Abstract
MicroRNAs (miRNAs) regulate specific immune mechanisms but their genome-wide regulation of T-lymphocyte activation is largely unknown. We performed a multidimensional functional genomics analysis to integrate genome-wide differential mRNA, miRNA, and protein expression as a function of human T-lymphocyte activation and time. We surveyed expression of 420 human miRNAs in parallel with genome-wide mRNA expression. We identified a unique signature of 71 differentially expressed miRNAs, 57 of which were previously not known as regulators of immune activation. The majority of miRNAs are upregulated, mRNA expression of these target genes is downregulated and this is a function of binding multiple miRNAs (combinatorial targeting). Our data reveal that consideration of this complex signature, rather than single miRNAs, is necessary to construct a full picture of miRNA-mediated regulation. Molecular network mapping of miRNA targets revealed the regulation of activation-induced immune signaling. In contrast, pathways populated by genes that are not miRNA targets are enriched for metabolism and biosynthesis. Finally, we specifically validated miR-155 (known) and miR-221 (novel in T-lymphocytes) using locked nucleic acid inhibitors. Inhibition of these 2 highly upregulated miRNAs in CD4+ T cells were shown to increase proliferation by removing suppression of 4 target genes linked to proliferation and survival. Thus, multiple lines of evidence link top functional networks directly to T-lymphocyte immunity underlining the value of mapping global gene, protein and miRNA expression.
Keywords: Adaptive immunity, lymphocyte activation, functional genomics, microRNA, high-throughput genomics, molecular networks, T-lymphocytes, immune regulation
Introduction
T-lymphocytes regulate the adaptive immune response by serving as antigen-specific effector cells. Activation via T cell receptor (TCR) engagement and CD28 co-stimulation is characterized by gene upregulation (1) and is a highly regulated process requiring coordination of multiple signaling pathways for proliferation, cytokines, and differentiation. After antigen clearance, some effector cells must be reduced or eliminated by mechanisms like activation-induced cell death (AICD) (2). Thus, activation must be regulated in a coordinated fashion to achieve a balance between proliferation, memory and quiescence.
MicroRNAs (miRNAs) have emerged as post-transcriptional regulators of gene expression in a variety of biological processes (3–7). The mode of miRNA regulation is protein repression via complementary sequence recognition in the 3′ UTR of the target mRNA and/or degradation of the target transcript (8–11). A recent paper indicates the major effect of miRNAs is to decrease mRNA levels (12).
MiRNAs can potentially regulate hundreds of proteins (13) and modulate concentration of proteins over a narrow range in a dose-dependent manner (14, 15). MiRNAs are involved in hematopoietic cell function and development, as summarized in (16–64). A few miRNAs have been linked to specific T-lymphocyte mechanisms: 181a (37), 181c (39), 155 (28), 150 (18), 146 (20), and 142 (40) via regulation of T cell sensitivity to antigen stimulation, regulating transcription factors, and AICD. However at the global level, little is known about the impact of activation-induced miRNAs on mRNA and protein expression in human T-lymphocytes, particularly in the context of mapping miRNA-regulated molecular networks.
Here we show that differentially upregulated miRNAs regulate T-lymphocyte activation by targeting highly differentially expressed genes involved in networks critical for cell activation, proliferation and survival. We used a multi-dimensional approach to integrate genome-wide miRNA, mRNA and protein expression. We surveyed expression for 420 human miRNA sequences at 0, 24, 48 and 72 hours after activation. In parallel, we profiled global mRNA and protein expression. We found 71 significantly differentially expressed miRNAs of which 57 have not been previously linked to T-lymphocyte function. Testing several established miRNA target prediction algorithms, we demonstrated globally that targets of multiple upregulated miRNAs (combinatorial targeting) have decreased mRNA expression with activation. In validation, we showed that inhibition of 2 highly upregulated miRNAs in CD4+ T cells increased proliferation by removing suppression of 4 target genes involved in proliferation and survival. Thus, our studies provide novel evidence for a large number of functional molecular networks populated by downregulated targets of highly upregulated miRNAs.
Materials and Methods
T-lymphocyte isolation
Blood draw for this study was accepted by our institution’s ethical commission and all subjects gave their written consent according to review board guidelines. CD2+ T-lymphocytes were purified from Ficoll-Hypaque density separated peripheral blood mononuclear cells (PBMC) of 7 healthy human donors. MACS CD2+ micro-magnetic beads were used for the positive isolation of CD2+ T-cells, using a MACS separator with LS columns (Miltenyi Biotec). CD4+ T-lymphocytes were isolated from PBMC of 3 human blood donors by negative selection with MACS CD4+ T cell negative isolation kit II (Miltenyi Biotec) according to manufacturer’s protocol.
Lymphocyte activation, RNA and protein isolation
Freshly isolated CD2+ T-lymphocytes were resuspended in RPMI-1640 medium (Hyclone) supplemented with 10% (vol/vol) FBS and 2 mM glutamine, with penicillin (100 U/ml) and streptomycin (100 U/ml) and activated with CD3/CD28 Dynal beads (Invitrogen) according to manufacturer’s protocol. Cell activation was confirmed by flow cytometry for the following activation markers: CD134 (OX40), CD150 (SLAM), CD25 (IL-2Rα), CD69, and CD71 (transferin receptor) and intracellular cytokines: IFNγ, IL-10, IL-2, IL-4, and TNFα for activated T cells as shown previously(1). The cells were harvested and stabilized in RNALater (Ambion) at 0, 24, 48 and 72 hours post-activation. Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion), which also allows for the isolation of the total proteome fraction.
miRNA profiling
TaqMan stem-loop RT–PCR method(65) was performed on an ABI 7900HT Real-Time PCR system for 420 human miRNA primer/probes on 0.5 μg total RNA from 0, 24, 48, and 72 hour CD2+ T-lymphocyte samples. A two-tailed Student’s t-test with a p-value threshold of 0.05 and FDR adjusted p-value (q-value) threshold of 0.1, where q-value= p-value * number tested/rank, between 0 and 48 h was used on the normalized data to identify differentially expressed miRNAs. Q-value of 0.1 implies that 10% of significant tests will result in false positives. MiRNA expression following LNA nucleoporation in CD4+ T cells was measured with TaqMan MicroRNA Assays (Applied Biosystems) for hsa-miR-155 and 221 in accordance with manufacturers’ protocols. U6 was used as an internal control.
Microarray profiling
1.5 μg of total RNA per sample was converted into labeled cDNA using the GeneChip® WT Sense Target Labeling kit (Affymetrix). Labeled cDNA was hybridized to Affymetrix Human Exon 1.0 ST arrays. Data for mRNA transcript profiles were generated in the form of CEL files using standard protocols.
Microarray Data
Data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE14352 and can be viewed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14352
Differential gene expression analysis
Raw data expression values from CEL files were normalized by RMA algorithm provided through Affymetrix Power Tools and summarized in AltAnalyze software (http://altanalyze.org), retaining only probesets that align to a single Ensembl gene. Gene-expression values were calculated based on the mean expression of all “core” probesets detected above background (DABG p-value thresholds for p<0.01, FDR 0.01). Fold changes and p-value (two-tailed t-test assuming unequal variance) were calculated for each timepoint comparisons. Statistics were performed for specified pair-wise comparisons between all timepoints of activation. In parallel, we performed our own analysis of differential gene expression to corroborate AltAnalyze results. CEL files for each donor from the 1.0ST HuEx Arrays were normalized by RMA using a custom CDF downloaded from the University of Michigan (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp). Differential expression was measured with the Limma package (http://www.bioconductor.org/packages/2.6/bioc/html/limma.html) using a two-class model. All calculations were performed in R/Bioconductor. Genes were filtered using a fold change filter of 1.5 (0.58 in log2) and an FDR filter of 0.01. Genes that were detected as significantly differentially expressed by both AltAnalyze and Limma analysis were then selected for further analysis.
