Abstract
Rate variation in genes from all three genomes has been observed frequently in plant lineages with a parasitic and mycoheterotrophic mode of life. While the loss of photosynthetic ability leads to a relaxation of evolutionary constraints in genes involved in the photosynthetic apparatus, it remains to be determined how prevalent increased substitution rates are in nuclear DNA of non-photosynthetic angiosperms. In this study we infer rates of molecular evolution of 18S rDNA of all parasitic and mycoheterotorphic plant families (except Lauraceae and Polygalaceae) using relative rate tests. In several holoparasitic and mycoheterotrophic plant lineages extremely high substitution rates are observed compared to other photosynthetic angiosperms. The position and frequency of these substitutions have been identified to understand the mutation dynamics of 18S rRNA in achlorophyllous plants. Despite the presence of significantly elevated substitution rates, very few mutations occur in major functional and structural regions of the small ribosomal molecule, providing evidence that the efficiency of the translational apparatus in non-photosynthetic plants has not been affected.
Keywords: 18S rDNA, Mycoheterotrophy, Parasitism, Substitution rates, Relative rate test
Introduction
In flowering plants, a fully heterotrophic mode of life is an exceptional trait. Little over 1% of all described species derive all of their carbon from other organisms (Kuijt 1969; Heide-JØrgensen 2008). Based on the partners involved in the interaction, two groups of heterotrophic plants can be distinguished: parasitic and mycoheterotrophic plants. Parasitic plants directly penetrate host plants via their haustoria to obtain water and (in)organic solutes (Nickrent et al. 1998). These plants include hemiparasites and holoparasites, a division based on the presence or absence of chlorophyll during at least one part of their life cycle, respectively. Parasitic plants are mainly restricted to eudicots, with the exception of three magnoliid genera: Hydnora, Prosopanche (Hydnoraceae) and Cassytha (Lauraceae). A parasitic lifestyle has evolved at least 11 times independently in angiosperms, occurring in 20 families and close to 4,500 extant species (Nickrent et al. 1998; Barkman et al. 2007; Heide-JØrgensen 2008). The phylogenetic positions of many parasitic lineages are still unknown at low taxonomic level (APG 2003).
In contrast to parasitic plants, mycoheterotrophic plants derive carbon from fungi (Leake 2005). With a few exceptions, most mycoheterotrophic plant species exploit mycorrhizal fungi that are simultaneously mycorrhizal with neighboring photosynthetic plants. Because all carbon in this tripartite relationship is ultimately derived from autotrophic plants, mycoheterotrophic plants are considered to be epiparasitic on green plants (Bidartondo et al. 2002; Leake 2004). However, some mycoheterotrophic orchids are associated with litter- and wood-decay fungi (Ogura-Tsujita et al. 2009; Selosse et al. 2010). Over 400 fully mycoheterotrophic plant species have been reported in eudicots (Ericaceae, Gentianaceae and Polygonaceae) and monocots (Burmanniaceae, Corsiaceae, Iridaceae, Orchidaceae, Petrosaviaceae, Thismiaceae and Triuridaceae) (Leake 1994; Heide-JØrgensen 2008; Merckx et al. 2009). In addition, over 20,000 flowering plant species are thought to be at least partially mycoheterotrophic, mostly initial mycoheterotrophs in the Orchidaceae (Leake 2005; Selosse and Roy 2009).
With the loss of photosynthetic ability, genes required for the photosynthetic apparatus will undergo random mutations under relaxed natural selection (Conopholis americana, Wimpee et al. 1991; Cuscuta, Funk et al. 2007; McNeal et al. 2009; Revill et al. 2005; Epifagus virginiana, dePamphilis and Palmer 1990; Wolfe et al. 1992; dePamphilis et al. 1997; Lathraea clandistina, Delavaut et al. 1995; Orobanche hederae, Thalouarn et al. 1994). Although some chloroplast genes are retained and functional in several holoparasitic plants (Bungard 2004), many holoparasites have a reduced plastid genome due to excessive gene loss and increased substitution rates in the remaining genes (Wolfe et al. 1992). In chloroplast genome analysis of non-photosynthetic plants different explanations have been proposed why a reduced set of genes have been retained and need to be translated within the chloroplast as opposed to replacement by a product from cytosolic orthologues: (1) import of proteins with hydrophobic membranes from the cytosol back into the organelle would be impossible; (2) rate of synthesis of specific proteins can be regulated within a individual plastids preventing deadly side-effects of oxidative stress or lethal effects of accumulating toxic products; (3) synthesis and assembly of components of the photosynthetic complexes are tightly regulated within plastids (see review by Barbrook et al. 2006). The plastid genome of fully mycoheterotrophic plants remains largely unstudied, but rbcL data suggest that the genome is prone to a similar relaxation of purifying selection (Caddick et al. 2002; Barrett and Freudenstein 2008). These observations demonstrate that non-photosynthetic plants are under reduced selective constraints, which affect the structure and function of genes involved in photosynthetic reactions.
Somewhat surprisingly, extreme variation in rates of evolution has also been observed in the nuclear and/or mitochondrial genes of some fully mycoheterotrophic (Merckx et al. 2006, 2009; Petersen et al. 2006) and parasitic plants (Nickrent and Starr 1994; Nickrent et al. 1998; Davis et al. 2004; Chase 2004; Barkman et al. 2007). The causes for increased substitution rates in nuclear and mitochondrial DNA of achlorophyllous plants are still poorly documented and understood. Several hypotheses related to the loss of photosynthesis, effective population size, generation time and host specificity have been proposed to explain this phenomenon in parasitic plants. However, none of these hypotheses could unequivocally explain the rate increases in all parasitic plants (dePamphilis and Palmer 1990; Nickrent and Starr 1994; dePamphilis et al. 1997; Young and dePamphilis 2005).
In this study, we infer the variation of substitution rates of nuclear 18S rDNA of nearly all angiosperm families by comparing branch length variation of 37 fully mycoheterotrophic species, 33 parasitic plant species, and related autotropic lineages with relative rate tests (Robinson-Rechavi and Huchon 2000; Wilcox et al. 2004). Estimating rate variation across taxa, representing most groups of angiosperms, gives us the opportunity to address the following questions: How frequent can increased substitution rates be observed in nuclear loci of hemiparasitic, holoparasitic and mycoheterotrophic plants? Are individual achlorophyllous taxa or whole heterotrophic plant families prone to substitution rates? Is loss of chlorophyll in angiosperms associated with longer branches? Can we provide evidence for another hypothesis, which could cause the increased substitution rates in 18S rDNA of mycoheterotrophic and parasitic plants? What are the exact nucleotide positions of the substitution in 18S rRNA and are they interfering with functional sites?
Materials and methods
Taxon sampling
In total, 18S rDNA sequences of 178 angiosperm species were used for this study, representing the 45 orders of the APGII classification (APGII 2003). All families with parasitic and fully mycoheterotrophic species are represented in our study by one or more taxa, with the exception of Polygalaceae (Epirixanthes—fully mycoheterotrophic) and Lauraceae (Cassytha—hemiparasitic). To evaluate the substitution rates of mycoheterotrophic and parasitic taxa, 18S rDNA sequences of related autotrophic plants were obtained from Genbank and included as reference points to measure substitution rates. The outgroups used for the heterotrophic lineages, according to recent phylogenetic studies are showed in Table 1. All species sampled with voucher information and Genbank accession numbers are listed in the “Appendix”.
Table 1.
