Abstract
NK cell activity is regulated by the integration of positive and negative signals. One important source of these signals for human NK cells is the KIR family which includes both members that transduce positive and those that generate negative signals. KIR3DL1 inhibits NK cell activity upon engagement by its ligand HLA-Bw4. The highly homologous KIR3DS1 is an activating receptor, which has implicated in the outcome of a variety of pathological situations. However, unlike KIR3DL1, direct binding of KIR3DS1+ cells to HLA has not been demonstrated. We analyzed four key amino acid differences between KIR3DL1*01502 and KIR3DS1*013 to determine their role in KIR binding to HLA. Single substitutions of these residues dramatically reduced binding by KIR3DL1. In the reciprocal experiment, we found that the rare KIR3DS1 allotype KIR3DS1*014 binds HLA-Bw4 even though it differs from KIR3DS1*013 at only one of these positions (138). This reactivity was unexpectedly dependent on residues at other variable positions, as HLA-Bw4 binding was lost in receptors with KIR3DL1-like residues at both positions 199 and 138. These data provide the first evidence for the direct binding of a KIR3DS1+ cells to HLA-Bw4, and highlights the key role for position 138 in determining ligand specificity of KIR3DS1. They also reveal that KIR3DS1 reactivity and specificity is dictated by complex interactions between the residues in this region, suggesting a unique functional evolution of KIR3DS1 within the activating KIR family.
Introduction
The Killer Immunoglobulin-like Receptor (KIR) family of Natural Killer (NK) receptors consists of 14 members including receptors with both inhibitory and activating potential (1). This family shows a high degree of variation among populations, including variation in gene content, at the level of allelic polymorphism and the frequency and level of expression (2–4). Several of the KIR are known to bind to Human Leukocyte Antigen (HLA) class I molecules. KIR2DL1 and KIR2DL2/3 recognize HLA-C2 and C1 respectively (5), KIR3DL2 recognizes A*3 and A*11 allotypes (6), KIR2DL4 is reported to bind to HLA-G (7), while KIR3DL1 recognizes HLA-A or -B molecules expressing the Bw4 epitope (8). Activating members of the KIR family, which show a high degree of sequence similarity in the extracellular portion to inhibitory members, also bind to HLA, albeit with much lower affinity (9–11).
Recognition of HLA-Bw4 by KIR3DL1 allotypes is well characterized, with sequence variation in the KIR3DL1 molecule, the Bw4 molecule and the sequence of peptide presented by the Bw4 molecule all influencing the interaction (12–15). There is also growing evidence that KIR3DS1 may recognize HLA-Bw4. Several genetic studies have correlated the presence of both KIR3DS1 and HLA-Bw4 with the outcome of disease (16). In particular in HIV infection, it has been shown that the epistatic interaction of KIR3DS1 and a subgroup of Bw4 carrying isoleucine at position 80 (Bw4-80I) delays progression to AIDS, reducing viral load early as well as leading to a reduction in opportunistic infections later in disease (17, 18). Functional evidence to support the interaction of KIR3DS1 and HLA-Bw4, at least in the context of HIV infection, comes from the report that KIR3DS1+ NK cells can suppress HIV replication and lyse HIV+ target cells in a Bw4-dependent manner (19). However, several attempts to characterize the direct interaction of KIR3DS1 with Bw4 have been unsuccessful – traditional Bw4+ target cell lines are not killed in a KIR3DS1-dependent manner and HLA-Bw4 tetramers do not bind to KIR3DS1-expressing cells, even those tetramers that present HIV-derived peptides (13, 20, 21).
The highly polymorphic KIR3DL1 gene locus is unique in the KIR family in encoding both inhibitory (KIR3DL1) and activating (KIR3DS1) allotypes. The 3D KIR contain three extracellular Ig domains termed D0 (membrane distal), D1 and D2 (membrane proximal). Each of these domains is encoded by a separate exon, exons 3, 4, and 5 respectively of the KIR3DL1 gene (22). The D1 and D2 domains of KIR3DL1 are homologous to the 2 Ig domains of type I 2D KIR (2DL1, 2, and 3) which contain a D1 and D2 but have a pseudoexon in place of exon 3. Type II 2D KIR (2DL4 and 2DL5) contain a D0 and D2 configuration. The structure of the interaction of type I 2D KIR with their HLA-C ligands has been resolved and shown to be dependent on residues from both D1 and D2 (23). While no structure of a 3D KIR, either alone or in association with HLA has been resolved, a sequence comparison with the D1 and D2 domains of KIR3DL1 suggests that many of the predicted residues for 2D KIR are preserved in the apparent interaction of KIR3DL1 with Bw4 (23). The role of D0 is even less well defined, but this domain has been shown to be necessary in the binding of KIR3DL1 to HLA-B*5101 (24). A recent model of the interaction of KIR3DL1 with HLA-Bw4 proposes that D0 combines with D1 and D2 to form the binding interface, with D0 making direct contact with the HLA molecule (25).
