Abstract
Two novel synthetic peptides accelerate bone formation and can be delivered using a collagen matrix. The aim of this study was to investigate the effects on bone repair in a unicortical defect model. Treatment of mesenchymal cells produced an increase in alkaline phosphatase activity, showed nodule formation by the cells, and increased the expression of genes for runx2, osterix, bone sialoprotein, and osteocalcin. A collagen sponge soaked with peptide promoted repair of bone defects, whereas the control was less effective. The results from this study demonstrated that mesenchymal cells treated with peptide in vitro differentiate towards osteogenesis, and, that peptides delivered in vivo using a collagen sponge promote the repair of unicortical defects.
Protocol
1. In vivo Biopanning to Isolate Phage That Target Long Bones
Phage DNA was sequenced and Peptides with a corresponding sequence were synthesized. Peptides were synthesized with gly-gly-gly-ser-lysine linker (the linker is needed to couple fluorescent chromophores that help to track/trace the peptides).
A 10 week old Balb/c mouse was anesthetized, heart was exposed.
The Ph.D-12 Phage display peptide library kit (New England Biolabs, Beverly, MA) was used for in vivo biopanning, 10 x 1010 pfu, and injected into the heart.
After approximately 5 minutes, the mouse was euthanized. The femurs were exposed and dissected, the ends of the femurs were cut off, and the bone marrow was removed.
The inside and outside of the left femur were rinsed 10 times with PBS, and the femur was added to ER2537 bacteria (referred to as non-eluted phage, skip to step 6).
The intramedullary canal of the right femur was rinsed with 2%-dried skim milk in a syringe with a 21-gauge needle ten times to remove non-bound phage. Bound phage was eluted from the intramedullary canal with HCl-glycine buffer pH 2.2, neutralized with 1M Tris, added to ER2537 bacteria, and cultured for 4.5 hours to amplify the phage (referred to as eluted).
Bacteria were removed by centrifugal sedimentation for 10 min at 4°C, and phage was precipitated overnight at 4°C by the addition of PEG/NaCl solution. The phage was collected by centrifugation and suspended and precipitated twice by addition of 1/6 volume of PEG/NaCl. The final pellet was resuspended in TBS with 0.02% NaN3.
The amplified eluates from both the eluted and non-eluted phage were injected into separate mice, and phage was isolated as above. Only the left femur was removed from the mouse that was injected with the non-eluted phage, and the femur placed directly into the bacteria (see step 4 above). The eluted phage mouse was isolated only from the right femur and the phage released by HCl-glycine buffer before adding to the bacteria (see step 5 above).
This in vivo biopanning procedure was repeated three times before sequencing of DNA using-98 primer (provided with the Phage display kit).
Pooled phage were amplified in bacterial cultures, purified, quantitated and then were injected in a fourth mouse. Sequencing was repeated.
To demonstrate peptide specificity to bone and marrow, phage eluted from kidney and liver were analyzed as described above. In all four rounds there was minor amount of phage located in these organs (less than 10%).
Peptides were synthesized with a gly-gly-gly-ser-lys sequence, with and without biotin to be used for cell and tissue staining, as well as for the in vivo work.
2. Mesenchymal Cell Culture for Osteogenic Cell Differentiation Experiments
A cloned mouse pluripotential bone-marrow stroma (D1) cell line isolated in our lab1 was used for in vitro experiments. The cells were cultured in Dulbecco's modified essential medium, DMEM, low glucose (Gibco cat#11885-084) supplemented with 10% fetal bovine serum (Gibco cat# 16000-044), fifty milligrams of sodium ascorbate per milliliter (Sigma cat# A7631), and 100 U/ mL penicillin G and 100mg/ mL streptomycin (Gibco cat# 15140-122).
Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 water jacketed incubators and sub-cultured until cells were approximately 90% confluent.
Cell were trypsinized, centrifuged to pellet, counted, and seeded at a concentration of 5x103 cells/cm2 in tissue culture treated plasticware.