List of mycoheterotrophic/parasitic taxa and outgroups investigated in this study
| Mycoheterotroph/parasite | BRT (constrained) | BRT (unconstrained) | References |
|---|---|---|---|
| Outgroup | |||
| Hemiparasites | |||
| Convolvulaceae | |||
| Cuscuta gronovii | 0.332/0.390/0.471 | 0.334/0.401/0.466 | McNeal et al. (2007) |
| Convolvulus arvensis | 0.164/0.202/0.252 | 0.167/0.216/0.260 | |
| Krameriaceae | |||
| Krameria ixine | 0.117/0.155/0.208 | 0.137/0.173/0.221 | Simpson et al. (2004) |
| Guiacum sanctum | 0.096/0.137/0.176 | 0.118/0.152/0.201 | |
| Orobanchaceae | |||
| Pedicularis racemosa | 0.128/0.164/0.206 | 0.120/0.157/0.192 | Olmstead et al. (2001) |
| Orthocarpus erianthus | 0.143/0.184/0.231 | 0.142/0.179/0.219 | |
| Paulownia tomentosa | 0.123/0.154/0.194 | 0.115/0.151/0.189 | |
| Lamium amplexicaule | 0.139/0.171/0.214 | 0.128/0.165/0.202 | |
| Santalales (Loranthaceae) | |||
| Nuytsia floribunda | 0.114/0.152/0.195 | 0.106/0.139/0.181 | Der and Nickrent (2008) |
| Tupeia antarctica | 0.102/0.139/0.184 | 0.094/0.130/0.174 | Malécot and Nickrent (2008) |
| Santalales (Misodendraceae) | |||
| Misodendrum linearifolium | 0.193/0.242/0.308 | 0.194/0.245/0.298 | |
| Santalales (Olacaceae) | |||
| Erythropalum scandens | 0.092/0.124/0.161 | 0.081/0.116/0.150 | |
| Olax aphylla | 0.156/0.207/0.261 | 0.130/0.172/0.216 | |
| Santalales (Opiliaceae) | |||
| Opilia amentacea | 0.179/0.227/0.293 | 0.161/0.205/0.267 | |
| Lepionurus sylvestris | 0.170/0.215/0.269 | 0.149/0.196/0.250 | |
| Santalales (Santalaceae) | |||
| Lepidoceras chilense | 0.170/0.208/0.270 | 0.166/0.210/0.262 | |
| Santalum album | 0.126/0.166/0.231 | 0.138/0.175/0.221 | |
| Santalales (Schoepfiaceae) | |||
| Schoepfia arenaria | 0.158/0.197/0.242 | 0.133/0.184/0.229 | |
| Santalales (Viscaceae) | |||
| Arceuthobium verticilliflorum | 0.365/0.438/0.512 | 0.363/0.445/0.521 | |
| Dendrophtora domingensis | 0.233/0.288/0.351 | 0.244/0.300/0.368 | |
| Gunnera manicata | 0.131/0.167/0.212 | 0.112/0.150/0.189 | |
| Hamamelis virginiana | 0.127/0.166/0.206 | 0.132/0.169/0.222 | |
| Plumbago auriculata | 0.142/0.182/0.219 | 0.144/0.186/0.238 | |
| Holoparasites | |||
| Apodanthaceae | |||
| Pilostyles thurberi | 0.189/0.245/0.296 | 0.189/0.235/0.282 | Nickrent et al. (2004) |
| Celastrus scandens | 0.162/0.213/0.272 | 0.158/0.202/0.246 | |
| Balanaphoraceae | |||
| Ombrophytum subterraneum | 0.386/0.457/0.547 | 0.383/0.456/0.555 | Stevens (2001) |
| Gunnera manicata | 0.131/0.167/0.212 | 0.112/0.150/0.189 | |
| Hamamelis virginiana | 0.127/0.166/0.206 | 0.132/0.169/0.222 | |
| Plumbago auriculata | 0.142/0.182/0.219 | 0.144/0.186/0.238 | |
| Cynomoriaceae | |||
| Cynomorium coccineum | 0.189/0.236/0.285 | 0.182/0.226/0.285 | Nickrent et al. (2005) |
| Gunnera manicata | 0.131/0.167/0.212 | 0.112/0.150/0.189 | |
| Hamamelis virginiana | 0.127/0.166/0.206 | 0.132/0.169/0.222 | |
| Plumbago auriculata | 0.142/0.182/0.219 | 0.144/0.186/0.238 | |
| Cytinaceae | |||
| Bdallophyton americanum | 0.256/0.303/0.346 | 0.248/0.308/0.374 | Nickrent (2007) |
| Cytinus hypocistis | 0.322/0.406/0.467 | 0.343/0.410/0.479 | |
| Muntingia calabura | 0.136/0.172/0.217 | 0.147/0.187/0.233 | |
| Euphorbiaceae | |||
| Rafflesia keithii | 0.930/1.091/1.274 | 0.942/1.085/1.267 | Davis et al. (2007) |
| Rhizanthes infanticida | 0.982/1.136/1.310 | 0.977/1.132/1.317 | |
| Euphorbia pulcherrima | 0.114/0.156/0.203 | 0.107/0.145/0.182 | |
| Hydnoraceae | |||
| Hydnora africana | 0.293/0.359/0.410 | 0.364/0.432/0.497 | Nickrent et al. (2002) |
| Prosopanche americana | 0.351/0.425/0.495 | 0.397/0.487/0.566 | |
| Aristolochia macrophylla | 0.074/0.099/0.130 | 0.103/0.164/0.215 | |
| Lennoaceae | |||
| Pholisma arenarium | 0.242/0.290/0.346 | 0.236/0.276/0.320 | Olmstead and Ferguson (2001) |
| Hydrophyllum fendleri | 0.173/0.215/0.270 | 0.162/0.204/0.243 | |
| Mitrastemonaceae | |||
| Mitrastemon yamamotoi | 0.351/0.420/0.508 | 0.363/0.439/0.542 | Davis et al. (2007) |
| Vaccinium macrocarpon | 0.143/0.183/0.225 | 0.133/0.170/0.207 | |
| Orobanchaceae | |||
| Boschniakia rossica | 0.126/0.169/0.214 | 0.127/0.165/0.199 | Olmstead et al. (2001) |
| Conopholis americana | 0.140/0.178/0.221 | 0.132/0.173/0.210 | |
| Epifagus virginiana | 0.135/0.173/0.214 | 0.130/0.168/0.204 | |
| Harveya speciosa | 0.155/0.195/0.241 | 0.158/0.195/0.238 | |
| Lathraea clandestina | 0.154/0.194/0.242 | 0.146/0.190/0.229 | |
| Orobanche fasciculata | 0.178/0.220/0.276 | 0.171/0.217/0.256 | |
| Paulownia tomentosa | 0.123/0.154/0.194 | 0.115/0.151/0.189 | |
| Lamium amplexicaule | 0.139/0.171/0.214 | 0.128/0.165/0.202 | |
| Mycoheterotrophs | Family | ||
| Burmanniaceae | |||
| Apteria aphylla | 0.221/0.279/0.349 | 0.155/0.212/0.304 | Merckx et al. (2006) |
| Burmannia oblonga | 0.263/0.311/0.358 | 0.188/0.238/0.288 | |
| Burmannia sphagnoides | 0.198/0.254/0.303 | 0.136/0.187/0.249 | |
| Campylosiphon purpurascens | 0.147/0.188/0.246 | 0.083/0.122/0.160 | |
| Cymbocarpa refracta | 0.201/0.254/0.326 | 0.138/0.192/0.256 | |
| Dictyostega orobanchoides | 0.186/0.227/0.298 | 0.105/0.157/0.210 | |
| Gymnosiphon aphyllus | 0.178/0.232/0.286 | 0.115/0.159/0.215 | |
| Gymnosiphon divaricatus | 0.163/0.215/0.281 | 0.099/0.144/0.207 | |
| Hexapterella gentianoides | 0.141/0.187/0.243 | 0.075/0.118/0.166 | |
| Burmannia bicolor | 0.329/0.395/0.479 | 0.269/0.325/0.377 | |
| Dioscorea rockii | 0.139/0.189/0.239 | 0.083/0.123/0.169 | |
| Corsiaceae | |||
| Arachnitis uniflora * | 0.599/0.705/0.805 | 0.584/0.698/0.849 | Fay et al. (2006) |
| Luzuriaga latifolia | 0.102/0.140/0.183 | 0.037/0.075/0.107 | |
| Ericaceae | |||
| Hemitomes congestum* | 0.149/0.199/0.256 | 0.147/0.187/0.238 | Cullings (2000) |
| Monotropa uniflora | 0.191/0.242/0.293 | 0.151/0.198/0.248 | |
| Pityopus californicus* | 0.165/0.210/0.252 | 0.184/0.233/0.283 | |
| Pterospora andromedea | 0.157/0.200/0.241 | 0.156/0.191/0.232 | |
| Sarcodes sanguinea | 0.151/0.196/0.237 | 0.139/0.185/0.231 | |
| Arctostaphylos uva-ursi | 0.104/0.142/0.182 | 0.097/0.135/0.174 | |
| Pyrola picta | 0.120/0.159/0.197 | 0.114/0.150/0.191 | |
| Gentianaceae | |||
| Exacum paucisquamum* | 0.251/0.294/0.352 | 0.229/0.269/0.315 | Yuan et al. (2003) |
| Sebaea oligantha* | 0.195/0.241/0.303 | 0.177/0.217/0.255 | |
| Voyria caerulia* | 0.241/0.294/0.361 | 0.218/0.268/0.320 | |
| Voyria corymbosa* | 0.275/0.327/0.387 | 0.257/0.303/0.352 | |
| Voyria aurantiaca* | 0.255/0.299/0.366 | 0.227/0.273/0.319 | |
| Voyriella parviflora* | 0.170/0.221/0.284 | 0.152/0.198/0.238 | |
| Sebaea grandis | 0.187/0.244/0.308 | 0.174/0.219/0.263 | |
| Iridaceae | |||
| Geosiris sp. | 0.095/0.134/0.175 | 0.067/0.095/0.129 | Reeves et al. (2001) |
| Aristea glauca | 0.122/0.157/0.199 | 0.096/0.123/0.155 | |
| Orchidaceae | |||
| Corallorhiza maculata | 0.165/0.205/0.246 | 0.112/0.171/0.221 | Freudenstein et al. (2004) |
| Aplectrum hyemale | 0.161/0.200/0.246 | 0.108/0.167/0.217 | Molvray et al. (2000) |
| Erythrorchis cassythoides | 0.288/0.335/0.385 | 0.226/0.297/0.361 | |
| Cyrtosia septentrionalis | 0.277/0.329/0.378 | 0.231/0.293/0.367 | |
| Lecanorchis multiflora | 0.265/0.305/0.361 | 0.216/0.270/0.335 | |
| Vanilla aphylla | 0.273/0.313/0.366 | 0.219/0.279/0.346 | |
| Neottia nidus - avis | 0.201/0.245/0.293 | 0.167/0.217/0.272 | |
| Eburophyton austinae | 0.196/0.236/0.285 | 0.154/0.205/0.262 | |
| Rhizanthella gardneri | 0.436/0.490/0.555 | 0.378/0.443/0.529 | |
| Diuris sulphurea | 0.223/0.265/0.310 | 0.175/0.225/0.286 | |
| Wullschlaegelia calcarata | 0.175/0.222/0.265 | 0.137/0.189/0.239 | |
