Abstract
Murine Tcra and Tcrd gene segments are organized into single genetic locus (Tcra/Tcrd locus) which undergoes V(D)J recombination in CD4−CD8− double negative (DN) thymocytes to assemble Tcrd genes and in CD4+CD8+ double positive (DP) thymocytes to assemble Tcra genes. Recombination events are regulated by two developmental stage-specific enhancers, Eδ and Eα. Effects of Eα on Trca/Tcrd locus chromatin have been well documented, but effects of Eδ have not. In this regard, Eα acts over long distances to activate many Vα and Jα segments for recombination in DP thymocytes. However, in DN thymocytes it is unclear whether Eδ functions over long distances to regulate Vδ gene segments, or only functions locally to regulate Dδ and Jδ gene segments. Here we analyzed germline transcription, histone modifications and recombination on wild-type and Eδ-deficient alleles in adult and fetal thymocytes. We found that Eδ functions as a local enhancer whose influence is limited to no more than about 10 kb in either direction (including Dδ, Jδ and TRDV5 gene segments) in adult DN thymocytes. However we identified a unique long-distance role for Eδ promoting accessibility and recombination of fetal Vδ gene segment TRDV4, over a distance of 55 kb, in fetal thymocytes. TRDV4 recombination is specifically repressed in adult thymocytes. We found that this repression is enforced by a developmentally regulated loss of histone acetylation. Constitutively high levels of a suppressive modification, histone H3 lysine 9 dimethylation, may contribute to repression as well.
Introduction
The development of αβ and γδ T lymphocytes depends on the somatic assembly of T cell receptor (TCR) genes by V(D)J recombination (1). Among the four TCR genes, Tcrd and Tcra are uniquely organized into a single, complex genetic locus (the Tcra/Tcrd locus) that spans 1.6 megabases of murine chromosome 14 (2). Tcra/Tcrd locus recombination events occur according to a strict developmental program during thymocyte maturation, with Tcrd genes assembled in CD4−CD8− double negative (DN) thymocytes and Tcra genes assembled in those thymocytes that progress to the CD4+CD8+ double positive (DP) stage. Developmental programming is thought to be mediated by changes in chromatin structure that make certain recombination signal sequences (RSSs) accessible to the recombination activating gene (RAG) complex and by changes in locus organization that allow specific pairs of RSSs to undergo synapsis for recombination (3). How such changes are effected is only partially understood.
Two developmental stage-specific enhancers, Eδ and Eα, regulate the switch from Tcrd to Tcra rearrangement (4–7). Eδ is active in DN thymocytes; it promotes high level germline transcription from nearby Dδ and Jδ promoters and supports normal levels of fully Vδ-Dδ-Jδ rearranged genes (6, 8). However inefficient and incomplete (Vδ-Dδ, Dδ-Dδ, Dδ-Jδ) Tcrd gene assembly occurs even on Eδ-deleted alleles, implying that Tcrd recombination may be partially supported by additional cis-elements. Eα becomes active subsequently, in DP thymocytes; it provokes transcription from the T early α promoter upstream of the Jα segments, at a distance of 70 kb (9, 10). Eα- and T early α-dependent germline transcription across the Jα segments is essential for Jα accessibility and for Vα-to-Jα recombination at this stage (11, 12).
A pool of about 100 V gene segments is distributed across 1.5 megabases of DNA upstream of Dδ and Jα gene segments and supplies V genes for both the TCRδ and TCRα repertoires. A small subset of these V gene segments undergoes recombination to Dδ gene segments; sixteen have been classified as Vδ but a smaller number (TRDV2-2, TRDV5 and the four-membered TRAV15/TRDV6 family) tend to dominate the adult TCR δ repertoire (2, 13–15). In addition, one Vδ (TRDV4) rearranges specifically in fetal thymocytes. In contrast, the vast majority of the V gene segments may undergo recombination to Jα gene segments during subsequent Tcra gene assembly (14, 16).
Although the regulation of Tcra/Tcrd V gene segment usage is crucial for the development of distinct TCRδ and TCRα repertoires, the molecular basis for this regulation is only partly understood (1). Several Vδ gene segments are positioned relatively proximal to Dδ, Jδ and Cδ gene segments. However, prominent adult Vδ gene segments are characterized by active germline transcription and an accessible chromatin configuration in adult DN thymocytes, independent of their position in the locus (15). Moreover, recombination biases imposed by RSSs themselves cannot explain gene segment usage (17). Thus there must be regulation at the level of RSS accessibility (15, 17). However, it is not clear whether Vδ gene segment accessibility depends solely on intrinsic features of Vδ gene promoters, or whether there are contributions from distant cis-regulatory elements as well. Regardless, it is clear that many additional Tcra/Tcrd locus V gene segments become transcriptionally active and accessible in DP thymocytes, particularly within the proximal 500kb of the V segment array (15). These changes do not occur on Eα-deficient alleles, demonstrating that Eα can influence V gene segment accessibility over very long distances.
Although the studies outlined above suggest that Eα can shape the Vα repertoire through long-distance effects on Vα promoters, surprisingly little is known regarding the role of Eδ in shaping the Vδ repertoire. Although germline transcription of TRDV5, located 10 kb downstream of Eδ, was found to be reduced on Eδ-deficient alleles (18), information about more distant Vδ gene segments has been lacking. Furthermore, no information is available regarding the chromatin modification profile of Eδ-deficient alleles. Thus it is unknown whether support of Tcrd gene assembly by Eδ reflects a broad role for Eδ in promoting accessibility of Vδ, Dδ and Jδ gene segments in DN thymocytes, or a selective and localized role in Dδ or Jδ accessibility. Indeed it has been generally assumed that Eδ functions over relatively short distances, based in part on the observation that Eδ cannot support Tcrd gene recombination when repositioned 100 kb distant from Dδ and Jδ gene segments in the normal location of Eα (19).
To investigate a role for Eδ in regulating Vδ gene segments in DN thymocytes, we analyzed germline transcription, histone modifications, and recombination events on wild-type and Eδ-deleted alleles in adult and fetal thymocytes. These studies led us to identify a specific, long-distance role for Eδ in promoting accessibility and recombination of the TRDV4 gene segment in fetal thymocytes. They have also provided some insights into the mechanisms by which accessibility is generated at Dδ and Jδ gene segments.
