Abstract
Glioblastoma multiforme (GBM) is a malignant brain tumor characterized by diffuse infiltration of single cells into the brain parenchyma, which is a process that relies in part on aberrant biochemical and biophysical interactions between tumor cells and the brain extracellular matrix (ECM). A major obstacle to understanding ECM regulation of GBM invasion is the absence of model matrix systems that recapitulate the distinct composition and physical structure of brain ECM while allowing independent control of adhesive ligand density, mechanics, and microstructure. To address this need, we synthesized brain-mimetic ECMs based on hyaluronic acid (HA) with a range of stiffnesses that encompasses normal and tumorigenic brain tissue and functionalized these materials with short Arg-Gly-Asp (RGD) peptides to facilitate cell adhesion. Scanning electron micrographs of the hydrogels revealed a dense, sheet-like microstructure with apparent nanoscale porosity similar to brain extracellular space. On flat hydrogel substrates, glioma cell spreading area and actin stress fiber assembly increased strongly with increasing density of RGD peptide. Increasing HA stiffness under constant RGD density produced similar trends and increased the speed of random motility. In a three-dimensional (3D) spheroid paradigm, glioma cells invaded HA hydrogels with morphological patterns distinct from those observed on flat surfaces or in 3D collagen-based ECMs but highly reminiscent of those seen in brain slices. This material system represents a brain-mimetic model ECM with tunable ligand density and stiffness amenable to investigations of the mechanobiological regulation of brain tumor progression.
Keywords: Brain, cell adhesion, ECM (extracellular matrix), Hyaluronic acid/hyaluronan
INTRODUCTION
Glioblastoma multiforme (GBM) is the most common and lethal primary brain tumor [1, 2]. The relentless progression of this disease is due in part to the infiltration of cells from the primary tumor into distant regions of the brain, which renders complete surgical resection of the tumor mass virtually impossible and almost inevitably leaves the patient susceptible to recurrence, typically leading to death within 1-2 years of diagnosis. This aggressive invasion is a hallmark of malignant gliomas and is widely considered to be a key obstacle to the development of successful therapies [3]. For example, anti-angiogenic agents that are initially effective at reducing tumor bulk may induce an “adaptive-evasive” response characterized by increased cellular invasiveness that largely offsets these agents’ overall survival benefit [4, 5].
It is well established that the extracellular microenvironment plays an essential role in defining the ‘context’ that allows tumors to grow and spread [6]. There are several indications that the unusual aggressiveness of GBM may be mediated by its unique pattern of interactions with the extracellular matrix (ECM) of the brain [7]. Specifically, GBM tumors display a strikingly different pattern of invasion of brain tissue than systemic malignant tumors that metastasize to the brain. GBM tumors readily invade brain tissue including white matter, which is highly resistant to invasion by non-neural metastatic tumors [8]. Conversely, GBM tumors do not invade into blood vessel walls, unlike systemic metastatic tumors [9], and very rarely metastasize outside of the brain to other organs. This pattern may be due in part to the fact that brain ECM is highly specialized and distinct from that of other tissues in the body [10, 11]. For instance, brain ECM has a very low content of fibrous proteins such as collagen, fibronectin, and vitronectin, as well as basement membrane proteins such as laminin, which are largely restricted to the vascular and perivascular spaces in the brain.
One major component of brain ECM is hyaluronic acid (HA) [12, 13], a non-sulfated glycosaminoglycan that occupies a large fraction of the extracellular volume of the brain. In addition to HA, brain ECM contains other glycosaminoglycans such as heparan sulfate, a number of chondroitin sulfate proteoglycans of the lectican family such as aggrecan, neurocan, and versican, and proteins such as tenascin and thrombospondin. There are several lines of evidence that HA plays a significant role in glioma progression, including histological analyses of tumor tissue, which have shown that malignant gliomas contain greater levels of HA than normal brain tissue [14]. Treatment of cells with soluble HA, or inclusion of HA in the ECM by adsorbing it to tissue culture plastic or incorporating it into reconstituted ECM gels such as Matrigel or fibrin, has been shown to promote migration and invasion of glioma cells [15-18]. This effect is partly mediated by signals transduced by direct interactions between HA and its cell-surface receptors CD44 and RHAMM [15, 16, 19-21]. Correspondingly, CD44 is frequently overexpressed in gliomas and is correlated with increased malignancy and reduced mean survival time [22-24]. CD44 triggers a wide variety of downstream signaling programs upon engagement of HA [25], including pro-survival signaling through the PI3K-Akt [26] and EGFR [27] pathways, and increased migration via activation of Rho family GTPases [28].
In addition to the biochemical aspects of HA-induced signaling described above, there are indications that HA may play a biophysical role in GBM tumor invasion. The high viscosity and water-swelling ability of HA contribute to brain architecture and porosity and influence directional cell movements during morphogenesis [13, 29]. Addition of HA to fibrin-based ECMs has been shown to stimulate invasion of glioma cells via modulation of the fibrin fiber architecture [17]. These findings assume broader significance when viewed in the context of the growing consensus that cancer cells exert forces on and remodel their ECM and that this biophysical crosstalk can influence tumor progression [30, 31]. For example, we previously showed that the morphology, proliferation, and motility of GBM tumor cells cultured on two-dimensional (2D) polyacrylamide substrates conjugated with full-length collagen or fibronectin are regulated by the stiffness of the ECM [32, 33]. In a complementary study, Beadle et al. showed that glioma cells upregulate nonmuscle myosin II in order to generate sufficient contractile force to squeeze cell nuclei through tight spaces in the brain, suggesting that cells adapt their intrinsic contractile and adhesive machinery to overcome mechanical and microstructural barriers to migration [34]. Previous attempts to extend glioma cell-ECM mechanobiology to three-dimensional (3D) matrices have largely been restricted to reconstituted ECM preparations such as collagen and Matrigel [35-38]. While these studies have provided tremendous insight into mechanisms of tumor invasion, their interpretation is complicated by the fact that the biochemistry and architecture of these matrices differ in important respects from those of brain parenchyma. In addition, control of material properties in these systems is often achieved by changing the concentration of the gel-forming proteins, which simultaneously varies ECM mechanics, microstructure, and adhesivity, making it impossible to unambiguously attribute observed differences in cell behavior to chemical or mechanical stimuli [39].
For all of these reasons, there is a strong need for HA-based ECMs that offer independent control over biochemical functionality and mechanical rigidity and are amenable to studies of GBM tumor invasion. Although HA-based scaffolds have been used extensively for tissue engineering [40-48], only a few studies have used HA-based ECMs to study cell-matrix interactions in GBM that underlie invasion [49, 50]. Furthermore, these few GBM-specific studies have not addressed the potential regulatory role of ECM stiffness and architecture in tumor invasion and therefore leave open the question of whether the biophysical context in which HA is presented influences its effects on tumor progression. To meet this need, we synthesized and characterized a series of HA-based hydrogels with independently tunable stiffness and biochemical ligand density and used them to probe the dependence of glioma cell adhesion, morphology, cytoskeletal architecture, and motility on the ligand functionalization and stiffness in 2D culture. We also used these materials to investigate patterns of 3D tumor invasion across a panel of glioma cell lines.
MATERIALS AND METHODS
HA-methacrylate synthesis
HA-methacrylate was synthesized as originally described by Smeds et al. [51] according to the protocol detailed by Marklein et al. [52] (Figure 1). Briefly, HA of nominal molecular weight 60 kDa (66 kDa – 90 kDa range, Lifecore Technologies) was dissolved at 1 wt% in deionized water, and a six-fold molar excess of methacrylic anhydride (Sigma) (relative to the HA disaccharide repeat unit) was added dropwise to the solution on ice. The pH of the two-phase reaction mixture was adjusted to 8.0 with 5 M NaOH, and the reaction was allowed to continue overnight at 5 °C with frequent re-adjustment of the pH. The product was isolated by adding the reaction mixture to a five-fold volumetric excess of cold acetone, followed by centrifugation to recover the precipitate, which was then re-dissolved in deionized water, flash frozen in liquid nitrogen, and lyophilized. The powder thus obtained was analyzed by 1H-NMR, and the degree of functionalization was estimated to be 60% (see Supplementary Methods and Supplementary Figure S1). In order to synthesize HA-methacrylate with a higher degree of functionalization, the reaction was initially carried out overnight as above, after which an additional 3-fold molar excess of methacrylic anhydride was added and the reaction further continued for 1 day at 5 °C. This yielded HA-methacrylate with 85% degree of methacrylation as estimated by 1H-NMR.