| Orchis quadripunctata | 0.237/0.274/0.324 | 0.177/0.236/0.295 | |
| Petrosaviaceae | |||
| Petrosavia stellaris | 0.083/0.131/0.204 | 0.052/0.112/0.162 | Cameron et al. (2003) |
| Japonolirion osense | 0.073/0.117/0.180 | 0.056/0.102/0.152 | |
| Thismiaceae | |||
| Afrothismia hydra | 0.469/0.554/0.657 | 0.431/0.522/0.638 | Merckx et al. (2006) |
| Afrothismia winkleri | 0.472/0.557/0.658 | 0.428/0.518/0.632 | |
| Haplothismia exannulata | 0.285/0.333/0.394 | 0.210/0.268/0.333 | |
| Thismia rodwayi | 0.332/0.385/0.459 | 0.253/0.327/0.428 | |
| Thismia aseroe | 0.475/0.551/0.630 | 0.379/0.467/0.569 | |
| Tacca chantieri | 0.148/0.195/0.248 | 0.095/0.135/0.185 | |
| Dioscorea rockii | 0.139/0.189/0.239 | 0.083/0.123/0.169 | |
| Triuridaceae | |||
| Kupea martinetugei | 0.378/0.472/0.557 | 0.341/0.416/0.500 | Stevens (2001) |
| Sciaphila ledermannii* | 0.133/0.185/0.237 | 0.077/0.128/0.177 | |
| Sciaphila densiflora | 0.131/0.188/0.246 | 0.076/0.132/0.175 | |
| Stemona japonica | 0.103/0.156/0.206 | 0.075/0.119/0.164 | |
95% confidence intervals and median values of constrained and non-constrained Bayesian relative rates test (BRT) are indicated between “/”. Mycoheterotrophic/parasitic taxa are indicated in bold. Lineages with significant elevated substitution rates are underlined. Choice of outgroups were based on previous phylogenetic studies. Newly obtained 18S rDNA sequences for this study are indicated with an asterisk
DNA extraction, amplification and sequencing
Ten and six 18S rDNA sequences of mycoheterotrophic and autotrophic plants, respectively, were newly obtained for this study (Table 1; Appendix). Total DNA was extracted from silica-dried material with the Puregene DNA extraction kit (Gentra Systems, Landgraaf, The Netherlands) following the manufacturer’s instructions. The 18S rDNA region was amplified using primers NS1, NS2, NS3, NS4, NS5 and NS8 (White et al. 1990). Each amplification reaction was performed in 25 μl reaction mix containing 5 μl DNA, 4 μl H2O, 2.5 μl 10× PCR buffer, 0.75 μl 25 mM MgCl2, 10 μl of 2.2 mM forward and reverse primers, 2.5 μl 2 mM dNTP and 0.2 μl Taq DNA polymerase. The polymerase chain reactions were run on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) for 30 cycles starting at 94°C for 1 min, followed by denaturation (94°C for 30 s), annealing (44°C for 30 s), extension (72°C for 1 min) and a final extension (72°C for 7 min). PCR products were cleaned using a Nucleospin Extraction II kit (Machery-Nagel, Düren, Germany) according to the manufacturer’s instructions. Sequencing reactions were done using the ABI PRISM Big Dye Terminator Cycle sequencing kit (Applied Biosystems) with the same primers as listed above. All the samples were sequenced on an ABI 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analyses
The sequences were assembled and edited using Staden et al. (1998). A preliminary sequence alignment was created with Clustal X (Thompson et al. 1997) followed by manual adjustments with MacClade 4.04 (Maddison and Maddison 2001) resulting in an unequivocal alignment with a length of 1708 nucleotide positions. The best fitting model of DNA substitution was chosen by performing hierarchical Likelihood-ratio tests in MrModeltest v3.06 (Posada and Crandall 1998). The Likelihood-ratio tests and Akaike Information Criterion selected the GTR+I+G model of evolution. To reduce calculation time a starting tree for the Bayesian analyses was generated under Maximum Likelihood using Garli v0.951 (Zwickl 2006) with the GTR+I+G model of evolution. An initial unconstrained Bayesian analysis of the 18S rDNA dataset retrieved a moderate resolved topology with conflicting nodes as compared to the relationships of APGII (results not shown). However, the conflicting nodes in our unconstrained phylogeny did not receive significant Bayesian posterior probabilities. In order to improve the topology of this single gene analysis according to the classification of APGII and to compare the branch lengths of the heterotrophic lineages with the according autotrophic relatives, the analysis was rerun with 19 constraints using the ‘topologypr’ command in MrBayes (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) (Fig. 1a, b). The constraints resolved some major informal groups (e.g. magnoliids, monocots, commelinids, eudicots, core eudicots, rosids including eurosids I and II and asterids, including euasterids I and II) as recovered by multi-gene analyses (APG 2003) and forced five families and one order containing mycoheterotrophic and parasitic lineages in monophyletic groups (Euphorbiaceae, Gentianaceae, Orchidaceae, Petrosaviaceae and Santalales). Amborella was chosen as outgroup for the analysis. Bayesian analyses were run on the K.U.Leuven UNIX cluster (‘VIC’), running four Markov chains sampling every 100 generations for four million generations. The first 10,000 sampled trees (25%) were regarded as “burnin” and discarded. Convergence of the Markov chains was checked using Tracer v.1.4 (Rambaut and Drummond 2007).
Fig. 1.
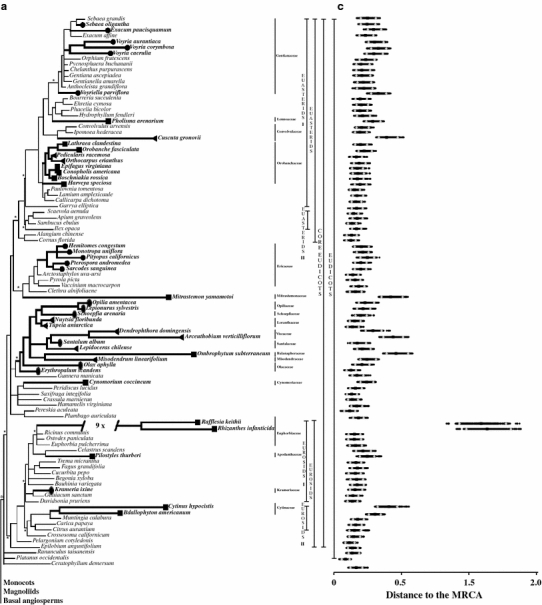
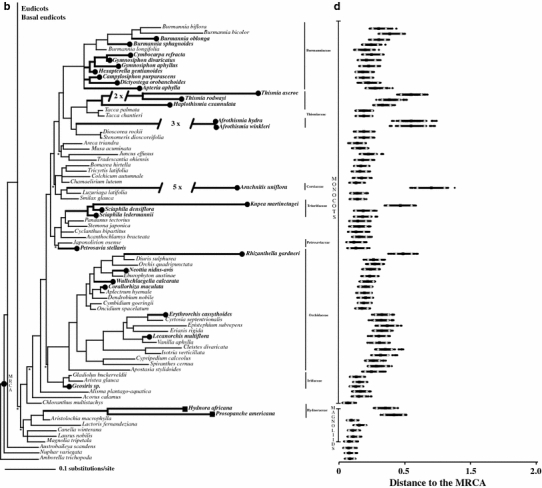
a, b The majority-rule consensus phylogram of the Bayesian analysis of the 18S rDNA data under topology constraints. The implemented constrains are indicated with asterisks. Branches in bold with a circle, a square, a diamond and an ellipse are, respectively, mycoheterotrophic, holoparasitic, hemiparasitic and facultative hemiparasitic lineages. c, d The Bayesian relative rates test shows the relative branch lengths of all ingroup taxa using Amborella as most recent common ancestor (MRCA). The squares indicate the mean branch lengths. The triangles delimit the 95% confidence intervals
Relative rate tests
Rates of molecular evolution of 18S rDNA data were estimated using a Bayesian relative rates test according to the method described by Wilcox et al. (2004). From 40,000 sampled trees of the Bayesian analysis with the topology constraints enforced, 25% burnin phase excluded, 500 phylograms were randomly selected. For these sampled trees, the distance from the most recent common ancestor (MRCA) of the ingroup to each of the terminal taxa was calculated with Cadence v1.0 (Wilcox et al. 2004). The MRCA of the ingroup is the ancestral node shared by all ingroup taxa. The values of these distances for every taxon were plotted in Excel (Excel 2004 for Mac version 11.4.1) in order to calculate the 95% confidence intervals. Based on the assumption that the confidence interval of a given taxon does not overlap with the confidence interval of another taxon, we can state that a significant difference in rate of molecular evolution between these two individual taxa has occurred (Wilcox et al. 2004). In order to determine the effect of the implementation of topology constraints on the estimation of substitution rates, the Bayesian relative rates test was repeated using phylograms sampled during the unconstrained Bayesian analysis.
To test whether grouped phylogenetic lineages accumulated nucleotide substitutions at significantly increased rates compared to an outgroup, we performed the relative rate test using the program package RRTree (Robinson-Rechavi and Huchon 2000). RRTree computes differences in molecular rates between non-coding DNA sequences using Kimura’s two parameter (K2P) (Kimura 1980) and Jukes and Cantor’s one parameter (JC) model. The K2P substitution model was selected for this study because it is the most complex model presently implemented in RRTree.