MudPIT proteomics
The Multi-dimensional Protein Identification Tool(66) (MudPIT) protocol was used as described previously(1). Protein fraction was denatured, alkylated, and trypsin digested. 50μg of digested protein sample at 0 and 48 hours was run in four technical replicates. Data were acquired using an LTQ LX linear ion trap mass spectrometer (ThermoFisher Scientific) interfaced in-line with 2D HPLC in a data-dependant manner where each analytical full scan (MS, 200–2,000m/z units range) was followed by three fragmentation scans (MS/MS) that targeted the three most abundant ions from the full scan. 40-micro second CID pulses of 35% intensity were used for precursor ion fragmentation. A default exclusion list (Xcalibur 2.0, ThermoFisher Scientific) of 180-second, 50 precursor ion members was used for data acquisition. Raw data were searched against the EBI database (12/01/2006 release) supplemented with a decoy database where each entry of the original protein contains its reversed sequence. Database searching used SEQUEST (v27) and outcomes were filtered using DTASelect version 2.0. Protein identifications were extracted and a measure of normalized amino acid coverage was used as label free quantification. Relative quantifications were done using spectral counts normalized to the median of the total spectral counts. Protein identifications across replicate experiments were pooled to represent a union for each category of 0 and 48 hours post-activation. Proteins identified in two or more technical replicates per category were kept for further analysis. Relative protein abundance was compared between the 0 and the 48-hour post-activation for proteins present in more than one category. Proteins identified in more than one technical replicate in a single category and not in any category were also considered for functional analysis as unique identifications. A two-tailed Student’s t-test was used for hypothesis testing and the significant differentially expressed proteins (p<0.05) were considered for functional analysis.
miRNA target analysis
For prediction of target genes of differentially expressed miRNAs, 3 publicly available algorithms were used: PITA, MiRanda, and TargetScan5.1. In the end, TargetScan predictions based on conservation scores were used to compute the 50th percentile targets in our expressed gene set.
Functional mapping
We used Ingenuity Pathway Analysis (https://analysis.ingenuity.com) to map molecular pathways and networks populated by predicted miRNA targets. Ingenuity Pathways Analysis Database is a constantly curated resource of published literature on gene functions and interactions. Canonical pathway and networks analysis was carried out by uploading the predicted downregulated genes targeted by the upregulated miRNAs. Significance of association between genes and pathway was measured by the Benjamini and Hochberg multiple testing corrected p-value that can be interpreted as an upper bound for the expected fraction of falsely rejected null hypotheses among all functions with p-values smaller than threshold of 0.05. Network node genes were based on especially high degree of links to other genes in the IPA database.
Electroporation
3×106 primary human CD4+ T-lymphocytes were electroporated in Nucleofector II instrument (AMAXA) using Human T cell Nucleofector kit (VPA-1002, Lonza) in duplicate with 50nM miRCURY LNA™ microRNA Power Inhibitor or scrambled negative control probes (Exiqon) against hsa-miR-221 and 155 according to the manufacturer’s protocol. After electroporation, cells were cultured in RPMI-1640 medium supplemented with 10% (vol/vol) FBS and 2 mM glutamine, and after 2 hours half the medium was replaced with fresh medium. At 24 h after electroporation, the cells were activated with CD3/CD28 Dynal beads (Invitrogen) for 48 hours.
Cell proliferation assay
Cell proliferation was measured using Ziva Cell Proliferation Assay (Jaden BioScience). Electroporated CD4+ T-lymphocytes were plated at 8×10^4/well in 96-well plate in duplicates for each condition and activated for 48 hours with CD3/CD28 beads. Cells were pulsed with 10uM BrdU/well 18 hours before harvesting. Forty-eight hours after activation, cells were harvested, expander beads removed and 10^4 cells/well were plated in 96-well Thermo Scientific Nunc Plate (Fisher). Cell proliferation was measured with a chemiluminescent substrate to detect the presence of an anti-BrdU antibody labeled with alkaline phosphatase on the Insight-Mi Luminometer (Jaden BioScience). The signal was fully developed and measured 60 minutes after the addition of the substrate.
Quantitative RT-PCR
300ng of total RNA from CD4+ T cells was transcribed into cDNA using qScript cDNA Supermix (Quanta Biosiences) according to manufacturer’s guidelines and gene expression for IRS2, IKBKE, FOS, and PIK3R1 was quantitated with PrimeTime qPCR assays (Integrated DNA Technologies) using PerfeCta qPCR FastMix kit (Quanta Biosiences) on the ABI 7900HT Fast Real-Time PCR instrument. Expression of 18s gene was set as endogenous control. For data analysis, the threshold cycle (67) value was determined and specific gene expression normalized to endogenous control using delta-delta Ct method. The normalized delta Ct from LNA transfected samples were then compared to the scrambled control to obtain delta-delta Ct values and used to calculate relative fold change compared to control. Experiments were performed in triplicate. The primer probe sequences for validation assays were as follows:
IKBKE Probe (0.5 nMoles): 5′-/56-FAM/TAC CTG ATC/ZEN/CCG GCT CTT CAC CA/3IABkFQ/-3′
IKBKE Primer1 (1 nMoles): 5′-CAT CTT GTC CAA ACA GCA CTG-3′
IKBKE Primer2 (1 nMoles): 5′-AAA ATATCA CGG AGA CCC AGG-3′
FOS Probe (0.5 nMoles): 5′-/56-FAM/TGC AGA CCG/ZEN/AGATTG CCA ACCT/3IABkFQ/-3′
FOS Primer1 (1 nMoles): 5′-CAT CAG GGATCT TGC AGG C-3′
FOS Primer2 (1 nMoles): 5′-GACTGA TAC ACT CCA AGC GG-3′
IRS2 Probe (0.5 nMoles): 5′-/56-FAM/AGG CCA CCA/ZEN/TCG TGA AAG AGT GAA G/3IABkFQ/-3′
IRS2 Primer1 (1 nMoles): 5′-TGA CAT CCT GGT GAT AAA GCC-3′
IRS2 Primer2 (1 nMoles): 5′-ACT TCT TGT CCC ACC ACT TG-3′
PIK3R1 Probe (0.5 nMoles): 5′-/56-FAM/CAC AAT GCT/ZEN/TTA CTT CGC CGT CCA C/3IABkFQ/-3′
PIK3R1 Primer1 (1 nMoles): 5′-CTG TAC AAGTTATAG GGCTCG G-3′
PIK3R1 Primer2 (1 nMoles): 5′-GAT GGC ACT TTT CTT GTC CG-3′
Statistics
All statistical analyses used the Student’s t-test of at least three independent experiments, unless stated otherwise. Differences with P-values <0.05 are considered significant.