3. Peptide Staining for Cultured Cells and Tissues
D1 or marrow cells were plated on fibronectin- or gelatin- coated chamber well slides containing DMEM and 10% fetal bovine serum for 24 hours.
Cells were washed with PBS. Cells/tissue were fixed with 4% paraformaldehyde for 30 minutes
Excess aldehyde were blocked using 0.5 M Glycine (pH 7.5) for 10 minutes.
Endogenous avidin and biotin receptors on the cell surface was blocked by the addition of 5% BSA+Avidin blocking solution for 1 hour, rinsed once with PBS for 5 minutes, then blocked with 5% BSA+Biotin blocking solution for 1 hour.
Then 100 μM biotinylated L7 and R1 peptides were added to wells for 30 minutes.
Cells were washed three times with PBS for 5 min each and 1:1000 dilution of Alexa Flour Streptavidin was added and incubated in the dark for 30 minutes.
Cells were washed and coverslips mounted with vectashield mounting media and examined under a fluorescence microscope.
4. Alkaline Phosphatase Activity Assay
Alkaline phosphatase colorimetric assay was performed on monolayers using an alkaline phosphatase kit (BioRad) as recommended by the manufacturer.
D1 cells were plated at a density 5 x 103 cells/cm2 on 6-well culture plates and treated with R1 peptide, ALP activity was quantified at days 4, 8, 12, 16 and 20.
To prepare the cell lysates, cell layers were washed three times with PBS buffer and then scraped into PBS followed by sonication and centrifugation to remove cellular debris.
Equal volumes of lysate (100 μL) was then mixed with 100 μL of the freshly prepared colorimetric substrate para-nitrophenyl phosphate, and incubated at 37°C for 30 minutes.
The enzymatic reaction was stopped by adding 100 μL of 0.2 N NaOH solutions. ALP activity was measured at 405 nm using ELISA reader (Bio-Rad).
Protein concentration of the cell lysates was measured with a BioRad Protein Assay Kit, and AP activity was then expressed as para-nitrophenol produced in nmol/min/mg of protein.
ALP activity was calculated according to the manufacturer's specifications (Sigma, Inc.).
Units of ALP activity were normalized to milligrams of total protein content with BCA assay (Pierce, Rockford, IL) of the cell lysate.
5. Mesenchymal Cell Differentiation using L7 and R1
Subconfluent mesenchymal (D1) cells were grown to approximately 90% confluency and treated with 5nM L7 and R1 peptide.
Cells were harvested at 0.5, 2, 6, 24 and 48 hrs, and the RNA was prepared using the Qiagen RNeasy kit using manufacturer's instructions.
Reverse transcriptase (RT) reactions were annealed (37°C, 10 minutes) and followed by first-strand cDNA synthesis (42°C, 30 minutes) and heat inactivation (95°C for 5 minutes). The resulting cDNA was stored frozen (-70°C) until assayed by real-time PCR.
Real Time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen). The reactions were performed with 25 μL of the SYBR Green kit master mix and forward and reverse primers (300 nmol/L) of bone specific genes for e.g. Runx2, Osterix, Osteocalcin, Bone sialoprotein, and wnt pathway genes such as b-catenin, LRP6, Wnt 5a, 7b, 10b, DKK1 and OPG and RANKL (see Table 1).
The 3 μL of cDNA sample was added to the final reaction mixture undiluted from the RT reaction. The 96-well real-time PCR format included six 10-fold dilutions in duplicate of the plasmid DNA standards. The wells of the plate were sealed with optical adhesive covers (BioRad) and centrifuged at low speed (300 g, 5 minutes) to ensure complete mixing.
Each sample was analyzed at least in duplicate with the iCycler instrument (BioRad Laboratories, Hercules, CA).
The PCR protocols used involved activation of AmpliTaq Gold DNA polymerase followed by 40 cycles of denaturation at 94°C for 30 seconds, respective annealing temperature for 30 seconds, and extension at 72°C for 30 seconds. The PCR threshold cycle number (CT) for each sample was calculated at the point where the fluorescence exceeded the threshold limit. The threshold limit was fixed along the linear logarithmic phase of the fluorescence curves at 10 to 20 standard deviations (SDs) above the average background fluorescence. Standard curve equations were calculated by regression analysis of average CT versus the log10 of the cDNA relative to the total RNA present. Relative expression of the target genes was normalized to 18S for bone specific genes and b-actin was used for Wnt pathway gene.