Allotypic polymorphism of KIR3DL1 leads to variation in the cell surface staining phenotype with two specific anti-KIR3DL1 antibody clones – DX9 and Z27. Initially this was described as some allotypes (including *005) giving a low intensity staining pattern with both antibodies, while others (including *01502) give a high intensity pattern (26). However, as more allotypes have been described, it has become clear that different allotypes show a broad range of antibody staining levels. Evidence suggests that these differences are a reflection of the level of receptor expressed at the cell surface rather than variation in antibody affinity (27). In one extreme case, the common *004 allotype, which folds poorly, is completely retained intracellularly (28). The KIR3DS1 allotype is not recognized by the DX9 clone, but gives a very low intensity staining pattern with Z27 (13, 29). Both DX9 and Z27 antibodies can prevent binding of KIR3DL1 to its Bw4 ligand, suggesting an overlap between the antibody epitope and the HLA binding site.
KIR3DL1*054 is a recently described allotype of KIR3DL1, which shows a pattern of amino acid substitutions in the D1 domain not seen in other inhibitory allotypes but present in the activating KIR3DS1 (2). As the D1 domain is believed to contribute to both the HLA binding face and the antibody recognition of these molecules we wished to address the role of the amino acids shared between *054 and KIR3DS1 in the regulation of expression levels and in the recognition of HLA. Delineating the role of these variable positions in the binding capacity of these receptors will allow us to better understand the functional consequences of KIR3DL1 polymorphism and, perhaps more importantly, provide insight into the specific requirements of KIR3DS1 for HLA recognition, which to date has proved elusive.
Materials and Methods
Site-directed mutagenesis of KIR3DL1*01502 and KIR3DS1*013
FLAG-tagged (N-terminal) KIR3DL1*01502 or KIR3DS1*013 (referred to here as KIR3DS1) sequences were cloned into the pEF6V5HisTOPO vector. The transmembrane domain of KIR3DS1 was modified to mirror that of KIR3DL1, negating the need for DAP12 association. These sequences were then mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions using the primers outlined in Table I.
Table I.
Mutagenesis Primers
Primer sequences used for the mutagenesis of KIR3DL1 and KIR3DS1 constructs
| Change | Primer Sequence (Forward and Reverse) | ||
|---|---|---|---|
| Amino Acid | Nucleotide | ||
| KIR3DL1 *01502 | G138W | G475T | CTGCACAAAGAGTGGATCTCTAAGGACCCC |
| GGGGTCCTTAGAGATCCACTCTTTGTGCAG | |||
| P163S | C550T | CAATTTCTCCATCGGTTCCATGATGCTTGCCC | |
| GGGCAAGCATCATGGAACCGATGGAGAAATTG | |||
| L166R | T560G | CGGTCCCATGATGCGTGCCCTTGCAGGG | |
| CCCTGCAAGGGCACGCATCATGGGACCG | |||
| KIR3DS1 | W138G | T475G | CTGCACAAAGAGGGGATCTCTAAGGACCCC |
| GGGGTCCTTAGAGATCCCCTCTTTGTGCAG | |||
| S163P | T550C | CAATTTCTCCATCGGTCCCATGATGCGTGCCC | |
| GGGCACGCATCATGGGACCGATGGAGAAATTG | |||
| R166L | G560T | CGGTTCCATGATGCTTGCCCTTGCAGGG | |
| CCCTGCAAGGGCAAGCATCATGGAACCG | |||
| L199P | T659C | CATCGTGGTCACAGGTCCATATGAGAAACCTTCTC | |
| GAGAAGGTTTCTCATATGGACCTGTGACCACGATG | |||
| KIR3DS1 _S163P | R166L | G560T | GGTCCCATGATGCTTGCCCTTGCAGGG |
| CCCTGCAAGGGCAAGCATCATGGGACC | |||
To generate double mutants, mutanted constructs were used as template with primers for mutation of the second position as outlined above except in the case of KIR3DS1 S163P + R166L where, due to the small distance between the residues, a second set of primers was used. The full coding sequence of the resulting plasmids was sequenced to confirm the mutations.
Analysis of KIR3DL1 expression on HEK293T cell lines
HEK293T cells were transfected as previously described (2) using plasmids containing full-length KIR3DL1*01502, KIR3DL1*054, KIR3DS1 or mutants thereof. Expression of the KIR on transfected cells was analyzed by monoclonal antibody staining using FITC-conjugated anti-FLAG (Sigma), PE-conjugated anti-KIR3DL1 mAb clone DX9 (BioLegend) or clone Z27 (Beckman Coulter) and analyzed on a BD FACScan. All experiments included staining with isotype control antibodies to permit accurate positioning of electronic gates for analysis of KIR3DL1 expression.
Analysis of HLA tetramer binding to KIR
The following tetramers were obtained through the NIH Tetramer Facility: B*0801 folded with FLKEQGGL (from HIV nef), A*2402 folded with RYPLTFGW (from HIV nef), B*2705 folded with SRHHAFLFR (from human aggrecan). B*5701 tetramer folded with TSTLQEQIGW (from HIV gag) was a kind gift from Dr Galit Alter (Harvard Medical School, USA).
PE-conjugated HLA tetramers were incubated with transfected cell lines for 30 minutes at 4°C prior to washing and analysis on a BD FACScan. For blocking experiments, cells were pre-incubated with unconjugated Z27 antibody (Beckman Coulter; 10 μg/ml) or mouse IgG1 (Sigma, 10 μg/ml) for 10 minutes at 4°C prior to staining with HLA tetramers. Integrity of the HLA tetramers was verified by staining of a leukocyte immunoglobulin-like receptor subfamily B member 1 (LILRB1) -transfected Ba/F3 cell line (not shown).