Patterns of nucleotide substitutions in 18S rDNA of mycoheterotrophic, parasitic and autotrophic angiosperms
Conserved and variable nucleotides in the 18S rDNA dataset were identified using the chart option in MacClade 4.04 (Maddison and Maddison 2001). Each character with the corresponding character states for each angiosperm taxon was optimized on the consensus tree obtained from the Bayesian analyses. This approach provides a minimum estimate of change for each site. The values of the numbers of character substitutions or steps for every nucleotide site are shown on a histogram constructed with Excel 2004 (for Mac version 11.4.1). This pattern of conserved and variable nucleotides was examined within heterotrophic (strict mycoheterotrophic and holoparasitic taxa) and autotrophic species separately, both groups including 54 taxa. The autotrophic species included within this analysis are all sister group lineages of the heterotrophic taxa. Hemiparasitic and partly mycoheterotrophic taxa were removed from the analysis. In addition, the difference between both groups was calculated and values of steps per nucleotide site were depicted on a histogram. Positive and negative values represent character substitutions contributed by heterotrophic lineages and autotrophic species, respectively. Furthermore, nucleotide substitutions of each nucleotide site were associated with the secondary structures and functional regions proposed by Wuyts et al. (2000) and Caetano-Anollés (2002). Nucleotides within the 18S rDNA alignment were divided into 915 stem and 793 loop positions, according to secondary structures obtained by the European Small Subunit Ribosomal RNA database (Van de Peer et al. 2000; Wuyts et al. 2004). Stem regions are defined as those bases that typically participate in base pairing, with the remainder occurring in non-base pairing loop structures. In this study, base pairing was restricted to Watson–Crick nucleotide pairs and G–U pairs.
Results
Evolutionary rates
The constrained phylogram (Fig. 1a, b) reveals heterogeneity in branch lengths among the taxa investigated. Several mycoheterotrophic and parasitic plant species show extremely elevated substitution rates resulting in long terminal branches (e.g. Rafflesia keithii, Rhizanthes infanticida and Arachnitis uniflora). The results of the Bayesian relative rates test are shown in Fig. 1 (panels c, d) and listed in Table 1. The values of the confidence intervals for every mycoheterotrophic and parasitic lineage with the associated outgroups are given in Table 1. In order to analyze the effect of the implementation of constraints on the substitution rates of mycoheterotrophic and parasitic lineages, a second Bayesian relative rates test was conducted on the same dataset using trees from the unconstrained analysis. Overall, the confidence intervals of the substitution rates in the constrained and non-constrained phylogenetic analyses showed no pronounced differences, which mean that the implementation of topology constraints did not affect branch lengths (Table 1).
According to the Bayesian relative rate test the substitution rates of several hemiparasitic (3 out of 16), holoparasitic (8 out of 17) and mycoheterotrophic (8 out of 37) species are significantly higher compared to their autotrophic relatives (Table 1). In general, most holoparasitic plant families share substantial elevated substitution rates, which are on average 1.8 (Bdallophyton americanum—Cytinaceae) to 7.3-fold (Rhizanthes infanticida—Euphorbiaceae) higher compared to their autotrophic relatives. Only the holoparasitic families, Apodanthaceae, Cynomoriaceae, Lennoaceae and Orobanchaceae, do not have significantly increased substitution rates. Hemiparasitic plants are characterized by less pronounced substitution rates where only three species are placed on significant longer branches: average of 1.6 (Dendrophthora domingensis—Viscaceae) to 2.4-fold (Arceuthobium verticilliflorum—Viscaceae) higher compared to their autotrophic relatives. The (facultative) hemiparasites of the families Krameriaceae and Orobanchaceae and the remaining Santalalean lineages are not prone to increased substitution rates in 18S rDNA. In mycoheterotrophic plants only eight out of 37 fully mycoheterotrophic plants have significantly elevated substitution rates with an average increase between 1.7 (Haplothismia exannulata—Thismiaceae) and 5.0-fold (Arachnitis uniflora—Corsiaceae). The most pronounced increase of substitution rates among mycoheterotrophic species included all five taxa of Thismiaceae and the single taxon of Corsiaceae. Although the branch lengths of the two mycoheterotrophic species Rhizanthella gardneri (Orchidaceae) and Kupea martinetugea (Triuridaceae) are considerably shorter they are still significantly longer compared to their autotrophic sister groups. The Bayesian relative rates test reveals no significant variation in rates of molecular evolution in species of the mycoheterotrophic families Burmanniaceae, Ericaceae, Gentianaceae, Iridaceae and Petrosaviaceae. However, depending on whether Burmannia bicolor or Dioscorea rockii was used as autotrophic relative, the mycoheterotroph Burmannia oblonga, is either placed on a significant or non-significant long branch than its green relatives.
The RRTree test revealed similar results compared to the Bayesian relative rate test (Table 2). However, significant increased substitution rates were observed in five additional mycoheterotrophic (Exacum paucisquamum and the genus Voyria) and holoparasitic groups (Cynomorium coccineum, Pholisma arenarium and Pilostyles thurberi). Furthermore, mycoheterotrophic/parasitic clades (i.e. Triuridaceae and the Santalales clade), which contain both lineages with and without significantly increased substitution rates according to the Bayesian relative rates test, are indicated by significant P-values for the whole clade.
Table 2.
Values of the relative rate test for comparing molecular evolutionary rates between parasitic/mycoheterotrophic lineages (Lineage 1) and autotrophic relatives (Lineage 2)
| Family | Lineage 1 | Lineage 2 | K1 | K2 | K1–K2 | K1/K2 | P valuea |
|---|---|---|---|---|---|---|---|
| Apodanthaceae | Pilostyles b | Celastrus | 0.055 | 0.043 | 0.012 | 1.3 | 2.2 × 10 −2 |
| Burmanniaceae | Apteria | B. biflora, B. bicolor | 0.006 | 0.068 | −0.062 | 0.1 | 3.0 × 10−1 |
| Burmannia oblonga | B. biflora, B. bicolor | 0.059 | 0.068 | −0.009 | 0.9 | 4.5 × 10−2 | |
| Burmannia sphagnoides | B. biflora, B. bicolor | 0.033 | 0.053 | −0.020 | 0.6 | 8.0 × 10−5 | |
| Campylosiphon, Dictyostega | B. biflora, B. bicolor | 0.063 | 0.093 | −0.029 | 0.7 | 7.7 × 10−5 | |
| Cymbocarpa, Gymnosiphon clade, Hexapterella | B. biflora, B. bicolor | 0.044 | 0.068 | −0.024 | 0.6 | 1.0 × 10−7 | |
| Convolvulaceae | Cuscuta | Convolvulus, Ipomoea | 0.077 | 0.044 | 0.033 | 1.8 | 1.0 × 10 −7 |
| Corsiaceae | Arachnitis | Luzuriaga, Smilax | 0.105 | 0.020 | 0.084 | 5.2 | 1.0 × 10 −7 |
| Cynomoriaceae | Cynomorium b | Hamamelis, Peridiscus | 0.053 | 0.039 | 0.014 | 1.4 | 3.9 × 10 −3 |
| Cytinaceae | Bdallophyton, Cytinus | Muntingia | 0.072 | 0.041 | 0.031 | 1.8 | 1.0 × 10 −7 |
| Ericaceae | Ericaceae clade | Arctostaphylos, Pyrola, Vaccinium | 0.054 | 0.053 | 0.001 | 1.0 | 7.4 × 10−1 |
| Euphorbiaceae | Rafflesia, Rhizanthes | Euphorbia, Ostodes, Ricinus | 0.150 | 0.035 | 0.115 | 4.3 | 1.0 × 10 −7 |
| Gentianaceae | Exacum b | Sebaea grandis | 0.064 | 0.050 | 0.014 | 1.3 | 1.5 × 10 −3 |
| Gentianaceae | Sebaea oligantha | Sebaea grandis | 0.051 | 0.050 | 0.001 | 1.0 | 5.2 × 10−1 |
| Gentianaceae | Voyria clade b | Sebaea grandis, Orphium | 0.063 | 0.052 | 0.011 | 1.2 | 1.4 × 10 −3 |
| Gentianaceae | Voyriella | Sebaea grandis, Orphium | 0.049 | 0.050 | −0.001 | 1.0 | 8.3 × 10−1 |
| Hydnoraceae | Hydnora, Prosopanche | Aristolochia, Lactoris | 0.086 | 0.039 | 0.047 | 2.2 | 1.0 × 10 −7 |
| Iridaceae | Geosiris | Aristea, Gladiolus | 0.034 | 0.035 | −0.001 | 1.0 | 7.6 × 10−1 |
| Krameriaceae | Krameria | Guaiacum | 0.039 | 0.039 | 0.000 | 1.0 | 9.9 × 10−1 |
| Lennoaceae | Pholisma b | Hydrophyllum, Phacelia, Ehretia | 0.060 | 0.044 | 0.016 | 1.4 | 7.7 × 10 −4 |
| Mitrastemonaceae | Mitrastemon | Arctostaphylos, Pyrola, Vaccinium | 0.086 | 0.043 | 0.044 | 2.0 | 1.0 × 10 −7 |
| Orchidaceae | Corallorhiza | Aplectrum, Dendrobium | 0.048 | 0.045 | 0.003 | 1.1 | 1.2 × 10−1 |
| Orchidaceae | Erytrorchis | Cyrtosia, Eriaxis, Epistephium | 0.060 | 0.062 | −0.002 | 1.0 | 6.0 × 10−1 |
| Orchidaceae | Lecanorchis | Vanilla, Eriaxis, Epistephium | 0.050 | 0.054 | −0.004 | 0.9 | 2.7 × 10−1 |
