Abstract
HLA-DM (DM) catalyzes CLIP release, stabilizes MHC class II molecules and edits the peptide repertoire presented by class II. Impaired DM function may have profound effects on antigen presentation events in the thymus and periphery that are critical for maintenance of self-tolerance. The associations of the HLA-DQ2 (DQ2) allele with celiac disease and type 1 diabetes mellitus have been appreciated for a long time. The explanation for these associations, however, remains unknown. We previously found that DQ2 is a poor substrate for DM. Here, to further characterize DQ2-DM interaction, we introduced point mutations into DQ2 on the proposed DQ2/DM interface in order to restore the sensitivity of DQ2 to DM. The effects of mutations were investigated by measuring the peptide dissociation and exchange rate in vitro, CLIP and DQ2 expression on the cell surface and the presentation of α-II-gliadin epitope (residues 62-70) to murine, DQ2-restricted T cell hybridomas. We found that the three α chain mutations (α+53G, α+53R or αY22F) decreased the intrinsic stability of peptide-class II complex. More interestingly, the α+53G mutant restored DQ2 sensitivity to DM, likely due to improved interaction with DM. Our data also suggest that α-II-gliadin 62-70 is a DM-suppressed epitope. The DQ2 resistance to DM changes the fate of this peptide from a cryptic to an immunodominant epitope. Our findings elucidate the structural basis for reduced DQ2-DM interaction and have implications for mechanisms underlying disease associations of DQ2.
Introduction
Epidemiological studies have revealed the now well-established association between the HLA-DQ2 allele (A1*0501/B1*0201) and multiple autoimmune diseases, including celiac disease (CD) (1) and type 1 insulin-dependent diabetes (T1D) (2). CD is induced by the ingestion of gluten, a component derived from wheat, barley and rye. It is characterized by T and B cell-mediated autoimmunity that results in inflammation of the small intestine (3). About 90-95% of CD patients express the HLA-DQ2 allele, and most of the remaining patients carry the HLA-DQ8 allele (4). Although the strong associations between DQ2 and CD have been appreciated for a long time, the explanations for these genetic links remain unclear.
The process of peptide loading onto MHC II in APC endosomes is facilitated by several molecules: invariant chain (Ii), human leukocyte antigen DM (HLA-DM), and its modulator, HLA-DO (DO) (5, 6). The newly synthesized MHC class II αβ heterodimers are assembled with trimers of invariant chain in the ER (7, 8). This association prevents premature ligand binding to MHC II in the ER and guides the export of (αβ)3Ii3 complexes through the Golgi apparatus, targeting late endosomal and lysosomal vesicles (9-13). In these compartments, Ii is progressively degraded until only small Ii fragments called CLIP (class-II-associated invariant chain peptide) remain bound in the peptide-binding groove of MHC II (14, 15). The subsequent release of CLIP from MHC II and exchange for antigenic peptide are both mediated by DM (16-18). In addition, the peptide repertoire presented by MHC class II is modified by DM, a process referred to as “peptide editing”, whereby unstable peptides are removed by DM and peptides with high kinetic stability resistant to this process are selected for class II presentation (18). After endosomal editing by DM, MHC II molecules loaded with peptide are transported to the cell surface for inspection by CD4+ T cells.
In recent work, we found that DQ2 molecules interact with HLA-DM atypically (19). First, CLIP dissociation and peptide exchange of DQ2 are insensitive to DM (19). Also, compared to DQ1, DQ2 is resistant to DM-mediated conformational changes and the surface abundance of DQ2 is not affected by coexpression of DM (19). These results imply inefficient interaction between DQ2 and DM. This atypical interaction may contribute to the mechanism by which HLA-DQ2 confers susceptibility to autoimmune disease. To elucidate the structural basis of the DQ2 resistance to DM effect, we introduced several point mutations into DQ2 on the proposed DM-interacting site to identify one or more mutations that are able to restore DQ2 susceptibility to DM. We found that insertion of a glycine residue into the 53 position of DQ2 α chain not only alters the intrinsic stability of complexes with several DQ2-binding peptides, but also improves the DQ2-DM interaction. Our study suggests the interface residues important for DQ2-DM interaction and provides additional insight into understanding the association of DQ2 with autoimmunity.
Methods
Cell lines
The T x B hybrid cell lines, T2 (MHC II-/DM−) and T2DM (MHC II-/DM+) were obtained from the laboratory of Dr. Lisa Denzin, Sloan-Kettering Institute, Memorial Sloan-Kettering Cancer Center, New York. They were established as described (20, 21). The T cell hybridoma cells, HH8 and DB4, were generous gifts from Prof. James McCluskey, University of Melbourne, Parkville, Australia (22). Soluble DQ2 was expressed in S2 D. melanogaster insect cells using a baculovirus expression vector system (23).
Peptides
Biotin-labeled peptides, MHC Ia 49-63 (APWIEQEGPEYWDQE) and MB 65KD Hsp 243-255 (65-kD heat-shock protein of Mycobacterium bovis, KPLLIIAEDVEGEY) were synthesized by AnaSpec Inc (San Jose, CA). Unlabeled MHC Ia 49-63 was a kind gift from Dr. Alessandro Sette at La Jolla Institute for Allergy & Immunology, CA. The peptide α-gliadin 57-73 Q65E (QLQPFPQPELPYPQPQS) was a generous gift from Prof. James McCluskey, University of Melbourne, Parkville, Australia.
cDNA constructs and transfection
The plasmids expressing WT soluble DQ2α and β, pRmHa3-DQA1*0501 and pRmHa3-DQB1*0201, were obtained from the laboratory of Dr. Ludvig Sollid, University of Oslo, Norway. Site-directed mutagenesis was done by the GeneTailor Site-Directed Mutagenesis System from Invitrogen. The primer sets used to make mutants are summarized in Table 1. The plasmids pRmHa3-DQA1*0501 and pRmHa3-DQB1*0201 were co-transfected with the neomycin-resistance plasmid, pUChsneo, into S2 cells using calcium phosphate. The positively transfected cells were selected with neomycin (1.5mg/ml) for 4 weeks.
Table 1.
Primers used for site-directed mutagenesis
| mutants | Primers (the upper one is sense primer; the lower one is anti-sense primer) |
|---|---|
| αY22F | 5′-CTTACGGTCCCTCTGGCCAGTTCACCCATGAAT-3 ′ |
| 5′-CTGGCCAGAGGGACCGTAAGACTGGTACA-3′ | |
| αQ50F | 5′-GGTGTTTGCCTGTTCTCAGATTCTTTAGATTTGA-3 ′ |
| 5′-TCTGAGAACAGGCAAACACCAGACAGTCTC-3′ | |
| α+53G | 5′-CCTGTTCTCAGACAATTTAGAGGATTTGACCCGC-3 ′ |
| 5′-TCTAAATTGTCTGAGAACAGGCAAACACCAG-3′ | |
| α+53R | 5′-CCTGTTCTCAGACAATTTAGAAGGTTTGACCCGC-3 ′ |
| 5′-TCTAAATTGTCTGAGAACAGGCAAACACCAG-3′ | |
| βT98E | 5′-TGCAGCGGCGAGTGGAGCCCGAAGTGACCATCT-3 ′ |
| 5′-GGGCTCCACTCGCCGCTGCAAGGTCGTGCG-3′ | |
| βT98K | 5′-TGCAGCGGCGAGTGGAGCCCAAGGTGACCATCT-3 ′ |
| 5′-GGGCTCCACTCGCCGCTGCAAGGTCGTGCG-3′ |
To stably express full length DQ2 in T2/T2DM cells, cDNA of WT DQ2α was amplified by PCR using sense primer 5′-CGCGATCCATGATCCTAAACAAAGCT-3′ and antisense primer 5′-ACGCGTCGACTCACAAGGGCCCTTGGTG-3′ to add a BamHI site to the 5′ end and a SalI site to the 3′ end. The digested DQ2α was cloned into the retrovirus vector, PBMN-ZIN-neo, which was cut with the same enzymes. Full length cDNA of WT DQ2β was amplified with the primer set: 5′-CCCAAGCCTATGTCTTGGAAAAAGGCT-3′ and 5′-ATTTGCGGCCGCTCAGTGCAGGAGCCCTTT-3′ to add a HindIII site and EcoRI site to 5′ and 3′ end. The PCR product was cut with HindIII and EcoRI, and ligated into the retrovirus vector, PBMN-LZRS-puro, that was digested with the same enzymes. Mutants were generated with the same primer sets and system mentioned above.
The α chain of DQ2 was first transfected into Phoenix Retroviral System, and the supernatant was collected to infect T2 or T2DM cells. Forty-eight hours after infection, the positively infected cells were treated with neomycin (1mg/ml) for one week. The DQ2β chain was then introduced into cells expressing α chain by the same approach. Finally, cells expressing DQ2 dimer on the surface were enriched by PE-conjugated anti-DQ2 Ab (Ia3, BD bioscience) and anti-PE MACS microbeads (Miltenyi biotec).