Results
Activated T-lymphocytes demonstrate a unique activation-induced miRNA signature
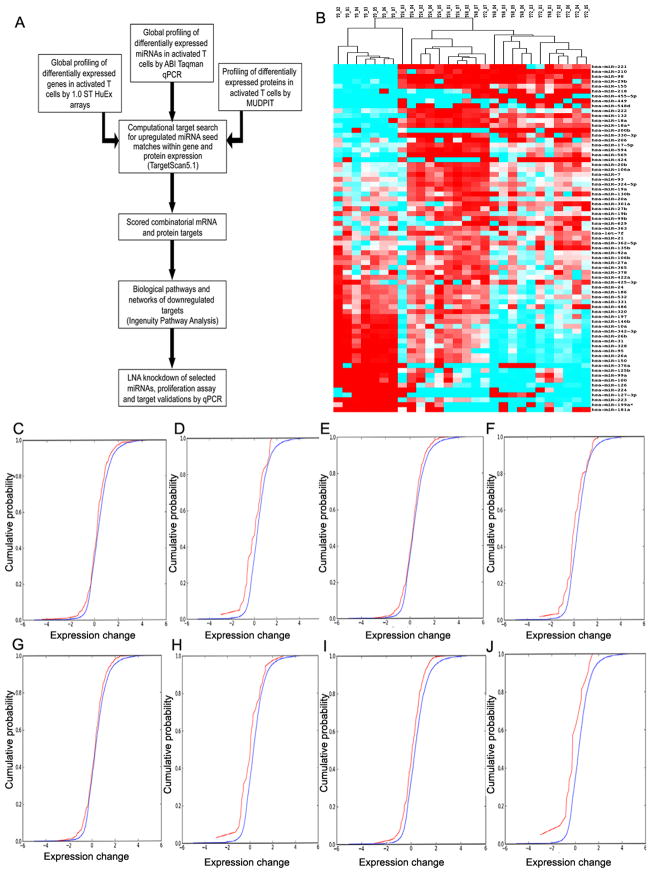
We used a multi-dimensional approach to integrate genome-wide miRNA, mRNA and protein expression (Fig. 1A). We activated human T-lymphocytes via CD3/CD28 co-stimulation and harvested cells at 0, 24, 48 and 72 hours. This activation strategy modeled allogeneic activation(68, 69). We surveyed miRNA expression using real-time qPCR for 420 human miRNA sequences. Specific miRNAs were differentially expressed in T-lymphocytes as a function of activation. We identified 71 differentially expressed miRNAs (p<0.05, q<0.1) between 0 and 48 hours, of which 51 were upregulated (Table I). We chose 48 hours as a key timepoint in T-lymphocyte activation based on peak cell proliferation, cytokine production and expression of activation markers (1). These changes in miRNA expression are robust across all donors (Fig. 1B). The top 12 up-regulated miRNAs were miR-221, 210, 98, 29b, 155, 218, 455-3p, 449, 548d, 222, 132 and 18a. The top down-regulated miRNAs were miR-181a, 223, 224, 150, 146b, 126, 127-3p, 376a, 100, 99a, 125b and 26a (Table II).
Figure 1.
Combinatorial targeting by multiple upregulated miRNAs during T cell activation demonstrates decreased mRNA levels after activation with increased miRNA binding. (A) Schematic of our experimental approach. (B) miRNA signature of T cell activation: Heat-map of 71 statistically significant (p<0.05, q<0.1) differentially expressed miRNAs at 0 and 48 hours. Heatmap shows expression at 0, 24, 48 and 72 hours across 7 donors. Red represents positive change, cyan represents negative change, white represents no change. (C): A Cumulative Distribution Function (CDF) plot of relative fold change between 0 and 48 hours of combined PITA, TargetScan/conservation and TargetScan/context, top 50th percentile predictions in each, 2+ miRNAs targeting each gene. Target genes in red, non-target genes in blue. (D): Same as (A) with 4+ miRNAs targeting each gene (E): PITA, top 50th percentile predictions, 4+ miRNAs targeting a given gene. Target genes in red, non-target genes in blue. (F): PITA, top 50th percentile predictions with 7+ miRNAs targeting a given gene. Target genes in red, non-target genes in blue. (G): TargetScan/context score, top 50th percentile predictions, 4+ miRNAs targeting a given gene. Target genes in red, non-target genes in blue. (H): TargetScan/context score, top 50th percentile predictions, 7+ miRNAs targeting a given gene. Target genes in red, non-target genes in blue. (I): TargetScan/conservation score, top 50th percentile predictions, 4+ miRNAs targeting a given gene. Target genes in red, non-target genes in blue. (J): TargetScan/conservation score, top 50th percentile predictions, 7+ miRNAs targeting a given gene. Target genes in red, non-target genes in blue.
Table 1.
Differentially upregulated (>2 fold) miRNAs in activated T cells detected 0vs48
| miRNA | p-value | * q-value | Fold Change |
|---|---|---|---|
| hsa-miR-221 | 2.77E-08 | 3.31E-06 | 7881.6 |
| hsa-miR-210 | 2.12E-04 | 2.54E-03 | 1846.7 |
| hsa-miR-98 | 8.96E-04 | 6.69E-03 | 237.6 |
| hsa-miR-29b | 1.97E-03 | 1.27E-02 | 194.7 |
| hsa-miR-155 | 4.19E-08 | 3.34E-06 | 70.5 |
| hsa-miR-218 | 4.80E-03 | 2.55E-02 | 45.4 |
| hsa-miR-455-5p | 1.34E-03 | 9.13E-03 | 32.2 |
| hsa-miR-449 | 1.01E-02 | 4.75E-02 | 20.5 |
| hsa-miR-548d | 9.97E-03 | 4.76E-02 | 18.8 |
| hsa-miR-222 | 2.89E-07 | 1.73E-05 | 17.1 |
| hsa-miR-132 | 6.22E-05 | 1.06E-03 | 15.3 |
| hsa-miR-18a | 2.05E-03 | 1.29E-02 | 15.3 |
| hsa-miR-18a* | 7.19E-09 | 1.72E-06 | 15.0 |
| hsa-miR-200b | 2.90E-02 | 9.76E-02 | 13.1 |
| hsa-miR-330-3p | 1.39E-02 | 5.61E-02 | 11.1 |
| hsa-miR-206 | 1.28E-02 | 5.68E-02 | 10.0 |
| hsa-miR-17-5p | 5.48E-06 | 2.18E-04 | 9.8 |
| hsa-miR-424 | 1.32E-02 | 5.44E-02 | 9.6 |
| hsa-miR-20b | 2.34E-06 | 1.12E-04 | 9.0 |
| hsa-miR-106a | 6.51E-06 | 2.22E-04 | 8.4 |
| hsa-miR-7 | 7.05E-05 | 1.05E-03 | 8.3 |
| hsa-miR-93 | 3.08E-05 | 7.37E-04 | 7.6 |
| hsa-miR-324-5p | 1.37E-05 | 4.09E-04 | 6.4 |
| hsa-miR-19a | 2.89E-04 | 2.76E-03 | 6.3 |
| hsa-miR-130b | 2.51E-04 | 2.50E-03 | 6.1 |
| hsa-miR-20a | 2.39E-05 | 6.34E-04 | 6.0 |
| hsa-miR-301a | 2.24E-04 | 2.55E-03 | 6.0 |
| hsa-miR-27b | 2.81E-02 | 9.74E-02 | 5.9 |
| hsa-miR-19b | 2.24E-04 | 2.43E-03 | 5.8 |
| hsa-miR-99b | 2.35E-04 | 2.45E-03 | 5.1 |
| hsa-miR-629 | 4.60E-04 | 4.07E-03 | 4.7 |
| hsa-miR-363 | 2.35E-02 | 8.27E-02 | 4.2 |
| hsa-let-7f | 1.30E-02 | 5.65E-02 | 3.8 |
| hsa-miR-21 | 3.68E-03 | 2.00E-02 | 3.7 |
| hsa-miR-362-5p | 1.77E-03 | 1.18E-02 | 3.7 |
| hsa-miR-135b | 6.35E-04 | 5.06E-03 | 3.5 |
| hsa-miR-92a | 5.26E-05 | 9.67E-04 | 3.4 |
| hsa-miR-106b | 6.54E-03 | 3.33E-02 | 3.4 |
| hsa-miR-27a | 3.41E-03 | 1.90E-02 | 3.1 |
| hsa-miR-365 | 1.04E-02 | 4.80E-02 | 2.7 |
| hsa-miR-378 | 2.17E-02 | 7.98E-02 | 2.7 |
| hsa-miR-422a | 1.64E-04 | 2.18E-03 | 2.6 |
| hsa-miR-425-3p | 8.26E-03 | 4.11E-02 | 2.5 |
| hsa-miR-24 | 6.69E-05 | 1.07E-03 | 2.3 |
Shown are significantly differentially upregulated miRNAs, with fold change greater than 2-fold.
q-value is the FDR threshold for the corresponding p-value
Table II.