6. In vivo Preparation of Gelfoam Composite
Using aseptic routines, gelfoam cylinders (Pharmacia & Upjohn cat# 09-0353-01), 3 mm in diameter by 8 mm in length, were created using self-designed instrument.
Twenty-four hours before surgery, the gelfoam cylinders were soak-loaded with either L7 or R1 peptide (20 μM in PBS) or PBS buffer without peptide. The gelfoam cylinders were transferred to sterile 12 well culture dishes and stored overnight at -4°C.
At the time of surgery, the gelfoam composite in culture dishes were moved out of the refrigerator and put on ice until surgery.
7. Bone Defects
All animal procedures were approved by the Animal Care and Use Committee of the Health System at the University of Virginia, prior to being performed. Ten nine month-old adult syngeneic male Fischer 344 rats (350-400 g) were anesthetized intraperitoneally with a mixture of ketamine (50 g per gram body weight) and xylazine (5 g per gram body weight).
A bilateral surgical approach was made to the anteromedial aspect of tibia. Rats at this age or older show insignificant increases in cross-sectional area, cortical bone area, bone mass, bone density, and axial length of the anteromedial aspect of tibia. A large, oblong unicortical defect was created which measured 3 mm wide by 8 mm long using a pneumatic burr (Hall High Speed Drill; Zimmer) under saline irrigation.
The bone defect was treated with gelfoam either with or without peptides. All implanted scaffolds, measuring 3 mm width by 8 mm length by 1.5 mm depth, were inserted by gentle tapping. No external or internal limb fixation devices were used postoperatively, and animals allowed to ambulate without restriction immediately following surgery.
8. Histology and Morphometry
Bone regeneration was evaluated by gross morphology and digital images taken.
Rats were euthanized and the tibias were collected at 3, 5, and 12 weeks to quantify the amount of new bone generated in response to injury.
After checking the bone defect carefully, the harvested tibia were fixed in 10% neutral buffered formalin, decalcified in RDO rapid decalcifier for 72 hours, then processed routinely and embedded in paraffin, and sectioned longitudinally.
Bone regeneration was evaluated by gross and light microscopic examination.
Ten animals were used for each condition (i.e., empty defect, gelfoam alone, and gelfoam plus R1 peptide). The 8mm unicortical defect was represented across ~40 tissue sections, each of which was 8 μm thick. Of those 40 sections, we used a minimum of 6 sections to quantify the amount of new bone.
All the tissue sections were stained with hematoxylin/eosin (H&E), which labels osteoid matrix.
All slides were evaluated by microscopy.
9. Bone Repair
Overview
Bone repair is clearly a very important area of consideration in the field of musculoskeletal tissue regeneration. The peptides that target bone could have significant utility in the practice of Orthopaedic Surgery, particularly in the use of protheses that can be indwelling for years if not decades. The biomimetic properties of bone tropic factors could create a technological step forward in the field of tissue engineering, as well as for prosthetic surgery of bone. Inclusion of bone targeting factors in synthetic or natural polymers may assist specificity of cell adherence, thereby scavenging endogenous cells within tissues that are undergoing repair and regeneration. An additional goal was to establish if our unique bone targeting peptides identified by in vivo biopanning of a phage display library would bind to cells isolated from bone and bone marrow, and have the potential to modulate osteogenesis in vitro and bone regeneration in vivo.
In our experiments, we delivered the peptides into femoral defects in rats using natural and synthetic polymer matrices and bone regeneration examined by histology, biochemistry, gene expression, micro-CT and biomechanics in order to elucidate details of peptide activity on bone regeneration. Further development of this approach can lead to the discovery and development of biologically active compounds that target bone and potentiate repair through mechanisms that are well characterized biologically at the cellular and molecular levels.