In vitro peptide loading assay
Soluble DQ2 was engineered to have a CLIP peptide tethered to the β chain via a thrombin cleavable linker and an Acid-Base Zipper replaced the transmembrane domains. Soluble DQ2 protein was expressed in S2 cells and purified with anti-Flag affinity M2 column. sDQ2 was predigested with thrombin for 2 hours at room temperature to free the tethered CLIP peptide. Digested sDQ2 was then incubated with biotin-labeled peptides, MHC Ia 49-63 or MB 65kD Hsp 243-255, with or without DM, at pH 4.7, 37°C for 2 hours to allow peptide loading into DQ2. The reaction buffer contained 50mM NaOAc, 150mM NaCl, 1% BSA, 0.5% NP40, and 0.1% NaN3. After 2 hours of incubation, the reaction was stopped by neutralizing pH with a buffer containing 50mM Tris-HCl, 150mM NaCl, 1% BSA, 0.5% NP40, and 0.1% NaN3 with pH 8.6. DQ2-peptide complexes (100ul) were transferred to a 96-well plate and captured by anti-DQ2 Ab, SPVL3, for one hour at room temperature. The loaded biotin-peptide was detected by Streptavidin-Europium. The europium emission fluorescence was measured in a time-resolved fluorometer (EG&G Wallac, Gaithersburg, MD) after enhancer was added.
In vitro peptide dissociation assay
Thrombin-digested sDQ2 was loaded with the peptide biotin-MHC Ia 49-63 at 37°C overnight. MHC Ia/DQ complexes were separated from free peptide using Sephadex G50-Superfine spin columns. Unlabeled MHC Ia 49-63 peptide was added to a concentration five times higher than the biotin-labeled peptide to prevent rebinding of dissociated biotinylated MHC Ia 49-63. The dissociation was then allowed to proceed at 37°C with or without soluble HLA-DM for various times. At each time point, reactions were stopped by adding two volumes of ice-cold neutralization buffer (50mM Tris-HCl, 150mM NaCl, 1% BSA, 0.5% NP40, and 0.1% NaN3, pH 8.6). Neutralized reaction mixtures (100 μl) were transferred to SPVL3-coated, blocked flat-bottom 96-well microtiter plates. After 2 hours of capture at room temperature, plates were washed in PBS containing 0.05% Tween-20. DQ-bound biotinylated peptide was detected by addition of Streptavidin-Europium. After 1 hour of incubation and further washes, enhancement solution (100 μl) was added, and time-resolved fluorescence was detected using a fluorescence plate reader (EG&G Wallac, Gaithersburg, MD). The signals of each time point was normalized to the values of time zero which was considered as one and single-exponential decay curves were fitted with Prism5 to calculate half-lives (t1/2).
Flow cytometry
Surface DQ2 was detected by anti-DQ2 Ab, SPLV3, Ia3 or 2E11.12 and the secondary Ab, PE-conjugated goat anti-mouse IgG (BD bioscience). In the double staining of DQ2 and CLIP, surface DQ2 was detected with PE-conjugated Ia3 and CLIP was stained by FITC-conjugated anti-CLIP Ab (CerCLIP, BD bioscience). For intracellular staining of DM, cells were first fixed and permeabilized using the Cytofix/Cytoperm kit (BD bioscience) and then stained with PE-conjugated anti-DM Ab (MaP.DM1, BD bioscience). Cells were analyzed using a FACScan flow cytometer (BD Biosciences) and data were analyzed using FlowJo software (Tree Star).
Processing of gluten
Gluten (750 mg, G5004, Sigma) was added into 50 ml of 0.01M HCl (pH 2.0) and was digested with 30 mg of pepsin (P6887, Sigma) for 1 hr at 37°C. Na2HPO4 (175 mg) was added after pepsin digestion and pH was adjusted to 6. For 1 ml of gluten suspension, 7.8 ul of trypsin (50 mg/ml, T8802, Sigma) and 7.8 ul of chymotrypsin (50 mg/ml, C4129, Sigma) were added for further digestion at 37°C for 2h. Both trypsin and chymotrypsin were prepared in 25 mM Na2HPO4, pH 6.0. Enzymes were inactivated by heating for 5 mins at 95°C. After inactivation, samples were spun down quickly to pellet the undigested gluten. The supernatant was collected and dialyzed against water with a membrane with a molecular mass cut-off of 12-14 kDa (Spectrum Laboratories) at 4°C overnight. The resultant high-molecular-weight fraction of digested gluten (HMWF PT-gluten) was concentrated with Amicon Ultra Centrifugal Filters with a cut-off of 10 kDa. Finally, PT-gluten was incubated with 100 ug/ml of guinea pig transglutaminase (stock 10 mg/ml, T-5398; Sigma) at 37°C for 2h with 1mM CaCl2 for deamidation.
T cell proliferation assay
Irradiated (8,000 rads) T2 or T2DM cells expressing DQ2 WT or mutant (100,000) were incubated with the titrated α-gliadin 57-73 Q65E or PT-gluten and T cell hybridoma, DB4 or HH8, (100,000) for 16-24 hours. After incubation, medium was collected and the IL-2 in the supernatant was measured by mouse IL-2 ELISA kit (BD Bioscience).
Results
Rationale for choice of DQA*0501 and DQB*0201 mutations
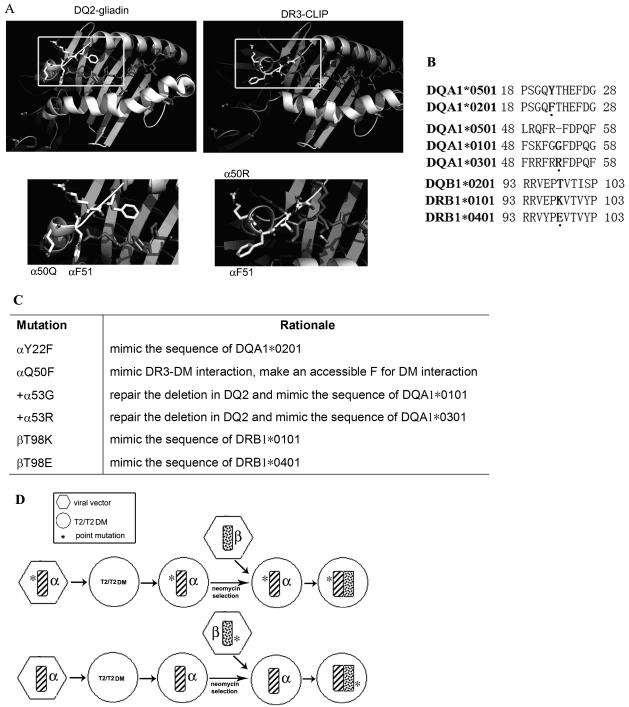
We have previously mapped the interface on DR3 that interacts with DM (24) as well as the interface on DM that interacts with DR3 (25). Based on these studies and available crystal structures, we know that the solvent accessible F51 in DRA1*0101 is critical for DM interaction. As indicated in Figure 1A (magnified view in bottom-right panel), F51 in DR3 α chain protrudes out and provides a putative interaction site for DM. In contrast, its structural homologue in DQ2 (DQA1*0501, DQB1*0201) is protected by a steric block (Fig. 1A, magnified view in bottom-left panel). To test whether this is the basis of reduced DM-DQ2 binding, we mutated the amino acid Q50 in DQA1*0501 to F. In addition, DQA1*0501 contains a deletion of residue 53, which is not observed in DQ1 (DQA1*0101, DQB1*0501) or DQ8 (DQA1*0301, DQB1*0302) (Fig. 1B, middle panel), both of which are susceptible to DM function. To study the role of α53 in DM interaction, the deletion was repaired with the homologous residues from DQA1*0101 (G) or DQA1*0301 (R). Either of these two mutations is expected to reorient the DM-contact region in DQ2 and improve DM-DQ2 interaction. Our previous studies of DR-DM interaction also show that DR1 and DR4 are both better substrates for DM than DR3, because their β chains contain polymorphisms that favor DM binding, with K (DRβ*0101) or E (DRβ*0401) at position β98 (Fig. 1B, bottom panel and R. Doebele and EDM, unpublished data). Based on these findings, point mutation T98K or T98E was introduced into the β chain of DQ2, representing the polymorphism in DR1 or DR4. DQ2.2 (DQA1*0201, DQB1*0202) is another HLA-DQ2 molecule that has high homology to DQ2.5 (DQA1*0501, DQB1*0201), but does not confer risk for celiac disease. A recent study has shown that DQα22 polymorphism controls the kinetic stability of DQ2-peptide complexes (26). Replacement of αY22 in DQ2.5 with the homologous residue of DQ2.2 (F) decreases the overall peptide binding stability and inhibits T cell proliferation in response to gliadin peptide (26). This mutant (αY22F) is also included in our study to investigate its influence on DM-DQ2 interaction. The mutations that were chosen are summarized in Figure 1C.
Figure 1.