| Orchidaceae | Neottia | Eburophyton | 0.060 | 0.055 | 0.006 | 1.1 | 1.2 × 10−1 |
| Orchidaceae | Rhizanthella | Diuris, Orchis | 0.088 | 0.050 | 0.038 | 1.8 | 2.0 × 10 −7 |
| Orchidaceae | Wullschlaegelia | Eburophyton, Diuris | 0.044 | 0.054 | −0.010 | 0.1 | 7.4 × 10−3 |
| Orobanchaceae | Orobanchaceae clade | Paulownia, Lamium, Callicarpa | 0.044 | 0.425 | 0.002 | 0.1 | 5.6 × 10−1 |
| Petrosaviaceae | Petrosavia | Japonolirion | 0.035 | 0.031 | 0.004 | 1.1 | 2.5 × 10−1 |
| Santalales | Santalales clade | Gunnera, Peridiscus, Plumbago | 0.051 | 0.039 | 0.012 | 1.3 | 6.1 × 10 −6 |
| Thismiaceae | Afrothismia clade | Dioscorea, Stenomeris, Tacca | 0.113 | 0.041 | 0.072 | 2.8 | 1.0 × 10 −7 |
| Thismiaceae | Haplothismia, Thismia clade | Dioscorea, Stenomeris, Tacca | 0.082 | 0.042 | 0.040 | 1.9 | 1.0 × 10 −7 |
| Triuridacaee | Kupea, Sciaphila clade | Pandanus, Stemona | 0.056 | 0.037 | 0.019 | 1.5 | 5.7 × 10 −6 |
P values of lineages with significant increased substitution rates are indicated in bold
K1 mean divergence between lineage 1 and the most recent common ancestor of lineages 1 and 2; K2 mean divergence between lineage 2 and the most recent common ancestor of lineages 1 and 2; K1/K2 rate ratio
aSignificance of the P values <0.05
bLineages with significant increased substitution rates which are not observed in the Bayesian relative rates test
Patterns of nucleotide substitution in 18S rDNA of heterotrophic and autotrophic angiosperms
Alignment of 18S rDNA angiosperm sequences showed an alternation of conserved and variable regions. Using the chart option in MacClade, patterns of nucleotide substitutions were generated, describing a mosaic of conserved and variable nucleotides across the angiosperms (Fig. 2). Due to missing data the first and last nucleotide positions of the alignment could not be compared among all sequences. This resulted in a substitution pattern starting and ending in helix structure number 8 and 49, respectively.
Fig. 2.
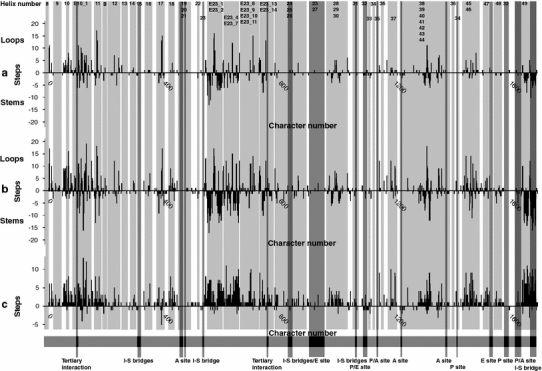
Histograms showing the pattern of nucleotide substitutions in 18S rDNA across angiosperm taxa. The histogram describes for each nucleotide site the amount of parsimony steps or character substitutions (y-axis; a, b loops above, stems below; c heterotrophic minus autotrophic). Helix numbering according to Wuyts et al. (2000) is shown at the top. The major functional and structural sites according to Caetano-Anollés (2002) are indicated at the bottom. a Pattern of nucleotide substitution of 54 autotrophic taxa. b Pattern of nucleotide substitution of 37 mycoheterotrophic and 17 holoparasitic taxa. c Difference between mycoheterotrophic/parasitic and autotrophic values of character substitutions per nucleotide site. Light gray and dark gray shading displays the different helix structures and functional regions, respectively
Variable nucleotides tend to occur both in loop and stem structures in both autotrophs and holoparasites/mycoheterotrophs, but highly variable regions (≥10 steps) are more abundant in loop structures (e.g. 11, 17, E23_2, E23_7, E23_12, E23_13 and 49), than in stem structures (e.g. E10_1, 11, 43 and 49) (Fig. 2a, b). Conserved regions with no or few mutations appear both in stem and loop structures (8, 9, 13, 14, 15, 16, 4, 19, 20, 21, 3, 22, 23, E23_8, E23_9, E23_10, E23_11, 26, 23, 27, 28, 30, 31, 2, 32, 33, 34, 35, 36, 38, 39, 40, 41, 42 and 34) (Fig. 2a, b). The loop and stem structures comprised 1943 and 1745 of total steps, respectively.
Patterns of substitutions were separately analyzed in autotrophic (Fig. 2a) and holoparasitic/mycoheterotrophic lineages (Fig. 2b), showing a similar distribution pattern: the positions of conserved and variable nucleotides are almost identical in the autotrophic and achlorophyllous histograms. However, more and highly variable nucleotides are presented on the latter histogram. Figure 2c confirms this result by calculating the difference of steps between mycoheterotrophic/holoparasitic taxa and autotrophic species. The distribution pattern showed that most of the substitutions occurred within achlorophyllous taxa, revealing that mainly heterotrophic taxa contribute to the observed mutations.
Discussion
Rate heterogeneity
Both Bayesian relative rate test and the distance based relative rate test showed similar results (Tables 1, 2), providing evidence for their accuracy and robustness.
The statistical tests showed remarkably long terminal branches for the heterotrophic taxa Rafflesia keithii, Rhizanthes infanticida, Arachnitis uniflora, Afrothismia hydra, A. winkleri and Thismia aseroe, compared to other angiosperms. Long terminal branches were not observed in chlorophyllous relatives of these taxa. These significantly increased substitution rates suggest that a non-photosynthetic mode of life could be a necessary condition for the occurrence of extremely increased substitution rates in 18S rDNA. However, our results show that the loss of photosynthesis in 29 mycoheterotrophic and nine holoparasitic representatives does not necessarily imply significantly increased substitution rates in 18S rDNA. Several non-photosynthetic lineages in Apodanthaceae, Burmanniaceae, Cynomoriaceae, Ericaceae, Gentianaceae, Iridaceae, Lennoaceae, Orobanchaceae, Orchidaceae, Petrosaviaceae and Triuridaceae lack significantly accelerated substitution rates, according to the Bayesian relative rates test. On the other hand all sampled mycoheterotrophic and holoparasitic Balanaphoraceae, Corsiaceae, Cytinaceae, Euphorbiaceae, Hydnoraceae, Mitrastemonaceae and Thismiaceae are characterized by elevated substitution rates (Fig. 1; Tables 1, 2). Previous studies investigating the substitution rates in mycoheterotrophic and parasitic plants confirm the observation that there is no evolutionary trend between the presence or absence of chlorophyll and elevated substitution rates (mycoheterotrophs: Cameron and Chase 2000; Molvray et al. 2000; Merckx et al. 2006, 2009 and parasites: Nickrent and Starr 1994; Nickrent et al. 1998).
Currently it is still unclear why some heterotrophic plants accumulate much more substitutions in their ribosomal DNA than autotrophic plants, but numerous potential hypotheses explaining causes of accelerated substitution rates have been proposed. The long-term effects of a small effective population size resulting in a genetic bottleneck effect (Wu and Li 1985), the influence of a short generation time and the correlated higher number of mutation-generating reproductive events (Wu and Li 1985), an increased tolerance of mutations due to a relaxation of selective constraints, variations in mutation rate (Sniegowski et al. 2000), DNA repair efficiency (Modrich and Lahue 1996) and speciation rates (Barraclough and Savolainen 2001) are possible factors that trigger high substitution rates in mycoheterotrophic and parasitic plants. None of these hypotheses could unequivocally explain higher substitution rates in parasitic plants (Nickrent and Starr 1994; dePamphilis et al. 1997; Young and dePamphilis 2005). However, in mycoheterotrophic plants, the effective population hypothesis may have played an important role in many cases of elevated substitution rates. The genera Kupea (Triuridaceae), Afrothismia (Thismiaceae) and Arachnitis (Corsiaceae) have very limited distribution ranges: Kupea martinetugei has only been reported from two localities in Cameroon (Cheek et al. 2003), Afrothismia is restricted to extremely scattered populations in the tropical rain forests in Africa (Maas-van de Kamer 1998; Franke et al. 2004) and Arachnitis is restricted in its distribution to disjunct areas in the southern part of South America (Ibisch et al. 1996). A very limited geographic distribution or reduced effective population size may indeed explain the significantly increased substitution rates of 18S rDNA in these mycoheterotrophic taxa. In parasitic lineages, however, Nickrent and Starr (1994) postulated that the effective population hypothesis could not be the only cause of higher substitution rates. A very restricted distribution pattern has been observed in the holoparasites Rafflesia (Euphorbiaceae), Rhizanthes (Euphorbiaceae) and Prosopanche (Hydnoraceae), which have accumulated significantly more substitutions compared to their autotrophic relatives. In contrast, Arceuthobium (Viscaceae) species have large and extremely widespread populations and show also significantly increased evolutionary rates. However, it remains to be studied whether populations of non-photosynthetic plants with wide distribution ranges are still mating (see Taylor et al. 2004).