Materials and Methods
Mice
The following mouse strains were used: 129 (Jackson Laboratory), Rag2−/− (20) (Jackson Laboratory) on a 129 background, Eδ−/− (6) and Eδ−/−Rag2−/−. Eδ−/− mice were kindly provided by Barry Sleckman (Washington University, St. Louis MO). Strains carrying Eδ-deleted alleles were on a mixed background but carried the 129 Tcra/Tcrd locus. Adult thymi were harvested from three week old recombinase-deficient mice or four to six week old recombinase-sufficient mice. Fetuses were harvested from timed pregnancies, with the day of detection of a vaginal plug designated F0.5. All mice were used in accordance with protocols approved by the Duke University Animal Care and Use Committee.
Chromatin immunoprecipitation (ChIP)
Chromatin was prepared as described (21, 22). Thymocytes (0.5–10.0 × 106) were washed with Wash Buffer I (10 mM sodium butyrate, 5mM Na3EDTA, 0.15 mM spermine, 0.5 mM spermidine, 0.1 mM PMSF, 0.1 mM benzamidine, pH7.0, in PBS without Mg2+ and Ca2+) followed by Wash Buffer II (Wash Buffer I without Na3EDTA). Washed cells were then lysed in 200–400 μl of 80 mM NaCl, 10 mM Tris-HCl pH8.0, 10 mM sodium butyrate, 6 mM MgCl2, 1 mM CaCl2, 250 mM sucrose, 0.15 mM spermine, 0.5mM spermidine, 0.02% (vol/vol) NP40, 0.1 mM PMSF and 0.1 mM benzamidine for 5 min on ice. Nuclei were pelleted and washed once with 10 mM NaCl, 10 mM Tris-HCl pH8.0, 10 mM sodium butyrate, 3 mM MgCl2, 1 mM CaCl2 and 250 mM sucrose. Digestion was then performed to generate mainly mononucleosomes with a minor fraction of dinucleosomes, by incubation for 5 min at 37°C in 200 μl of the same buffer containing 5 U micrococcal nuclease (Worthington). The reaction was stopped by addition of 300 μl 10 mM Tris-HCl pH8.0, 5 mM EDTAand 10 mM sodium butyrate. Sonication was then conducted on ice for eight cycles of 15 seconds on and 20 seconds off using a Sonicator 3000 (Misonix) with the output set to 3.0. After centrifugation for 10 min at 18,000×g, the supernatant was transferred to fresh tube and Triton X-100 was added to a final concentration of 1% (vol/vol). Chromatin was then precleared with Protein A-Sepharose/salmon sperm DNA slurry (Millipore) and was subsequently incubated overnight at 4°C with anti-acetylated H3 (Millipore 06-599), anti-acetylated H4 (Millipore 06-598), anti-dimethylated H3K4 (Millipore 07-030), anti-trimethylated H3K4 (Millipore 04-745), anti-dimethylated H3K9 (Abcam ab1220), anti-trimethylated H3K27 (Millipore, 07-449) or control rabbit IgG (R&D Systems, ab-105-c). Protein A-Sepharose/salmon sperm DNA slurry was added for an additional 1 hr incubation, after which immunoprecipitates were washed vigorously and DNAs were purified. Immunoprecipitated and input DNAs were quantified by real time PCR using a Roche LightCycler and a FastStart DNA Master SYBR Green I kit (Roche). For immunoprecipitations using anti-acetylated H3, anti-acetylated H4, anti-dimethylated H3K4 and anti-trimethylated H3K4, analysis of β2-microglobulin (B2m) was used to normalize ratios of bound/input in different samples. Analysis of MageA2 was used to normalize immunoprecipitations using anti-dimethylated H3K9. Primers sequences are provided in Supplementary Table 1. PCR conditions were as follows: 5 min at 95°C followed by 45 cycles of 1 s at 95°C, 5 s at 62°C, 7 s at 72°C.
Germline transcription
RNA was extracted from unfractionated thymocytes using TRIzol (Invitrogen) according to the manufacturer’s instructions. Contaminating genomic DNA was removed by incubation with 1U DNase I (New England Biolabs) for 10 min at 37°C. SuperScript reverse transcriptase (Invitrogen) and random hexamer primers were used to synthesize cDNA according to the manufacturer’s instructions. PCR conditions were described previously (15). After agarose gel electrophoresis and transfer to nylon membranes, PCR products were detected by hybridization with a 32P-labeled oligonucleotide probe. Primer and probe sequences are listed in Supplementary Table 1. Real-time PCR was carried out as described above; primers are listed in Supplementary Table 1.
PCR and Southern blot analysis of rearrangements
Total or sorted DN3 thymocytes were lysed by incubation in 10 mM Tris-HCl pH8.0, 150 mM NaCl, 10 mM EDTA, 0.4% (wt/vol) SDS and 0.1 mg/ml proteinase K overnight at 37°C. Genomic DNA was prepared by phenol/chloroform extraction and ethanol precipitation. PCR conditions were as follows: 3 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, 1 min at 72°C; 5 min at 72°C. After agarose gel electrophoresis and transfer to nylon membranes, PCR products were detected by hybridization with 32P-labeled oligonucleotide probes. Primer and probe sequences are provided in Supplementary Table 1.
PCR analysis of RSS cleavage
Thymocyte genomic DNA obtained from recombinase-sufficient mice was analyzed by real-time PCR using primer pairs that define an RSS-spanning and a neighboring amplicon. Quantification was performed using standard curves constructed from amplification of thymocyte genomic DNA obtained from recombinase-deficient mice. Loss of the RSS amplicon in recombinase-sufficient thymocytes can result from its rearrangement or its deletion due to rearrangement of upstream RSSs; loss of the neighboring amplicon reflects its deletion only. Unrearranged RSS was calculated as residual RSS amplicon/residual neighbor amplicon to specifically reflect RSS loss due to RSS rearrangement. Primer sequences are provided in Supplementary Table 1. PCR conditions were as described for ChIP analysis.