Rationale for choice of DQA1*0501 and DQB1*0201 mutations. (A) Ribbon diagrams (based on crystal structures) of the peptide loading grooves of HLA-DQ2/gliadin (left) and HLA-DR3/CLIP (right). The structures indicated by highlighted boxes are magnified and shown in the bottom panels. (B) The top panel is amino-acid alignment of α allele of DQ2.5 (DQA1*0501) and α allele of DQ2.2 (DQA1*0201). The middle one is the alignment of DQA1*0501 and A1*0101 and A1*0301. Alignment of DQB1*0201, B1*0101 and B1*0401 is shown in the bottom panel. Amino acids of interest are bolded and highlighted by the dots underneath. (C) Summary of rationale for choice of DQA1*0501 and DQB1*0201 mutations. (D) Strategies used to construct T2/T2DM cell lines expressing DQ2 WT or mutant. DQ2 α chain was introduced into T2/T2DM cells using retroviral transduction, followed by subsequent retrovirus-mediated β chain transfer. Cells expressing surface DQ2 dimer were isolated by MACS. *denotes the site-directed mutation.
Both DQ2α and β chain of WT and mutants were sequentially transfected into T2DM− (T2) and T2DM+ (T2DM) cells using retroviral transduction (Fig. 1D). Neither T2 nor T2DM expresses endogenous class II due to a large homozygous deletion in the MHC. Cells expressing DQ2 dimers on the surface were enriched by MACS microbeads. We first screened the mutants by FACS to identify those associated with reduced surface CLIP/DQ2 ratio in the presence of DM, indicating a better DM-DQ2 interaction. Both β chain mutations (βT98K and βT98E) showed comparable CLIP/DQ2 ratio to WT (data not shown). Therefore, we focused on the α chain mutations in the following studies.
Expression of the soluble form of WT and mutant DQ2 molecules
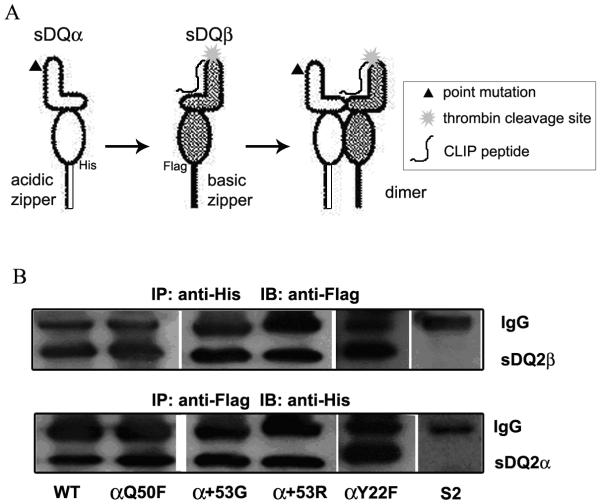
WT and mutant soluble DQ2 (sDQ2) molecules, lacking transmembrane and cytoplasmic domains, were expressed in S2 cells. These molecules were engineered with a CLIP peptide tethered to the β chain N-terminus using a linker containing a thrombin cleavage site and with C-terminal epitope tags followed by complementary acidic and basic zippers (Fig. 2A). sDQ2 proteins were affinity purified from cell culture supernatants, and dimerization was confirmed by co-precipitation of FLAG-tagged β chain with His-tagged α chain (Fig. 2B, top panel), and vice versa (Fig. 2B, bottom panel). Lysate from untransfected S2 cells was included as negative control.
Figure 2.
(A) Schematic representation of site-directed mutations introduced in recombinant soluble DQ2 molecules. The mutated α chain was co-transfected with WT β chain into S2 insect cells. Complementary acidic and basic zippers with upstream epitope tags (His or Flag) were added at the C-terminal ends of the chains. A CLIP peptide was tethered to the N-terminus of the β chain using a linker with an internal thrombin cleavage site. (B) Dimerization of DQ2 WT or mutant molecules was tested by co-immunoprecipitation. sDQ2 α chain was precipitated by anti-His tag antibody (Ab) and the co-precipitation of β chain was detected by Ab against Flag epitope (top panel). Cell lysate from untransfected S2 cells was included as a negative control. The reverse co-precipitation (precipation of β chain by anti-Flag and western blot with anti-His for α chain) is shown in the bottom panel.
Intrinsic peptide exchange was increased by α+53G, α+53R or αY22F
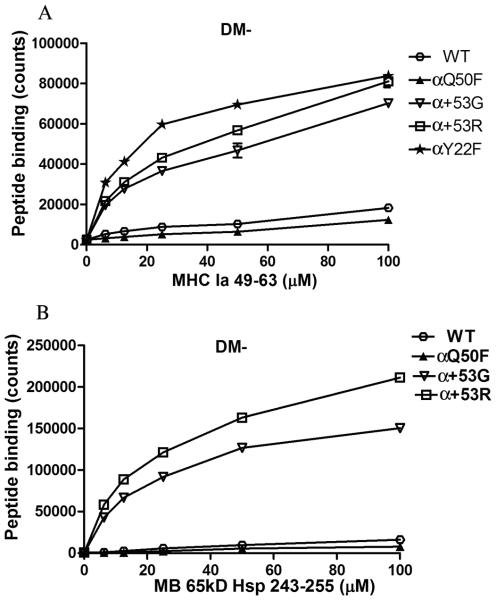
To investigate the effect of mutations on intrinsic peptide exchange, we performed a peptide loading assay using affinity purified sDQ2 molecules and biotin-labeled peptides that have high binding affinity to DQ2. sDQ2 (200 nM) cleaved by thrombin was incubated with increasing amounts of biotinylated peptide (6.25-100 μM) at pH 4.7, 37°C for 2 hours, allowing peptide loading into sDQ2. sDQ2-peptide complexes were then captured by anti-DQ2 Ab, SPVL3 and the biotinylated peptide was detected by streptavidin-europium. Peptide exchange in the absence of DM was significantly increased in α+53G, α+53R or αY22F compared to WT and αQ50F for MHC Ia 49-63 (Fig. 3A). The intrinsic peptide exchange of these three α chain mutants was increased about 3~6 fold compared to that of WT. Similar results were seen for α+53G and α+53R when another high binding affinity peptide, MB 65KD 243-255, was tested (Fig. 3B). The spontaneous peptide exchange of αQ50F is comparable to WT. This finding indicates that the mutants of α+53G, α+53R or αY22F decrease the intrinsic stability of DQ2-CLIP complexes, facilitating exchange.
Figure 3.
Increased intrinsic peptide exchange in α+53G, α+53R or αY22F. WT or mutated sDQ2 (200 nM) was first cleaved by thrombin to free the tethered CLIP peptide and then incubated with increasing amounts of biotinylated high-affinity peptide (6.25-100 uM), MHC Ia 49-63 (A) or MB 65KD 243-255 (B). The sDQ2-peptide complexes were captured by anti-DQ2 Ab, SPVL3 and the loaded peptides were detected by streptavidin-europium. Background from peptide alone at each dose was subtracted. Each condition was run in duplicate and each experiment was repeated at least three times. Mean and SD from one representative experiment was shown.
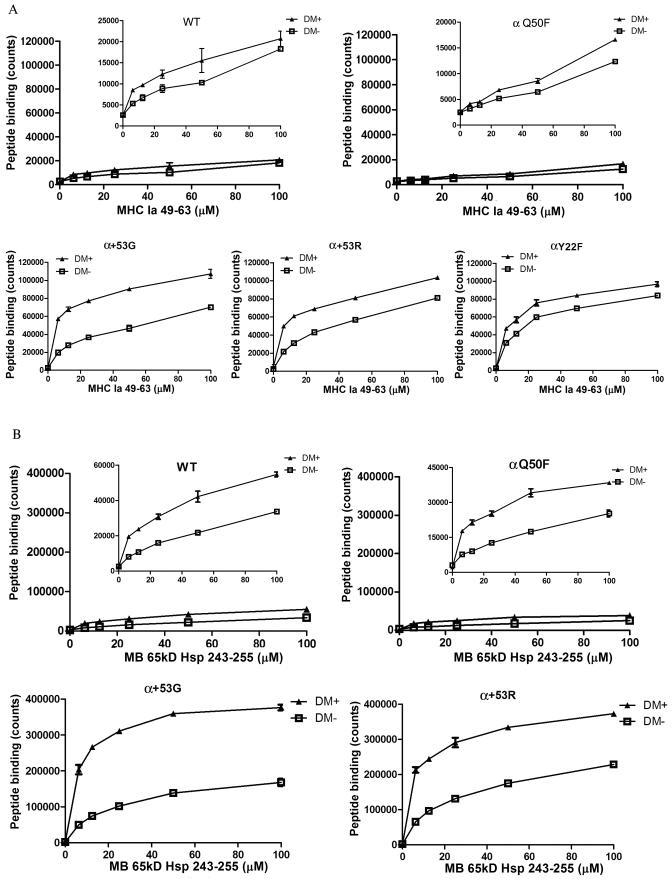
DM-mediated peptide exchange was also increased in α+53G, α+53R or αY22F
To measure the net effect of DM on peptide exchange, sDQ2 (200 nM) was incubated with increasing amounts of peptide (6.25-100 uM) in the absence or presence of 2.4 μM of DM. The results with or without DM were plotted together: the distance between the two curves represents the DM effect. As indicated in Figure 4A, DM enhanced the exchange of CLIP for MHC Ia 49-63 in sDQ2 WT and all the mutants, but to different extents. For WT and αQ50F, the addition of DM led to a minimal increase in peptide exchange, which was consistent with our previous finding that DQ2 was insensitive to DM function (19). For α+53G, α+53R or αY22F, however, the net DM effect was significantly higher. Among these three mutants, α+53G had the most evident difference between DM- and DM+. Similar patterns were observed for another peptide, MB 65KD 243-255 (Fig. 4B). Our previous studies have demonstrated that the intrinsic dissociation rate is positively correlated to DM-mediated dissociation rate (27). Therefore, the increased peptide exchanged in the presence of DM observed in α+53G, α+53R or αY22F could be at least partially explained by the faster intrinsic peptide release and the consequent improvement in interaction between DQ2 and DM. The extent of DM enhancement of peptide loading shown on Figures 4A and 4B also argues that α+53G is more sensitive to DM function than α+53R or αY22F.