An alternative hypothesis to explain the absence or presence of accelerated substitution rates in mycoheterotrophic plants is the absolute time when photosynthesis capacity was lost. Non-photosynthetic plants without significant increased substitution rates might have lost their chlorophyll only recently. However dating analysis on Burmanniaceae, which show no significant increase in substitution rates, suggests a Late Cretaceaus/Eocene origin of mycoheterotrophy, rejecting this hypothesis in that case (Merckx et al. 2008). Probably no single mentioned hypothesis will be able to explain unequivocally the substitution patterns observed in mycoheterotrophic and parasitic plants, but most likely a combination of several mechanisms is affecting divergent rDNA sequences.
Nucleotide variability and functionality of 18S rDNA
The number of substitutions per site in 18S rDNA differs greatly among sites. Alignment of 18S rDNA sequences reveals the presence of conserved areas with few variable substitutions interspersed (Nickrent and Soltis 1995; Van de Peer et al. 1993, 1996). We observed similar results in our dataset, showing a mosaic pattern with long stretches of conserved nucleotides and short variable regions in 18S rDNA (Fig. 2). Both stem and loop structures contain variable and conservative nucleotides. A higher proportion of highly variable nucleotides (≥10 steps) and a higher number of steps occurred in loops compared to stem structures, suggesting differences in selective constrains between both structures. This result confirms the pattern found in eukaryotes, where loops evolve considerably faster than stems (Smit et al. 2007).
More important is the similar distribution pattern between heterotrophic and related autotrophic plants (Fig. 2a, b). Despite a high number of variable nucleotides in mycoheterotrophic and holoparasitic plants (Fig. 2c), mutations seem to be strictly positioned to specific nucleotide sites, suggesting fixed selective constraints in the 18S rRNA molecule. These variable nucleotide sites occur in regions that are known to be highly variable within eukaryotes (Wuyts et al. 2000, 2001). The variable regions include mainly peripherally located eukaryotic-specific structures attached to a conserved core structure. Consequently, most conserved nucleotide sites are near the ribosome centre with increased nucleotide variability towards the ribosome surface (Wuyts et al. 2001). Nucleotides at the periphery of the ribosome, which are prone to substitutions, were located in structure number 6, 10, E10_1, 11, 17, 18, E23, 29, 43, 45 and 46. These variable nucleotides are under less functional constraints compared to centrally located sites with a catalytic or binding activity, which have less freedom to mutate. In Fig. 2, functional structures involved in peptidyl transferase activity, the translational cycle and interaction between rRNA subunits delimited by Caetano-Anollés (2002) are presented. As expected, substitutions are rare in those functional sites even within the faster evolving heterotrophic taxa. Based on these observation we can conclude that no major interference with the functions of the ribosome occurs and that the small ribosomal subunit retains its functionality, despite extremely high substitution rates in achlorophyllous taxa detected.
Acknowledgments
The authors thank Martin Bidartondo, Jonathan Kissling, Sainge Moses and Paul Maas for sending plant material and DNA extractions. This work was supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT Vlaanderen, no. 71488). VM is supported by the Fund for Scientific Research Flanders (FWO Vlaanderen). General financial support was provided by the K.U.Leuven (OT/05/35).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix
List of sampled species, Genbank accession numbers and voucher information arranged into the major Angiosperm groups. Newly obtained 18S rDNA sequences for this study are indicated with an asterisk. Sequences obtained from Genbank do not list voucher information.
Taxon-Genbank accession; Voucher.
Outgroup
Amborella trichopoda Baill.-U42497
Basal Angiosperms
Austrobaileya scandens C.T. White-AF206858, Nuphar variegata Engelm. ex Clinton-AF206972
Magnoliids
Aristolochia macrophylla Lam.-AF206855, Canella winterana Gaertn.,-AF206879, Chloranthus multistachys P’ei-AF206885, Hydnora africana Thunb.-L25681, Lactoris fernandeziana Phil.-LFU42783, Laurus nobilis Cav.-AF197580, Magnolia tripetala L.-AF206956, Prosopanche americana (R.Br.) Kuntze-L24047
Monocots
Acanthochlamys bracteata P.C.Kao-AY952411, Acorus calamus L.-L24078, Afrothismia hydra Sainge & T.Franke-DQ786083, Afrothismia winkleri Sainge & T.Franke-EU420992, Alisma plantago - aquatica L.-AF197585, Aplectrum hyemale Torr.-U59937, Apostasia stylidioides Rchb.f.-AF135207, Arachnitis uniflora* Phil.-HQ448758; Cocucci 2122, Aristea glauca Klatt-AF206854, Areca triandra Roxb.-AY952409, Apteria aphylla (Nutt.) Barnh. ex Small-DQ786035, Bomarea hirtella Herb.-AF206871, Burmannia bicolor Mart.-DQ786072, Burmannia biflora L.-DQ786070, Burmannia longifolia Becc.-AF309398, Burmannia oblonga Ridl.-DQ786064, Burmannia sphagnoides Becc.-AF309400, Campylosiphon purpurascens Benth.-EU420996, Chamaelirium luteum Miq.-AF206884, Cleistes divaricata Ames-AF135205, Colchicum autumnale L.-U42072, Corallorhiza maculata Greene-U59940, Cyclanthus bipartitus Poit.-AF168837, Cymbidium goeringii (Rchb.f.) Rchb.f.-AJ271248, Cymbocarpa refracta Miers-DQ786038, Cypripedium calceolus L.-AF069208, Cyrtosia septentrionalis (Rchb.f.) Garay-AF135198, Dendrobium nobile Lindle.-AB027309, Dictyostega orobanchoides (Hook.) Miers-DQ786056, Dioscorea rockii Prain & Burkill-DQ786090, Diuris sulphurea R.Br.-AF135196, Eburophyton austinae A. Heller-U59949, Epistephium subrepens Hoehne-AF135200, Eriaxis rigida Rchb.f.-AF135201, Erythrorchis cassythoides (A. Cunn. ex Lindl.) Garay-AF135199, Geosiris sp. Bail-EU816707, Gladiolus buckerveldii (L. Bolus) Goldblatt L54062, Gymnosiphon aphyllus Blume-AF309402, Gymnosiphon divaricatum (Benth.) Benth. & Hook.-DQ786043, Haplothismia exannulata Airy Shaw-DQ786082, Hexapterella gentianoides Urb.-DQ786057, Isotria verticillata Raf.-AF135204, Japonolirion osense Nakai-AF206942, Juncus effusus L.-AF206944, Kupea martinetugei Cheek & Williams-EU816706, Lecanorchis multiflora J.J.Sm.-AF135203, Luzuriaga latifolia Poir.-AF233091, Musa acuminata Colla-AF069226, Neottia nidus - avis (L.) Rich.-U59948, Oncidium sphacelatum Lindl.-U59939, Orchis quadripunctata Cirillo ex Ten.-AF135206, Pandanus tectorius Parkinson ex J.P.du Roi-AY952391, Petrosavia stellaris Becc.-AF206987, Rhizanthella gardneri R.S.Rogers-AF135197, Sciaphila densiflora Schltr.-EU816704, Sciaphila ledermannii* Engl.-HQ448766; Merckx 128, Smilax glauca Walter-AF207022, Spiranthes cernua Rich.-AF135195, Stemona japonica Franch. & Sav.-AF207028, Stenomeris dioscoreifolia Planch.-DQ786087, Tacca chantrieri André-DQ786086, Tacca palmata Blume-EU421000, Thismia aseroe Becc.-AF309404,Thismia rodwayi F.Muell.-AF309403, Tradescantia ohiensis Raf.-AF069213, Tricyrtis latifolia Maxim.-AF207046, Vanilla aphylla Wight-AF135202, Wullschlaegelia calcarata Benh.-EU816708
Basal Eudicots
Ceratophyllum demersum L.-U42517, Gunnera manicata Linden-U43787, Platanus occidentalis L.-U42794, Ranunculus taisanensis Hayata-D29780
Core Eudicots
Arceuthobium verticilliflorum Engelm.-L24042, Crassula marnieriana Huber & Jacobsen-U42525, Cynomorium coccineum L.-AF039069, Dendrophthora domingensis Eichler-X16601, Erythropalum scandens Blume-DQ790111, Hamamelis virginiana L.-AF094551, Lepidoceras chilense (Molina) Kuijt-EF464459, Lepionurus sylvestris Blume-DQ790101, Misodendrum linearifolium DC.-L24397, Nuytsia floribunda R.Br.-DQ790103, Olax aphylla R.Br.-L24405, Ombrophytum subterraneum (Asplund) B.Hansen-L24406, Opilia amentacea Wall.-L24407, Pereskia aculeata Mill.-AF206986, Peridiscus lucidus Benth.-AY372815, Plumbago auriculata Lam.-U42795, Santalum album L.–L24416, Saxifraga integrifolia Hook.-U42810, Schoepfia arenaria Britton–X16606, Tupeia antarctica Cham. & Schltdl.-L24425
Rosids
Bauhinia variegata L.-AF525295, Begonia oxyloba Welw. ex. Hook.f.-AY968392, Bdallophyton americanum (R. Br.) Harms-AY739089, Carica papaya L.-U42514, Celastrus scandens L.-AY674581, Citrus aurantium L.-U38312, Crossosoma californicum Nutt.-U42529, Cucurbita pepo L.-AF206895, Cytinus hypocistis L.-AY739092, Davidsonia pruriens F.Muell.-AF206897, Epilobium angustifolium L.-AF206907, Euphorbia pulcherrima Willd. ex Klotzsch-L37582, Fagus grandifolia Ehrh.-AF206910, Guaiacum sanctum L.-U42824, Krameria ixine L.-AF206948, Muntingia calabura L.-U42539, Ostodes paniculata Blume-AB268104, Pelargonium cotyledonis L’Hér-AF206982, Pilostyles thurberi A.Gray-AY739081, Rafflesia keithii Meijer-L24041, Rhizanthes infanticida H.Bänziger & B. Hansen-L24048, Ricinus communis L.-AB233559, Trema micrantha Blume-AF207044