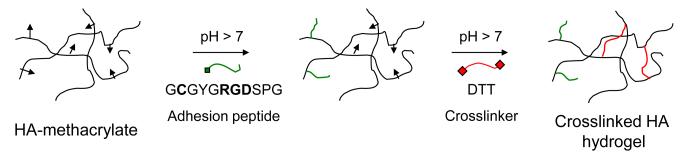
Figure 1.
Schematic description of HA functionalization and crosslinking. HA-methacrylate was functionalized with an RGD peptide using the Michael-type addition reaction between the methacrylate groups on the polymer and the cysteine thiol groups on the peptide. The same addition reaction with the methacrylate groups was used to induce crosslinking via reaction with dithiothreitol (DTT) to form HA hydrogels.
HA gelation and rheology
In order to form crosslinked HA gels, HA-methacrylate polymers with 60% and 85% degrees of functionalization (hereafter denoted as HA-60 and HA-85, respectively) and the crosslinker DL-dithiothreitol (DTT) were dissolved in Dulbecco’s Modified Eagle Medium (DMEM) and mixed to create gels with varying polymer weight fractions and thiol:HA repeat unit ratios. Oscillatory shear rheology was performed using an Anton Paar Physica MCR 301 rheometer in the parallel-plate configuration using a 25 mm diameter top plate. The gel was crosslinked in situ between the rheometer plates for 1 h at 37 °C in a humidified chamber. Typically, 60 μL of the gel solution was used with a gap width of 0.1 mm between the plates. Amplitude sweeps at constant frequency were performed to determine the linear viscoelastic range of deformation for each sample, after which frequency sweeps were performed at a strain amplitude within the linear range. Typically, frequencies from 0.05 Hz to 100 Hz were tested at a strain amplitude of 0.5%.
Scanning Electron Microscopy (SEM)
SEM sample preparation and imaging were performed as described previously [35]. Briefly, HA-DTT hydrogels were cast upon silicon wafers pre-treated with hydrophobic solution (OMS OptoChemicals) to promote gel adhesion. After swelling in phosphate-buffered saline (PBS) overnight, the hydrogels were fixed in 2% glutaraldehyde followed by 1% osmium tetroxide, both for 1 h at room temperature in 0.1 M sodium cacodylate buffer at pH 7.2. The samples were then dehydrated in ethanol, dried using the critical-point technique (AutoSamdri 815, Tousimis, Rockville, MD), and sputter-coated with approximately 2 nm of gold and palladium (Tousimis). SEM images were acquired using a Hitachi S-5000 scanning electron microscope.
Swelling studies
Hydrogels prepared as above were swollen in DMEM for 3 days at room temperature. The estimated equilibrium swelling ratio Q was defined as the ratio of the mass of the swollen hydrogel to that of the dry polymer obtained by lyophilization. The method of Canal and Peppas [53] was then used to estimate the mesh size ξ of the polymeric network (see Supplementary Methods).
Cell culture
U373-MG and U87-MG human glioblastoma and rat C6 glioma cells were cultured as previously described [32] in DMEM (Invitrogen) supplemented with 10% Calf Serum Advantage (JR Scientific, Inc.), 1% penicillin-streptomycin, 1% MEM non-essential amino acids and 1% sodium pyruvate (Invitrogen).
Preparation of 2D HA-RGD gels
For 2D studies, HA-RGD gels were prepared on glass-bottom 6-well plates (MatTek). The glass surface was briefly exposed to hydrophobic solution (OMS OptoChemicals) to promote adhesion of the HA gel. HA-60 and DTT were dissolved in DMEM, mixed in the appropriate ratios, and pipetted onto the glass surface. The gel precursor solution was then covered with a glass coverslip, which had been plasma-treated to increase its hydrophilicity. After incubation at 37°C for 2 h, the top coverslip was carefully removed to expose a flat HA hydrogel of approximate (nominal) thickness 200 μm. Hydrogels were then soaked in DMEM overnight at 37 °C to remove unreacted DTT.
In order to explore the effect of varying surface densities of RGD peptide (Ac-GCGYGRGDSPG-NH2, Anaspec), hydrogels of elastic modulus G′ ~ 35 kPa (8 wt% HA-60, 0.5 thiol:HA repeat unit ratio) were prepared as above. 25 μL of RGD peptide dissolved in DMEM at concentrations ranging from 0.1 mg/mL to 5 mg/mL were incubated on the hydrogel surface for 2 h at room temperature. Hydrogels were then rinsed and soaked in DMEM overnight at 37 °C to remove unreacted peptide.
To vary the mechanical properties of hydrogels, formulations with varying HA weight percent and crosslinking density were used, based on rheology data (Figure 2): G’ ~ 150 Pa (3 wt% HA-60, 0.1 thiol:HA repeat unit ratio), G’ ~ 1 kPa (3 wt% HA-60, 0.25 thiol:HA repeat unit ratio), and G’ ~ 5 kPa (5 wt% HA-60, 0.25 thiol:HA repeat unit ratio). HA-60 was functionalized with 1 mM RGD peptide by vortexing for 2 h at room temperature prior to DTT crosslinking and gelation in order to ensure that crosslinking did not compete with peptide conjugation. Glass surfaces coated with full-length human fibronectin (FN) as described previously [32] were used as controls.
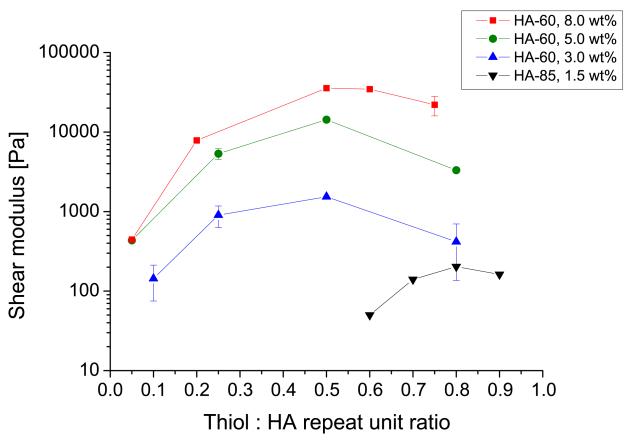
Figure 2.
Mechanical characterization of HA gels. Shear elastic moduli of the DTT-crosslinked HA hydrogels were measured by oscillatory rheometry. Each curve represents HA gels containing a particular HA-methacrylate polymer (HA-60 or HA-85 with 60% and 85% degree of methacrylation, respectively) and weight fraction, at varying ratios of thiols used for crosslinking. Error bars represent standard deviation; N ≥ 2 replicates.
Measurement of cell number
Cell density on hydrogel substrates was measured using the WST-1 assay (Roche), a colorimetric assay which infers cell number from mitochondrial metabolic activity. For these assays, 2,500 cells/cm2 were seeded with phenol red-free media and allowed to grow for 4 days prior to measurement. Samples were incubated following manufacturer’s instructions with 10% WST-1 reagent in the cell medium for 30 minutes at 37 °C. Absorbance at 440 nm was measured and converted to cell number using a concurrently obtained standard curve based on WST-1 measurements of defined numbers of cells. Data shown are the means of two independent experiments, each done in triplicate (N = 6).
Fluorescence microscopy and morphometric analysis
In order to evaluate cell adhesion and spreading on 2D HA-RGD hydrogels, U373-MG human GBM cells were seeded on hydrogel surfaces at an initial density of 5,000 cells/cm2 for 24 h. Cells were then fixed with 4% paraformaldehyde in PBS, permeabilized using 0.1% Triton-X100 in PBS, and blocked using 5% goat serum in PBS. To visualize vinculin, cells were treated with a mouse anti-vinculin primary antibody (Sigma) and AlexaFluor-546 goat anti-mouse secondary antibody (Invitrogen). Actin filaments were visualized with AlexaFluor-488 labeled phalloidin (Invitrogen). Cell nuclei were visualized using 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen). All fluorescence imaging was performed on a Nikon TE2000E2 epifluorescence microscope as described previously [32, 33]. Morphometric analysis of projected cell area and circularity (calculated as 4π × area/perimeter2, which varies from 1 for a circle to 0 for a straight line) was performed by thresholding the actin fluorescence images to define the cell boundaries and applying automated particle shape analysis (ImageJ).