OP9-DL1 culture
Monolayers of OP9-DL1 cells (23) were cultured in MEM Alpha (GIBCO) supplemented with 20% fetal bovine serum (Atlanta Biologicals), 1×Penicillin-Streptomycin (GIBCO) and 1mM sodium pyruvate (GIBCO). Antibodies PE-Cy 5 anti-CD4 (553654), PE-Cy 5 anti-CD8a (553034), PE-Cy 5 anti-CD3e (553065), FITC anti-CD25 (553072) and PE anti-CD44 (553134) were purchased from BD-Pharmingen. Anti-CD117 magnetic beads (18757) were purchased from Stemcell Technologies. CD117+ thymocytes were enriched by AutoMACS in Possels mode. Sorted DN1 (CD4−CD8−CD3−CD117+CD44+CD25−) thymocytes from adult 129 or Eδ−/− mice were then cultured on an OP9-DL1 monolayer in IMDM (GIBCO) supplemented with 5% fetal bovine serum, 1×penicillin/streptomycin, 1 mM sodium pyruvate, 55 μM 2-mercaptoethanol (GIBCO), 5 ng/ml rmFlt-3 Ligand (R&D Systems, 427-FL) and 5 ng/ml IL-7 (R&D Systems, 407-ML). After 14 days of culture in the presence or absence of 3 ng/ml trichostatin A (TSA, Sigma), DN3 cells (CD25+CD44−) were sorted for analysis.
Results
Eδ behaves as a local enhancer in adult DN thymocytes
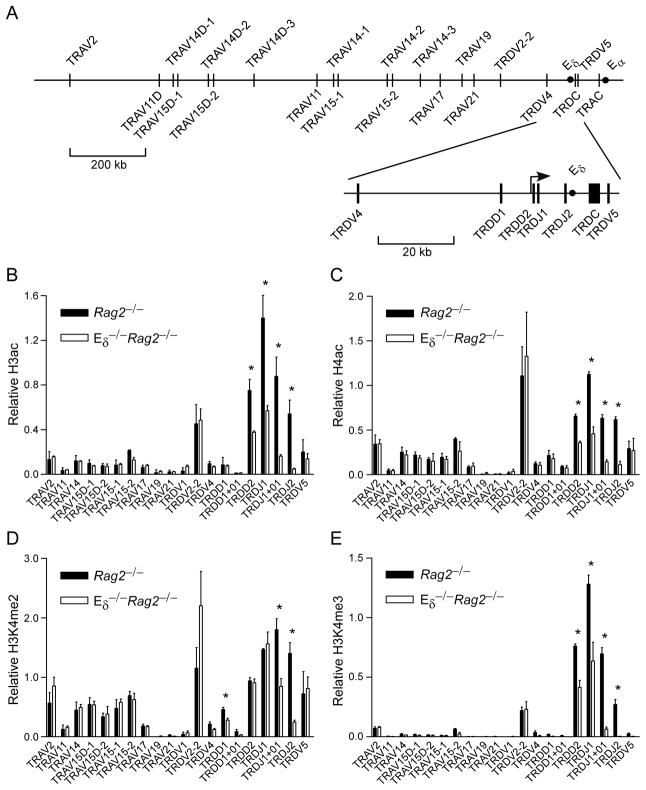
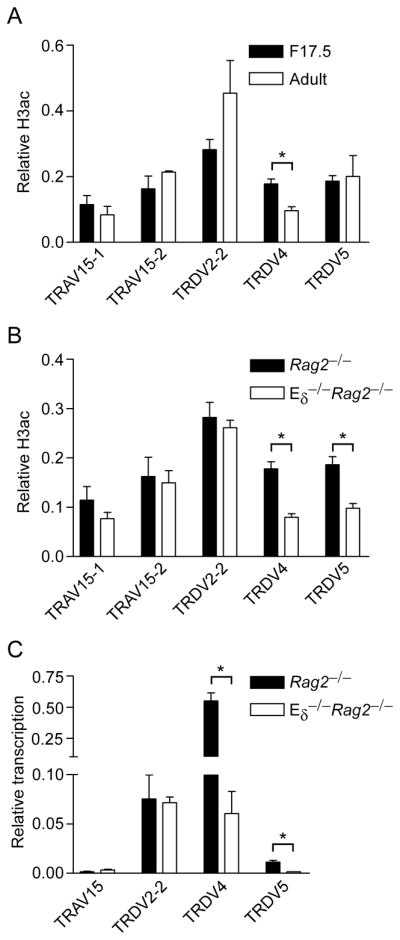
In our previous studies we observed that the Vδ gene segments that dominate the adult TCRδ repertoire (TRDV2-2, TRDV5, TRAV15D-1, TRAV15D-2, TRAV15-1 and TRAV15-2) (Fig. 1A) displayed elevated histone H3 acetylation (H3ac) and H4 acetylation (H4ac) and germline transcription in DN thymocytes of adult Rag2−/− mice (15). To investigate whether Vδ chromatin structure depends on Eδ, we used ChIP to monitor four histone modifications in thymocytes from Rag2−/− and Eδ−/− Rag2−/− mice. On wild-type alleles, we found H3ac and H4ac to be high (>50% of B2m) from TRDD2 (D2) to TRDJ2 (J2) (Fig. 1B, C). As noted previously (15), H3ac and H4ac were present at moderate (5-50% of B2m) to high levels at six dominant Vδ gene segments (TRDV2-2, TRDV5 and TRAV15 family) and at certain other V segments (TRAV2 and TRAV14 family). By the criteria noted above, we also detected moderate acetylation at TRAV17, TRDV4 and TRDD1. On Eδ-deficient alleles, H3ac and H4ac were reduced in the D2-to-J2 region, but we observed no substantial changes at V gene segments (Fig 1B, C).
Figure 1.
Influence of Eδ on Tcra/Tcrd locus histone modifications in adult DN thymocytes. A. Map of the Tcra/Tcrd locus depicting the relative positioning of gene segments analyzed in this study. Enhancers Eδ and Eα (circles), and the D2 promoter (bent arrow) are also depicted. Not all Tcra/Tcrd gene segments are shown. H3ac (B), H4ac (C), H3K4me2 (D) and H3K4me3 (E) were measured by ChIP using chromatin prepared from Rag2−/− and Eδ−/−Rag2−/− thymocytes. TRDD1+01 and TRDJ1+01 are sites situated 1kb downstream of D1 and J1, respectively. The data represent the mean ± SEM of three independent chromatin preparations for each genotype. Values of bound/input were expressed relative to those for B2m (normalized to one) in each sample. Note that PCR for TRAV11 detects both TRAV11 and TRAV11D; PCR for TRAV14 detects six members of the TRAV14 family (TRAV14D-1, D-2, D-3,-1,-2,-3). The significance of differences between Eδ−/− and wild-type were evaluated by two-tailed Student’s t-test: *, P<.01.