Differentially downregulated miRNAs in activated T cells detected 0 vs. 48
| miRNA | p-value | q-value | Fold Change |
|---|---|---|---|
| hsa-miR-197 | 1.72E-02 | 6.75E-02 | −1.2 |
| hsa-miR-146b | 1.67E-02 | 6.67E-02 | −1.6 |
| hsa-miR-10a | 1.95E-02 | 7.52E-02 | −1.6 |
| hsa-miR-342-3p | 1.88E-04 | 2.37E-03 | −1.7 |
| hsa-miR-26b | 2.90E-02 | 9.90E-02 | −1.8 |
| hsa-miR-31 | 4.91E-04 | 4.05E-03 | −1.8 |
| hsa-miR-328 | 9.47E-05 | 1.33E-03 | −1.9 |
| hsa-miR-95 | 2.58E-03 | 1.54E-02 | −2.1 |
| hsa-miR-26a | 2.96E-03 | 1.72E-02 | −2.4 |
| hsa-miR-150 | 4.30E-05 | 8.56E-04 | −3.0 |
| hsa-miR-125b | 3.33E-03 | 1.89E-02 | −3.4 |
| hsa-miR-99a | 5.04E-03 | 2.62E-02 | −3.8 |
| hsa-miR-100 | 2.23E-03 | 1.37E-02 | −3.9 |
| hsa-miR-376a | 2.19E-02 | 7.93E-02 | −4.1 |
| hsa-miR-126 | 1.19E-03 | 8.34E-03 | −5.2 |
| hsa-miR-127-3p | 2.31E-02 | 8.23E-02 | −7.2 |
| hsa-miR-224 | 1.31E-02 | 5.60E-02 | −7.9 |
| hsa-miR-223 | 4.05E-05 | 8.80E-04 | −8.7 |
| hsa-miR-199a* | 9.15E-03 | 4.46E-02 | −31.0 |
| hsa-miR-181a | 2.07E-02 | 7.72E-02 | −71.8 |
Shown are significantly differentially downregulated miRNAs.
Based on current literature, of these 71 differentially expressed miRNAs, only 14 have a documented function in T-lymphocytes: miR-150 (18, 19), miR-155 (18, 25), miR-181a(37), miR-106a(31, 33), the miR-17~92 cluster (30, 32), miR-24 (6, 47), miR-21 (18, 53), miR-223 (41) and miR-let-7f (18) (Tables I and II). Five additional miRNAs are linked to development, or aberrant activation of hematopoietic cells (Supplemental Table S1). For example, our results indicate that miR-150 is downregulated in T cells upon activation, consistent with studies in murine lymphocytes (19). In contrast, miR-155 is upregulated in activated human and murine T-lymphocytes (26). Importantly, we identified 57 differentially expressed miRNAs currently undocumented in T-lymphocyte activation.
Predicted targets of differentially upregulated miRNAs are globally downregulated
Starting with the 71 differentially expressed miRNAs, we mapped the gene targets of the 51 differentially upregulated. Predictive algorithms rely on multiple parameters: seed complementarity, thermodynamics and biochemical properties of binding and evolutionary conservation. Unfortunately, these algorithms suffer from high false positive and negative rates. Combining predictions from different algorithms may be useful, but there is little overlap in top targets predicted by different algorithms (70, 71). We tested 50th percentile predictions for these 51 differentially upregulated miRNAs using four algorithms: PITA (72), MiRanda (73) and TargetScan5.1 context or conservation scores (74). We measured the change (delta) in mRNA expression between 0 and 48 hours for all genes above background (expression ≥6.5; log2 scale). We then plotted differences in distribution of deltas between predicted targets and non-targets. MiRanda failed to produce any expression correlations with our data and was not used further. Predictions with TargetScan and PITA based on testing the effects of single miRNA binding also revealed no shift in mRNA signals with activation. In contrast, plotting the gene expression of targets predicted to bind multiple upregulated miRNAs (e.g. ≥4 or ≥7) revealed that combinatorial miRNA binding decreases mRNA expression (Fig. 1C–D). Combinatorial targeting benchmarked at the 50th percentile with both PITA and TargetScan gave the best predictions (Fig. 1E–J). As shown in Fig. 1I and J, TargetScan conservation predictions with combinatorial binding of 4 or more miRNAs show the best results correlating increased miRNA binding with decreased target gene expression.
T-lymphocyte activation is marked by global gene upregulation including miRNA processing machinery
We showed previously that T-lymphocyte activation is dominated by widespread differential gene upregulation (1). We therefore analyzed differential gene expression in parallel with miRNA expression. Genome-wide mRNA transcript analysis revealed 3798 differentially expressed mRNA transcripts between 0 and 48 hours (p-value < 0.01, FDR 1%): 3362 upregulated (89%) and 436 downregulated. Upregulation of the miRNA processing/biogenesis genes included: XPO5, EIF2C2/AGO2, SIP1/GEMIN2, 4, 5, 6, and 7, RANGAP1, YBX1 and ADARB1 (Table III).
Table III.
miRNA processing machinery genes detected at 48 hours post activation
| Gene# | Definition | Function | Fold change T48 vs T0 | miRNAs |
|---|---|---|---|---|
| XPO5 | Exportin-5 | Mediates the nuclear export of microRNA precursors. | 4.30 | miR-218 miR-24 |
| RANGAP1 | Ran GTPase-activating protein 1 | mRNA processing and transport | 1.60 | ND |
| EIF2C2/AGO2 | Protein argonaute-2 | Provides endonuclease activity to RNA- induced silencing complexes (RISC). Cleaves siRNA/mRNA heteroduplexes bound to RISC. | 3.84 | ND |
| SIP1/GEMIN2 | Survival of motor neuron protein- interacting protein 1 | Core component of the survival of motor neuron (SMN) complex, which plays an essential role in spliceosomal snRNP assembly in the cytoplasm and is required for pre-mRNA splicing in the nucleus. | 2.23 | ND |
| GEMIN4 | Gem-associated protein 4 | Component of the SMN complex, which is required for pre-mRNA splicing in the nucleus. | 1.59 | miR-155 |
| GEMIN5 | Gem-associated protein 5 | Component of the SMN complex, which is required for pre-mRNA splicing in the nucleus. | 2.13 | ND |
| GEMIN6 | Gem-associated protein 6 | Component of the SMN complex, which is required for pre-mRNA splicing in the nucleus. | 2.11 | ND |
| GEMIN7 | Gem-associated protein 7 | Component of the SMN complex, which is required for pre-mRNA splicing in the nucleus. | 1.90 | ND |
| YBX1 | Nuclease-sensitive element-binding protein 1 | Participates in different steps of mRNA biogenesis, including mRNA transcription, processing, and transport from the nucleus into the cytoplasm, binds to splice sites in pre-mRNA, and regulates splice site selection. | 1.53 | ND |
| ADARB1 | Double-stranded RNA- specific editase 1 | Binds to short interfering RNAs (siRNA) without editing them and suppresses siRNA-mediated RNA interference. | 1.50 | miR-218 |
The FDR adjusted p values of every gene in this table is <0.007. Shown are genes implicated in miRNA processing/biogenesis that were significantly differentially expressed between 0 and 48 hours, with relative fold changes.