KIR3DS1 reporter assay
Stable Jurkat cell lines expressing the extracellular domain of KIR3DS1 fused to the CD3 zeta chain were used in an NFAT reporter assay as described in (13). A reporter cell line for KIR3DS1*014 was generated by mutating KIR3DS1 using the primer pair outlined in Table 1 and stimulated with plate-bound Z27 antibody, the HLA Class I deficient 721.221 cell line or HLA-B*5701 and HLA-B*5801 transfectants. Lysates were harvested and luciferase activity was measured using Promega’s Dual-Luciferase® Reporter Assay System according to the manufacturer’s instructions. Luciferase activity was normalized relative to unstimulated controls.
Results
Novel Allelic Variant Of KIR3DL1 Is Poorly Recognized By Two Anti-3DL1 Antibodies Despite Maintained Cell Surface Expression
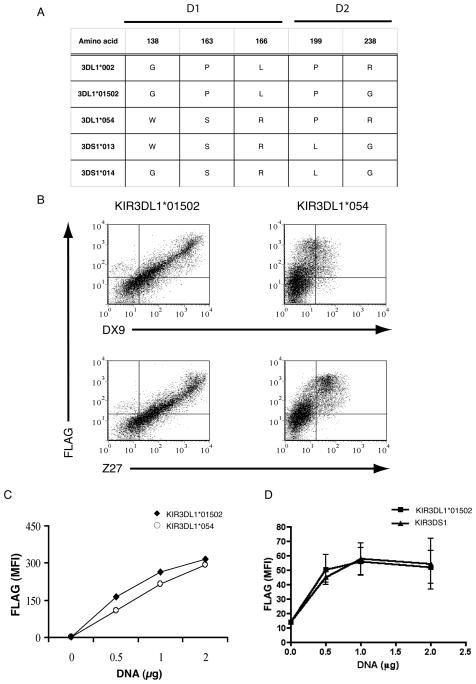
The recently described KIR3DL1 allotype *054 is identical to the highly expressed KIR3DL1*002 allotype except for the D1 domain, encoded by exon 4, where it is identical to KIR3DS1 (Figure 1A). We have recently described that a HIVpositive donor expressing this rare allotype gives a very low intensity staining profile with the DX9 antibody (2). This is possibly a reflection of reduced cell surface expression, poor antibody affinity or a consequence of viral infection. To examine the cell surface expression and antibody affinity of this KIR3DL1 allotype, we transfected HEK293T cells with a construct consisting of the N-terminal FLAG-tagged *054 sequence. These cells were then stained with the anti-KIR3DL1 antibodies DX9 and Z27 and with anti-FLAG and compared to the profile observed for another highly expressed KIR3DL1 allotpye *01502, which has an additional variation at position 238 (Figure 1A). The intensity of FLAG staining observed with the *054 construct was comparable to that seen with the highly expressed *01502 allotype confirming that the *054 allotype is well expressed at the cell surface (Figure 1B, C). However staining with both Z27 and DX9 antibodies was much dimmer than that seen with *01502 allotype, indicating that while both these antibodies recognize *054 they do so with reduced affinity. Thus, while variation in antibody staining for other allotypes of KIR3DL1 has been ascribed to variation in cell surface expression, in the case of *054 reduced intensity of antibody staining is a reflection of reduced affinity of both anti-3DL1 antibodies for this allotype.
Figure 1.
Allelic Variant *054 Is Poorly Recognized By Two Anti-3DL1 Antibodies Despite Maintained Cell Surface Expression. (A) A comparison of selected KIR3DL1 and KIR3DS1 allotypes at key D1 and D2 residues (B) HEK293T cells were transfected with 2 μg of plasmid containing FLAG tagged KIR3DL1*01502 (left panel) or KIR3DL1*054 (right panel) and stained with anti-FLAG and anti-KIR3DL1 DX9 (top panel) or Z27 (bottom panel) antibodies. (C) HEK293T cells were transfected with increasing concentrations of plasmid expressing FLAG tagged KIR3DL1*01502 (closed diamonds) or KIR3DL1*054 (open circles) and stained with anti-FLAG antibody. (D) HEK293T cells were transfected with increasing concentrations of plasmid expressing FLAG tagged KIR3DL1*01502 (closed squares) or KIR3DS1 (closed triangles) and stained with anti-FLAG antibody. MFI of FLAG was calculated by gating on the FLAG positive cells. The results are representative of findings made in at least two independent experiments.
The recognition of KIR3DS1 by Z27 also results in a very low intensity staining pattern, while this allotype is not recognized by DX9. Although it has been suggested that this is due to reduced antibody recognition, and not low expression of KIR3DS1, this has not been formally addressed. To examine this, we compared the intensity of FLAG staining of HEK293T cells transfected with FLAG-tagged KIR3DS1 and KIR3DL1*01502 constructs. We found no difference in the level of FLAG staining seen (Figure 1D), proving that these two receptors are expressed at a similar level at the cell surface and implicating poor recognition of KIR3DS1 by Z27 in the observed low intensity staining.
