 Figure 4. LdcI assay results. A plot of the rate of LdcI activity (in nmoles cadaverine produced min-1 μg-1 LdcI) is shown in the presence of 100 μM GTP, GDP, pppGpp, ppGpp and in the absence of nucleotide. The stringent response nucleotides (p)ppGpp are capable of significantly inhibiting LdcI activity. The error bars represent the standard deviation of at least six independent measurements.
Figure 4. LdcI assay results. A plot of the rate of LdcI activity (in nmoles cadaverine produced min-1 μg-1 LdcI) is shown in the presence of 100 μM GTP, GDP, pppGpp, ppGpp and in the absence of nucleotide. The stringent response nucleotides (p)ppGpp are capable of significantly inhibiting LdcI activity. The error bars represent the standard deviation of at least six independent measurements.
Discussion
In the lysine decarboxylase assay, TNBS is reacted with the primary amines of L-lysine and cadaverine to form TNP-lysine and TNP-cadaverine adducts (Figure 1). Due to the presence of the carboxylic acid group on TNP-lysine, this adduct remains soluble in water while the TNP-cadaverine, lacking the carboxylic acid group, is capable of partitioning into toluene11. This type of assay can be utilized more broadly on other types of amino acids where the loss of a carboxylic acid group occurs during the reaction. This occurs during the decarboxylation of L-ornithine by the inducible ornithine decarboxylase SpeF to form the polyamine putrescine7 and the decarboxylation of L-arginine by the inducible arginine decarboxylase AdiA to form the polyamine agmatine4.
The LdcI assay described here provides a relatively fast method for the determination of the activity of the purified protein in vitro. The major advantages of this assay are:
i)Use of multiple replicates per experiment improves the precision of each measurement;
ii)The assay may be conducted over a wide range of buffer conditions (different pH, salt, reducing agent etc.) without modification of the protocol;
iii)The assay may be modified for measuring the in vivo activity of LdcI by determining the amount of cadaverine excreted during cell growth.
The major limitations of this assay are:
i)The sensitivity of the experiments are limited by the linear range of absorbance of the TNP-adducts;
ii)The multiple processing steps increase the magnitude of experimental errors;
iii)Not all amino acid decarboxylases are amenable to this type of protocol. For example, the decarboxylation of L-glutamic acid by the inducible glutamic acid decarboxylases GadA/GadB generates γ-amino-butyric acid2, the TNBS-adduct of which will be soluble in water due to the presence of the side-chain carboxylic acid group.
The biochemical investigation of the acid stress response of E. coli is an expanding area of research and will allow us to better understand the molecular basis of stress response in E. coli and related γ-proteobacteria that have similar acid stress response systems such as Salmonella enterica serovar Typhimurium12 and Vibrio cholera13. The discovery that LdcI activity is inhibited by the stringent response regulator ppGpp has provided us with a previously unknown insight into the regulation of this protein.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank Dr. Dr. Michael Cashel (National Institutes of Health, Bethesda, MA, USA) for sending us bacterial strains, plasmids, and necessary protocols. We thank Dr. John Glover (Department of Biochemistry, University of Toronto) for use of the SpecraMax plate reader. UK is the recipient of a National Sciences and Engineering Research Council of Canada (NSERC) Postgraduate Scholarship, a Canadian Institutes of Health Research Strategic Training Program in the Structural Biology of Membrane Proteins Linked to Disease, and a University of Toronto Open Fellowship. This work was supported by a grant from the Canadian Institutes of Health Research (MOP-67210) to WAH.
References
- Gale EF, Epps HM. The effect of the pH of the medium during growth on the enzymic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem J. 1942;36:600–618. doi: 10.1042/bj0360600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Williams C, Miller C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol. 2003;185:6556–6561. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo DL, Boeker EA, Byers B, Waron H, Fischer EH. Purification and physical properties of inducible Escherichia coli lysine decarboxylase. Biochemistry. 1974;13:662–670. doi: 10.1021/bi00701a005. [DOI] [PubMed] [Google Scholar]
- Snider J. Formation of a distinctive complex between the inducible bacterial lysine decarboxylase and a novel AAA+ ATPase. J Biol Chem. 2006;281:1532–1546. doi: 10.1074/jbc.M511172200. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20922–20927. [PubMed] [Google Scholar]
- Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000;8:1–6. doi: 10.1016/s0969-2126(00)00085-x. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. In: Escherichia coli and Salmonella : cellular and molecular biology. Curtiss R, Neidhardt FC, editors. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends in Microbiology. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Phan AP, Ngo TT, Lenhoff HM. Spectrophotometric assay for lysine decarboxylase. Anal Biochem. 1982;120:193–197. doi: 10.1016/0003-2697(82)90336-0. [DOI] [PubMed] [Google Scholar]
- Park YK, Bearson B, Bang SH, Bang IS, Foster JW. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- Merrell DS, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- Kanjee U, Gutsche I, Alexopoulos E, Zhao B, Bakkouri MEl, Thibault G, Liu K, Ramachandran S, Snider J, Pai EF, Houry WA, A W. Linkage between the Bacterial Acid Stress and Stringent Responses Revealed by the Structure of the Inducible Lysine Decarboxylase. EMBO Journal. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]