Figure 4.
Increased DM-dependent peptide exchange was observed in α+53G, α+53R or αY22F. Peptide exchange of cleaved sDQ2-CLIP (200 nM) with high-affinity peptides (6.25-100 uM), MHC Ia 49-63 (A) or MB 65KD 243-255 (B), was measured in the presence (2.4 uM) or absence of DM. Counts of WT and αQ50F were rescaled and shown in the inserted graphs. Each condition was run in duplicate. Shown is mean and SD from one representative experiment of at least three independent experiments.
A short time-course (0~4 h) experiment was also performed to directly compare the peptide exchange efficiency of WT and the α+53G mutant in the presence or absence of DM. As shown in Supplemental Figure 1, the peptide exchange of CLIP for MHC Ia peptide was increased in the α+53G mutant at every time point tested (Supp. Fig. 1).
α+53G is a better substrate of DM
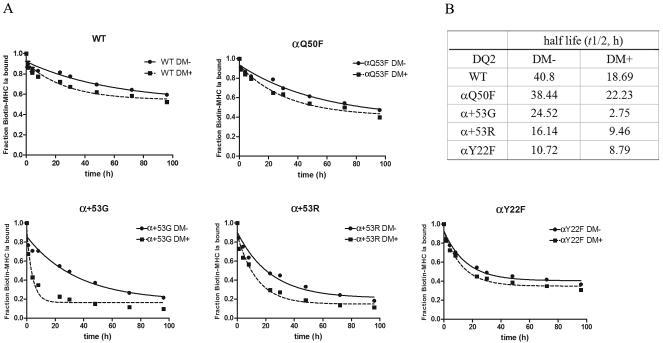
To investigate the possibility that the specifically improved DM-DQ2 interaction contributes to the enhanced DM-mediated peptide exchange, we measured the dissociation rates of biotin-MHC Ia 49-63 in the absence or presence of DM. With or without DM, the peptide dissociation rate was faster in α+53G, α+53R or αY22F than WT and αQ50F (Fig. 5A). This data confirms that these three mutants decrease the intrinsic stability of these complexes and this effect contributes to faster peptide dissociation in the presence of DM. Interestingly, although αY22F had the quickest intrinsic dissociation rate, the lowest t1/2 in the presence of DM was detected in α+53G (Fig. 5B). After DM was added, the peptide dissociation from α+53G increased nearly 12 times. Although DM also accelerated peptide release from WT and other mutants, the effect was modest (only 1~2 times). These findings argue that the acceleration of peptide dissociation observed in α+53G is not solely the result of moderately lowered intrinsic stability. Better DQ2-DM interaction is likely caused by this mutation. These two combined effects make α+53G the best substrate of DM among WT and other mutants.
Figure 5.
MHC Ia 49-63 dissociation from WT or mutant DQ2 molecules in vitro. Thrombin-digested sDQ2 molecules were preloaded with biotin-MHC Ia 49-63 overnight. The unbound peptide was removed, and dissociation of labeled peptide was followed in the absence (filled circles) or presence (filled squares) of DM (1.25 μM) for different periods of time. The signals at each time point were normalized to the starting counts for each molecule, and single-exponential decay curves were fit in Prism5 to calculate half lives.
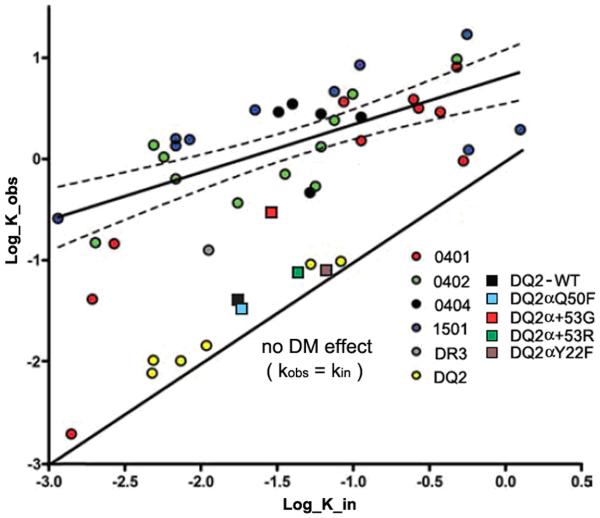
To further assess the DM susceptibility of these five complexes, we calculated the dissociation rate constants, k, of each complex in the presence or absence of DM, and plotted them in Figure 6 together with our previous data in which peptide dissociation rates with or without DM were measured for a panel of DR- or DQ2-peptide complexes with different intrinsic stability. Previously, for most DR-peptide complexes we studied, a positive correlation between the intrinsic dissociation rate constant and DM-mediated dissociation rate constant was observed with a correlation of r = 0.69 (27). All the six DQ2-peptide complexes (yellow dots), however, did not conform to this best-fitting line of correlation (19). Instead, they were all distributed along the line with a slope of 1 through the origin, which indicates no DM effect (kin = kobs). These results imply that DQ2 is resistant to DM function. The square representing αQ50F (light blue) was partly overlapped with WT (black square), suggesting this mutant behaves similarly to WT. Both α+53R and αY22F significantly increased the intrinsic off-rate (kin); thus, the squares representing these two mutants (green for α+53R, purple for αY22F) shifted to the right along the x-axis. Interestingly, WT and all three mutants (αQ50F, α+53R or αY22F) still followed the no-DM-effect line, suggesting the susceptibility to DM is not altered. α+53G (red square), however, behaved differently from the other mutants and WT. It slightly shifted to the right along the x-axis due to the moderately lowered intrinsic stability. At the same time, it obviously shifted up along the y-axis and became closer to the line indicating the correlation, reflecting improved DM susceptibility.
Figure 6.
Relationship between intrinsic and DM-catalyzed dissociation. Dissociation rate constants, k, calculated from the t1/2 by the equation t1/2= ln2/k. To compare current results to previous work on 36 complexes, dissociation in the presence of sDM (k_obs) are corrected for the differences in the concentration of DM used with the equation k_obs = k_in + [DM]km. k_in is the dissociation rate in the absence of DM. [DM] is the concentration of DM used for each reaction. km represents the change of dissociation rate induced by DM, which is calculated by the equation km = k_obs’-k_in. k_obs’ is the original dissociation rate in the presence of DM before normalization that is calculated from the equation t1/2= ln2/k. The values of k_in and k_obs are plotted on a log10 scale in units of hours−1. The x-axis shows dissociation in the absence of DM (k_in); the y-axis shows dissociation in the presence of sDM (k_obs). The solid best-fit straight line through the data has a slope of 0.47 (95% confidence interval, 0.3–0.64; thin dashed curves): the extremely stable DR*0401 complex and the complexes studied in this report were excluded from the correlation analysis. The solid line of slope 1 through the origin indicates no DM effect; the vertical distance of each data point from this line is a measure of DM susceptibility (27). The dissociation rates of biotin-MHC Ia 49-63 from DQ2 WT or mutants were calculated and plotted as described above. Data from DQ2 WT and mutants are represented by squares of different colors. The concentration of DM used for DQ2 WT and mutants was 1.25 μM.