D1 Residues Differentially Contribute To The DX9 And Z27 Epitope
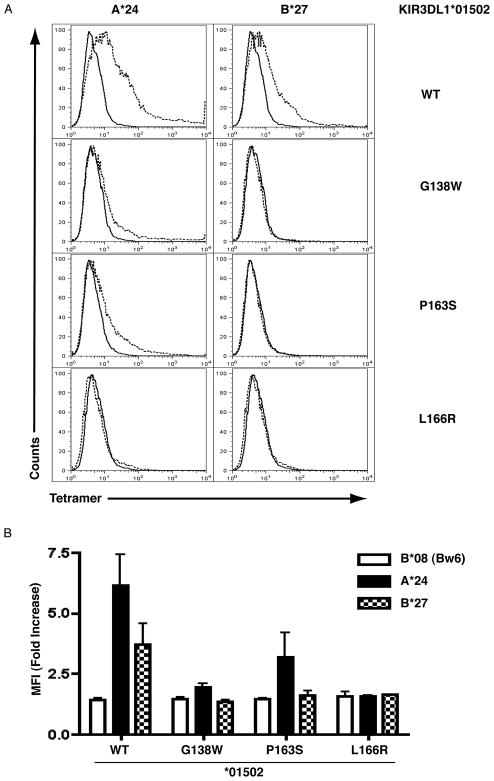
We next examined the role of the D1 amino acid residues in controlling antibody affinity by mutating these amino acid residues in *01502 to the corresponding amino acid found in *054 and 3DS1. Protein levels were titrated by transfection of HEK293T cells with increasing concentration of DNA. Mutation of *01502 at a single residue 138 (glycine to tryptophan, G138W) dramatically reduces recognition by both DX9 and Z27 antibodies (Figure 2A), implicating this residue in the epitope recognized by both antibody clones. The intensity of staining was still greater than that seen with the *054 allotype suggesting that while position 138 plays an important role in the formation of the binding face, position 163, 166 or 238 also contribute. Indeed the combined mutation of residues 163 and 166 also caused a reduction in the affinity of both antibodies for *01502 (data not shown). When we examined the role of 163 and 166 individually, we found that mutation of 166 had a profound effect on the recognition by DX9 but did not appear to contribute substantially to the Z27 epitope (Figure 2B). Single mutation of the residue at position 163 did not influence binding of either antibody. All mutant constructs were carefully analyzed for evidence of intracellular retention/variation in levels of cell surface expression using anti-FLAG antibody. We did not see any consistent difference relative to *01502. To reduce the noise due to slight variations in transfection efficiency, we normalized DX9 and Z27 binding intensity relative to the levels of FLAG staining observed. This ratio gives a reflection of the antibody affinity without influence from slight variations in the level of receptor expression.
Figure 2.
Allelic Polymorphism In KIR3DL1 Uncovers Amino Acid Variations In D1 Domain That Differentially Influences DX9 And Z27 Recognition. (A) HEK293T cells were transfected with increasing concentration of plasmid expressing FLAG tagged KIR3DL1*01502 (squares), KIR3DL1*054 (upright triangle) or KIR3DL1*01502 G138W (inverted triangle) and stained with anti-FLAG, DX9 and Z27 antibodies. The mean fluorescent intensity of the positive population is shown. (B) As above with plasmids containing FLAG-tagged KIR3DL1*01502 (squares), *01502 P163S (upright triangles) and *01502 L166R (inverted triangles). Data are expressed as DX9 or Z27 MFI relative to the anti-FLAG MFI of the total cell population to normalize for variation in cell surface expression due to transfection efficiency [e.g. for *01502 1 μg DNA: FLAG MFI 26, DX9 MFI 129, Ratio 5.0]. (C) HEK293T cells were transfected with increasing concentration of plasmid expressing FLAG tagged KIR3DS1 (closed squares), KIR3DS1 W138G (triangle), KIR3DS1 S163P (open squares), KIR3DS1 R166L (diamonds) or KIR3DS1 L199P (circles) and analyzed as in (A). (D) A comparison of the staining profile of transfected HEK293T cells stained with anti-FLAG and DX9 (top panel) and Z27 (bottom panel) antibodies. Results are representative of findings made in at least two independent experiments.
As mutation of *01502 at residues 138, 166 and 163+166 results in reduced antibody recognition, we wished to examine the role of these amino acid in the binding face of KIR3DS1. These positions were altered in KIR3DS1 to express the residues found in inhibitory KIR3DL1 allotypes. We observed differences in antibody recognition by DX9 and Z27 antibodies (Figure 2C), in the absence of any change in the degree of cell surface expression. Mutation of residue 138 from tryptophan to the glycine residue, which is highly conserved across all KIR receptors (apart from KIR3DS1), results in increased recognition by the Z27 antibody but did not confer recognition by the DX9 antibody. This suggests that this amino acid is an important determinant of the weak reactivity of Z27 seen for 3DS1, but reversion of this position to the common KIR3DL1 residue is not sufficient to allow DX9 recognition. Together with the dramatic loss of DX9 reactivity in the reciprocal mutation of *01502 (*01502 G138W, Figure 2A) these data suggest that while the presence of glycine at position at 138 is necessary for strong DX9 recognition, it is not sufficient. In contrast, substitution at position 166 resulted in recognition (albeit weak) by DX9 but did not significantly affect the affinity of Z27 antibody (Figure 2C), demonstrating the independent effects of these two positions on DX9 and Z27 reactivity. As KIR3DS1 and several revertants do not show any affinity for the DX9 antibody, regardless of good cell surface expression, the DX9 MFI expressed relative to FLAG drops to less than one.