Staining
The peptides were synthesized with biotin-labels in order to determine binding and localization to mesenchymal cells. Fluorescein-isothiocyanate (FITC)-labeled avidin was used to localize microscopically the binding of peptides on cells grown in vitro. The peptides bind to mesenchymal cells in culture as well as to bone and marrow in mouse rib tissue sections in vitro (Figure 1).
Von Kossa Staining
A mesenchymal cell (D1) was cloned from bone marrow and used to determine the osteogenic effect of peptides on cells in vitro. The cells were treated with peptides L7 and R1 to determine changes in cell morphology and gene expression. At day 4, the cells had started to aggregate, and the aggregates enlarged with time to form nodules, a cell behavior pattern similar to bone cells in culture ( Figure 2 ).
The addition of peptide L7 or R1 to the cells enhanced staining of the cultures with von Kossa. Cells were then treated with different concentrations of peptide L7 or R1 for up to 16 days in culture. It was determined that 5 nM peptide was most effective in vitro . Alkaline phosphatase activity in the cultures that were treated with peptide increased from between 2 and 3.5 fold. The fold increase depended on the concentration of peptide and the time of treatment in culture.
Real-time PCR
Cells treated with L7 and R1 peptides were analyzed using Real-Time PCR for gene expression for two known bone cell transcription factors (Osterix and Runx2) and three bone matrix proteins (bone sialoprotein, osteocalcin and collagen type I). Treatment with R1 showed increases in Osterix and Runx2 gene expression. Collagen type 1 gene expression remained unchanged. Treatment with L7 showed an increase in BSP and osteocalcin gene expression as well as expression of the type I collagen gene. This demonstrates that both peptides have an effect on genes that are related to the onset and progression of osteogenesis in mesenchymal cell cultures.
The treatment of in vitro cell cultures with the peptides R1 and L7 has revealed an anabolic effect related to bone formation using mesenchymal cells. An increase in the ratio of osteoprotegerin to RANKL indicates a decrease in the recruitment of the bone resorbing cells that are known as osteoclasts. Treatment of cells with the peptides also leads to an increase in the expression of genes that are associated with bone formation and diminishes the factors that inhibit osteogenesis.
Histology
To determine the quantity of bone, we exposed the slices of bone to μLtraviolet light thereby producing autofluorescence. This provides greater contrast between bone and non-bone tissues for the purpose of quantitation 2. The sections were photographed using both bright field and UV light (Figure 3) a Nikon and UV light by a Nikon digital imaging system at the same magnification (x10 objective). The resulting digital images were analyzed with Metavue imaging software (Molecular Devices). We chose a fixed, rectangular region of interest (ROI) that contained the 8 mm unicortical defect. The injury site was always represented inside this ROI by manually placing the box in the correct position on each image. The H&E-positive pixels were partially automated using the magic wand tool set to a color tolerance. This tolerance setting resulted in highlighted pixels with a range of green that corresponded precisely with the histological appearance of osseous tissue in the H&E-stained sections. The total number of H&E-positive pixels for each section was recorded. The pixel counts from individual sections were averaged for each tibial sample and the differences within and among treatment groups were calculated based on these averages. The areas for all the samples were then compared based on the amount of new cortical bone ( Figure 4 ).
The presence of bone in the sections was quantified to determine bone repair in the defects treated with gelfoam+peptide by comparison to gelfoam alone. Defects that were filled with gelfoam+R1 peptide showed the largest amount of cortical repair with a 10, 7 and 2-fold increase at 3, 5, 12 weeks respectively. The differences in cortical bone repair in the gelfoam+peptide compared with gelfoam alone were most noticeable at 3 and 5 weeks demonstrating that the peptide promotes cortical bone regeneration ( Figure 5 ).
10. Anabolic Effect of Osteogenic Peptides
The OPG/RANKL ratio is greater than the RANKL/OPG ratio, which suggests that the peptides may have a direct effect on osteoblasts and regulate bone resorption.