H3 lysine 4 dimethylation (H3K4me2) is a chromatin mark that is distributed across active genes; H3 lysine 4 trimethylation (H3K4me3) better correlates with transcriptional activity, is generally associated with the 5′ portion of active transcription units (24) and notably serves as a docking site for RAG2 protein during V(D)J recombination (25–27). Profiles of these histone modifications followed the trend of Eδ-dependence near Dδ and Jδ gene segments and Eδ-independence at more distal locations (Fig. 1D, E). However, unlike H3ac, H4ac and H3K4me3, we found that H3K4me2 was Eδ-independent at D2 and J1. Interpretation of this finding is difficult since H3K4me2 abundance varies in a complex way that is determined in part by its conversion to H3K4me3 at the 5′ end of active transcription units (24).
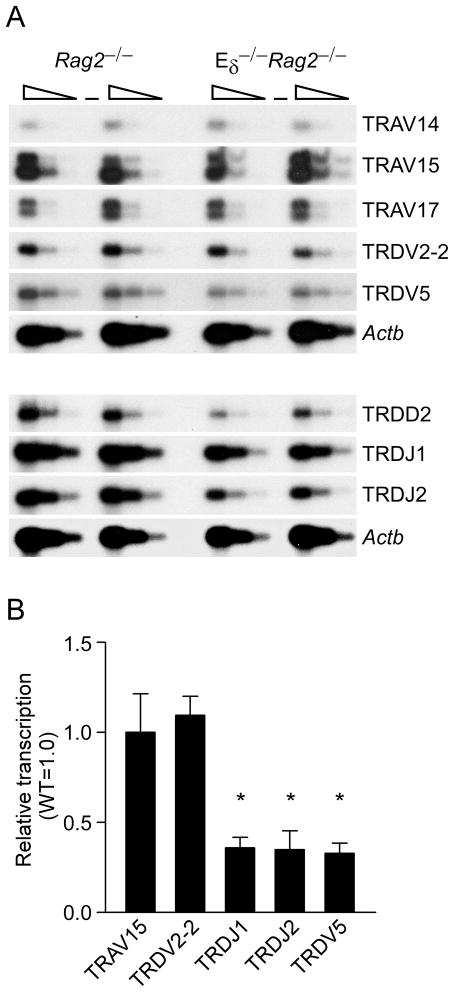
To extend these observations, we analyzed germline transcription of Vδ, Dδ and Jδ gene segments by semiquantitative RT-PCR (Fig. 2A). The results matched the chromatin data quite closely, as germline transcription at D2, TRDJ1 (J1) and J2 was reduced substantially on Eδ-deficient as compared to wild-type alleles, whereas (with the exception of TRDV5) germline transcription of V gene segments was unaffected. These conclusions were confirmed by quantitative real-time PCR, which detected approximately 70% reductions in transcription at J1, J2 and TRDV5, but not at the more distant TRAV15 and TRDV2-2 gene segments (Fig. 2B). We note that among the histone modifications tested, transcription correlated least well with H3K4me2, as reduced transcription at D2 and J1 occurred without concomitant reductions in this modification (Fig. 1D). However as noted previously, the abundance of H3K4me2 can be difficult to interpret. Reduced transcription at TRDV5 was consistent with previous data (18); however, we did not detect differences in H3ac, H4ac or H3K4me2 at this site on Eδ-deficient alleles. Nevertheless, although H3K4me3 at TRDV5 was very low on wild-type alleles, it was still 5–10-fold over the IgG control in different experiments (data not shown), and, consistent with the transcription data, averaged 8.5-fold over Eδ−/− alleles (Fig. 1E). Taken together, the chromatin and transcription data support the notion that Eδ functions as a local enhancer whose influence is limited to no more than about 10 kb in either direction in adult DN thymocytes.
Figure 2.
Influence of Eδ on Tcra/Tcrd locus germline transcription in adult DN thymocytes. A. Germline transcription was measured by RT-PCR using serial three-fold dilutions of cDNA (wedges) prepared from four independent cDNA preparations from Rag2−/− and Eδ−/−Rag2−/− thymocytes. Two preparations for each genotype are analyzed in the top set of panels; two different preparations for each genotype are analyzed in the bottom set of panels. (−) no reverse transcriptase. The TRAV14 and TRAV15 primers detect all members of the TRAV14 and TRAV15 families. B. Real-time PCR of germline transcription using cDNA preparations from Rag2−/− and Eδ−/−Rag2−/− thymocytes. The data represent the mean ± SEM of five independent cDNA preparations for each genotype, all normalized to values for β-actin (Actb). The values for Eδ−/−Rag2−/− for each site were then expressed relative to those for Rag2−/−, which were normalized to one. The significance of differences between Eδ−/− and wild-type were evaluated by two-tailed Student’s t-test: *, P<.05.
Eδ regulates TRDV4 and TRDV5 chromatin in fetal thymocytes
The nearest Vδ gene segment upstream of Eδ is TRDV4, at a distance of 55 kb. TRDV4 recombination is restricted to the fetal thymus, likely the result of a specific suppression mechanism in adult as has been described for Vγ3 and Vγ4 (28–31). To ask whether Eδ regulates TRDV4 chromatin we analyzed H3ac at the TRDV4 promoter using chromatin prepared from fetal day 17.5 (F17.5) thymocytes of Rag2−/− and Eδ−/−Rag2−/− mice. Consistent with the unique developmental profile of TRDV4 usage, TRDV4 displayed an 85% increase in H3ac in fetal as compared to adult thymocytes, whereas none of the other Vδ gene segments tested displayed similar increases (Fig. 3A). Moreover, elevated H3ac at TRDV4 in fetal thymocytes was clearly Eδ-dependent (Fig. 3B). H3ac at TRDV5 was Eδ-dependent at this stage as well (Fig. 3B).
Figure 3.