The effect of mutations on CLIP phenotype
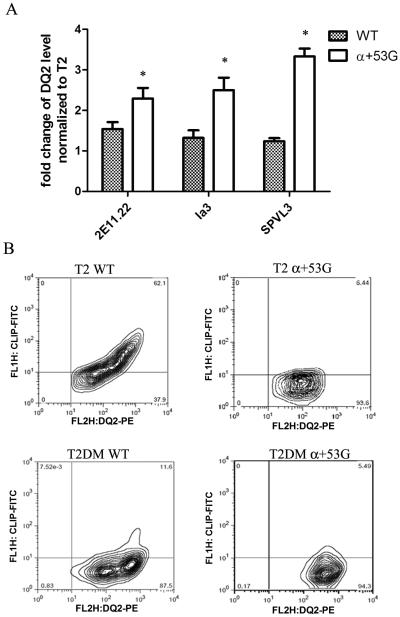
To evaluate the functional effect of α+53G, WT or α+53G in cells, the mutant cDNAs were stably expressed in T2 and T2DM cell lines, which are human T cell leukemia/B cell line hybrids that have no endogenous MHC class II molecules (Fig. 1D). In addition to its critical role in peptide exchange, DM also functions as a chaperone and conformational editor of MHC class II (28-30). Previous studies have suggested that DM can increase cell surface level of some class II molecules, in particular those with low affinity for CLIP (28, 31-34). Because of the poor interaction between DQ2 and DM, DQ2 resisted DM-mediated conformation change and cell surface abundance of DQ2 was not influenced by DM (19). Due to the better sensitivity of α+53G to DM function, we expected to see more DQ2 abundance on α+53G-expressing cells. To test this, we measured cell surface abundance of DQ2, using three different anti-DQ2 antibodies: 2E11.12, Ia3 and SPVL3, at saturating concentrations. Fold change in mean of MFI of DM+ compared with DM− cells were calculated and plotted in Fig. 7A. As we expected, coexpression of DM significantly increased surface expression of α+53G by ~2-3 times, with < 1.5 fold increase in WT (Fig. 7A).
Figure 7.
(A) Cells containing α+53G exhibited higher cell surface expression of DQ2 in the presence of DM. DQ2 was stained by three different clones of antibodies, 2E11.12, SPVL3 and Ia3 and PE-conjugated goat anti-mouse secondary Ab. The means of MFI in T2DM were normalized to those of T2 for both WT and α+53G. Mean and SD from three independent stainings are shown. * P< 0.05 (B) The percentage of DQ2+CLIP+ cells was decreased in α+53G-expressing cells. DQ2 and CLIP on cell surface were measured by PE-conjugated anti-DQ2 Ab (Ia3) and FITC-conjugated anti-CLIP Ab (CerCLIP), respectively.
To assess the effect of α+53G on CLIP accumulation at the cell surface, transfected T2 and T2DM cells were stained for DQ2 and CLIP with PE-conjugated anti-DQ2 Ab (Ia3) and FITC-conjugated anti-CLIP Ab (CerCLIP). The percentage of cells positive for both DQ2 and CLIP in α+53G-expressing T2 cells was significantly lower than that of WT (6.44% versus 62.1%, Fig. 7B, the top two panels), which likely is due to the low stability of α+53G/CLIP complexes, allowing exchange of CLIP for other peptides. When DM is present, it catalyzes CLIP exchange for other peptides in endocytic compartments. The percentage of double positive cells (DQ2+CLIP+) in WT decreased from 62.1% to 11.6% in the presence of DM. The sensitivity of WT DQ2 to DM effect is probably caused by the high expression of DM in transfected cell lines, the absence of the normal competition between DQ2 and DR3 for interaction with DM that occurs in cells expressing the physiological (DR3/DQ2) haplotype (35) and the resulting increased molar ratio of DM to DQ2. The percentage of double positive cells in α+53G-expressing cells was nearly equivalent after transfection of DM (from 6.44% to 5.49%). This is likely explained at least in part (see discussion) by the rapid dissociation of α+53G-CLIP complexes in the absence of DM, which leaves no window to measure DM/α+53G interaction using CLIP. (Fig. 7B, the bottom two panels).
The influence of α+53G on gliadin presentation to T cell hybridomas
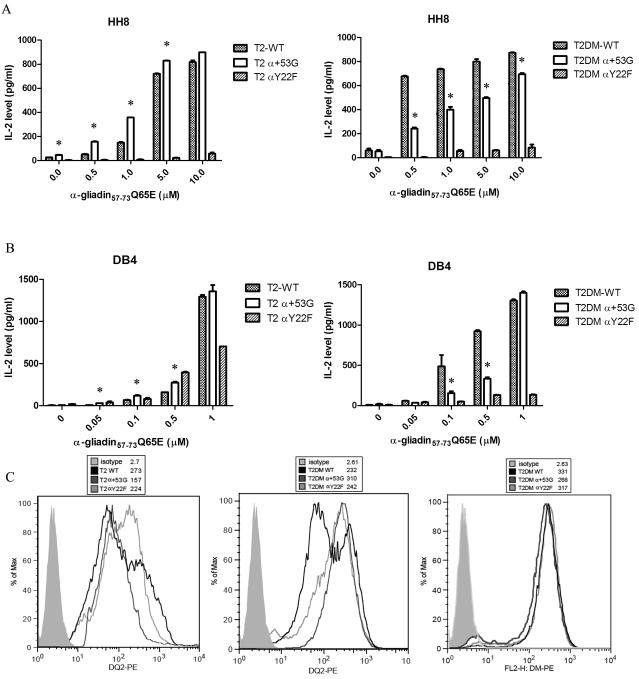
Altered intrinsic stability and/or DM interaction may influence the T cell stimulatory capacity of antigenic peptides. We tested the presentation of α-gliadin 57-73 Q65E (QLQPFPQPELPYPQPQS) by irradiated T2 and T2DM expressing DQ2 WT, α+53G or αY22F to DQ2-restricted mouse T cell hybridomas, HH8 or DB4. The peptide α-gliadin 57-73 Q65E contains the partially overlapping α-I (QLQPFPQPELPY) and α-II (PQPELPYPQPQL) gliadin epitopes, both of which have been identified as dominant epitopes recognized by T cells present in the small intestine after in vivo gluten challenge in HLA-DQ2+ patients with celiac disease (36). Both hybridomas express the markers of hCD4, mCD4 and mCD3, and are specific for the α-II gliadin epitope (residues 62-70) {(22), and unpublished data from Prof. James McCluskey’s laboratory}. In the presence of low concentration of peptide (0.5-5uM for HH8; 0.05-0.5uM for DB4), T cell activation was more efficient with T2-α+53G (Fig. 8A&B, left panels). This finding likely reflects the more efficient CLIP replacement by exogenous gliadin peptide on the surface of α+53G-expressing T2 cells. In contrast, decreased hybridoma activation was observed when cells were co-cultured with T2DM-α+53G (Fig. 8A&B, right panels) with low concentration of peptide. A likely explanation is the more effective “editing” of DQ2/peptide complexes in these cells, restricting peptide surface exchange. Interestingly, the difference between WT and α+53G was overcome when high concentration of peptide was used (10uM for HH8; 1uM for DB4), indicating that comparable peptide exchange on cell surface can be achieved in the presence of abundant peptides. To exclude the possibility that the differential hybridoma activation induced by WT and α+53G was caused by the differences in DQ2 expression level, we measured cell surface levels of WT and α+53G DQ2 by FACS. As shown in Figure 8C, α+53G expression on the T2 surface was lower than WT, whereas α+53G was expressed at higher levels than WT in T2DM cells. Thus, surface expression levels do not explain the pattern of hybridoma activation induced by α+53G. The αY22F mutant was not able to efficiently activate either T cell hybridoma, perhaps because the αY22F mutation substantially decreases the stability of peptide-class II complexes (26), and accelerates release of gliadin peptide from surface DQ molecules. In all the T2DM cells, over 95% of total cells were DM positive and DM levels in WT, α+53G and αY22F-expressing cells were comparable (Fig. 8C).
Figure 8.
α+53G enhances gliadin peptide presentation to T cell hybridoma without DM and inhibits peptide presentation with DM coexpression. Irradiated T2/T2DM cells expressing DQ2 WT, α+53G or αY22F were co-cultured with DQ2-restricted T cell hybridomas, HH8 (A) or DB4 (B), in the presence of increasing amount of gliadin peptide (0.01-10 uM for HH8; 0.05-1 uM for DB4). IL-2 level was measured by ELISA. Each condition was done in replicates and each experiment was repeated independently at least twice. Mean and SD from one representative experiment was shown. (C) Comparison of DQ2 and DM level in T2/T2DM cells expressing WT, α+53G or αY22F. Surface DQ2 expression was detected by PE-conjugated anti-DQ2 Ab (Ia3). Intracellular DM was stained by PE-conjugated anti-DM Ab (Map.DM1) after cells were fixed and permeabilized. The solid light gray histogram represents isotype control; black line is WT; dark gray line is α+53G; light gray line is αY22F. Mean of MFI is indicated in parentheses. * P< 0.05
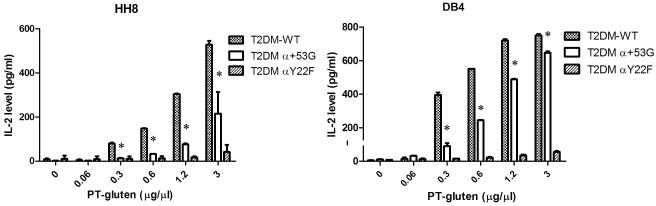
Suppressed procession and presentation of gluten by α+53G-expressing T2-DM cells
HMWF PT-gluten is the high molecular weight (>10kDa) fraction of processed gluten protein fragments, generated by pepsin and trypsin/chymotrypsin digestion and treated with transglutaminase. It has been shown that the presentation of PT-gluten requires internal processing (37) and we also saw that T cell proliferation was dramatically inhibited when APCs were fixed (data not shown). It is known that DM plays an important role in selection of endosomally-generated peptides for presentation. To better understand the effect of DQ2 mutants on this functional interaction with DM, we measured T cell hydridoma activation in response to T2-DM cells fed with PT-gluten. Interestingly, activation of both HH8 and DB4 was significantly compromised when α+53G was coexpressed with DM (Fig. 9). DM+ cells with DQ2 WT more efficiently presented the epitope α-gliadin 57-73 Q65E than DM-cells (Supp. Fig. 2A&B, left panels). This difference between DM− and DM+ was modest (for HH8) or even reversed (for DB4) for cells containing α+53G (Supp. Fig. 2A&B, right panels). Considering the improved sensitivity of α+53G to DM effect as demonstrated above, these findings suggest that the presentation of epitope α-gliadin 57-73 Q65E is suppressed by DM in endocytic compartments.