Synthetic peptides may have advantages over larger molecules such as speed of preparation, molecular stability, long shelf lives and potentially therapeutic applications. Significant progress has been made in attempts to modulate bone loss and regeneration by controlling cytokines and growth factors. These osteotropic peptides may offer attractive alternatives to existing therapies that stimulate bone anabolism and bone regeneration.
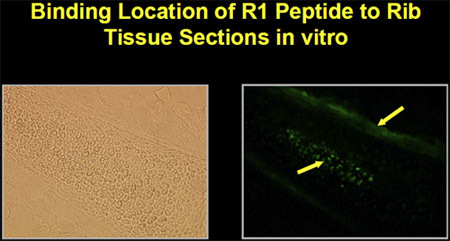 Figure 1. Peptide binding to marrow and bone in mouse rib tissue section in vitro.
Figure 1. Peptide binding to marrow and bone in mouse rib tissue section in vitro.
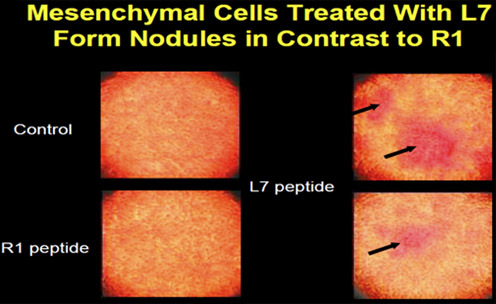 Figure 2. Mesenchymal cells (D1) form nodules after peptide treatment.
Figure 2. Mesenchymal cells (D1) form nodules after peptide treatment.
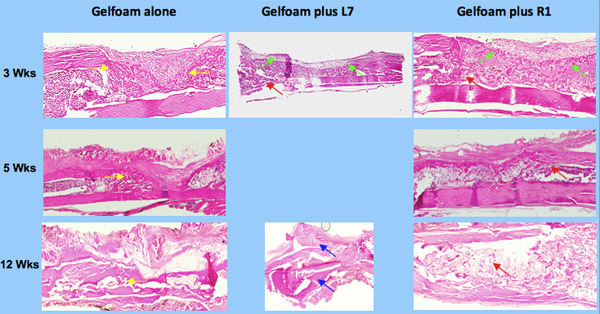 Figure 3. Histology of bone repair using osteogenic peptides, L7 and R1.
Figure 3. Histology of bone repair using osteogenic peptides, L7 and R1.
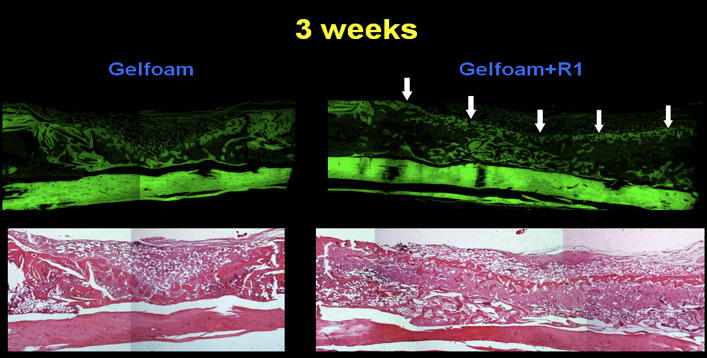 Figure 4. Bone sections stained with H & E under UV light which causes autofluorescence. This provides greater contrast between bone and non-bone tissues for quantitation purposes.
Figure 4. Bone sections stained with H & E under UV light which causes autofluorescence. This provides greater contrast between bone and non-bone tissues for quantitation purposes.
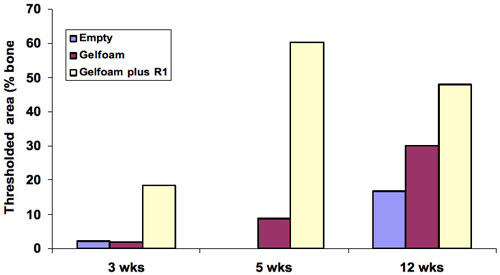 Figure 5. Quantitation of bone in the histology sections showed the largest amount of cortical repair was at 5 weeks with a 10-fold change, 7 and 2-fold at 3 and 5 weeks demonstrating that the peptide promotes cortical bone regeneration.