Influence of Eδ on Tcra/Tcrd locus histone acetylation and germline transcription in fetal thymocytes. H3ac was measured by ChIP using chromatin prepared from (A) F17.5 and adult Rag2−/− DN thymocytes and (B) F17.5 Rag2−/− and Eδ−/−Rag2−/− thymocytes. The data represent the mean ± SEM of three to five independent chromatin preparations for each genotype and developmental stage. Values of bound/input were expressed relative to B2m (normalized to one) in each sample. C. Germline transcription was measured by quantitative real-time PCR using cDNA prepared from F17.5 Rag2−/− and Eδ−/−Rag2−/− thymocytes. The data represent the mean ± SEM of two independent cDNA preparations for each genotype, all normalized to values for Actb. *, P<.05 by two-tailed Student’s t-test.
To confirm the conclusions from chromatin analysis, we measured germline transcription of Vδ gene segments in fetal thymocytes of Rag2−/− and Eδ−/−Rag2−/− mice (Fig. 3C). Consistent with the chromatin data, TRDV4 and TRDV5 both displayed substantial reductions in germline transcripts on Eδ-deficient alleles, whereas there were no significant changes in germline transcription of TRDV2-2 and TRAV15. These results demonstrate that Eδ supports TRDV4 and TRDV5 promoter activity and chromatin structure in fetal thymocytes.
Eδ regulates TRDV4 and TRDV5 recombination fetal thymocytes
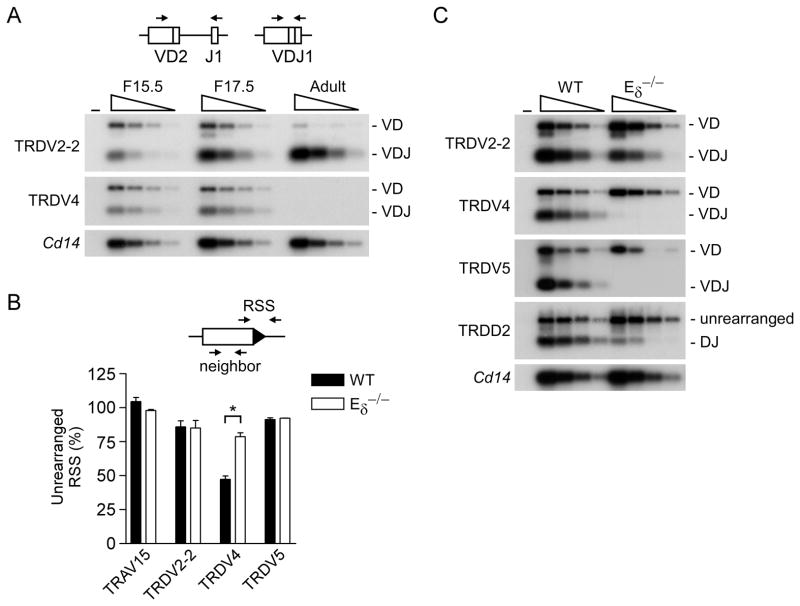
Based on the above results we predicted that Eδ would play a critical role in TRDV4 recombination in fetal thymocytes. To begin to test this, we first compared TRDV4 and TRDV2-2 recombination on wild-type alleles in F15.5, F17.5 and adult thymocytes (Fig. 4A). The Tcrd gene is unique among antigen receptor genes in that its recombination is not strictly ordered. Thus VD, DD and DJ recombination products are all detectable and fully rearranged VDJ alleles can presumably be assembled through multiple pathways (32). PCR using a V-specific primer and a J1 primer can amplify two species indicative of Tcrd recombination events, a smaller product arising from fully rearranged templates (VDJ1) and a larger product arising from partially rearranged templates (VD2) (Fig. 4A). Prior work has shown that D1 and D2 are both incorporated into Tcrd rearrangements in adult thymocytes whereas D2 is selectively incorporated in fetal thymocytes (13, 33). Our PCR strategy does not report on D1 usage, as amplicon sizes would be virtually indistinguishable regardless of its inclusion. Using this strategy, we detected complete (VDJ1) and partial (VD2) rearrangements of TRDV4 in wild-type F15.5 thymocytes. These rearrangements increased slightly in F17.5 thymocytes, but were undetectable in adult thymocytes (Fig. 4A). In contrast, we detected low levels of complete and partial TRDV2-2 rearrangements in F15.5 thymocytes and complete rearrangements increased in F17.5 and even moreso in adult thymocytes (Fig. 4A). Thus, TRDV4 rearrangement was restricted to the fetal period.
Figure 4.
Influence of Eδ on Tcrd gene rearrangement in fetal thymocytes. A. J1 rearrangement in 129 mice. Three-fold serial dilutions (wedges) of genomic DNA from thymocytes of F15.5, F17.5 and adult 129 mice were analyzed by PCR followed by Southern blot. PCR was performed using TRDV2-2 or TRDV4 primers in combination with a J1 primer; a 32P-labeled internal J1 oligonucleotide was used as a probe. Cd14 PCR insured use of similar amounts of DNA. (−), no DNA. The data are representative of three independent experiments. B. RSS cleavage in genomic DNA of wild-type (WT) 129 and Eδ−/− fetal thymocytes. Real-time PCR was used to quantify percent unrearranged RSS relative to a neighboring amplicon. TRAV15 primers detect all members of the TRAV15 family. The data represent the mean ± SEM of four and two independent genomic DNA preparations, respectively, from WT and Eδ−/− F17.5 thymocytes. *, P<.05 by two-tailed Student’s t-test. C. Tcrd rearrangement in fetal thymocytes of wild-type (WT) 129 and Eδ−/− mice. Genomic DNA samples from F17.5 thymocytes were analyzed by PCR using the indicated primers in conjunction with a J1 primer, followed by Southern blot using a 32P-labeled internal J1 oligonucleotide probe. The data are representative of two to three independent experiments.