Asterids
Alangium chinense (Lour.) Harms-AF206843, Anthocleista grandiflora Gilg-AJ236026, Apium graveolens L.-AF206852, Arctostaphylos uva - ursi Spreng.-L49272, Boschniakia rossica (Cham. & Schltdl.) Standl.-U59951, Bourreria succulenta Jacq.-U38319, Callicarpa dichotoma Raeusch.-AJ236048, Chelonanthus purpurascens* (Aubl.) Struwe, S.Nilsson & V.A.Albert-HQ448759; BINCO-FG43, Clethra alnifolia L.-AF419793, Convolvulus arvensis L.-AJ236013, Conopholis americana (L.) Wallr.-U59954, Cornus florida L.-X17370, Exacum paucisquamum* (C.B.Clarke) Klack-HQ448760; Yuan CN2k1-31, Cuscuta gronovii Wild. ex Roem. & Schult –L24747, Ehretia cymosa Thonn.-U59938, Epifagus virginiana (L.) W.P.C.Barton-U59955, Exacum affine Balf.f.-AJ236023, Garrya elliptica Dougl. ex Lindl.-U42540, Gentiana asclepiadea* L.-HQ448773; 19801931, Gentianella amarella* (L.) H. Sm.-HQ448761; Fay 14626, Harveya speciosa Bernh.-U59950, Hemitomes congestum* A.Gray-HQ448762; Bidartondo s.n., Hydrophyllum fendleri A. Heller-AJ236019, Ilex opaca Soland.-AF206938, Ipomoea hederacea Jacq.-U38310, Lamium amplexicaule L.-L49287, Lathraea clandestine L.-U59941, Mitrastemon yamamotoi Makino -AY739090, Monotropa uniflora L.-L25680, Orobanche fasciculata Nutt.-U59961, Orthocarpus erianthus Benth.-U38316, Orphium frutescens* E.Mey.-HQ448763; J. Kissling and L. Zeltner 44, Paulownia tomentosa Steud.-AJ236039, Pedicularis racemosa Dougl. ex Hook.-U59959, Phacelia bicolor Torr. Ex S.Watson-L49292, Pholisma arenarium Nutt.-U59935, Pityopus californicus* (Eastw.) Copel.-HQ448764; Bidartondo s.n., Pterospora andromedea Nutt.-U59943, Pycnosphaera buchananii* N.E.Br.-HQ448765; Bingham MG9370, Pyrola picta Sm.-U59936, Sambucus ebulus L.-AJ236005, Sarcodes sanguinea Torr.-U59945, Scaevola aemula R.Br.-AJ236008, Sebaea grandis* Steud.-HQ448767; Dessein et al. 752, Sebaea oligantha* Schinz-HQ448768; Merckx 103, Vaccinium macrocarpon Ait.-AF419808, Voyria aurantiaca* Splitq.-HQ448769; Maas et al. 9610, Voyria caerulea* Aubl.-HQ448770; Maas et al. 9636, Voyria corymbosa* Splitq.-HQ448771; Maas et al. 9611, Voyriella parviflora* Miq.-HQ448772; Maas et al. 9678.
References
- APG An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APGII. Bot J Linn Soc. 2003;141:399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x. [DOI] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic orgasisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Barkman TJ, McNeal JR, Lim S-H, Coat G, Croom HB, Young ND, dePamphilis CW. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Savolainen V. Evolutionary rates and species diversity in flowering plants. Evolution. 2001;55:677–683. doi: 10.1554/0014-3820(2001)055[0677:ERASDI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Freudenstein JV. Molecular evolution of rbcL in the mycoheterotrophic coralroot orchids (Corallorhiza Gagnebin, Orchidaceae) Mol Phylogenet Evol. 2008;47:665–679. doi: 10.1016/j.ympev.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Bidartondo MI, Redecker D, Hijri I, Wiemken A, Bruns TD, Domínguez L, Sérsic A, Leake JR, Read DJ. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature. 2002;419:389–392. doi: 10.1038/nature01054. [DOI] [PubMed] [Google Scholar]
- Bungard RA. Photosythetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays. 2004;26:235–247. doi: 10.1002/bies.10405. [DOI] [PubMed] [Google Scholar]
- Caddick LR, Rudall PJ, Wilkin P, Hedderson TAJ, Chase MW. Phylogenetics of Dioscoreales based on combined analyses of morphological and molecular data. Bot J Linn Soc. 2002;138:123–144. doi: 10.1046/j.1095-8339.2002.138002123.x. [DOI] [Google Scholar]
- Caetano-Anollés G. Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res. 2002;30:2575–2587. doi: 10.1093/nar/30.11.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron KM, Chase MW. Nuclear 18S rDNA sequences of Orchidaceae confirm the subfamilial status and circumcription of Vanilloideae. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 457–471. [Google Scholar]
- Cameron KM, Chase MW, Rudall PJ. Recircumscription of the monocotyledonous family Petrosaviaceae to include Japonolirion. Brittonia. 2003;55:214–225. doi: 10.1663/0007-196X(2003)055[0214:ROTMFP]2.0.CO;2. [DOI] [Google Scholar]
- Chase MW. Monocot relationships: an overview. Am J Bot. 2004;91:1645–1655. doi: 10.3732/ajb.91.10.1645. [DOI] [PubMed] [Google Scholar]
- Cheek M, Williams SA, Etuge M. Kupea martinetugei, a new genus and species of Triuridaceae from western Cameroon. Kew Bull. 2003;58:225–228. doi: 10.2307/4119366. [DOI] [Google Scholar]
- Cullings K. Reassessment of phylogenetic relationships of some members of the Monotropoideae based on partial 28S ribosomal RNA gene sequencing. Can J Bot. 2000;78:1–2. [Google Scholar]
- Davis JI, Stevenson DW, Petersen G, Seberg O, Cambell LM, Freudenstein VF, Goldman DH, Hardy CR. A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values. Syst Bot. 2004;29:467–510. doi: 10.1600/0363644041744365. [DOI] [Google Scholar]
- Davis CC, Latvis M, Nickrent DL, Wurdack KJ, Baum DA. Floral gigantism in Rafflesiaceae. Science. 2007;315:1812. doi: 10.1126/science.1135260. [DOI] [PubMed] [Google Scholar]
- Delavaut P, Sakanyan V, Thalouarn P. Divergent evolution of two plastid genes, rbcL and atpB, in non-photosynthetic parasitic plant. Plant Mol Biol. 1995;29:1071–1079. doi: 10.1007/BF00014978. [DOI] [PubMed] [Google Scholar]
- dePamphilis CW, Palmer JD. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990;348:337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- dePamphilis CW, Young ND, Wolfe AD. Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rates variation. Proc Natl Acad Sci USA. 1997;94:7367–7372. doi: 10.1073/pnas.94.14.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der JP, Nickrent DL. A molecular phylogeny of Santalaceae (Santalales) Sys Bot. 2008;33:107–116. doi: 10.1600/036364408783887438. [DOI] [Google Scholar]
- Fay MF, Chase MW, RØnsted N, Devey DS, Pillon Y, Pires JC, Petersen G, Seberg O, Davis J. Phylogenetics of Liliales: summarized evidence from combined analyses of five plastid and one mitochondrial loci. Aliso. 2006;22:559–565. [Google Scholar]
- Franke T, Sainge MN, Agerer R. A new species of Afrothismia (Burmanniaceae; tribe: Thismieae) from the Western foothills of Mount Cameroon. Blumea. 2004;49:451–456. [Google Scholar]
- Freudenstein JV, van den Berg C, Goldman DH, Kores PJ, Molvray M, Chase MW. An expanded plastid DNA phylogeny of Orchidaceae and analysis of jacknife branch support strategy. Am J Bot. 2004;9:149–157. doi: 10.3732/ajb.91.1.149. [DOI] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of the two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide-JØrgensen HS. Parasitic flowering plants. Leiden: Brill; 2008. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ibisch PL, Neinhuis C, Rojas NP. On the biology, biogeography, and taxonomy of Arachnitis Phil. Nom. Cons. (Corsiaceae) in respect to a new record from Bolivia. Willdenowia. 1996;26:321–332. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kuijt J. The biology of parasitic flowering plants. Berkeley: University of California Press; 1969. [Google Scholar]
- Leake JR. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994;127:171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x. [DOI] [PubMed] [Google Scholar]
- Leake JR. Myco-heterotrophic/epiparasitic plant interactions with ectomycorrhizal and arbuscular mycorrhizal fungi. Curr Opin Plant Biol. 2004;7:422–428. doi: 10.1016/j.pbi.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Leake JR. Plants parasitic on fungi: unearthing the fungi in myco-heterotrophs and debunking the ‘saprophytic’ plant myth. Mycologist. 2005;19:113–122. [Google Scholar]
- Maas-van de Kamer H (1998) Burmanniaceae. In: Kubitzki K (ed) Families and genera of vascular plants, Monocotyledons, Lilianae (exept Orchidaceae). Springer, Berlin, pp 154–164
- Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Version 4.01. Sunderland: Sinauer Associates; 2001. [Google Scholar]
- Malécot V, Nickrent D. Molecular phylogenetic relationships of Olacaceae and related Santalales. Sys Bot. 2008;33:97–106. doi: 10.1600/036364408783887384. [DOI] [Google Scholar]