ND = miRNA not detected
Predicted targets of upregulated miRNAs populate networks associated with immunity, cell survival, and proliferation
Using TargetScan conservation predictions we identified 1640 candidate miRNA targets of which 214 were downregulated (Supplemental Table S2). Thus, half of all 436 downregulated genes are targets of upregulated miRNAs. Functional pathway and network enrichment analysis was done for the 182 of 214 downregulated targets that mapped to known functional pathways and the 200 that mapped to molecular networks.
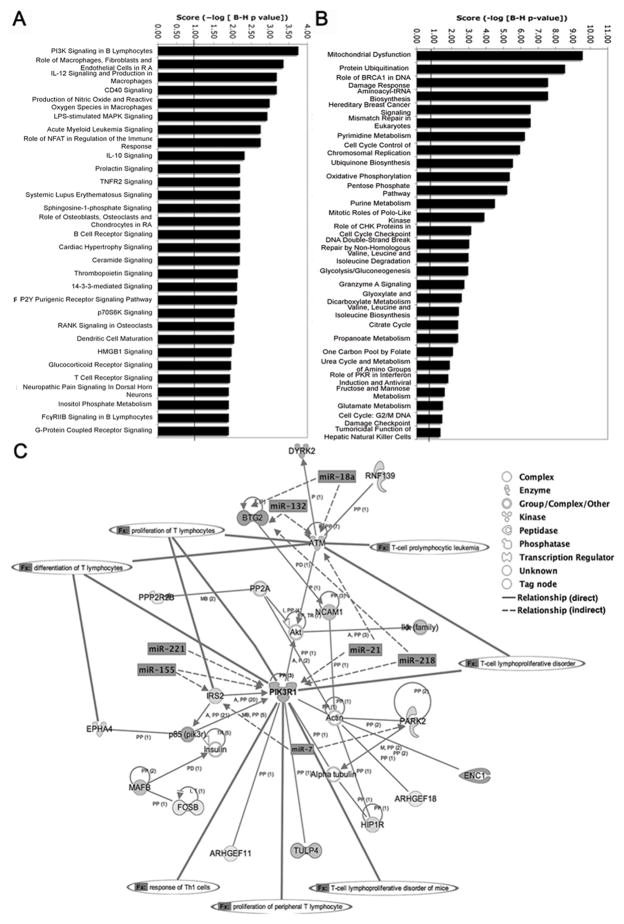
Pathway analysis revealed statistically significant enrichment for 71 canonical pathways (multiple test correction p-value<0.05), with the top 30 pathways shown in Fig. 2A. Represented were primarily immune signaling pathways including IL-12, PI3K, IL-10, CD40, NFAT, S1P and TCR. Cell survival, growth and proliferation pathways included prolactin, TNFR2, ceramide, thrombopoietin, and p70S6K signaling. In sharp contrast, differentially expressed genes that were not predicted targets of miRNAs were highly enriched for metabolism and biosynthesis pathways (Fig. 2B).
Figure 2.
Functional analysis of predicted targets of upregulated miRNAs reveals networks associated with immunity, cell survival, and proliferation. (A) Top 30 overrepresented canonical pathways for downregulated gene targets of upregulated miRNAs. Pathways are sorted by score (−log [multiple testing corrected p-value]). A higher score indicates that the pathway is more significantly associated with genes of interest. The line represents statistically significant threshold limit. (B) Significantly overrepresented canonical pathways for non-target differentially expressed genes 0vs48 hours. (C) Top direct interaction network representing 21 downregulated target focus genes, overlaid with predicted major miRNA targeting and functions associated with nodal genes.
Molecular networks were constructed from the miRNA targets with downregulated expression. Network eligibility was based on connectivity to other genes with known interactions. Highly connected genes represent network “nodes” or hubs, where closely connected genes are functionally similar. The top network was comprised of 21 genes significantly enriched for functions linked to T-lymphocyte activation, proliferation and survival (Fig. 2C). The hub genes in this network are PIK3R1 with 6 connections and ATM, PARK2, HIP1R and NCAM1 with 3 connections each. Members of this network are predicted targets for 17 upregulated miRNAs. The central node gene PIK3R1 belongs to the phosphoinositide 3-kinase family that phosphorylates phosphatidylinositol-(4,5)-phosphate (PIP2) to phosphatidylinositol-(3,4,5)-phosphate (PIP3) to regulate cell proliferation, and cytokine production (75). PIK3R1 is a predicted target of 4 miRNAs (miR-155, 21, 218, and 221). Thus, downregulated gene targets of upregulated activation-induced miRNAs are associated with proliferation and cell survival signaling networks. We identified 12 other target gene networks (Supplemental Table S3).
Predicted targets of downregulated miRNAs are upregulated with activation
With respect to the impact of the 20 downregulated miRNAs, we predicted 1347 gene targets out of the total of 3798 activation-induced genes. 487 genes were only targeted by downregulated miRNAs. In contrast, the majority (860) were also targeted by upregulated miRNAs, matching our observations on the apparent importance of combinatorial targeting. We predicted that targets of only downregulated miRNAs should have upregulated mRNA levels. Indeed, 410 (84%) were upregulated >1.5 fold
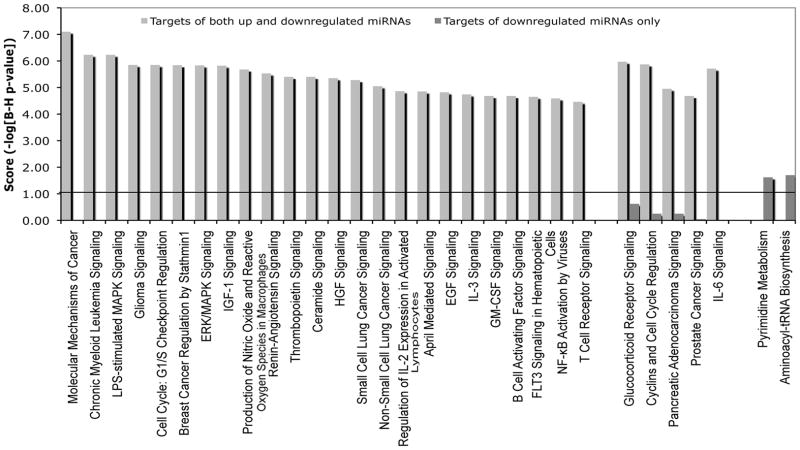
We mapped the functional pathways enriched for these two classes of targets. The pathways linked to only downregulated miRNAs are predominantly cell metabolism and biosynthesis. Since miRNAs targeting these genes are downregulated with activation, the presumed regulation of their targets is removed or at least significantly decreased. The functional role of these genes in metabolism and biosynthesis, much like the genes we found were not targeted by miRNAs, supports the observation that activation-induced miRNAs target a functionally distinct class of genes. In contrast, the pathways linked to combinatorial targeting by both up and downregulated miRNAs are enriched for signaling in immunity, growth and cell proliferation (Fig. 3).