Measurement of cell motility
Live-cell imaging was performed with a Nikon TE2000E2 microscope equipped with an incubator chamber for control of temperature, humidity, and CO2, based on our published protocols [32, 33, 35]. Briefly, U373-MG cells were seeded at 5,000 cells/cm2 and allowed to adhere to the hydrogel surfaces for at least 4 h prior to imaging. Phase contrast images were then obtained with a 10x objective every 30 minutes over an 18-hour period for at least 10 positions per sample and three samples per condition, in at least three independent experiments. The migration speed for each 30-minute time-step was determined by dividing the displacement of the nucleus (Manual Tracking plugin in ImageJ) by 30 minutes; these step-speeds were then averaged over at least 4 h to obtain the average motility speed for each cell. At least 120 cells per condition were tracked, excluding cells that came into contact with other cells or moved outside the boundaries of the field of view.
3D invasion of glioma tumor spheroids
In order to study 3D invasion of glioma cells in HA-RGD hydrogels, multicellular tumor spheroids were created using the hanging drop method as described previously [35, 54]. Briefly, glioma cells were suspended in growth medium, and 13 μL droplets of cell suspension containing ~500 cells were plated on the lid of a petri dish, which was then inverted over a dish containing growth medium for 4 days to yield multicellular spheroids. HA-RGD gels were prepared as described above with 2 mM RGD coupled to HA prior to gelation. To ensure that the tumor spheroid would be implanted entirely within the gel, a layer of hydrogel 5 mm in diameter and approximately 900 μm thick was first prepared on the surface and allowed to gel for 2 h. Another layer of HA-RGD hydrogel was then pipetted on the pre-formed gel surface and tumor spheroids of ~ 250 μm diameter were collected using a micropipette and immediately implanted in the hydrogel during gelation. After additional 2 h gelation, the hydrogels were incubated in full growth media for 7 days. To visualize 3D invasion, live-cell microscopy was performed as described above after 4 days in culture.
Data analysis and statistics
2D glioma cell morphology and migration speed data were obtained from two or more independent experiments, each done in duplicate. Significant differences among cell numbers inferred from WST-1 measurements were determined using one-way ANOVA followed by the Tukey-Kramer HSD (honestly significant difference) test for multiple comparisons. Significant differences among non-normally distributed data sets (cell spreading, circularity, and migration speed) were determined using the non-parametric Kruskal-Wallis test followed by Dunn’s test for multiple comparisons. In both cases, statistically significant differences among discrete groups are indicated on graphs by uppercase letters (A, B, C, etc.). Non-normally distributed data sets are shown as box-whisker plots, with boxes indicating 25th, 50th, and 75th percentiles, whiskers indicating 5th and 95th percentiles, and + marks indicating 1st and 99th percentiles.
RESULTS
HA-methacrylate synthesis and chemical characterization
To fabricate HA hydrogels (Figure 1), we first functionalized HA with methacrylate groups using a simple, one-step reaction with methacrylic anhydride. We chose this strategy because the methacrylate groups may be used for conjugation and crosslinking via a rapid Michael-type addition with thiol-containing molecules, a reaction which may be carried out in aqueous medium at physiological temperature and pH and, if needed, in the presence of proteins [55] or cells [56, 57]. We synthesized two variants of HA-methacrylate, HA-60 and HA-85, which respectively have 60% and 85% methacrylation of the HA backbone as determined by 1H-NMR (Supplementary Figure S1). To create hydrogels of defined mechanical stiffness, we crosslinked HA-methacrylate with DL-dithiothreitol (DTT) via Michael-type addition in defined stoichiometries (described below). To incorporate additional cell-adhesive functionality, we used Michael-type addition to graft cysteine-containing RGD peptides to the methacrylated HA backbone.
Mechanical characterization of HA hydrogels
To determine if we could synthesize HA hydrogels spanning a range of elastic properties relevant to brain ECM, we used parallel-plate rheology to measure shear elastic moduli of HA hydrogels prepared from HA-60 or HA-85 at various weight fractions and DTT crosslink ratios (Figure 2). As expected, elastic modulus increased with the HA weight fraction for a given degree of methacrylation, consistent with the notion that hydrogel elasticity increases with density [58]. For a given HA-methacrylate polymer and weight fraction, the elastic modulus (G’) also depended strongly on the ratio of thiols to HA repeat units, which is expected to govern the crosslink density. Optimal crosslinking (maximal elastic modulus) was obtained at a thiol:HA repeat unit ratio of 0.5 for HA-60 and 0.8 for HA-85, which in each case corresponds approximately to a 1:1 ratio of thiol groups to methacrylate groups. The reduction in elastic modulus at high crosslinker concentrations has been reported in other systems [56] and is consistent with a mechanism in which single-thiol ligation between DTT and the HA backbone is favored over dual-thiol ligation/crosslinking when thiols are in excess of methacrylate groups. To test this hypothesis, we directly measured the free thiol content of the hydrogels after gelation using Ellman’s test (Supplementary Figure S2A). Indeed, the residual thiol content increased significantly above a 1:1 ratio of thiols to methacrylates, indicating the presence of singly ligated DTT molecules. We confirmed that these free thiols are quenched to levels that would not be expected to interfere with cell physiology by repeating Ellman’s test after overnight incubation in culture medium at 37 °C (Supplementary Figure S2B). Thus, by strategically choosing HA weight fraction, degree of methacrylation, and degree of crosslinking, we can generate hydrogels with elastic moduli ranging from 50 Pa to 35 kPa, which brackets stiffness ranges previously observed in brain tissue [59-61].
Microstructural characterization of HA hydrogels
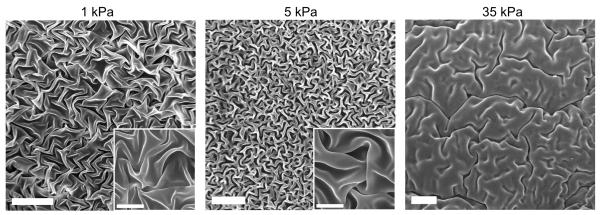
As described earlier, brain ECM lacks the highly fibrillar architecture associated with many connective tissues and commonly used reconstituted ECM-mimetic hydrogels such as collagen I and fibrin. To characterize the microstructure of our HA gels, we used scanning electron microscopy (SEM), which revealed dense, folded sheets of polymer free of fibrillar structures (Figure 3). Hydrogels with greater HA weight fractions and thus higher stiffnesses exhibited a denser architecture. A similarly smooth, sheet-like appearance was reported in previous studies of HA hydrogels [62-64]. These studies also observed that the sheet-like architecture was interrupted with micron-scale pores; the absence of pores in our images may be due to differences in dehydration protocols or other sample preparation details.
Figure 3.
Scanning Electron Microscopy (SEM) imaging of dehydrated HA gels (higher magnification images in insets). The gel microstructure consists of dense, folded sheets of polymer free of fibrillar structures and does not contain micron-sized pores. The apparent density of the polymer network increases with increasing polymer weight fraction and stiffness. Scale bar = 20 μm; inset scale bar = 5 μm.
Due to these sample preparation artifacts and spatial resolution limits, accurate quantification of mesh sizes from SEM images is challenging. We therefore exploited the equilibrium swelling behavior of our HA hydrogels to determine the theoretical mesh size of the polymer network (Table 1; see Supplementary Methods for details). This analysis revealed mesh sizes ranging from 50-150 nm, consistent with previously reported values for HA-based hydrogels used in tissue engineering applications [65]. Based on the SEM images and mesh size calculations, we conclude that the HA hydrogels are nanoporous, similar to hydrogels prepared from poly-ethylene glycol [66] or agarose [67]. The relationship between experimentally-determined equilibrium shear modulus G’ and swelling ratio followed an inverse power-law with an exponent of −3.6 (see Supplementary Figure S3), similar to an exponent of −4 previously reported for disulfide-crosslinked HA hydrogels [68].
Table 1.