Figure 9.
The activation of T cell hybridoma by PT-gluten is repressed by α+53G in the presence of DM. Irradiated T2DM cells expressing DQ2 WT, α+53G or αY22F were co-cultured with DQ2-restricted T cell hybridoma, HH8 (left) or DB4 (right), in the presence of increasing amount of PT-gluten (0.06-3 μg/μl). IL-2 level was measured by ELISA. Each condition was done in replicate and each experiment was repeated independently at least twice. Mean and SD from one representative experiment was shown. * P< 0.05
Discussion
It is generally accepted that DQ2 is a strong, inherited risk factor for several autoimmune diseases, especially CD (38, 39). Currently, the susceptibility of DQ2 to CD is understood to reflect the preferential binding of gliadin peptide to the DQ2 molecule (19, 23, 40). One of the structural characteristics of DQ2 that makes it able to accommodate the proline-rich gliadin peptide is the single residue deletion at position α53 (23, 40). A hydrogen bond formed between the amide carboxylic oxygen of the residue α53 and the amide hydrogen of the P1 residue is a general feature of most MHC II-peptide interaction (41, 42). Due to the lack of an α53 residue, proline can be accommodated by DQ2 at the P1 pocket without energetic cost. It is possible that insertion of an extra G or R in α53 influences the accommodation of proline at the P1 pocket of DQ2, although the gliadin-derived peptide α-gliadin 57-73 Q65E still binds sufficiently to stimulate the T hybridomas tested. Also, according to the crystal structure of the DQ2-α–I-gliadin complex, a hydrogen bond is formed between the amide hydrogen of α52R of DQ2 and the amide carboxylic oxygen of the P-2 residue (23), which may also be affected by the addition of G or R at α53. These two factors may explain the accelerated spontaneous release of DQ2-binding peptides from these two mutants (Fig. 5 and Fig. 6). As for the mutant αY22F, recently reported by Dr. Ludvig Sollid’s group (26), it disrupts a water-mediated hydrogen bond formed between α22Y and the bound peptide, leading to a decrease in the overall peptide-binding stability. These three α chain mutations also have increased peptide exchange in the absence of DM (Fig. 3), which is caused, we think, mainly by the increased off-rate of CLIP.
A positive correlation between intrinsic peptide/class II and DM-mediated dissociation rate has been reported (27). Most DR-peptide complexes in our previous studies showed the predicted behavior (Fig. 6); however, all six DQ2-peptide complexes with varied levels of intrinsic stability (yellow dots) were away from the best-fitting straight correlation line. In fact, they were all around the line with a slope of 1 through the origin, which indicates no DM effect (kobs = kin). These findings imply that DQ2 is resistant to DM effect, probably due to the poor interaction between these two molecules. If the mutants of DQ2 only reduce the intrinsic peptide dissociation rate, they should shift to the right along the x-axis, but still follow the no-DM-effect line, which is what we saw with α+53R or αY22F (Fig. 6). The mutant α+53G, however, had a shift both to the right and up, which makes it closer to the line of correlation, similar to other class II molecules that interact with DM (Fig. 6). This result indicates that the faster peptide dissociation from α+53G in the presence of DM is not only because of the accelerated spontaneous peptide release, but also because the increased susceptibility of α+53G to DM effect.
The DM-interacting site on DQ2 has not been mapped yet. Our previous mutational analysis revealed that αF51 in DR3 plays a critical role in DM interaction (24). This residue is located on the interface of DR3 that interacts with DM and may function as a lever to move the extended strand including residues 51-53 (24). Because the structural homologue of αF51 in DQ2 is sterically blocked (Fig. 1A), we mutated αQ50 to F to provide an interaction site for DM. This mutation did not improve the DQ2 susceptibility to DM (Fig. 4 &5). In fact, the recombinant αQ50F protein is more sensitive to freezing and thawing than WT and other mutants (data not shown), probably because the mutation alters the local charge and influences the protein stability. The point deletion at α53 is also a special characteristic of DQA1*0501 that is not found in DQA1*0101 and DQA1*0301, which are both sensitive to DM function. Although the insertion of G or R both accelerated intrinsic peptide release, only α+53G has improved DQ2 susceptibility to DM. This difference probably is due to the different charge of G and R. The repulsion between the inserted α53R and the endogenous α52R in DQ2 may cause local conformational change that does not occur when uncharged G is inserted.
The point deletion at the position 53 of DQ α chain is also found in other DQ alleles, including all the DQA1*05 alleles, DQA1*0601, A1*0602, A1*0401, and A1*0404. The relationship between some of these alleles and autoimmune diseases has been reported. For example, DQ7.6 (DQA1*0601:DQB1*0301) is associated with asthma in Chinese population (43) and pauciarticular juvenile chronic arthritis without anti-nuclear antibodies (44). The allele of DQA1*0401 has been found associated with primary chronic progressive multiple sclerosis among the Ashkenazi patients (45). It is possible that the interaction between these alleles and DM is also defective, similar to what we have observed in DQ2, and it will be interesting to determine whether our finding that the insertion of glycine restores the DM susceptibility also applies to these DQ alleles.
Our previous data show that DQ2 purified from B cells is associated with two different groups of CLIP peptides, the traditional CLIP peptides (CLIP1) and the unusual CLIP peptides (CLIP2), which bind to DQ2 in overlapped, but distinct binding registers (19). One of the major functions of DM is to replace CLIP with antigenic peptides in endosomal compartments (16-18). Because the available anti-CLIP antibody (CerCLIP) can only recognize the traditional CLIP1, but not the unusual CLIP2 peptide, which is the predominant one associated with DQ2, FACS staining with CerCLIP is not able to reveal the complete CLIP profile binding to WT or mutants. It is thus possible that the mutation of α+53G changes the ratio of CLIP1 and CLIP2 peptide presenting on the cell surface. MALDI-TOF analysis of CLIP peptides eluted from DQ2 will be needed to assess this possibility.
Because DM edits the peptide repertoire presented by MHC class II to CD4+ cells, the sensitivity to DM function may have a profound effect on T cell-mediated immune response. Even in the presence of DM, the peptide repertoire bound to wt DQ2 is not edited. Due to the sensitivity of α+53G to DM-mediated peptide editing, α+53G is loaded in endosomal compartments with the help of DM with peptides of high kinetic stability, which are eventually presented on the cell surface. The high stability of α+53G-peptide complexes only allows limited peptide exchange on the cell surface when exogenous gliadin peptide is provided. As a result, the presentation of the α-gliadin epitope to T cell hybridomas is inhibited (Fig. 8A&B, right panels). In the absence of DM editing, the lower intrinsic stability of α+53G-peptide complexes facilitates peptide exchange at the cell surface, and the subsequent activation of T cell hybridomas by gliadin peptide is thus increased (Fig. 8A&B, left panels).