Figure 3.
Functions of genes targeted by downregulated miRNAs. Top 30 overrepresented canonical pathways for gene targets of upregulated as well as downregulated miRNAs and downregulated miRNAs only. Pathways are sorted by score (−log [multiple testing corrected p-value]). A higher score indicates that the pathway is more significantly associated with genes of interest. The line represents statistically significant threshold limit. The pathways linked to only downregulated miRNAs are predominantly cell metabolism and biosynthesis, much like the genes not targeted by miRNAs. In contrast, the pathways linked to combinatorial targeting by both up and downregulated miRNAs are enriched for signaling in immunity, growth and cell proliferation.
Correlating global protein expression to predicted miRNA targets
Our hypothesis was that upregulated miRNAs regulate the immune response and should repress target proteins during T-lymphocyte activation. Target protein repression can be accomplished by either inhibiting translation or enhancing mRNA degradation. While it has been shown that most translational repression is coupled to miRNA-mediated mRNA degradation (12, 14), we considered the possibility that some miRNA targets might be specifically repressed at the translational level without decreases in corresponding mRNAs. A focus exclusively on the downregulation of mRNAs would miss such targets. Therefore, a high-throughput shotgun proteomics protocol (66) was used to analyze global protein expression between 0 and 48 hours.
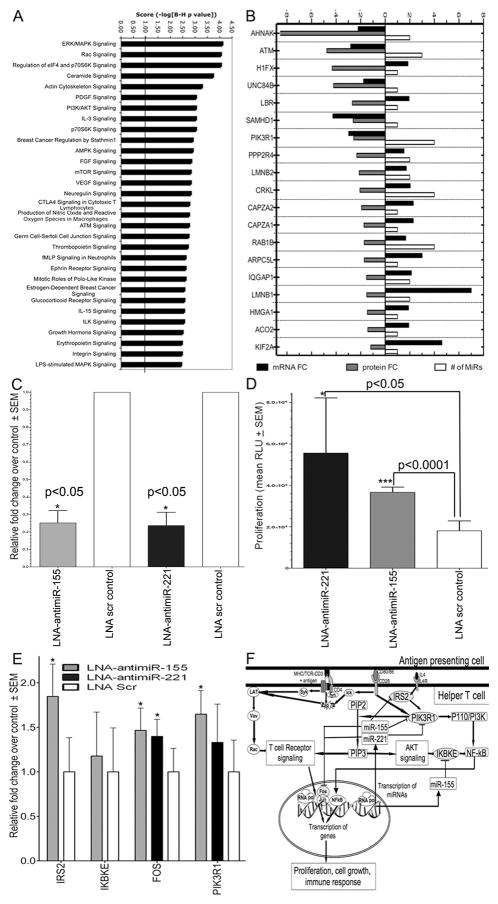
A total of 589 differentially expressed proteins, and another 876 proteins expressed uniquely at T0 or T48 were identified. Correlating the predicted gene targets of miRNAs to expressed proteins, we identified 234 protein-mRNA transcript targets of upregulated miRNAs (Supplemental Table S4). 81 of these proteins had decreased expression at 48 hours. Interestingly, 70 of these protein targets have upregulated mRNA expression. Thus, these proteins are regulated by post-transcriptional mechanisms not coupled to mRNA decay. Functional analysis of these 81 downregulated protein targets revealed significant enrichment for signaling pathways in immune response, cell cycle, growth and proliferation (Fig. 4A). In contrast, the 153 upregulated protein targets were enriched for only 4 pathways: RAN signaling; Glycolysis/Gluconeogenesis; Phenylalanine, Tyrosine and Tryptophan Biosynthesis; and Alanine and Aspartate Metabolism.
Figure 4.
Inhibition of miR-155 (known) and miR-221 (novel), 2 highly upregulated miRNAs, in CD4+ T cells increased proliferation by removing suppression of 4 target genes linked to proliferation and survival. (A) Top 30 overrepresented canonical pathways for downregulated protein targets of upregulated miRNAs. Pathways are sorted by score. (B) Expression of 19 predicted downregulated protein targets associated with top 3 networks. (C) Real-Time qPCR expression of miR-155 and miR-221 in CD4+ T cells nucleoporated with 50nM LNA-antimiR-155 (tan), LNA-antimiR-221 (black), or LNA-scrambled control showing decreased miRNA expression after LNA-antimiR transfection relative to scrambled control, set as 1. Shown are fold changes relative to scrambled control, normalized to U6 snRNA. (D) Increased cell proliferation following knockdown of miR-221 (black) or miR-155 (tan), compared to scrambled control by Cell Proliferation Assay. (E) Real-Time qPCR analysis of predicted targets in CD4+ T cells transfected with LNA-antimiR-155 (tan), LNA-antimiR-221 (black), and LNA scrambled control, presented relative to the expression in scrambled control sample, set as 1. (F) Proposed model of miR-155 and 221 negative-feedback regulation of proliferation following T cell activation. CD3/CD28 co-stimulation induces signaling cascades that result in transcription of genes that promote proliferation, activation and immune response. Upregulation of miR-155 and 221 serves to diminish proliferation and immune response by targeting key proliferative genes such as PIK3R1, IRS2 and IKBKE as well as transcription factor FOS. Error bars in (C–E) represent mean ± SEM of triplicate experiments from 3 donors (n=3). *p < 0.05, ***p < 0.0001 (t-test) compared to scrambled control.
Because networks represent integration of multiple associations, we examined the top 3 networks to identify 19 downregulated proteins as predicted targets of 1 or more miRNAs (Fig. 4B). Within these 19 genes was AHNA, targeted by miR-200b and miR-7, and critical for calcium entry during immune T-lymphocyte activation (76). AHNA was 2.2-fold down at the mRNA level and 8-fold down at the protein level. ATM, targeted by miR-132, miR-18a, and miR-21, regulates cell cycle, promotes normal lymphocyte development and protects from neoplastic transformation (77). ATM was 2.8-fold down by mRNA and 4.8-fold down by protein. PIK3R1 is an adaptor kinase involved in TCR signaling and CD28 co-stimulation with 3-fold mRNA and 2.6-fold protein downregulation. It is a predicted target of 4 upregulated miRNAs (Fig. 2C) and is identified as a downregulated network hub by both gene expression and proteomics.
Several network proteins demonstrated increased mRNA but decreased protein levels (Fig. 4B). LMNB1, a predicted target of miR-218 and miR-7, showed the highest mRNA upregulation by 7-fold but was 1.5-fold down by protein. Inhibition of T-lymphoblast proliferation is associated with downregulation of LMNB1 protein (78). CRKL is targeted by 4 miRNAs and involved in signal transduction through WIP, JNK and Zap70(79). IQGAP1, targeted by 2 miRNAs, regulates lymphocyte cytoskeleton rearrangement in the immune synapse (80). Thus, these multiple lines of evidence linking the top functional networks directly to T-lymphocyte immunity underlines the value of such mapping based on global gene, protein and miRNA expression.
Knockdown of miR-221 and miR-155 increases T-lymphocyte proliferation by removing negative regulation of target genes
The premise of target predictions and functional network mapping is that upregulated miRNAs regulate genes that populate critical networks in T-lymphocyte activation. To validate our approach we knocked down two of the highest upregulated miRNAs: miR-221 and miR-155. Validations were done using purified CD4+ T-lymphocytes to simplify the cell subset composition and reflect our recent finding that CD4+ T-lymphocytes are selectively activated and proliferatively expanded in the early posttransplant period (81). We confirmed that miR-221 and 155 were significantly upregulated in CD4+ T cells at 48 hours of activation (data not shown). While the function of miR-155 has been widely studied in T cells (26, 41), miR-221 associated with cell cycle progression (44), has not been studied in T-lymphocytes.