Position 199 Contributes To The Z27 Epitope
KIR3DS1 also contains a unique amino acid at position 199 in the D2 domain, expressing a leucine in place of a proline residue. Mutation of this residue in KIR3DS1 does not alter either cell surface expression or DX9 reactivity but did abrogate recognition by the Z27 antibody (Figure 2D). This result is somewhat unexpected as Z27 shows greater affinity for inhibitory allotypes (expressing a proline at this position) than for KIR3DS1. However in the context of the KIR3DS1 sequence the presence of proline at this position completely abrogated recognition, suggesting that the exact nature of the epitope recognized by Z27 differs between inhibiting and activating forms of this 3D KIR. Indeed when the sequence was further altered to more closely resemble the sequence in inhibitory allotypes, with residue 138 also reverted, Z27 recognition was restored to a level greater than that seen with native KIR3DS1 sequence, but less than that seen with single mutation of position 138 (Figure 2D). The double 138+199 variant was also recognized by DX9, showing that while substitution at either 138 or 199 alone was not sufficient to generate a protein recognized by DX9, the combination of these two positions results in antibody recognition. Interestingly the *054 allotype which is identical to KIR3DS1 in the D1 domain (including W138), but contains a proline at position 199 is recognized, albeit weakly, by DX9. *054 differs from KIR3DS1 at several positions both in the D0 and D2 domains, implicating at least one of these residues in the generation of the DX9 epitope.
Single Amino Acid Substitutions In D1 Of KIR3DL1 Abrogate HLA Binding
HLA-Bw4 binding has been detected with all tested inhibitory KIR3DL1 subtypes; binding to KIR3DS1 however cannot be observed under similar conditions. This mirrors the reactivity seen with the DX9 antibody suggesting that the key amino acids involved in both antibody and HLA recognition are the same. To test this possibility, we examined the ability of 3DL1*01502 constructs to bind to HLA tetramers, including two HLA-Bw4 tetramers (B*2705 and A*2402). Expression of 3DL1*01502in HEK293T cells was found to lead to binding of Bw4 tetramers (Figure 3A), which could be inhibited by blocking with the Z27 antibody (data not shown). Binding to the Bw6 tetramer B*0801 was used as a negative control. Substitution at position 138 (G138W) dramatically reduced the binding of KIR3DL1 to the tested tetramers (Figure 3B). Similarly mutation of position 166 also resulted in a loss of tetramer binding while single mutation of position 163 resulted in a loss of binding to B*2705-SRHHAFLFR tetramer but only reduced the binding to A*2402-RYPLTFGW. Thus it appears that all three D1 residues that differ between KIR3DL1 and KIR3DS1 play a role in HLA recognition. As KIR3DL1*054 contains all three of these substitutions (G138W, P163S, L166R), we predicted that this allotype would not recognize HLA-Bw4 and indeed binding to HLA-B*5701 was lost (data not shown).
Figure 3.
Single Amino Acid Substitutions In The D1 Domain Of KIR3DL1 Abrogates HLA Binding. (A) HEK293T cells were transfected with 2 μg of plasmid containing FLAG tagged KIR3DL1*01502, *01502 G138W, *01502 P163S and *01502 L166R and stained with PE-conjugated HLA tetramers. Dashed line represents staining with HLA-A*2402 (left panel) or HLA-B*2705 (right panel), full line represents staining with control HLA-B*0801. (B) The mean fluorescence intensity of the tetramer stained cells is expressed as fold change over the fluorescence of unstained cells. Data shown are the averages of two independent experiments, error bars represent standard deviation.
Single Amino Acid Reversion Of Position 138 Of KIR3DS1 Permits HLA Binding
Mutation of *01502 suggests that all three 3DS1-specific D1 positions contribute to HLA binding, in particular positions 138 and 166. To examine the influence of these amino acid positions in the context of the KIR3DS1 sequence we expressed KIR3DS1 and single mutants in HEK293T cells and examined HLA reactivity. Despite the evidence from KIR3DL1 that the presence of glycine at position 138 and leucine at position at 166 was necessary for HLA recognition (Figure 3), we found that KIR3DS1 with a single substitution at position 138 was sufficient to confer HLA binding. This binding is seen with both HLA-Bw4 tetramers A*24 and B*57 but specificity for HLA-Bw4 is maintained, as no binding to the HLA-Bw6 tetramer B*08 was detected (Figure 4A). KIR3DS1 transfectants did not bind to any of the tested tetramers (data not shown). The binding to both HLA-A*24 and HLA-B*57 could be blocked by preincubation with the Z27 antibody. Single reversion of either 163 or 166 in KIR3DS1 was not sufficient to allow tetramer binding (data not shown).
Figure 4.