Epitopes of an intact antigen that elicit potent T cell activation are classified as immunodominant epitopes. By contrast, epitopes that fail to trigger a T cell response, but can bind to class II, are called cryptic epitopes. It has been proposed that DM determines the immunodominant and cryptic fate of CD4+ T cell epitopes (34, 46-48). DM removes cryptic peptides from the loading groove of class II in endosomal compartments and thereby antagonizes or completely eliminates the presentation of cryptic epitopes. The cryptic fate of α-gliadin epitopes was initially implied by work with DR-transfected murine fibroblasts (DM negative cells) that presented naturally processed A-gliadin protein (AGL, whole gliadin derived from unbleached flour) (49) to T cell clones more efficiently than EBV-transformed cell lines (DM+) (50). The naturally processed peptide containing the α-gliadin II epitope is not efficiently removed by DM in endosomal compartments, at least in part because of the poor interaction between WT DQ2 and DM, leading to presentation of this epitope (Supp. Fig. 2, left panels). However, the DQ2 WT DM+ cells have modestly higher levels of surface DQ2 WT than the DM-null DQ2 WT cells (Fig. 7A), perhaps due to variation in the (highly oligoclonal) transfectant lines, and/or to some DM chaperone effect at the high DM:DQ2 molar ratio, although in DR3/DQ2/DM+ lymphoblastoid lines, DM is not an effective chaperone for DQ2 (19). This increase likely enhances presentation of the epitope and contributes to the increased activation of T cell hybridomas by the DQ2+DM+ cells. The difference in DQ2 surface level between DM− and DM+ cells is even more evident for α+53G, likely due to chaperone effects, Fig. 7A), However, the T cell activation is not significantly increased or even suppressed by coexpression of DM (Supp. Fig. 2, right panels). This likely reflects the sensitivity of α+53G to DM editing, which results in crypticity of the α-gliadin 57-73 Q65E epitope in the context of the α+53G mutant. Thus, an attractive hypothesis is that the resistance of WT DQ2 to DM changes the fate of α-gliadin 57-73 Q65E from a cryptic epitope to an immunodominant epitope, influencing the generation of a gliadin-induced immune response in patients expressing the DQ2 allele. The two hydridomas, HH8 and DB4, used in this study both specifically recognize the α-II-gliadin epitope (residues 62-70) from α-gliadin57-73 Q65E {(22), and unpublished data from Prof. James McCluskey’s lab}.We are currently analyzing other disease-related gliadin epitopes (51) to determine the generality of our finding. As α+53G mutation effects both the peptide binding behavior and the DM interaction of the protein, we are studying other DQ2 mutants for an ideal one that only improves DM interaction without influencing the peptide binding groove. We predict that presentation of the gliadin epitopes will be suppressed such a mutant as their affinity for the DQ2 binding groove is lower than that of CLIP (19), which is sensitive to DM when the low affinity of DM for DQ2 is overcome with high levels of DM (19).
We have previously hypothesized that disease-susceptibility of certain MHC class II alleles may be highly related to alterations in events surrounding peptide loading and editing in endosomes, with consequences for antigen presentation events in the thymus and periphery that facilitate loss of self-tolerance and the development of autoimmune disease (52-55). It has been demonstrated that in mice lacking H2-DM molecules on certain H2 backgrounds, such as H-2b, class II is predominantly occupied by CLIP (56-58). A very narrow spectrum of class II-associated peptides leads to a reduction in the number of CD4+ T cells to approximately 30 to 50% of that in normal mice (56-59), and though positive selection occurs, it clearly restricts the diversity of the repertoire (58, 60). However, the population of CD4+ T cells from these DM-knockout mice is tolerized by CLIP/class II complexes rather than a broad range of self-peptides during negative selection. As a consequence, large proportion of H2-DM-CD4+ T cells show broad (“self”) reactivity to antigen presenting cells from a DM+ mouse of the same (I-Ab) class II haplotype (56-59). In the case of DQ2, the predominance of (unexchanged) CLIP peptides associated with DQ2 likely affects positive selection in the thymus, likely reducing their number and diversity. Further, these peripheral DQ2-restricted T cells are likely to be potentially self-reactive, having been negatively selected primarily on DQ2/CLIP. This potential for autoreactivity would be revealed if/when conditions allow other self-peptides to be presented, as may happen at sites of inflammation where self-peptides are generated extracellularly by proteases and where the pH of the microenvironment drops, favoring peptide exchange (61, 62). In addition, in peripheral antigen presenting cells, reduced DQ2/DM interaction and sufficient DQ2/CLIP affinity allows DQ2/CLIP to accumulate on the cell surface; these complexes more easily undergo peptide exchange than tightly bound peptides that have been subject to and survived DM editing.
In this study, we investigated the structural basis for the poor DQ2-DM interaction and identified one mutant, α+53G, which is able to restore DQ2 sensitivity to DM editing. Our results provide important mechanistic insights into the unique features of DQ2 interaction with the antigen presentation machinery. This may ultimately lead to improved understanding of the association of DQ2 with autoimmunity and may suggest new therapeutic approaches for DQ2-associated autoimmune disorders.
Supplementary Material
Acknowledgement
We thank Dr. Lisa Denzin, Dr. Ludvig Sollid, Prof. James McCluskey, Tracy Holmes II from Dr. Chaitan Khosla’s lab, and John Sidney from Dr. Alessandro Sette for their help with plasmids, cell lines and peptides.
Footnotes
This work is supported by funding from the NIH, 5R21DK079163-02 (to E.D.M.), Deans Postdoctoral Fellowship, 1048364-106- KAQEL, (to T.H.), Immunology Training Grant, NIH/NIAID, 5T32AI07290-24, (to T.H.), NIAID grants, 1F32AI089080-01, (to T.H.), Deans Postdoctoral Fellowship, Aaron Fund 1048085-153-KAUMZ (to H.M) and NIH Molecular and Cellular Immunobiology Training grant 5 T32 AI07290-24 and 5 T32 AI07290-26 (to H.M.).
- DM
- HLA-DM
- DQ2
- HLA-DQ2 (DQA1*0501, DQB1*0201)
- DO
- HLA-DO
- Ii
- invariant chain
- CLIP
- class-II-associated invariant chain peptide
- s
- soluble
- HMWF PT-gluten
- high-molecular-weight fraction of pepsin- and trypsin/chymotrypsin-digested gluten
References
- 1.Lundin KE, Gjertsen HA, Scott H, Sollid LM, Thorsby E. Function of DQ2 and DQ8 as HLA susceptibility molecules in celiac disease. Hum Immunol. 1994;41:24–27. doi: 10.1016/0198-8859(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 2.Pociot F, McDermott MF. Genetics of type 1 diabetes mellitus. Genes Immun. 2002;3:235–249. doi: 10.1038/sj.gene.6363875. [DOI] [PubMed] [Google Scholar]
- 3.Green PH, Cellier C. Celiac disease. N Engl J Med. 2007;357:1731–1743. doi: 10.1056/NEJMra071600. [DOI] [PubMed] [Google Scholar]
- 4.Johnson TC, Diamond B, Memeo L, Negulescu H, Hovhanissyan Z, Verkarre V, Rotterdam H, Fasano A, Caillat-Zucman S, Grosdidier E, Winchester R, Cellier C, Jabri B, Green PH. Relationship of HLA-DQ8 and severity of celiac disease: comparison of New York and Parisian cohorts. Clin Gastroenterol Hepatol. 2004;2:888–894. doi: 10.1016/s1542-3565(04)00390-8. [DOI] [PubMed] [Google Scholar]
- 5.Busch R, Doebele RC, Patil NS, Pashine A, Mellins ED. Accessory molecules for MHC class II peptide loading. Curr Opin Immunol. 2000;12:99–106. doi: 10.1016/s0952-7915(99)00057-6. [DOI] [PubMed] [Google Scholar]
- 6.Busch R, Rinderknecht CH, Roh S, Lee AW, Harding JJ, Burster T, Hornell TM, Mellins ED. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 7.Jones PP, Murphy DB, Hewgill D, McDevitt HO. Detection of a common polypeptide chain in I--A and I--E sub-region immunoprecipitates. Mol Immunol. 1979;16:51–60. doi: 10.1016/0161-5890(79)90027-0. [DOI] [PubMed] [Google Scholar]
- 8.Machamer CE, Cresswell P. Biosynthesis and glycosylation of the invariant chain associated with HLA-DR antigens. J Immunol. 1982;129:2564–2569. [PubMed] [Google Scholar]
- 9.Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 10.West MA, Lucocq JM, Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature. 1994;369:147–151. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- 11.Tulp A, Verwoerd D, Dobberstein B, Ploegh HL, Pieters J. Isolation and characterization of the intracellular MHC class II compartment. Nature. 1994;369:120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Y, Xu X, Wandinger-Ness A, Dalke DP, Pierce SK. Separation of subcellular compartments containing distinct functional forms of MHC class II. J Cell Biol. 1994;125:595–605. doi: 10.1083/jcb.125.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters PJ, Neefjes JJ, Oorschot V, Ploegh HL, Geuze HJ. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 1991;349:669–676. doi: 10.1038/349669a0. [DOI] [PubMed] [Google Scholar]