The impact of inhibiting miR-221 and 155 on T-lymphocyte proliferation was measured by transfecting cells with specific inhibitors or scrambled controls followed by activation. We obtained >60% knockdown of both miRNAs in 3 donors (n=3) as measured by qPCR (Fig. 4C). Significantly increased proliferation resulted from inhibiting either miR-155 or 221 as compared to scrambled control (Fig. 4D).
Among the predicted targets of miR-221 and miR-155, we chose 4 genes for validation by real-time qPCR: PIK3R1, FOS, IRS2, and IKBKE (Fig. 4E). FOS and IKBKE have been previously validated as targets of miR-221 (82) and 155 (83), respectively. At 48 hours after activation the expression of FOS and PIK3R1 is statistically increased by knocking down these two miRNAs. While changes in IKBKE levels were not statistically significant, IRS2, a predicted target of miR-155, increased expression after miR-155 knockdown. Upregulation of target genes following knockdown of miR-221 and 155 is consistent with the evidence above for mRNA repression mediated by miR-221 and/or 155 during T-lymphocyte activation (Fig. 4F).
Discussion
We investigated genome-wide miRNA, mRNA and protein expression following human T-lymphocyte activation. T-lymphocyte activation relies on signaling cascades that create a balance between activation, memory and quiescence. This balance is modulated by mechanisms regulating gene expression including post-transcriptional miRNA regulation. Here we show a unique miRNA signature with a total of 71 differentially expressed miRNAs with 51 upregulated between 0 and 48 hours. This signature comprises 57 miRNAs with no documented roles in T-lymphocyte function. Additionally, our data validated previous findings for a number of miRNAs with known functions in T-lymphocytes: upregulation of miR-155 can regulate the susceptibility of human and murine CD4+ T cells to nTreg cell-mediated suppression (26), the miR-17~92 cluster inhibits T cell activation (30, 32), miR-106a is implicated in IL-10 regulation(31, 33), miR-24 can inhibit cell proliferation by targeting cell-cycle genes(6, 47), and miR-21, upregulated by STAT3, prevents CD4+ T cell apoptosis and is implicated in lymphocyte oncogenesis (18, 53). Also, consistent with the importance of activation-induced miRNA expression, we observed upregulation of 10 miRNA biogenesis/processing machinery genes.
MiRNAs are known inhibitors of gene expression. The challenge is to map miRNAs to specific gene targets and the molecular networks they regulate. To address this challenge we investigated the predictive values of 4 widely used computational algorithms. First, if the results from all the algorithms are compared using a single miRNA hit/seed approach, the predicted targets are poorly correlated between methods to the extent that different methods will report very different results. Second, single hit/seed predictions did not correlate with mRNA repression. In contrast, combinatorial targeting (multiple seeds per target) gave the best predictions. Targetscan conservation predictions with combinatorial binding of 4 or more miRNAs showed the correlation between increased miRNA binding with decreased target gene expression.
By integrating activation-induced miRNA, mRNA and protein expression changes with target predictions, we tested our hypothesis that target genes are involved in regulating immune activation, cell proliferation and survival. Indeed, functional analysis demonstrated that downregulated miRNA targets populated signaling pathways highly enriched for immune response, proliferation, and survival. In contrast, activation-induced genes not predicted to be miRNA targets demonstrated significant enrichment for pathways of metabolism, DNA stability and cell cycle. These novel results reveal that predicted targets of activation-induced miRNAs are functionally distinct from non-target genes and presumably evolved with distinct selection pressures for such regulation. We also hypothesized that some targets might be specifically regulated by post-transcriptional mechanisms not coupled to mRNA decay. Based on our proteomics, we detected a number of such downregulated protein targets despite increased mRNA expression. Thus, inhibition of protein translation is not always coupled to corresponding mRNA degradation.
By investigating connectivity between predicted targets, we identified highly connected genes that function as network nodes, with closely connected genes being functionally similar. The top nodal gene PIK3R1 of one such network is a predicted target of miR-221 and 155, and downregulated at both mRNA and protein levels. This gene is an adaptor kinase involved in TCR signaling and CD28 co-stimulation, and regulates cell growth, proliferation, and T cell cytokine production (75). Thus, functional network analysis underlines the value of the mapping done here based on global gene, protein and miRNA expression.
In validation, we focused on the functional roles of two top upregulated miRNAs in our data: miR-155, widely studied in T cells, and miR-221, not previously associated with T cell function. Knockdown of either miRNA produced a significant increase in proliferation of activated CD4+ T cells, confirming that these two miRNAs actually have anti-proliferative roles during activation. We identified 4 potential targets of miR-155 and/or 221, and mapped a functional network critical to T cell activation (Fig. 4F). In addition to identifying PIK3R1 as a new target gene for miR-155 and 221, we discovered that the transcription factor FOS is also a target of both miR-155 and 221. We identified two more targets of miR-155: the novel IRS2, an adaptor for tyrosine kinases, and a previously verified miR-155 target IKBKE, that regulates NF-κB activation (83). Knockdown of miR-221 and/or 155 increased target mRNA expression for PIK3R1, FOS, and IRS2.
In conclusion, we propose a model where in the course of T-lymphocyte activation by TCR engagement and CD28 co-stimulation, there is a significant upregulation of miRNAs that are critical to this process. These activation-induced miRNAs create a negative-feedback loop to inhibit cell proliferation and regulate cell survival by targeting a series of molecular networks that we have mapped. In parallel, there is also a subset of miRNAs downregulated by activation and 84% of their predicted target genes are shown to be upregulated at the mRNA level. Moreover, there is a functionally-specific class of genes linked closely to the immune response that evolved to be the natural targets of these miRNAs in T-lymphocytes, with functions revealed by molecular networking mapping that are clearly distinct from the activation-induced genes that are not miRNA targets.
Supplementary Material
Acknowledgments
We acknowledge Priscilla Crisler for human blood drawing and Drs. William Soo Hoo and Connie Kohne (Jaden BioScience) for expert assistance in developing the cell proliferation assay.
This work was supported by National Institutes of Health Grants: U19 A1063603 and R01 AI081757.
Abbreviations used in this article
- TCR
T cell receptor
- AICD
Activation-Induced Cell Death
- miRNA
microRNA
- PBMC
Peripheral Blood Mononuclear Cells
- qPCR
quantitative real-time PCR
- MudPIT
The Multi-dimensional Protein Identification Tool
Footnotes
Author Contributions
YAG, SMK, and DRS conceived and designed the experiments and wrote the manuscript; YAG, SMK and AAN performed the experiments; YAG, AAN, and TH analyzed the data; CC, DC, SRH, and JRYIII contributed reagents/materials/analysis tools.
Databank submission of data: The entire set of CEL files from this study is available as Series GSE14352 at the NCBI Gene Expression Omnibus (GEO) site.