Equilibrium swelling data and mesh size estimates for HA-DTT hydrogels.
| HA-methacrylate polymer |
Polymer wt% |
Thiol:HA repeat unit ratio |
Average shear modulus, G’ [Pa] |
Average swelling ratio, Q |
Average mesh size, ξ [nm] |
|---|---|---|---|---|---|
| HA-60 | 3.0 | 0.10 | 144 | 76 | 93 |
|
| |||||
| HA-60 | 3.0 | 0.25 | 903 | 78 | 96 |
|
| |||||
| HA-60 | 5.0 | 0.25 | 5343 | 36 | 49 |
|
| |||||
| HA-60 | 8.0 | 0.50 | 35616 | 49 | 81 |
|
| |||||
| HA-85 | 1.5 | 0.60 | 50 | 172 | 155 |
|
| |||||
| HA-85 | 1.5 | 0.70 | 140 | 164 | 150 |
|
| |||||
| HA-85 | 1.5 | 0.80 | 203 | 128 | 123 |
Glioma cell behavior as a function of RGD peptide density
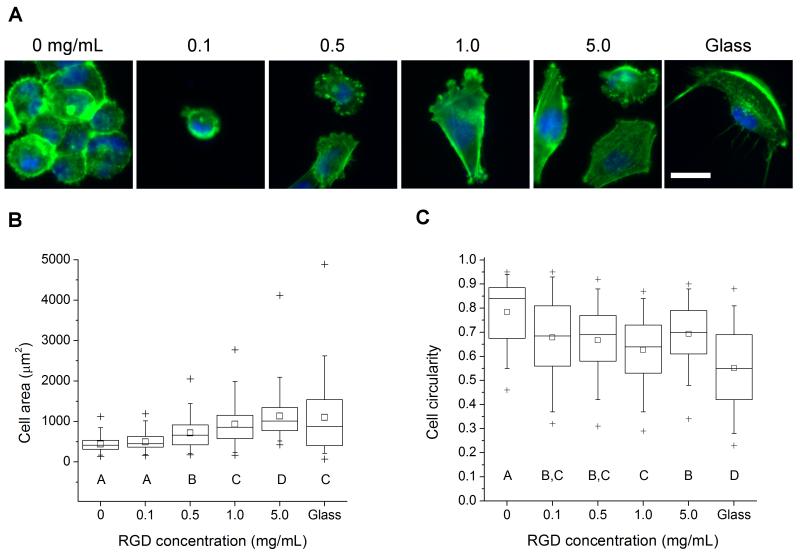
We next explored our ability to systematically vary ECM ligand density in HA gels, as well as the sensitivity of glioma cell adhesion and cytoarchitecture to these variations. We began by asking whether the density of RGD peptide coupled to the surface of hydrogels of fixed stiffness influences cell morphology and cytoskeletal organization (Figure 4A). We selected hydrogels of shear modulus 35 kPa, because we had shown previously that polyacrylamide matrices tuned to this stiffness range and conjugated with high densities of full-length ECM proteins are capable of supporting cell spreading [32]. The extent of cell adhesion increased modestly with increasing RGD density (Supplementary Figure S4). A graded increase in RGD density led to pronounced effects on cellular morphology, with a progressive increase in cell area (Figure 4B) and decrease in circularity (Figure 4C). Moreover, substrate ligand density strongly influenced cytoskeletal organization, with actin stress fibers only observable in cells spread on surfaces coated with RGD concentrations above 0.5 mg/mL. This relationship between ECM ligand density, cell spreading, and actin cytoskeletal architecture is broadly consistent with many previous studies in non-HA matrix systems [69-71]. Thus, we can exercise control over ECM ligand density comparable to existing synthetic hydrogel systems, and we observe the expected variations in cell morphology.
Figure 4.
Glioma cell morphology on 35 kPa HA gels with varying surface density of RGD peptide. (A) Morphology and cytoarchitecture of cells adhered to variable-RGD density gels after 24 hr incubation, as visualized by epifluorescence imaging of F-actin (green) and nuclear DNA (blue). Scale bar = 50 μm. (B) Quantification of projected cell spreading area. (C) Quantification of cell shape, as measured by circularity (see methods). N ≥ 55 cells for each condition. Statistically distinct groups (p < 0.05) determined by Dunn’s test are marked by A, B, C, D (see methods).
Glioma cell behavior as a function of HA hydrogel stiffness
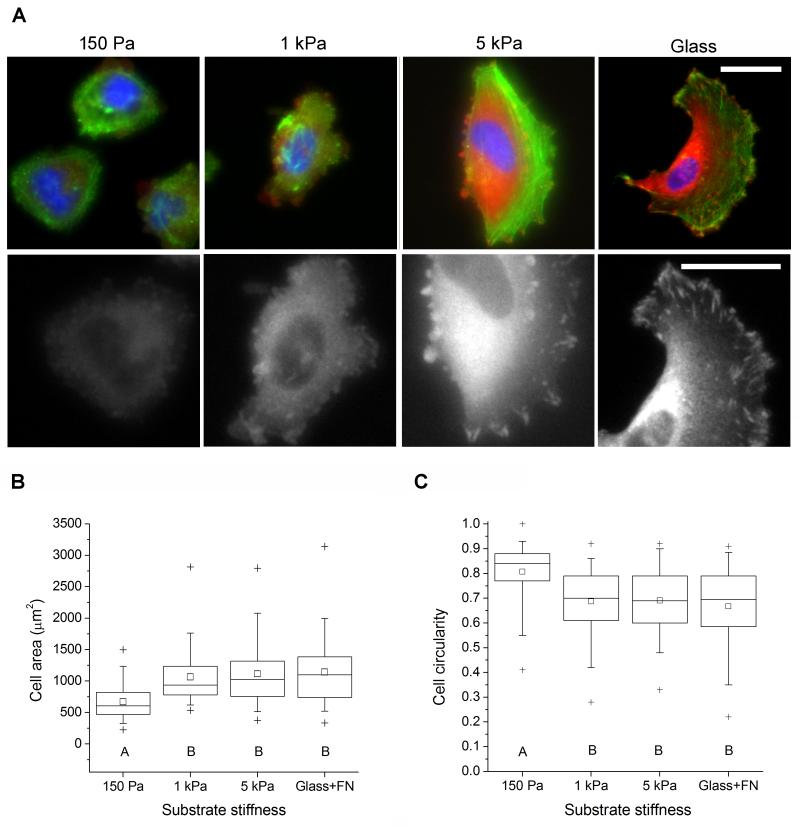
Having established that we can vary ECM ligand density on HA gels and can thereby control cell spreading and cytoarchitecture, we asked whether we could exercise similar control by modulating ECM mechanical rigidity. We functionalized HA-60 with 1 mM RGD prior to DTT crosslinking to prepare a series of hydrogels with constant RGD density but variable elastic modulus. The extent of initial cell attachment did not depend on hydrogel stiffness (Supplementary Figure S5), implying that RGD ligand densities were functionally similar across all substrates. Cell morphology depended strongly on ECM stiffness (Figure 5A), with cells on the softest hydrogel (G’ = 150 Pa) primarily exhibiting rounded morphologies and diffuse distributions of F-actin and the focal adhesion protein vinculin. In contrast, cells on 1 kPa and 5 kPa hydrogels spread and acquired a polarized shape, as evidenced by significant increases in average cell area (Figure 5B) and decreases in circularity (Figure 5C). Cells on 5 kPa hydrogels also exhibited hallmark characteristics of well-spread cells including broad lamellipodia, distinct actin-based stress fibers, and elongated vinculin-positive focal adhesions. These results are qualitatively similar to our previous observations of U373-MG human GBM cells on polyacrylamide ECMs covalently modified with either full-length fibronectin [32] or collagen I [33], providing additional validation for HA as a platform for studying ECM stiffness-mediated regulation of GBM and independent experimental support for ECM stiffness as a key governing parameter of GBM cell behavior.
Figure 5.
Glioma cell adhesion to variable-stiffness RGD-functionalized HA gels. (A) Morphology of cells adhered to variable-stiffness gels after 24 hr incubation. Top row: immunofluorescence imaging of vinculin (orange), F-actin (green), and nuclear DNA (blue). Bottom row: Isolated vinculin signal at higher magnification. Scale bar = 50 μm. (B) Quantification of projected cell area. (C) Quantification of circularity. N ≥ 85 cells for each condition. Statistically distinct groups (p < 0.01) determined by Dunn’s test are marked by A, B (see methods).