- 14.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 15.Sette A, Ceman S, Kubo RT, Sakaguchi K, Appella E, Hunt DF, Davis TA, Michel H, Shabanowitz J, Rudersdorf R, et al. Invariant chain peptides in most HLA-DR molecules of an antigen-processing mutant. Science. 1992;258:1801–1804. doi: 10.1126/science.1465617. [DOI] [PubMed] [Google Scholar]
- 16.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 17.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 18.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 19.Fallang LE, Roh S, Holm A, Bergseng E, Yoon T, Fleckenstein B, Bandyopadhyay A, Mellins ED, Sollid LM. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J Immunol. 2008;181:5451–5461. doi: 10.4049/jimmunol.181.8.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 21.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 22.de Kauwe AL, Chen Z, Anderson RP, Keech CL, Price JD, Wijburg O, Jackson DC, Ladhams J, Allison J, McCluskey J. Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4+ T cells. J Immunol. 2009;182:7440–7450. doi: 10.4049/jimmunol.0900233. [DOI] [PubMed] [Google Scholar]
- 23.Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U S A. 2004;101:4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 25.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 26.Fallang LE, Bergseng E, Hotta K, Berg-Larsen A, Kim CY, Sollid LM. Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat Immunol. 2009;10:1096–1101. doi: 10.1038/ni.1780. [DOI] [PubMed] [Google Scholar]
- 27.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 28.Koonce CH, Wutz G, Robertson EJ, Vogt AB, Kropshofer H, Bikoff EK. DM loss in k haplotype mice reveals isotype-specific chaperone requirements. J Immunol. 2003;170:3751–3761. doi: 10.4049/jimmunol.170.7.3751. [DOI] [PubMed] [Google Scholar]
- 29.Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 30.Lovitch SB, Pu Z, Unanue ER. Amino-terminal flanking residues determine the conformation of a peptide-class II MHC complex. J Immunol. 2006;176:2958–2968. doi: 10.4049/jimmunol.176.5.2958. [DOI] [PubMed] [Google Scholar]
- 31.Albert LJ, Ghumman B, Watts TH. Effect of HLA-DM transfection on hen egg lysozyme presentation by T2.Ak cells. J Immunol. 1996;157:2247–2255. [PubMed] [Google Scholar]
- 32.Fallas JL, Tobin HM, Lou O, Guo D, Sant’Angelo DB, Denzin LK. Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation. J Immunol. 2004;173:1549–1560. doi: 10.4049/jimmunol.173.3.1549. [DOI] [PubMed] [Google Scholar]
- 33.Honey K, Forbush K, Jensen PE, Rudensky AY. Effect of decreasing the affinity of the class II-associated invariant chain peptide on the MHC class II peptide repertoire in the presence or absence of H-2M. J Immunol. 2004;172:4142–4150. doi: 10.4049/jimmunol.172.7.4142. [DOI] [PubMed] [Google Scholar]
- 34.Lich JD, Jayne JA, Zhou D, Elliott JF, Blum JS. Editing of an immunodominant epitope of glutamate decarboxylase by HLA-DM. J Immunol. 2003;171:853–859. doi: 10.4049/jimmunol.171.2.853. [DOI] [PubMed] [Google Scholar]
- 35.Price P, Witt C, Allcock R, Sayer D, Garlepp M, Kok CC, French M, Mallal S, Christiansen F. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 36.Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med. 2000;6:337–342. doi: 10.1038/73200. [DOI] [PubMed] [Google Scholar]
- 37.Qiao SW, Bergseng E, Molberg O, Xia J, Fleckenstein B, Khosla C, Sollid LM. Antigen presentation to celiac lesion-derived T cells of a 33-mer gliadin peptide naturally formed by gastrointestinal digestion. J Immunol. 2004;173:1757–1762. doi: 10.4049/jimmunol.173.3.1757. [DOI] [PubMed] [Google Scholar]
- 38.Sollid LM, Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910–922. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- 39.Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169:345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergseng E, Xia J, Kim CY, Khosla C, Sollid LM. Main chain hydrogen bond interactions in the binding of proline-rich gluten peptides to the celiac disease-associated HLA-DQ2 molecule. J Biol Chem. 2005;280:21791–21796. doi: 10.1074/jbc.M501558200. [DOI] [PubMed] [Google Scholar]
- 41.Sant AJ, Beeson C, McFarland B, Cao J, Ceman S, Bryant PW, Wu S. Individual hydrogen bonds play a critical role in MHC class II: peptide interactions: implications for the dynamic aspects of class II trafficking and DM-mediated peptide exchange. Immunol Rev. 1999;172:239–253. doi: 10.1111/j.1600-065x.1999.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 42.Stratikos E, Wiley DC, Stern LJ. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II alpha-chain. J Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- 43.Guo X, Ni P, Li L. Association between asthma and the polymorphism of HLA-DQ genes. Zhonghua Jie He He Hu Xi Za Zhi. 2001;24:139–141. [PubMed] [Google Scholar]
- 44.Donn RP, Thomson W, Pepper L, Carthy D, Farhan A, Ryder C, Southwood T, Holt L, Ollier W. Antinuclear antibodies in early onset pauciarticular juvenile chronic arthritis (JCA) are associated with HLA-DQB1*0603: a possible JCA-associated human leucocyte antigen haplotype. Br J Rheumatol. 1995;34:461–465. doi: 10.1093/rheumatology/34.5.461. [DOI] [PubMed] [Google Scholar]
- 45.Kwon OJ, Karni A, Israel S, Brautbar C, Amar A, Meiner Z, Abramsky O, Karussis D. HLA class II susceptibility to multiple sclerosis among Ashkenazi and non-Ashkenazi Jews. Arch Neurol. 1999;56:555–560. doi: 10.1001/archneur.56.5.555. [DOI] [PubMed] [Google Scholar]
- 46.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinderknecht CH, Belmares MP, Catanzarite TL, Bankovich AJ, Holmes TH, Garcia KC, Nanda NK, Busch R, Kovats S, Mellins ED. Posttranslational regulation of I-Ed by affinity for CLIP. J Immunol. 2007;179:5907–5915. doi: 10.4049/jimmunol.179.9.5907. [DOI] [PubMed] [Google Scholar]
- 48.Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, Lazarski CA. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 49.Kagnoff MF, Austin RK, Johnson HC, Bernardin JE, Dietler MD, Kasarda DD. Celiac sprue: correlation with murine T cell responses to wheat gliadin components. J Immunol. 1982;129:2693–2697. [PubMed] [Google Scholar]
- 50.Franco A, Appella E, Kagnoff MF, Chowers Y, Sakaguchi K, Grey HM, Sette A. Peripheral T cell response to A-gliadin in celiac disease: differential processing and presentation capacities of Epstein-Barr-transformed B cells and fibroblasts. Clin Immunol Immunopathol. 1994;71:75–81. doi: 10.1006/clin.1994.1054. [DOI] [PubMed] [Google Scholar]
- 51.Marti T, Molberg O, Li Q, Gray GM, Khosla C, Sollid LM. Prolyl endopeptidase-mediated destruction of T cell epitopes in whole gluten: chemical and immunological characterization. J Pharmacol Exp Ther. 2005;312:19–26. doi: 10.1124/jpet.104.073312. [DOI] [PubMed] [Google Scholar]
- 52.Thomson G, Robinson WP, Kuhner MK, Joe S, MacDonald MJ, Gottschall JL, Barbosa J, Rich SS, Bertrams J, Baur MP, et al. Genetic heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin-dependent diabetes mellitus. Am J Hum Genet. 1988;43:799–816. [PMC free article] [PubMed] [Google Scholar]
- 53.Mellins ED. The role of the MHC in autoimmunity: an overview. J Rheumatol Suppl. 1992;33:63–69. [PubMed] [Google Scholar]
- 54.Rinderknecht CH, Lu N, Crespo O, Truong P, Hou T, Wang N, Rajasekaran N, Mellins ED. I-Ag7 is subject to post-translational chaperoning by CLIP. Int Immunol. 22:705–716. doi: 10.1093/intimm/dxq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinderknecht CH, Roh S, Pashine A, Belmares MP, Patil NS, Lu N, Truong P, Hou T, Macaubas C, Yoon T, Wang N, Busch R, Mellins ED. DM influences the abundance of major histocompatibility complex class II alleles with low affinity for class II-associated invariant chain peptides via multiple mechanisms. Immunology. 131:18–32. doi: 10.1111/j.1365-2567.2010.03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fung-Leung WP, Surh CD, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H2-M-deficient mice. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 57.Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 58.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 59.Surh CD, Lee DS, Fung-Leung WP, Karlsson L, Sprent J. Thymic selection by a single MHC/peptide ligand produces a semidiverse repertoire of CD4+ T cells. Immunity. 1997;7:209–219. doi: 10.1016/s1074-7613(00)80524-5. [DOI] [PubMed] [Google Scholar]
- 60.Bevan MJ. In thymic selection, peptide diversity gives and takes away. Immunity. 1997;7:175–178. doi: 10.1016/s1074-7613(00)80520-8. [DOI] [PubMed] [Google Scholar]
- 61.Ng AW, Bidani A, Heming TA. Innate host defense of the lung: effects of lung-lining fluid pH. Lung. 2004;182:297–317. doi: 10.1007/s00408-004-2511-6. [DOI] [PubMed] [Google Scholar]
- 62.Radu CG, Nijagal A, McLaughlin J, Wang L, Witte ON. Differential proton sensitivity of related G protein-coupled receptors T cell death-associated gene 8 and G2A expressed in immune cells. Proc Natl Acad Sci U S A. 2005;102:1632–1637. doi: 10.1073/pnas.0409415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.