References
- 1.Grigoryev YA, Kurian SM, Nakorchevskiy AA, Burke JP, Campbell D, Head SR, Deng J, Kantor AB, Yates JR, 3rd, Salomon DR. Genome-wide analysis of immune activation in human T and B cells reveals distinct classes of alternatively spliced genes. PLoS One. 2009;4:e7906. doi: 10.1371/journal.pone.0007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderson MR, Tough TW, Davis-Smith T, Braddy S, Falk B, Schooley KA, Goodwin RG, Smith CA, Ramsdell F, Lynch DH. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med. 1995;181:71–77. doi: 10.1084/jem.181.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 8.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 9.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 10.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 11.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 12.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 16.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, Fu X, Liu H, Lu L, Wu Y. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R81. doi: 10.1186/ar3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 22.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl HF, Fauti T, Ullrich N, Bopp T, Kubach J, Rust W, Labhart P, Alexiadis V, Becker C, Hafner M, Weith A, Lenter MC, Jonuleit H, Schmitt E, Mennerich D. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS One. 2009;4:e7158. doi: 10.1371/journal.pone.0007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 30.Cox MB, Cairns MJ, Gandhi KS, Carroll AP, Moscovis S, Stewart GJ, Broadley S, Scott RJ, Booth DR, Lechner-Scott J. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One. 2010;5:e12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C, Laschober GT, Lepperdinger G, Sampson N, Berger P, Herndler-Brandstetter D, Wieser M, Kuhnel H, Strasser A, Rinnerthaler M, Breitenbach M, Mildner M, Eckhart L, Tschachler E, Trost A, Bauer JW, Papak C, Trajanoski Z, Scheideler M, Grillari-Voglauer R, Grubeck-Loebenstein B, Jansen-Durr P, Grillari J. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9:291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki K, Kohanbash G, Hoji A, Ueda R, McDonald HA, Reinhart TA, Martinson J, Lotze MT, Marincola FM, Wang E, Fujita M, Okada H. miR-17–92 expression in differentiated T cells - implications for cancer immunotherapy. J Transl Med. 2010;8:17. doi: 10.1186/1479-5876-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci U S A. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 35.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 37.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 38.de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Q, Guo ZY, Li W, Wen WH, Meng YL, Jia LT, Wang J, Yao LB, Jin BQ, Wang T, Yang AG. Human activated CD4(+) T lymphocytes increase IL-2 expression by downregulating microRNA-181c. Mol Immunol. 2010 doi: 10.1016/j.molimm.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, Zhang GM, Feng ZH. miR-142-3p restricts cAMP production in CD4+CD25− T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP, Jr, Sloand EM, Kajigaya S, Young NS. Hematopoietic-specific microRNA expression in human cells. Leuk Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 43.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 44.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasahara Y, Rachid R, Byrne MJ, de la Fuente MA, Abraham RT, Ramesh N, Geha RS. Mechanism of recruitment of WASP to the immunological synapse and of its activation following TCR ligation. Mol Cell. 2002;10:1269–1281. doi: 10.1016/s1097-2765(02)00728-1. [DOI] [PubMed] [Google Scholar]
- 46.Jindra PT, Bagley J, Godwin JG, Iacomini J. Costimulation-dependent expression of microRNA-214 increases the ability of T cells to proliferate by targeting Pten. J Immunol. 2010;185:990–997. doi: 10.4049/jimmunol.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgantas RW, 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 50.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, Calin GA, Liu CG, Andreeff M, Croce CM. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 51.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009;106:15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, Willemze R, Tensen CP, Vermeer MH. MicroRNA-21 Expression in CD4+ T Cells Is Regulated by STAT3 and Is Pathologically Involved in Sezary Syndrome. J Invest Dermatol. 2010 doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 54.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, Weiss MJ. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, Chen Z, Zhang S. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem. 2008;104:805–817. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 56.Sonoki T, Iwanaga E, Mitsuya H, Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia. 2005;19:2009–2010. doi: 10.1038/sj.leu.2403938. [DOI] [PubMed] [Google Scholar]
- 57.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 58.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, Liso A, Diverio D, Mancini M, Meloni G, Foa R, Martelli MF, Mecucci C, Croce CM, Falini B. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, Chen J, Rowley JD, Zeleznik-Le NJ. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 62.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, Calin GA, Liu CG, Bloomfield CD, Andreeff M, Croce CM. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, Goodnow CC, Vinuesa CG. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 64.Weitzel RP, Lesniewski ML, Haviernik P, Kadereit S, Leahy P, Greco NJ, Laughlin MJ. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. 2009;113:6648–6657. doi: 10.1182/blood-2008-09-181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 67.Meloni MA, Galleri G, Carta S, Negri R, Costanzo G, De Sanctis V, Cogoli A, Pippia P. Preliminary study of gene expression levels in human T-cells exposed to cosmic radiations. J Gravit Physiol. 2002;9:P291–292. [PubMed] [Google Scholar]
- 68.Zhang J, Bardos T, Li D, Gal I, Vermes C, Xu J, Mikecz K, Finnegan A, Lipkowitz S, Glant TT. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J Immunol. 2002;169:2236–2240. doi: 10.4049/jimmunol.169.5.2236. [DOI] [PubMed] [Google Scholar]
- 69.Chambers CA. The expanding world of co-stimulation: the two-signal model revisited. Trends Immunol. 2001;22:217–223. doi: 10.1016/s1471-4906(01)01868-3. [DOI] [PubMed] [Google Scholar]
- 70.Gamazon ER, Im HK, Duan S, Lussier YA, Cox NJ, Dolan ME, Zhang W. Exprtarget: an integrative approach to predicting human microRNA targets. PLoS One. 2010;5:e13534. doi: 10.1371/journal.pone.0013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 72.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 73.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soond DR, Bjorgo E, Moltu K, Dale VQ, Patton DT, Torgersen KM, Galleway F, Twomey B, Clark J, Gaston JS, Tasken K, Bunyard P, Okkenhaug K. PI3K p110delta regulates T-cell cytokine production during primary and secondary immune responses in mice and humans. Blood. 2010;115:2203–2213. doi: 10.1182/blood-2009-07-232330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matza D, Badou A, Jha MK, Willinger T, Antov A, Sanjabi S, Kobayashi KS, Marchesi VT, Flavell RA. Requirement for AHNAK1-mediated calcium signaling during T lymphocyte cytolysis. Proc Natl Acad Sci U S A. 2009;106:9785–9790. doi: 10.1073/pnas.0902844106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matei IR, Guidos CJ, Danska JS. ATM-dependent DNA damage surveillance in T-cell development and leukemogenesis: the DSB connection. Immunol Rev. 2006;209:142–158. doi: 10.1111/j.0105-2896.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 78.Schultze FC, Petrova DT, Oellerich M, Armstrong VW, Asif AR. Differential proteome and phosphoproteome signatures in human T-lymphoblast cells induced by sirolimus. Cell Prolif. 2010;43:396–404. doi: 10.1111/j.1365-2184.2010.00690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peterson AC, Marks RE, Fields PE, Imamoto A, Gajewski TF. T cell development and function in CrkL-deficient mice. Eur J Immunol. 2003;33:2687–2695. doi: 10.1002/eji.200324294. [DOI] [PubMed] [Google Scholar]
- 80.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443:462–465. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- 81.Grigoryev YA, Kurian SM, Avnur Z, Borie D, Deng J, Campbell D, Sung J, Nikolcheva T, Quinn A, Schulman H, Peng SL, Schaffer R, Fisher J, Mondala T, Head S, Flechner SM, Kantor AB, Marsh C, Salomon DR. Deconvoluting post-transplant immunity: cell subset-specific mapping reveals pathways for activation and expansion of memory T, monocytes and B cells. PLoS One. 2010;5:e13358. doi: 10.1371/journal.pone.0013358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ichimura A, Ruike Y, Terasawa K, Shimizu K, Tsujimoto G. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol Pharmacol. 2010;77:1016–1024. doi: 10.1124/mol.109.063321. [DOI] [PubMed] [Google Scholar]
- 83.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.