Reversion Of W138 Of KIR3DS1 To The G138 Of KIR3DL1 Allows Binding Of HLA. (A) HEK293T cells were transfected with 2 μg of plasmid containing FLAG tagged KIR3DS1 W138G and stained with PE-conjugated HLA tetramers. Cells were pre-incubated with Z27 or mouse IgG1 (10 μg/ml) for ten minutes prior to staining. (B) Jurkat cell lines expressing KIR3DS1*013 or KIR3DS1*014 extracellular domain fused to the CD3 zeta chain were stimulated with Z27 antibody or 721.221 cell line transfectants at a 1:1 stimulator to responder ratio. Luciferase activity was measured after 18 hours as a measure of NFAT activity and normalized to unstimulated control. Data shown is representative of n=2 (KIR3DS1*013) or n=4 (KIR3DS1*014) independent experiments.
To confirm the recognition of HLA-Bw4 by KIR3DS1 W138G we tested the interaction using a chimeric reporter assay. Jurkat cells stably expressing KIR3DS1 or KIR3DS1 W138G as chimeric receptors with CD3 zeta chain were stimulated with 721.221 cell lines expressing HLA-B*5701 or HLA-B*5801 (both Bw480I allotypes). Although both KIR3DS1 cell lines responded with a clear increase in NFAT upon stimulation with plate-bound Z27 antibody, only KIR3DS1 W138G showed recognition of HLA-Bw4 (Figure 4B).
Complex Interaction Of 3DS1-Specific Residues Combine To Control HLA Recognition
Mutation of position 138 in KIR3DS1 (tryptophan to glycine) resulted in increased affinity of the Z27 antibody, however reverting both 138 and 199 together reduced the affinity back to baseline (Figure 2D). This suggests that the interaction of these amino acid residues in the D1 and D2 domains of the protein determine antibody reactivity, at least in part. To address the possibility that this was also true for HLA binding, we generated a panel of KIR3DS1 mutations in which combinations of position 138, 163, 166 and 199 were reverted to the residue found in KIR3DL1. These constructs were transfected into the HEK293T cells and their ability to bind Bw4 tetramers was examined as before. Reversion of W138G alone was sufficient to allow tetramer binding and combining this with reversion of position 166 (arginine to leucine) gave similar levels of binding (Figure 5A). The combination of 138 + 163 resulted in increased tetramer staining, despite the dominant role of position 166 in KIR3DL1 binding. Interestingly, when reversion of position 138 was combined with position 199 (leucine to proline), HLA reactivity was lost (Figure 5A). Triple revertants of 138+199 with either 163 or 166 similarly gave no HLA reactivity. Only when all the residues in the cluster were reverted to the KIR3DL1 sequence was HLA binding restored in the presence of proline 199 (Figure 5B).
Figure 5.
Complex Interaction Of KIR3DS1-Specific Residues Combine To Control HLA Recognition. HEK293T cells were transfected with 2 μg of plasmid containing FLAG tagged KIR3DS1 or the indicated double, triple or quadruple mutant and stained with PE-conjugated HLA tetramers. The mean fluorescence intensity of the tetramer stained cells is expressed as fold change over the fluorescence of unstained cells. Data shown are the averages of two independent experiments, error bars represent standard deviation.
Discussion
KIR3DL1 is a highly polymorphic KIR with greater than 60 alleles described to date. Allotypic variation is known to influence the pattern of antibody reactivity, HLA-Bw4 binding characteristics and peptide specificity of these KIR3DL1 variants. The KIR3DL1 allele *054 is identical to the common *002 allele at all positions except those in exon 4, where it is identical to KIR3DS1. This new allele is likely to have arisen by a gene conversion event where exon swapping between an existing KIR3DL1 allele (*002) and KIR3DS1 occurred. In our previous study we identified this allele in only one person of Hispanic origin (2), and there are no additional reports in the ImMunoGene Tics (IMGT) database. We have previously shown that NK cells from a *054 donor display a very low intensity staining profile with an anti-KIR3DL1 antibody, which suggested that this allotype may be expressed at a low level on the cell surface, possibly due to intracellular retention as has been shown for other allotypes (2). However, we have shown here that the *054 allotype is well expressed at the cell surface and that the dim staining profile is a reflection of reduced antibody affinity. This is similar to the poor recognition by Z27 of KIR3DS1, which our findings here show is also well expressed at the cell surface. Much of the reduced recognition of *054 relative to *01502 is due to the dramatic amino acid change at position 138 – from the small glycine residue to the bulky tryptophan, which considerably reduced the affinity of both antibody clones. Although the staining pattern of DX9 mirrors that of Z27 for inhibitory KIR3DL1 allotypes, it has been appreciated that the exact epitopes for these two antibodies differ with mutation at residue 200 playing a role in the DX9 but not the Z27 recognition (30). Here we identify a role for position 138 in both antibody epitopes, while DX9 is more heavily dependent on position 166, and Z27 on position 163. Sharma et al reported similar results for the role of position 163 and 166 in a mutagenesis study of KIR3DL1*015, but failed to detect any effect of position 138 (25). While this discrepancy is puzzling, the role of position 138 in both the DX9 and Z27 epitope is further supported in the analysis of 3DS1 mutants in which single residues were reverted to those found in KIR3DL1 allotypes. Interestingly, while in the context of KIR3DS1 position 138 and 199 cooperate to form a DX9 epitope, the double revertant showed weaker Z27 staining than that seen with the single 138. This is surprising given that KIR3DL1 allotypes that are well recognized by Z27 have these residues at position 138 and 199, highlighting the role of positions 163 and 166 in the formation of the binding face. Thus the variation among donors in the profile of antibody staining of KIR3DL1 is attributable to allelic polymorphism leading to variation in the frequency of gene expression (31), mature protein at the cell surface (26, 27) as well as variation in antibody affinity.