Glioma motility on variable-stiffness HA hydrogels
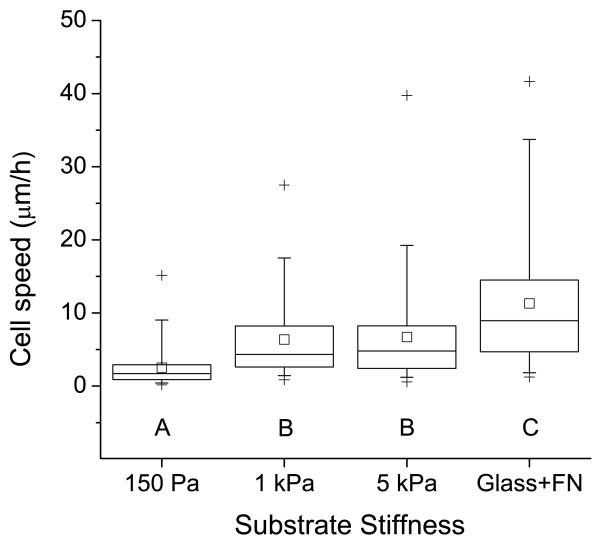
Since the architecture of cell-ECM adhesions and the actin cytoskeleton are strongly influenced by HA-RGD hydrogel stiffness (Figure 5), and since ECM stiffness has been observed to strongly influence cell motility in other material systems [72-74], we next asked whether modulation of HA stiffness would alter motility. We measured the speed of random cell migration on variable-stiffness HA hydrogels with equivalent RGD ligand functionality (Figure 6 and Supplementary Movies S1 to S4). Cell migration speed increased dramatically with increasing substrate stiffness, with maximum cell speeds on 5 kPa hydrogels nearly the same as those observed on fibronectin-coated glass. Cells on 150 Pa hydrogels were unable to form stable lamellipodia and migrate productively, whereas cells on 5 kPa hydrogels exhibited typical mesenchymal motility with broad lamellipodial protrusions followed by cell-body extension and rear-edge retraction. Intermediate-stiffness (1 kPa) hydrogels contained subsets of cells exhibiting both phenotypes. This relationship between ECM stiffness and cell migration speed is consistent with our studies of U373-MG cell migration on fibronectin-coated polyacrylamide ECMs [32] and, more generally, with our computational models of motility of highly polarized cells on defined-stiffness ECMs [75].
Figure 6.
Regulation of glioma cell motility by matrix stiffness. The plot depicts the average speed of random motility of U373-MG cells cultured on RGD-functionalized HA gels of constant peptide density and varying stiffness. N ≥ 120 cells for each condition. Statistically distinct groups (p < 0.01) determined by Dunn’s test are marked by A, B, C (see methods).
Glioma net cell growth on variable-stiffness HA hydrogels
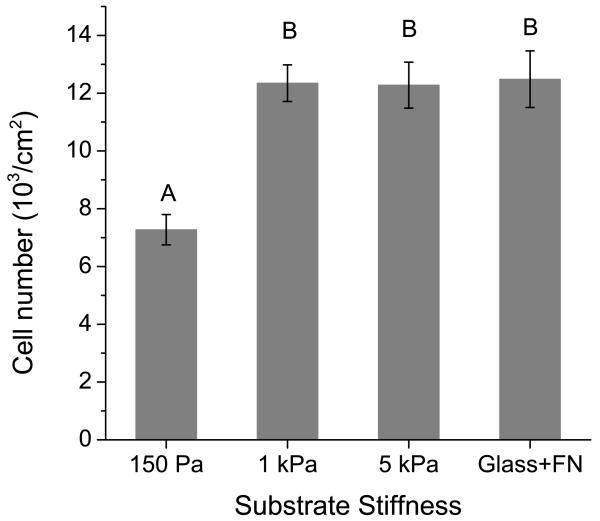
Cell spreading has been strongly correlated with proliferation in a variety of other cell-matrix systems [76], and our own previous studies have shown that increasing ECM stiffness stimulates the proliferation of human glioma cells [32]. To investigate the relationship between ECM stiffness and cell growth in the HA hydrogel ECM system, we cultured cells on HA hydrogels of constant RGD density and varying stiffness and then used a spectrophotometric assay of mitochondrial metabolic activity (WST-1) to estimate cell number (Figure 7). After 4 days of culture, significantly fewer cells were present on 150 Pa gels than on 1 kPa and 5 kPa hydrogels or fibronectin-coated glass, all three of which were statistically indistinguishable from one another. It is important to note that these data are not strictly interpretable in terms of differences in cell proliferation and could in principle be due to stiffness-dependent differences in cell detachment and apoptosis. However, we did not observe significant cell detachment on any substrate, and our live-cell motility imaging suggested that the vast majority of rounded cells on the 150 Pa gels were capable of extending processes, with some observed to divide. Taken together with our earlier observation that initial cell adhesion is independent of ECM stiffness (Supplementary Figure S5), these results are consistent with our previously reported stiffness-dependent increase in cell proliferation on polyacrylamide hydrogels [32] and provide further evidence that glioma cells proliferate in a stiffness-dependent manner.
Figure 7.
Cell number on variable-stiffness HA-RGD substrates. U373-MG cells were cultured on matrices of the specified rigidity and constant ligand density for 4 days. Cell number was then measured using the WST-1 metabolic assay. Error bars represent standard error of the mean; N = 6 replicates. Statistically distinct groups (p < 0.01) determined by Tukey’s test are marked by A, B (see methods).
Glioma invasion in 3D through HA-RGD hydrogels
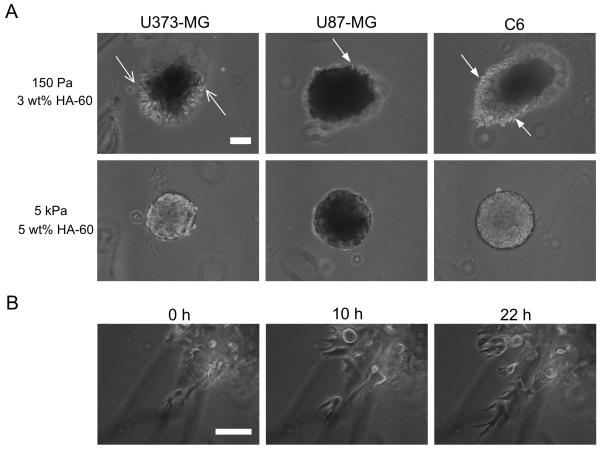
Brain ECM is a 3D structure, and so we next asked whether our HA-based hydrogels are capable of supporting the 3D invasion of tumor cells and whether that invasion was sensitive to hydrogel composition. We used the tumor spheroid invasion paradigm that has been used successfully to study glioma invasion in 3D biomimetic ECMs [35-37], and studied invasion across a panel of human and rat glioma cell lines. While all cell lines invaded the soft (150 Pa, 3 wt% HA-60) HA-RGD hydrogel after 3 days in culture (Figure 8A), there were clear differences in the extent and pattern of invasion observed across cell lines. U87-MG and U373-MG human GBM cells invaded to a lesser extent than rat C6 cells, and whereas U373-MG cells appeared to disperse from the spheroid and invade singly, U87-MG and C6 cells retained tight spheroid borders with cells invading actively at the edges (Figure 8A and Supplementary Movies S5 to S7). Interestingly, the mode of motility was distinct from our previous observations in 2D (Supplemental Movies S1-S4) and fibrous 3D collagen-based matrices [35, 77], in that invading cells exhibited non-mesenchymal motility devoid of lamellipodia at the leading edge (Figure 8B and Supplementary Movies S5 to S7). Instead, cells exhibited highly dynamic leading processes that underwent cycles of protrusion, retraction, and branching followed by sudden forward movement of the cell body. This mode of motility is remarkably similar to that observed for glioma cells and neural progenitors migrating in brain slice cultures [34, 78], which is postulated to be an adaptation necessary for migration through the sub-micron pores in the tightly packed extracellular spaces of the brain [79]. Strikingly, invasion was completely abrogated in the denser hydrogel (5 kPa, 5 wt% HA-60) (Figure 8A), although the spheroids continued to grow in volume indicating ongoing cell proliferation and mass expansion. The abolition of invasion in these hydrogels could be due to density-dependent alterations in HA-based adhesive signaling, hyaluronidase secretion, matrix secretion, or other factors. The high HA density may also pose an effectively insurmountable steric barrier to migration, qualitatively consistent with our previous observations in collagen-agarose composite hydrogels where increasing the density of agarose inhibited invasion of U373-MG tumor spheroids [35] due to increasing steric barriers and disruption of collagen remodeling. Thus, 3D invasion of glioma spheroids in HA-RGD hydrogels recapitulates key morphological features of the invasive phenotype of glioma in vivo.
Figure 8.
3D invasion of glioma spheroids through HA-RGD hydrogels. (A) Variation of extent and patterns of invasion with cell type and matrix density. U373-MG cells dispersed and invaded as single cells (open arrows) whereas U87-MG and C6 cells retained spheroid borders with cells invading at the edges (filled arrows). No cells invaded the dense 5 kPa hydrogel. (B) Time-lapse images of U373-MG cells invading the 150 Pa hydrogel. Cells exhibited distinctly non-mesenchymal motility, with dynamically extending and branching leading processes, following by abrupt movement of the cell-body forward. Scale bar = 100 μm.