Figure 5. Quantitation of bone in the histology sections showed the largest amount of cortical repair was at 5 weeks with a 10-fold change, 7 and 2-fold at 3 and 5 weeks demonstrating that the peptide promotes cortical bone regeneration.
Disclosures
The production of this video was sponsored by New England Biolabs, Inc, which produces some of the reagents used in this article.
Acknowledgments
National Institutes of Health, AR53579, Osteogenesis: Potentiation with Bone Tropic Peptides National Institutes of Health, AR056422, Anabolic Effect of Osteogenic Peptides National Institutes of Health, T32 AR050960, Musculoskeletal Tissue Repair and Regeneration DOD, PC051219 Nucleolin: A Novel Mediator of Prostate Cancer and Bone Marrow Endothelial Cell Adhesion Department of Defense, PC061508, Prostate Cancer Metastasis Cell-Bone Marrow adhesion Mediators
References
- Diduch DR, Coe MR, Joyner C, Owen ME, Balian G. Two cell lines from bone marrow that differ in terms of collagen synthesis, osteogenic characteristics, and matrix mineralization. J. Bone Joint Surg. Am. 1993;75:92–105. doi: 10.2106/00004623-199301000-00012. [DOI] [PubMed] [Google Scholar]
- Stoddart MJ, Furlong PI, Simpson AE, Davies CM, Richards RG. Viability in ex vivo cultured cancellous. Eur. Cells. Mater. 2007;13:69–70. doi: 10.22203/ecm.v012a02. [DOI] [PubMed] [Google Scholar]
- Lehr JE, Pienta KJ. Preferential adhesion of prostate cancer cells to a human bone marrow endothelial cell line. J. Natl. Cancer Inst. 1998;90:118–123. doi: 10.1093/jnci/90.2.118. [DOI] [PubMed] [Google Scholar]
- Schuster N, Götz C, Faust M, Schneider E, Prowald A, Jungbluth A, Montenarh M. Wild-type p53 inhibits protein kinase CK2 activity. J. Cell Biochem. 2001;81:172–183. doi: 10.1002/1097-4644(20010401)81:1<172::aid-jcb1033>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Winter G. Isolation of a peptide antagonist to the thrombin receptor using phage display. J. Mol. Biol. 1994;244:361–369. doi: 10.1006/jmbi.1994.1736. [DOI] [PubMed] [Google Scholar]
- Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Rio GD, Krajewski S, Lombardo CR, Rao R, Ruoslahti E, Bredesen DE, Pasqualini R. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- Goodson RJ, Doyle MV, Kaufman SE, Rosenberg S. High-affinity urokinase receptor antagonists identified with bacteriophage peptide display. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7129–7133. doi: 10.1073/pnas.91.15.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Ruoslahti E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J. Cell Biol. 1995;130:1189–1196. doi: 10.1083/jcb.130.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J. Clin. Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajotte D, Ruoslahti E. Membrane dipeptidase is the receptor for a lung-targeting peptide identified by in vivo phage display. J. Biol. Chem. 1999;274:11593–11598. doi: 10.1074/jbc.274.17.11593. [DOI] [PubMed] [Google Scholar]
- Wang B. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes RA, Cooper CR, Beck GL, Pruitt F, Brown ML, Balian G. Bone Stromal Cells as Therapeutics Targets in Osseous Metastasis. In: Meadows G, Kluwer KH, editors. Cancer Growth and Progression. "Integration/Interaction of Oncologic Growth". Boston, MA: Academic Publishers; 2005. pp. 369–386. [Google Scholar]
- Santos JL, Pandita D, Rodrigues J, Pêgo AP, Granja PL, Tomás H, Balian G. Receptor-Mediated Gene Delivery in Mesenchymal Stem Cells using PAMAM Dendrimers conjugated with Osteotropic Peptides. Molecular Pharmaceutics. 2010 doi: 10.1021/mp9002877. Forthcoming. [DOI] [PubMed] [Google Scholar]