In addition to the difference seen between KIR3DL1 and KIR3DS1 in antibody affinity, the more relevant difference in these molecules is the lack of HLA binding to KIR3DS1. Our analysis of KIR3DL1*01502 suggests that, similar to the DX9 epitope, the key residues involved in HLA recognition are 138 and 166, with a lesser role for 163. Single mutation of either 138 or 166 completely abrogated binding to two different Bw4 tetramers, while mutation of position 163 lead to the loss of binding to B*2705 and a reduction in A*2402 binding. The critical individual role of these multiple D1 variants suggests that the rare KIR3DL1 allotype *054, which varies from all other 3DL1 allotypes at these three positions, would not be capable of binding HLA-Bw4. Indeed this proved to be the case, as no binding was detectable to HLA-B*57 tetramer (data not shown). This rare allotype is likely to represent a recently arisen variant of KIR3DL1, generated via an exon 4 swap with KIR3DS1 that shares with KIR3DS1 a lack of broad HLA-Bw4 reactivity. It differs from KIR3DS1 in the potential HLA binding face in possessing proline rather than leucine at position 199 (encoded by exon 5). Based on the evidence we present here that position 199 cooperates with the residues of D1 to determine HLA binding, we predict that *054 will not share the (as yet undefined) specificity of KIR3DS1. Rather KIR3DL1*054 may represent a KIR3DL1 allotype with no affinity for HLA-Bw4, although more extensive analysis would be required to confirm this.
Analysis of the D1 dimorphic residues in the context of KIR3DL1 binding to HLA-Bw4 suggests than any one of D1 changes could be responsible for the lack of detectable HLA binding by KIR3DS1. It was therefore unexpected when reversion of position 138 alone in KIR3DS1 allowed binding to HLA. This variant of KIR3DS1 bound to both A*2402 and B*5701 (both Bw4 80I molecules), but not B*2705 (Bw4 80T) tetramer. This is of particular interest as an allotype of KIR3DS1 (3DS1*014) has been described which carries a glycine in place of tryptophan at this position (32, 33). Thus we have characterized KIR3DS1*014 as an activating 3D receptor with the potential for robust recognition of multiple Bw4 allotypes. Phylogentic analysis of KIR3DL1 and KIR3DS1 sequences by Norman et al (33) reveals that the extracellular domains of KIR3DS1 lineage diverged from KIR3DL1 lineages >3.9 million years ago. This analysis suggests that KIR3DS1*014 arose earlier than KIR3DS1*013, and likely represents an intermediate between the ancestral HLA-binding inhibitory receptors and KIR3DS1*013. KIR3DS1*013 was subsequently subject to positive selection and now represents the overwhelming majority of KIR3DS1 sequences globally. This suggests that there was a strong evolutionary pressure to maintain the KIR3DS1*013 allotype with its restricted Bw4 recognition rather than an activating KIR3DS1*014 with broad Bw4 specificity.
KIR3DS1*014 has been reported in only three populations (English, Tanzanian and Nigerian) and here only at low allele frequency (0.003, 0.006 and 0.003 respectively). Interestingly, 2/3 of these populations are Africian, which show a higher frequency of HLA-Bw4 carriage (and in particular HLA-Bw4 80I), (34) which, as we show here, acts as a ligand for this receptor. It is therefore attractive to speculate that this activating receptor may have in the past provided a selective advantage in these regions, where e.g. high ligand frequency and unique environment of pathogens may have allowed those individuals with KIR3DS1*014 to mount a more effective immune response. The low frequency of this receptor that is now present in these populations may therefore represent a move towards loss of this activating receptor in the absence of that selective pressure, a feature seen in the evolution of other activating KIR (discussed below).
The lack of a well defined ligand for KIR3DS1*013 remains a significant challenge in the field. The ability to detect evidence of an interaction of KIR3DS1 with HLA-Bw4 only in the presence of HIV infection suggests that this recognition may be controlled at the level of peptide presented. Alternatively, factors including the level of HLA class I expression and the presence of additional cofactors or co-receptors may determine the nature of the interaction. Among other members of the KIR family, KIR3DL2 shows a strong dependence on the presented peptide, with binding to A*3 and A*11 appearing to rely on the presence of EBV-derived sequences (6). The interaction of KIR2DS1 with HLA-C2 is influenced by the level of HLA expression, with binding seen only after an increase in expression as a consequence of EBV infection (9).