Quantitative real-time PCR across Vδ RSSs was then used to assess RSS loss and thus the extent of Vδ rearrangement in fetal thymocytes. The results indicated that TRDV4 dominates Vδ rearrangement events in the fetal period, as it was rearranged on 52% of wild-type alleles (Fig. 4B). In contrast, TRDV2-2 was rearranged on 14% and TRDV5 on only 9% of wild-type alleles (Fig. 4B). TRAV15 rearrangement was not detectable using this approach, indicating that this Vδ family makes at best a very minor contribution to the fetal Vδ repertoire. Analysis of Eδ-deficient fetal thymocytes revealed TRDV4 rearrangement to be reduced by 60% (to 21% of alleles) (Fig. 4B). In contrast, reductions in TRDV2-2 and TRDV5 rearrangement were notapparent. Thus, Eδ is critical for high frequency TRVD4 rearrangement in the fetal thymus. To confirm and extend these findings, we performed PCR with V-specific and J1 primers to assess the effects of Eδ-deficiency on the production of complete (VDJ1) and partial (VD2) fetal Tcrd rearrangement events (Fig. 4C). Complete rearrangements involving TRDV4 were abolished on Eδ-deficient alleles. Rather, the residual TRDV4 rearrangements on Eδ-deficient alleles were exclusively in the form of TRDV4-D2 rearrangement intermediates. A similar recombination defect was observed for TRDV5 (Fig. 4C). Since there was no apparent quantitative defect in TRDV5 recombination as assessed by RSS loss (Fig. 4B), this result may indicate a selective defect in TRDV5-D2-to-J1 rearrangement in Eδ-deficient fetal thymocytes. However, because the frequency of TRDV5 recombination is low even in wild-type fetal thymocytes, the assay for RSS loss may not have had sufficient accuracy to distinguish TRDV5 usage on wild-type and Eδ-deficient alleles (Fig. 4B).
In contrast to TRDV4 and TRDV5, we detected a very mild defect in TRDV2-2 rearrangement, with complete rearrangements slightly reduced and partial rearrangements slightly increased (Fig. 4C). This result is consistent with the fact that TRDV2-2 RSS loss is similar on wild-type and Eδ-deficient alleles (Fig. 4B). Given that D2-to-J1 rearrangement is dramatically impaired on Eδ-deficient alleles (Fig. 4C), we conclude that TRDV2-2 rearrangement occurs primarily by way of a VD intermediate on these alleles, and that the VD-to-J step may be mildly impaired. In contrast TRDV4 rearrangement is substantially impaired at the V-to-D step, and TRDV4 and TRDV5 rearrangements are both blocked at the VD-to-J step.
Developmental regulation of TRDV4 recombination by histone acetylation
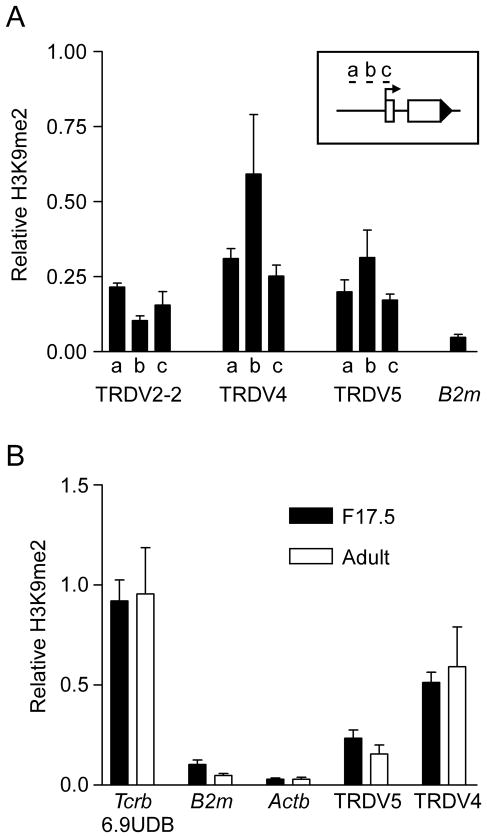
Our data indicated that Eδ can stimulate H3ac at TRDV4 in fetal thymocytes but cannot do so in adult thymocytes (Figs. 1B, 3A). We wondered whether potential effects of Eδ at TRDV4 in adult thymocytes were counteracted by suppressive histone modifications specifically targeted to TRDV4. In this regard, the suppressive histone modification H3 lysine 27 trimethylation (H3K27me3) was shown to be elevated at proximal VH gene segments in adult but not fetal pro-B cells (34) and to promote distal VH gene recombination in adults (35). However, we detected only low levels of H3K27me3 at TRDV4 in adult DN thymocytes (data not shown). We did detect the suppressive histone modification H3 lysine 9 dimethylation (H3K9me2) at levels that were substantially higher at TRDV4 than at TRDV2-2 or TRDV5 (Fig. 5A). Surprisingly, however, H3K9me2 at TRDV4 was similar in fetal and adult thymocytes (Fig. 5B), indicating that TRDV4 repression in adults is not a consequence of developmentally regulated H3K9me2.
Figure 5.
H3K9 dimethylation of Vδ gene segments in adult and fetal thymocytes. A. H3K9me2 of TRDV2-2, TRDV4 and TRDV5 was measured by ChIP using chromatin prepared from adult Rag2−/− thymocytes. Each V gene segment was analyzed at three sites (a, b, c, as diagrammed. Values of bound/input were expressed relative to MageA2 (normalized to one). The data represent the mean ± SEM of three independent chromatin preparations. B. H3K9me2 was compared in chromatin prepared from F17.5 and adult Rag2−/− thymocytes. The data represent the mean ± SEM of three to four independent chromatin preparations. TRDV4 and TRDV5 were analyzed at sites “b”. 6.9UDB is a positive control site within the Tcrb locus (39); B2m and Actb served as negative control sites.