DISCUSSION
In this study, we synthesized and characterized HA-based hydrogels with independently tunable stiffness and ligand functionality and used this brain-mimetic ECM platform to study the dependence of GBM tumor cell morphology, motility, and growth on the adhesivity and mechanical rigidity of the ECM. We also demonstrated that these materials support 3D tumor invasion with migratory modes similar to those observed in tissue. Our work addresses a significant need for well-defined model systems that capture the HA-rich milieu of brain ECM while permitting investigation of how brain tumor biology is regulated by ECM biochemical and biophysical properties. Further, our findings on the dependence of GBM tumor cell behavior on the mechanical features of HA hydrogels bear relevance to the pathophysiology of GBM progression in vivo.
In addition to providing a well-defined platform for studying glioma biology, our study also advances the field’s ability to manipulate HA for biomaterials applications, such as the engineering of soft tissues [46]. Although several investigators have previously reported on the development of HA-based hydrogels with tunable stiffness and ligand density [45, 52, 80, 81], key design relationships such as the effects of polymer weight fraction, degree of functionalization, and crosslink density on hydrogel structure and mechanics remain incompletely understood. Our data begin to fill this gap by outlining conditions for synthesizing hydrogels with moduli spanning three orders of magnitude (50 Pa to 35 kPa) that overlap significantly with the range of stiffnesses observed in soft tissues. By systematically varying the thiol:HA repeat ratio and measuring residual thiol concentrations (Supplementary Figure 2A), we also demonstrate that hydrogel elasticity can be optimized with respect to crosslinker (DTT) concentration; above this optimum, crosslinking moieties are more likely to react with single sites on the backbone rather than ligating two distal sites. Finally, our SEM imaging (Figure 3) demonstrates that these HA hydrogels are non-fibrillar and lack micron-scale pores. This not only mimics the native structure of brain ECM [10, 79] but also profoundly influences the mode of cell motility through these matrices, as we discuss in more detail below.
Our study may be placed in the context of previous in vitro, in vivo, and clinical studies that relate to the mechanobiological regulation of GBM. The utility of ultrasound elastography in guiding surgical tumor resection suggests that tumor tissue may be mechanically distinct from normal brain tissue [82]. Further, AFM measurements of the elasticity of rat brain tissue reveal significant mechanical heterogeneity within the hippocampus [59], and between white and gray matter in the cerebellum [60]. In this context, the mechanosensitive behavior of GBM tumor cells suggest that mechanical cues present in the tumor ECM may promote cell adhesion and migration, thereby facilitating the spread of the disease. We have reported previously on the mechanosensitive behavior of glioma cells on variable-stiffness 2D polyacrylamide substrates [32] and in 3D hydrogels composed of collagen and agarose [35]. Others have studied glioma cell migration as a function of the pore size and density of collagen matrices [38, 83]. Our findings that cell spreading, cytoskeletal assembly, migration speed, and 3D invasion are all strongly regulated by ECM structural and mechanical properties are consistent with these published reports and reinforce the notion that tumor invasion may be tightly regulated by the biophysical microenvironment.
Our study also adds to the field’s specific understanding of the role of HA in the regulation of GBM growth and spread. As described earlier, HA is enriched in tumor ECM [14], and adhesive signaling triggered by HA binding to its receptors (e.g., CD44, RHAMM) – which are also overexpressed in GBM [22, 23] – has been shown to contribute to tumor progression [25]. While several studies have reported that HA promotes glioma migration and invasion in vitro, these conclusions have largely been drawn from experiments in which HA has either been added as a soluble factor or incorporated as a matrix component in a background of collagen or Matrigel [15, 16]. By contrast, our results take advantage of a solid-state HA-based ECM platform that is comparatively free of confounding influences from other matrix elements and clearly show that in the absence of steric barriers GBM cell morphology and motility are regulated by HA stiffness (Figures 5 and 6), which suggests that the physical presentation of HA may strongly affect its adhesion-dependent biology. This in turn begs the question of whether this mechanosensitivity is actuated through integrins (via the RGD peptides) [84], CD44 and other HA receptors, or some combination of the two. This materials platform should permit elucidation of the relative roles of CD44-mediated and integrin-mediated signaling in glioma mechanobiology, which may have implications for other tissues in which HA receptor-based adhesion figures prominently, such as cartilage [85].
Although previous studies have reported the colonization of HA hydrogels by glioma cells [49, 50], our high-resolution time-lapse imaging of this process uniquely enables us to observe that glioma tumor spheroids competently invade soft HA-RGD hydrogels that are within the range of stiffness observed in brain tissue [59], but not significantly denser hydrogels (Figure 8A). Moreover, the motility exhibited by invading cells in 3D soft HA hydrogels is strikingly similar to that observed in migration of glioma cells in brain slice cultures [34]. This behavior is not observed in fibrillar ECMs composed of collagen I, where cells migrate in a largely mesenchymal mode through micron-sized pores following collagen fibers and tracks [35, 36]. Since the HA-RGD hydrogels have nanoscale pores (Figure 3 and Table 1) similar to the extracellular spaces in the brain [79], our data support the view that the distinct motility exhibited in these ECMs, where the cell body appears to be extruded through tight spaces, is specifically due to the constraints imposed by the dense extracellular environment.
Interestingly, we also observed differences in the extent of invasion across cell lines, with an order of invasiveness of C6 > U87-MG > U373-MG, which roughly corresponds to the observed proliferation rates of these cells in culture (data not shown). However, the observed differences in invasion may also be due to differences in HA-specific interactions across cell lines, such as varying expression of CD44 [86] or hyaluronidases [87]. Indeed, the ability of tumor cell lines to invade and colonize HA hydrogels has been correlated with the expression of hyaluronidase [88], which can decrease steric barriers to invasion by degrading the matrix [89]. The two human GBM cell lines used in this study (U373-MG and U87-MG) have been reported to express certain hyaluronidases at levels that are either very low [90] or close to zero [49]. Future studies should lend insight into the molecular mechanisms that underlie these cell-specific differences.
CONCLUSIONS
In summary, we have developed and characterized an HA-based ECM platform with independently tunable stiffness and ligand functionalization that mimics key biochemical and biophysical features of brain matrix, and demonstrated its utility in elucidating ECM mechanobiological regulation of glioma cell morphology, motility, and 3D invasion. We anticipate this material platform will be broadly useful for in vitro studies of cell-ECM interactions in the brain. Further, due to the excellent biocompatibility of HA [91], these materials may be used as an implantable scaffold to investigate matrix regulation of tumor invasion in vivo [92] as well as in regenerative medicine approaches to treat specific central nervous system pathologies [46].