The low affinity interaction of the KIR2DS molecules with their HLA-C ligands has been mapped to specific amino acid residues in the extracellular domain that differ from those found in the inhibitory counterparts. For KIR2DS1 this difference in binding affinity was mapped to position 70 of the KIR molecule, with substitution of the threonine of 2DL1 for the lysine of 2DS1 diminishing the binding of the long tailed KIR (35). Similarly, the binding of 2DS2 to HLA-C is of much lower affinity than that of the corresponding inhibitory KIR (2DL2/3). This receptor differs from KIR2DL2/3 by only 4 amino acids in the extracellular domain - substitution at position 45 (a tyrosine in 2DS2) for phenylalanine was found to enhance the affinity of the 2DS KIR for HLA-C ligand (10, 11). The restriction of HLA binding of KIR2DS4 (which recognizes some C1 and C2 allotypes as well as A*11) has been attributed to a gene conversion event with KIR3DL2 that resulted in a reduction of affinity of the parental molecule for HLA-C (36). Thus it appears that a common event in the evolution of the activating KIR from their older inhibitory counterparts is the introduction of specific amino acid changes that result in a reduction in affinity for HLA. In contrast, for KIR3DS1 there are at least 4 amino acid changes, over two different exons, which could be responsible for the lack of broad HLA binding by KIR3DS1. While analysis of single changes to the *01502 protein suggests that any one of these changes could be responsible for the “tuning down” of KIR3DS1 binding, the presence of additional variant positions that impact HLA recognition, suggest that the driving force by which KIR3DS1 was generated from KIR3DL1 may differ from that of the HLA-C recognizing 2D KIR. Studies of KIR3DS1 revertants in which combinations of two or more of these variant positions are mutated suggest that the role of individual amino acids in the recognition of HLA by KIR3DS1 may also differ from KIR3DL1. Thus, while proline at 199 appears to be necessary for KIR3DL1 recognition of HLA (25) it prevents HLA binding by KIR3DS1 W138G mutants. One intriguing interpretation of these data is that the HLA recognition by KIR3DS1 does not simply mirror that of KIR3DL1 but with lower affinity, as is the case of KIR2DS1 and KIR2DS2. Rather, the additional amino acid changes may reflect a reprogramming or restriction of the affinity for HLA-Bw4 thereby limiting reactivity. Analysis of the evolution of activating KIR from their inhibitory KIR suggests that this is a recurrent process with cycles of gain and loss in the absence of a strong, persistent evolutionary pressure to maintain activating KIR (37). This is consistent with data from disease association studies that suggest a protective role of activating KIR under certain circumstances, including viral infection, but a deleterious one under others (e.g. in autoimmune conditions). Thus, for KIR3DS1 the presence of multiple functionally important amino acid changes relative to KIR3DL1 may be the result of multiple pressures on KIR3DS1 to preserve a protective function, while restricting reactivity to prevent unwanted host damage that might be predicted with KIR3DS1*014. This is a particularly important consideration in our understanding of the role and evolution of activating KIR, as well as in the search for a functional interaction between KIR3DS1 and its ligand, given the growing number of disease outcomes that are influenced by the presence of the KIR3DS1 gene.
Acknowledgments
The authors thank Drs Peter Parham and Galit Alter for the generous gift of reagents. We are grateful to Kathleen Noer and Roberta Matthai for cell sorting. We also thank Dr Colm O’hUigin for critical reading of the manuscript.
This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations used in this paper
- KIR
Killer-cell immunoglobulin-like receptor
References
- 1.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 2.Thomas R, Yamada E, Alter G, Martin MP, Bashirova AA, Norman PJ, Altfeld M, Parham P, Anderson SK, McVicar DW, Carrington M. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 4.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 5.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, Conte R, Di Donato C, Parham P, Moretta L. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci U S A. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 12.Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, Long EO. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 14.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 16.Carrington M, Martin MP, van Bergen J. KIR-HLA intercourse in HIV disease. Trends Microbiol. 2008;16:620–627. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 18.Qi Y, Martin MP, Gao X, Jacobson L, Goedert JJ, Buchbinder S, Kirk GD, O’Brien SJ, Trowsdale J, Carrington M. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2:e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillespie GM, Bashirova A, Dong T, McVicar DW, Rowland-Jones SL, Carrington M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 22.Wilson MJ, Torkar M, Trowsdale J. Genomic organization of a human killer cell inhibitory receptor gene. Tissue Antigens. 1997;49:574–579. doi: 10.1111/j.1399-0039.1997.tb02804.x. [DOI] [PubMed] [Google Scholar]
- 23.Boyington JC, Brooks AG, Sun PD. Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol Rev. 2001;181:66–78. doi: 10.1034/j.1600-065x.2001.1810105.x. [DOI] [PubMed] [Google Scholar]
- 24.Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 25.Sharma D, Bastard K, Guethlein LA, Norman PJ, Yawata N, Yawata M, Pando M, Thananchai H, Dong T, Rowland-Jones S, Brodsky FM, Parham P. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 27.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 29.Pascal V, Yamada E, Martin MP, Alter G, Altfeld M, Metcalf JA, Baseler MW, Adelsberger JW, Carrington M, Anderson SK, McVicar DW. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 30.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J Exp Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crum KA, Logue SE, Curran MD, Middleton D. Development of a PCR-SSOP approach capable of defining the natural killer cell inhibitory receptor (KIR) gene sequence repertoires. Tissue Antigens. 2000;56:313–326. doi: 10.1034/j.1399-0039.2000.560403.x. [DOI] [PubMed] [Google Scholar]
- 33.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 34.Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 35.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 36.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, Parham P. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]