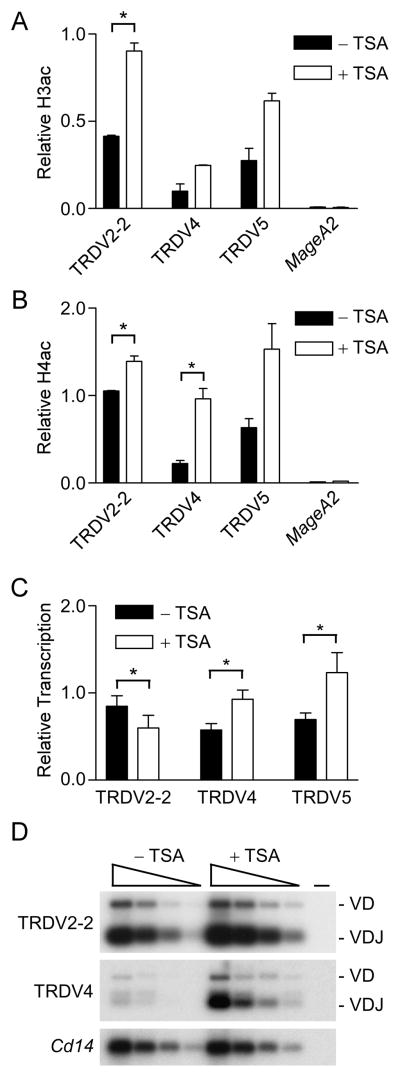
To assess whether TRDV4 recombination was suppressed in adult thymocytes by the observed loss of Eδ-dependent histone acetylation, we first asked whether TRDV4 histone acetylation could be modulated by inhibition of histone deacetylases (HDACs). Indeed, incubation of adult Rag2−/− thymocytes with HDAC inhibitor trichostatin A (TSA) increased H3ac and H4ac at TRDV2-2, TRDV4 and TRDV5 (Fig. 6A, B), although some of the differences noted fell just short of reaching statistical significance. In conjunction with these changes we detected increases in TRDV4 and TRDV5 germline transcription (Fig. 6C). We note that these increases could be more substantial, and the apparent decrease in TRDV2-2 transcription could be illusory, if TSA treatment were found to upregulate transcription of the control Actb gene that was used for normalization. We then tested the effects of TSA on TRDV4 rearrangement in 14 day cultures of DN thymocytes with OP9-DL1 stromal cells. TSA treatment stimulated more than a ten-fold increase in fully rearranged TRDV4 (Fig. 6D). In contrast, TSA had a relatively modest effect on rearrangement of TRDV2-2, likely because untreated adult thymocytes are already permissive for rearrangement of this Vδ gene segment. We conclude that the balance between histone acetyltransferase and HDAC activity differs at TRDV4 in fetal and adult thymocytes, and that this difference is causal in restricting TRDV4 usage to fetal thymocytes.
Figure 6.
Activation of Vδ gene segment chromatin and rearrangement using a HDAC inhibitor. H3ac (A) and H4ac (B) were measured by ChIP using chromatin prepared from adult Rag2−/− thymocytes that were cultured for 8 hrs with or without 3 ng/ml TSA. Values of bound/input were expressed relative to B2m (normalized to one) in each sample. The data represent the mean ± SEM of two to three independent chromatin preparations for each treatment. *, P<.05 by two-tailed Student’s t-test. C. Germline transcription was measured by quantitative real-time PCR using cDNA prepared from adult Rag2−/− thymocytes that were cultured for 8 hrs with or without 3 ng/ml TSA. The data represent the mean ± SEM of four independent cDNA preparations for each genotype, all normalized to values for Actb. *, P<.05 by two-tailed paired Student’s t-test. D. Adult 129 DN1 thymocytes were placed in culture for 14 days with or without 3 ng/ml TSA on OP9-DL1 stromal cells; DN3 thymocytes were sorted from cultured cells for preparation of genomic DNA. Three-fold serial dilutions (wedges) of genomic DNA were analyzed by PCR followed by Southern blot. PCR was performed using TRDV2-2 or TRDV4 primers in combination with a J1 primer; a 32P-labeled internal J1 oligonucleotide was used as a probe. Cd14 PCR insured use of similar amounts of DNA. (−), no DNA. The data is representative of three independent experiments.
Discussion
Although more than ten years has passed since Eδ was first shown to regulate Tcrd locus recombination events (6), a detailed characterization of the regulation of Tcrd locus chromatin accessibility has been lacking. In this study we provide new insights into the influences of Eδ on Tcrd locus chromatin, and how these influences support the recombination of Tcrd gene segments. We found that Eδ acts relatively locally to regulate chromatin structure and transcription in the 20 kb region extending from TRDD2 to TRDV5 in adult DN thymocytes. A relatively local influence of Eδ on Dδ and Jδ chromatin can readily explain the previously described recombination defect in Eδ-deficient mice, with reductions in complete VDJ-rearrangements and increases in VD and DD rearrangement intermediates (6). However, although Eδ regulates the nearby TRDV5, it appears to play no role in activating more distant Vδ gene segments in adult DN thymocytes. Thus, with the exception of TRDV5, Eδ does not function to define Tcra/Tcrd locus V gene segments as Vδ in these cells. Rather, this may be determined by unique features of individual V segment promoters. Notably, we found that in addition to regulating TRDV5, Eδ plays a distinct and unanticipated role in fetal thymocytes as a regulator of chromatin structure, transcription and recombination of fetal Vδ gene segment TRDV4. As a consequence, complete rearrangements of TRDV4 and TRDV5 were absolutely dependent on Eδ. The data for TRDV4 indicate that, at least in fetal thymocytes, Eδ can function over a distance of 55 kb.
A primary mechanism by which enhancers exert control over recombination is by activating transcription from germline promoters (11, 12). Germline transcription can then disrupt chromatin structure (36) and distribute histone modifications such as H3K4me3 that are crucial for recruitment and activation of the recombinase (25–27, 37). Tcrd gene recombination likely depends on the activities of distinct promoters that control transcription and accessibility of Vδ, Dδ and Jδ gene segments (8). In this context, the distinct recombination defects displayed by TRDV4, TRDV5 and TRDV2-2 in Eδ−/− fetal thymocytes are most easily explained by a hierarchy of promoter and accessibility defects on these alleles. We suggest that defective D2-to-J1 rearrangement predominantly reflects a major defect in J1 accessibility on Eδ-deficient alleles. We suspect, however, that there is only a modest defect in D2 accessibility; in this way, residual D2 accessibility can support levels of V-to-D2 rearrangement that vary largely as a function of V segment promoter activity and accessibility. Thus, for the Eδ-independent TRDV2-2, the frequency of V-to-D2 rearrangement on Eδ-deficient alleles is near that on wild-type alleles, whereas for the Eδ-dependent TRDV4, the frequency of V-to-D2 rearrangement is substantially reduced. Further, following initial TRDV2-2-to-D2 rearrangement, we suspect that potent Eδ-independent TRDV2-2 promoter activity can create substantial accessibility at both D2 and J1, providing the driving force for complete TRDV2-2-D2-J1 rearrangements on Eδ-deficient alleles. In contrast, for the Eδ-dependent TRDV4, reduced promoter activity on Eδ-deficient alleles is unable to support TRDV4-D2-to-J1 rearrangement following initial TRDV4-to-D2 rearrangement. Promoter and accessibility defects for TRDV5 are similar to those for TRDV4, thereby leading to a similar recombination profile. We note that our model for accessibility defects on Eδ−/− alleles is fully consistent with the unusually constellation of recombination intermediates previously detected on these alleles in adult thymocytes (6).