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J. Burdick and R. Marklein for valuable technical guidance, and G. Min at the Electron Microscopy Laboratory at UC Berkeley for assistance with SEM imaging. Y. K. gratefully acknowledges the support of Graduate Fellowships from the NSF and U.S. Department of Defense (NDSEG). S.K. gratefully acknowledges grant support from the Arnold and Mabel Beckman Foundation, the NSF (CMMI 0727420), and the NIH (1DP2OD004213, Director’s New Innovator Award, part of the NIH Roadmap for Medical Research; 1U54CA143836, Physical Sciences Oncology Center Grant).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64(4):458–78. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Pàez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Viñals F, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15(3):220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkel P, Ulbricht U, Bohlen P, Brockmann MA, Fillbrandt R, Stavrou D, et al. Inhibition of glioma angiogenesis and growth in vivo by systemic treatment with a monoclonal antibody against vascular endothelial growth factor receptor-2. Cancer Res. 2001;61(18):6624–8. [PubMed] [Google Scholar]
- 6.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1(1):46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36(6):1046–69. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Paganetti PA, Caroni P, Schwab ME. Glioblastoma infiltration into central nervous system tissue in vitro - involvement of a metalloprotease. J Cell Biol. 1988;107(6):2281–91. doi: 10.1083/jcb.107.6.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein JJ, Woodard CA. Glioblastoma cells do not intravasate into blood-vessels. Neurosurgery. 1995;36(1):124–32. doi: 10.1227/00006123-199501000-00016. [DOI] [PubMed] [Google Scholar]
- 10.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6(5):489–92. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 11.Gladson CL. The extracellular matrix of gliomas: modulation of cell function. J Neuropathol Exp Neurol. 1999;58(10):1029–40. doi: 10.1097/00005072-199910000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Laurent TC, Fraser JRE. Hyaluronan. FASEB J. 1992;6(7):2397–404. [PubMed] [Google Scholar]
- 13.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4(7):528–39. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 14.Delpech B, Maingonnat C, Girard N, Chauzy C, Maunoury R, Olivier A, et al. Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumor stroma. Eur J Cancer. 1993;29A(7):1012–7. doi: 10.1016/s0959-8049(05)80214-x. [DOI] [PubMed] [Google Scholar]
- 15.Koochekpour S, Pilkington GJ, Merzak A. Hyaluronic acid/CD44H interaction induces cell detachment and stimulates migration and invasion of human glioma cells in vitro. Int J Cancer. 1995;63(3):450–4. doi: 10.1002/ijc.2910630325. [DOI] [PubMed] [Google Scholar]
- 16.Radotra B, McCormick D. Glioma invasion in vitro is mediated by CD44-hyaluronan interactions. J Pathol. 1997;181(4):434–8. doi: 10.1002/(SICI)1096-9896(199704)181:4<434::AID-PATH797>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 17.Hayen W, Goebeler M, Kumar S, Riessen R, Nehls V. Hyaluronan stimulates tumor cell migration by modulating the fibrin fiber architecture. J Cell Sci. 1999;112(13):2241–51. doi: 10.1242/jcs.112.13.2241. [DOI] [PubMed] [Google Scholar]
- 18.Giese A, Loo MA, Rief MD, Tran N, Berens ME. Substrates for astrocytoma invasion. Neurosurgery. 1995;37(2):294–301. doi: 10.1227/00006123-199508000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Akiyama Y, Jung S, Salhia B, Lee SP, Hubbard S, Taylor M, et al. Hyaluronate receptors mediating glioma cell migration and proliferation. J Neurooncol. 2001;53(2):115–27. doi: 10.1023/a:1012297132047. [DOI] [PubMed] [Google Scholar]
- 20.Okada H, Yoshida J, Sokabe M, Wakabayashi T, Hagiwara M. Suppression of CD44 expression decreases migration and invasion of human glioma cells. Int J Cancer. 1996;66(2):255–60. doi: 10.1002/(SICI)1097-0215(19960410)66:2<255::AID-IJC20>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Turley EA, Noble PW, Bourguignon LYW. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277(7):4589–92. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the Hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70(6):2455–64. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranuncolo SM, Ladeda V, Specterman S, Varela M, Lastiri J, Morandi A, et al. CD44 expression in human gliomas. J Surg Oncol. 2002;79(1):30–6. doi: 10.1002/jso.10045. [DOI] [PubMed] [Google Scholar]
- 24.Ariza A, Lopez D, Mate JL, Isamat M, Musulen E, Pujol M, et al. Role of CD44 in the invasiveness of glioblastoma multiforme and the noninvasiveness of meningioma - an immunohistochemistry study. Hum Pathol. 1995;26(10):1144–7. doi: 10.1016/0046-8177(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 25.Bourguignon LYW. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin Cancer Biol. 2008;18(4):251–9. doi: 10.1016/j.semcancer.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourguignon LYW, Singleton PA, Zhu HB, Diedrich F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (Macrophage-Colony stimulating factor) production and breast tumor progression. J Biol Chem. 2003;278(32):29420–34. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 27.Tsatas D, Kanagasundaram V, Kaye A, Novak U. EGF receptor modifies cellular responses to hyaluronan in glioblastoma cell lines. J Clin Neurosci. 2002;9(3):282–8. doi: 10.1054/jocn.2001.1063. [DOI] [PubMed] [Google Scholar]
- 28.Bourguignon LYW, Zhu HB, Shao LJ, Chen YW. CD44 interaction with Tiam1 promotes Rac1 signaling and hyaluronic acid-mediated breast tumor cell migration. J Biol Chem. 2000;275(3):1829–38. doi: 10.1074/jbc.275.3.1829. [DOI] [PubMed] [Google Scholar]
- 29.Toole BP, Trelstad RL. Hyaluronate production and removal during corneal development in chick. Dev Biol. 1971;26(1):28. doi: 10.1016/0012-1606(71)90104-7. &. [DOI] [PubMed] [Google Scholar]
- 30.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–22. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Weaver V. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28(1):113–27. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69(10):4167–74. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen S, Dong MM, Kumar S. Isoform-specific contributions of alpha-actinin to glioma cell mechanobiology. PLoS One. 2009;4(12) doi: 10.1371/journal.pone.0008427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of Myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19(8):3357–68. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulrich TA, Jain A, Tanner K, MacKay JL, Kumar S. Probing cellular mechanobiology in three-dimensional culture with collagen–agarose matrices. Biomaterials. 2010;31(7):1875–84. doi: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman LJ, Brangwynne CP, Kasza KE, Filippidi E, Gordon VD, Deisboeck TS, et al. Glioma expansion in collagen I matrices: Analyzing collagen concentration-dependent growth and motility patterns. Biophys J. 2005;89(1):635–50. doi: 10.1529/biophysj.105.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hegedus B, Marga F, Jakab K, Sharpe-Timms KL, Forgacs G. The interplay of cell-cell and cell-matrix interactions in the invasive properties of brain tumors. Biophys J. 2006;91(7):2708–16. doi: 10.1529/biophysj.105.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HD, Guo TW, Wu AP, Wells A, Gertler FB, Lauffenburger DA. Epidermal Growth Factor-induced enhancement of glioblastoma cell migration in 3D arises from an intrinsic increase in speed but an extrinsic matrix- and proteolysis-dependent increase in persistence. Mol Biol Cell. 2008;19(10):4249–59. doi: 10.1091/mbc.E08-05-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 2011;3(4):267–78. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 40.Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(27):11298–303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leach J Baier, Bivens KA, Patrick CW, Jr, Schmidt CE. Photocrosslinked hyaluronic acid hydrogels: Natural, biodegradable tissue engineering scaffolds. Biotechnol Bioeng. 2003;82(5):578–89. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 42.Masters KS, Shah DN, Leinwand LA, Anseth KS. Crosslinked hyaluronan scaffolds as a biologically active carrier for valvular interstitial cells. Biomaterials. 2005;26(15):2517–25. doi: 10.1016/j.biomaterials.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Campoccia D, Doherty P, Radice M, Brun P, Abatangelo G, Williams DF. Semisynthetic resorbable materials from hyaluronan esterification. Biomaterials. 1998;19(23):2101–27. doi: 10.1016/s0142-9612(98)00042-8. [DOI] [PubMed] [Google Scholar]
- 44.Wang T-W, Spector M. Development of hyaluronic acid-based scaffolds for brain tissue engineering. Acta Biomater. 2009;5(7):2371–84. doi: 10.1016/j.actbio.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 45.Lei Y, Gojgini S, Lam J, Segura T. The spreading, migration and proliferation of mouse mesenchymal stem cells cultured inside hyaluronic acid hydrogels. Biomaterials. 2011;32(1):39–47. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burdick JA, Prestwich GD. Hyaluronic acid hydrogels for biomedical applications. Adv Mater. 2011;23(12):H41–H56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu XZ, Liu Y, Luo Y, Roberts MC, Prestwich GD. Disulfide cross-linked hyaluronan hydrogels. Biomacromolecules. 2002;3(6):1304–11. doi: 10.1021/bm025603c. [DOI] [PubMed] [Google Scholar]