- McNeal JR, Arumugunathan K, Kuehl JV, Boore JL, dePamphilis CW. Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae) BMC Biol. 2007;5:55. doi: 10.1186/1741-7007-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Arumugunathan K, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PloS One. 2009;4:e5982. doi: 10.1371/journal.pone.0005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx V, Schols P, Maas-van de Kamer H, Maas H, Huysmans S, Smets E. Phylogeny and evolution of Burmanniaceae (Dioscoreales) based on nuclear and mitochondrial data. Am J Bot. 2006;93:1684–1698. doi: 10.3732/ajb.93.11.1684. [DOI] [PubMed] [Google Scholar]
- Merckx V, Chatrou LW, Lemaire B, Sainge MN, Huysmans S, Smets E. Diversification of myco-heterotrophic angiosperms: Evidence from Burmanniaceae. BMC Evol Biol. 2008;8:178. doi: 10.1186/1471-2148-8-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx V, Bakker FT, Huysmans S, Smets E. Bias and conflict in phylogenetic inference of myco-heterotrophic plants: a case study in Thismiaceae. Cladistics. 2009;25:64–77. doi: 10.1111/j.1096-0031.2008.00241.x. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Molvray M, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 441–448. [Google Scholar]
- Nickrent DL. Cytinaceae are siter to Muntingiaceae (Malvales) Taxon. 2007;56:1129–1135. doi: 10.2307/25065907. [DOI] [Google Scholar]
- Nickrent DL, Soltis DE. A comparison of angiosperm phylogenies from nuclear 18S rDNA and rbcL sequences. Ann Miss Bot Gard. 1995;82:208–234. doi: 10.2307/2399878. [DOI] [Google Scholar]
- Nickrent DL, Starr EM. High rates of nucleotide substitutions in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. J Mol Evol. 1994;39:62–70. doi: 10.1007/BF00178250. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Colwell AE, Wolfe AD, Young ND, Steiner KE, dePamphilis CW. Molecular phylogenetic and evolutionary studies of parasitic plants. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II DNA sequencing. New York: Kluwer Academic; 1998. pp. 211–241. [Google Scholar]
- Nickrent DL, Blarer A, Qiu Y-L, Soltis DE, Soltis PS, Zanis M. Molecular data place Hydnoraceae with Aristolochiaceae. Am J Bot. 2002;89:1809–1817. doi: 10.3732/ajb.89.11.1809. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Blarer A, Qiu Y-L, Vidal-Russell R, Anderson F. Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol. 2004;4:40. doi: 10.1186/1471-2148-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Der JP, Anderson FE. Discovery of the photosynthetic relatives of the “Maltese mushroom” Cynomorium. BMC Evol Biol. 2005;5:38. doi: 10.1186/1471-2148-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura-Tsujita Y, Gebauer G, Hashimoto T, Umata H, Yukawa T. Evidence for novel and specialized mycorrhizal parasitism: the orchid Gastrodia confusa gains carbon from saprotrophic Mycena. Proc R Soc B. 2009;276:761–767. doi: 10.1098/rspb.2008.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead RG, Ferguson D (2001) A molecular phylogeny of the Boraginaceae/Hydrophyllaceae. BSA/ASPT/IOPB Meetings Abracts: 131
- Olmstead RG, dePamphilis CW, Wolfe AD, Young ND, Elisons WJ, Reeves PA. Desintegration of the Scrophulariaceae. Am J Bot. 2001;88:348–361. doi: 10.2307/2657024. [DOI] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Davis JI, Goldman DH, Stevenson DW, Campbell LM, Pires JC, Freudenstein JV, Hardy CR, Simmons MP. Mitochondrial data in monocot phylogenetics. In: Columbus JT, Friar EA, Porter JM, Prince LM, Simpson MG, editors. Monocots: comparative biology and evolution (excluding Poales) Claremont: Rancho Santa Ana Botanic Garden; 2006. pp. 52–62. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ (2007) Tracer v1. http://evolve.zoo.ox.ac.uk/software.html
- Reeves G, Chase MW, Goldblatt P, Rudall P, Fay MF, Cox AV, Lejeune B, Souza-Chies T. Molecular systematics of Iridaceae: evidence from four plastid DNA regions. Am J Bot. 2001;88:2074–2087. doi: 10.2307/3558433. [DOI] [PubMed] [Google Scholar]
- Revill MJW, Stanley S, Hibberd JM. Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta. J Exp Bot. 2005;56:2477–2486. doi: 10.1093/jxb/eri240. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Huchon D. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Roy M. Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci. 2009;14:64–70. doi: 10.1016/j.tplants.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Martos F, Perry B, Maj P, Roy M, Pailler T. Saprotrophic fungal symbionts in tropical achlorophyllous orchids: finding treasures among the ‘molecular scraps’? Plant Signal Behav. 2010;5:349–353. doi: 10.4161/psb.5.4.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson BB, Weeks A, Helfgott DM, Larkin LL. Species relationships in Krameria (Krameriaceae) based on ITS sequences and morphology: implications for character utility and biogeography. Sys Bot. 2004;29:97–108. doi: 10.1600/036364404772974013. [DOI] [Google Scholar]
- Smit S, Widmann J, Knight R. Evolutionary rates vary among rRNA structural elements. Nucleic Acids Res. 2007;35:3339–4708. doi: 10.1093/nar/gkm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal K, Bonfield J. The staden package. In: Miseners S, Krawetz S, editors. Computer Methods in Molecular Biology. New York: The Humana Press Inc.; 1998. pp. 115–130. [Google Scholar]
- Stevens PF (2001 onwards) Angiosperm Phylogeny Website. Version 9, June 2008 (and more or less continuously updated since then). http://www.mobot.org/MOBOT/research/Apweb/
- Taylor DL, Bruns TD, Hodges SA. Evidence for mycorrhizal races in a cheating orchid. Proc Biol Sci. 2004;271:35–43. doi: 10.1098/rspb.2003.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalouarn P, Theodet C, Russo N, Delevaut P. The reduced plastid genome of a non-photosynthetic angiosperm Orobanche hederae has retained the rbcL gene. Plant Physiol Biochem. 1994;32:233–242. [Google Scholar]
- Thompson JD, Gibson TJ, Pewniak F, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: flexible strategies for multiple sequence alignement aided by quality analyses tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Neefs J-M, De Rijk P, De Wachter P. Reconstructing evolution from eukaryotic small ribosomal subunit RNA sequences: calibration of the molecular clock. J Mol Evol. 1993;37:221–232. doi: 10.1007/BF02407359. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Van der Auwera G, De Wachter R (1996) The evolution of stramenopiles and alveolates as derived by ‘substitution rate calibration’ of small ribosomal subunit RNA J Mol Evol 42:201-210 [DOI] [PubMed]
- Van de Peer Y, De Rijk P, Wuyts J, Winkelmans T, De Wachter R. The European small subunit ribosomal RNA database. Nucleic Acids Res. 2000;28:175–176. doi: 10.1093/nar/28.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego, California: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Wilcox TP, Garcia de Leon FJ, Hendrickson DA, Hillis DM. Convergence among cave catfishes: long-branch attraction and a Bayesian relative rates test. Mol Phylogenet Evol. 2004;31:1101–1113. doi: 10.1016/j.ympev.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Wimpee CF, Wrobel RI, Garvin DK. A divergent plastid genome in Conopholis americana, an achlorophyllous parasitic plant. Plant Mol Biol. 1991;17:161–166. doi: 10.1007/BF00036822. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Mordon CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I, Li W-H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Van de Peer Y, De Wachter R. Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic Acids Res. 2001;29:5017–5028. doi: 10.1093/nar/29.24.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Perriere G, Van de Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:101–103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, dePamphilis CW. Rate variation in parasitic plants: correlated and uncorrelated patterns among plastid genes of different function. BMC Evol Biol. 2005;5:16. doi: 10.1186/1471-2148-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y-Y, Wohlhauser S, Möller M, Chassot P, Mansion G, Grant J, Küpfer P, Klackenberg J. Monophyly and relationships of the tribe Exaceae (Gentianaceae) inferred from nuclear ribosomal and chloroplast DNA sequences. MPE. 2003;28:500–517. doi: 10.1016/S1055-7903(03)00068-X. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin: The University of Texas; 2006. [Google Scholar]