- 48.Young JL, Engler AJ. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32(4):1002–9. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin SG, Jeong YI, Jung S, Ryu HH, Jin YH, Kim IY. The effect of hyaluronic acid on the invasiveness of malignant glioma cells : comparison of invasion potential at hyaluronic acid hydrogel and Matrigel. J Korean Neurosurg Soc. 2009;46(5):472–8. doi: 10.3340/jkns.2009.46.5.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coquerel B, Poyer F, Torossian F, Dulong V, Bellon G, Dubus I, et al. Elastin-derived peptides: matrikines critical for glioblastoma cell aggressiveness in a 3-D system. Glia. 2009;57(16):1716–26. doi: 10.1002/glia.20884. [DOI] [PubMed] [Google Scholar]
- 51.Smeds KA, Pfister-Serres A, Miki D, Dastgheib K, Inoue M, Hatchell DL, et al. Photocrosslinkable polysaccharides for in situ hydrogel formation. J Biomed Mater Res. 2001;54(1):115–21. doi: 10.1002/1097-4636(200101)54:1<115::aid-jbm14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 52.Marklein RA, Burdick JA. Spatially controlled hydrogel mechanics to modulate stem cell interactions. Soft Matter. 2010;6(1):136. [Google Scholar]
- 53.Canal T, Peppas NA. Correlation between mesh size and equilibrium degree of swelling of polymeric networks. J Biomed Mater Res. 1989;23(10):1183–93. doi: 10.1002/jbm.820231007. [DOI] [PubMed] [Google Scholar]
- 54.Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol Bioeng. 2003;83(2):173–80. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- 55.Elbert DL, Pratt AB, Lutolf MP, Halstenberg S, Hubbell JA. Protein delivery from materials formed by self-selective conjugate addition reactions. J Control Release. 2001;76(1-2):11–25. doi: 10.1016/s0168-3659(01)00398-4. [DOI] [PubMed] [Google Scholar]
- 56.Lutolf MP, Hubbell JA. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4(3):713–22. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 57.Khetan S, Katz JS, Burdick JA. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter. 2009;5(8):1601–6. [Google Scholar]
- 58.Anseth KS, Bowman CN, Brannon-Peppas L. Mechanical properties of hydrogels and their experimental determination. Biomaterials. 1996;17(17):1647–57. doi: 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- 59.Elkin BS, Azeloglu EU, Costa KD, Morrison B. Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J Neurotrauma. 2007;24(5):812–22. doi: 10.1089/neu.2006.0169. [DOI] [PubMed] [Google Scholar]
- 60.Christ AF, Franze K, Gautier H, Moshayedi P, Fawcett J, Franklin RJM, et al. Mechanical difference between white and gray matter in the rat cerebellum measured by scanning force microscopy. J Biomech. 2010;43(15):2986–92. doi: 10.1016/j.jbiomech.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 61.Cheng S, Clarke EC, Bilston LE. Rheological properties of the tissues of the central nervous system: a review. Med Eng Phys. 2008;30(10):1318–37. doi: 10.1016/j.medengphy.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Segura T, Anderson BC, Chung PH, Webber RE, Shull KR, Shea LD. Crosslinked hyaluronic acid hydrogels: a strategy to functionalize and pattern. Biomaterials. 2005;26(4):359–71. doi: 10.1016/j.biomaterials.2004.02.067. [DOI] [PubMed] [Google Scholar]
- 63.Brigham MD, Bick A, Lo E, Bendali A, Burdick JA, Khademhosseini A. Mechanically robust and bioadhesive collagen and photocrosslinkable hyaluronic acid semi-interpenetrating networks. Tissue Eng Part A. 2009;15(7):1645–53. doi: 10.1089/ten.tea.2008.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eng D, Caplan M, Preul M, Panitch A. Hyaluronan scaffolds: A balance between backbone functionalization and bioactivity. Acta Biomater. 2010;6(7):2407–14. doi: 10.1016/j.actbio.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 65.Collins MN, Birkinshaw C. Investigation of the swelling behavior of crosslinked hyaluronic acid films and hydrogels produced using homogeneous reactions. J Appl Polym Sci. 2008;109(2):923–31. [Google Scholar]
- 66.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys J. 2005;89(2):1374–88. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bellamkonda R, Ranieri JP, Bouche N, Aebischer P. Hydrogel-based 3-dimensional matrix for neural cells. J Biomed Mater Res. 1995;29(5):663–71. doi: 10.1002/jbm.820290514. [DOI] [PubMed] [Google Scholar]
- 68.Ghosh K, Shu XZ, Mou R, Lombardi J, Prestwich GD, Rafailovich MH, et al. Rheological characterization of in situ cross-linkable hyaluronan hydrogels. Biomacromolecules. 2005;6(5):2857–65. doi: 10.1021/bm050361c. [DOI] [PubMed] [Google Scholar]
- 69.Massia SP, Hubbell JA. An RGD spacing of 440nm is sufficient for integrin Alpha-V-Beta-3-mediated fibroblast spreading and 140nm for focal contact and stress fiber formation. J Cell Biol. 1991;114(5):1089–100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rezania A, Johnson R, Lefkow AR, Healy KE. Bioactivation of metal oxide surfaces. 1. Surface characterization and cell response. Langmuir. 1999;15(20):6931–9. [Google Scholar]
- 71.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113(10):1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 72.Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103(29):10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204(1):198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 75.Pathak A, Kumar S. From molecular signal activation to locomotion: an integrated, multiscale analysis of cell motility on defined matrices. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0018423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 77.Ulrich TA, Lee TG, Shon HK, Moon DW, Kumar S. Microscale mechanisms of agarose-induced disruption of collagen remodeling. Biomaterials. 2011;32(24):5633–42. doi: 10.1016/j.biomaterials.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kakita A, Goldman JE. Patterns and dynamics of SVZ cell migration in the postnatal forebrain: Monitoring living progenitors in slice preparations. Neuron. 1999;23(3):461–72. doi: 10.1016/s0896-6273(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 79.Thorne RG, Nicholson C. In vivo diffusion analysis with quantum dots and dextrans predicts the width of brain extracellular space. Proc Natl Acad Sci U S A. 2006;103(14):5567–72. doi: 10.1073/pnas.0509425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, et al. The effects of hyaluronic acid hydrogels with tunable mechanical properties on neural progenitor cell differentiation. Biomaterials. 2010;31(14):3930–40. doi: 10.1016/j.biomaterials.2010.01.125. [DOI] [PubMed] [Google Scholar]
- 81.Ghosh K, Pan Z, Guan E, Ge SR, Liu YJ, Nakamura T, et al. Cell adaptation to a physiologically relevant ECM mimic with different viscoelastic properties. Biomaterials. 2007;28(4):671–9. doi: 10.1016/j.biomaterials.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Unsgaard G, Rygh OM, Selbekk T, Müller TB, Kolstad F, Lindseth F, et al. Intra-operative 3D ultrasound in neurosurgery. Acta Neurochir (Wien) 2006;148(3):235–53. doi: 10.1007/s00701-005-0688-y. [DOI] [PubMed] [Google Scholar]
- 83.Yang Y-l, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010;31(21):5678–88. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 84.Katsumi A, Orr AW, Tzima E, Schwartz MA. Integrins in mechanotransduction. J Biol Chem. 2004;279(13):12001–4. doi: 10.1074/jbc.R300038200. [DOI] [PubMed] [Google Scholar]
- 85.Knudson W, Loeser RF. CD44 and integrin matrix receptors participate in cartilage homeostasis. Cell Mol Life Sci. 2002;59(1):36–44. doi: 10.1007/s00018-002-8403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Knupfer MM, Poppenborg H, Hotfilder M, Kuhnel K, Wolff JEA, Domula M. CD44 expression and hyaluronic acid binding of malignant glioma cells. Clin Exp Metastasis. 1999;17(1):71–6. doi: 10.1023/a:1026425519497. [DOI] [PubMed] [Google Scholar]
- 87.Novak U, Stylli SS, Kaye AH, Lepperdinger G. Hyaluronidase-2 overexpression accelerates intracerebral but not subcutaneous tumor formation of murine astrocytoma cells. Cancer Res. 1999;59(24):6246–50. [PubMed] [Google Scholar]
- 88.David L, Dulong V, Le Cerf D, Chauzy C, Norris V, Delpech B, et al. Reticulated hyaluronan hydrogels: a model for examining cancer cell invasion in 3D. Matrix Biol. 2004;23(3):183–93. doi: 10.1016/j.matbio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 89.Stern R. Hyaluronidases in cancer biology. Semin Cancer Biol. 2008;18(4):275–80. doi: 10.1016/j.semcancer.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 90.Enegd B, King JAJ, Stylli S, Paradiso L, Kaye AH, Novak U. Overexpression of hyaluronan synthase-2 reduces the tumorigenic potential of glioma cells lacking hyaluronidase activity. Neurosurgery. 2002;50(6):1311–8. doi: 10.1097/00006123-200206000-00023. [DOI] [PubMed] [Google Scholar]
- 91.Becker LC, Bergfeld WF, Belsito DV, Klaassen CD, Marks JG, Shank RC, et al. Final report of the safety assessment of hyaluronic acid, potassium hyaluronate, and sodium hyaluronate. Int J Toxicol. 2009;28:5–67. doi: 10.1177/1091581809337738. [DOI] [PubMed] [Google Scholar]
- 92.Liu YC, Shu XZ, Prestwich GD. Tumor engineering: orthotopic cancer models in mice using cell-loaded, injectable, cross-linked hyaluronan-derived hydrogels. Tissue Eng. 2007;13(5):1091–101. doi: 10.1089/ten.2006.0297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.