Although Eδ can activate TRDV4 in fetal thymocytes, it cannot do so in adult thymocytes. Previous work has demonstrated that the inability of fetal Vγ3 and Vγ4 gene segments to rearrange in adult thymocytes is due to suppression mediated by local promoter sequences (28, 30). TRDV4 usage may be regulated similarly. Notably, mice deficient in basic-helix-loop-helix transcription factor E2A display dysregulated rearrangement of Vγ3 and TRDV4 in adult thymocytes (29) implicating E2A as a suppressor of fetal Vγ and Vδ gene segments in adults. However it is not known whether this represents a direct effect of E2A on Vγ and Vδ promoters. Deletion of a TRDV4 promoter E box resulted in elevated TRDV4 promoter activity (38), but this result was obtained in transient transfection experiments and has not been further studied or confirmed in vivo. As in previous studies examining fetal Vγ3 gene rearrangement (30) we found that fetal TRDV4 rearrangement can be stimulated in adult DN thymocytes by artificially elevating histone acetylation through the use of HDAC inhibitor TSA. This suggests that the balance between histone acetyltransferase and HDAC activity at TRDV4 may regulate its developmental activation. Of particular interest, we found that the TRDV4 promoter displays unusually high levels of suppressive H3K9me2 in both adult and fetal thymocytes. Thus in adult thymocytes the TRDV4 histone modification profile is typical of a repressed gene, whereas in fetal thymocytes it appears to be a mosaic of histone modifications typical of both activation and repression. Prior data may be interpreted to indicate that fetal Vγ segment promoters are subject to a developmentally regulated suppressive mechanism in adult thymocytes (28–30). Our current data suggests that for TRDV4 at least a component of this suppression may be constitutive, and that even in fetal thymocytes, it may be primed for suppression in adults. Further studies will be required to better understand how the balance of additional suppressive and activating influences is modulated during development.
Supplementary Material
Acknowledgments
We thank Dr. Keji Zhao (NIH) for his ChIP protocol, Dr. Barry Sleckman (Washington University) for Eδ−/− mice, Dr. Motonari Kondo (Duke University) for OP9-DL1 cells and for use of the Auto-MACS, Dr. Mary Elizabeth Jones (Duke University) for advice with OP9-DL1 cultures, Dr. Yuan Zhuang for guidance on microdissection of fetal thymus, Eva Chung for help with the Auto-MACS, and Nancy Martin and Lynn Martinek of the Duke University Comprehensive Cancer Center Flow Cytometry Shared Resource for help with cell sorting. We also thank Dr. Shyam Unniraman and Han-yu Shih for critical review of the manuscript.
Abbreviations used in this paper
- ChIP
chromatin immunoprecipitation
- D2
TRDD2
- DN
double negative
- DP
double positive
- Eδ
Tcrd enhancer
- Eα
Tcra enhancer
- H3ac
histone H3 acetylation
- F15.5
fetal day 15.5
- F17.5
fetal day 17.5
- H4ac
histone H4 acetylation
- H3K4me2
histone H3 lysine 4 dimethylation
- H3K4me3
histone H3 lysine 4 trimethylation
- H3K27me3
histone H3 lysine 27 trimethylation
- H3K9me2
histone H3 lysine 9 dimethylation
- HDAC
histone deacetylase
- J1
TRDJ1
- J2
TRDJ2
- RSS
recombination signal sequence
- TSA
trichostatin A
Footnotes
This work was supported by National Institutes of Health Grant GM41052 (to M.S.K.)
References
- 1.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krangel MS, Carabana J, Abbarategui I, Schlimgen R, Hawwari A. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ locus. Immunol Rev. 2004;200:224–232. doi: 10.1111/j.0105-2896.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 3.Shih HY, Hao B, Krangel MS. Orchestrating T-cell receptor α gene assembly through changes in chromatin structure and organization. Immunol Res. 2010 doi: 10.1007/s12026-010-8181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauzurica P, Krangel MS. Temporal and lineage-specific control of T cell receptor α/δ gene rearrangement by T cell receptor α and δ enhancers. J Exp Med. 1994;179:1913–1921. doi: 10.1084/jem.179.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR α enhancer in αβ and γδ T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 6.Monroe RJ, Sleckman BP, Monroe BC, Khor B, Claypool S, Ferrini R, Davidson L, Alt FW. Developmental regulation of TCR δ locus accessibility and expression by the TCR δ enhancer. Immunity. 1999;10:503–513. doi: 10.1016/s1074-7613(00)80050-3. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez-Munain C, Sleckman BP, Krangel MS. A developmental switch from TCR δ enhancer to TCR α enhancer function during thymocyte maturation. Immunity. 1999;10:723–733. doi: 10.1016/s1074-7613(00)80071-0. [DOI] [PubMed] [Google Scholar]
- 8.Carabana J, Ortigoza E, Krangel MS. Regulation of the murine Dδ2 promoter by upstream stimulatory factor 1, Runx1, and c-Myb. J Immunol. 2005;174:4144–4152. doi: 10.4049/jimmunol.174.7.4144. [DOI] [PubMed] [Google Scholar]
- 9.Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP. Defect in rearrangement of the most 5′ TCR-Jα following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity. 1996;5:331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- 10.Hawwari A, Bock C, Krangel MS. Regulation of T cell receptor α gene assembly by a complex hierarchy of germline Jα promoters. Nat Immunol. 2005;6:481–489. doi: 10.1038/ni1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abarrategui I, Krangel MS. Regulation of T cell receptor-α gene recombination by transcription. Nat Immunol. 2006;7:1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 12.Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO J. 2007;26:4380–4390. doi: 10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber-Arden J, Wilbert OM, Kabelitz D, Arden B. Vδ repertoire during thymic ontogeny suggests three novel waves of γδ TCR expression. J Immunol. 2000;164:1002–1012. doi: 10.4049/jimmunol.164.2.1002. [DOI] [PubMed] [Google Scholar]
- 14.Bosc N, Lefranc MP. The mouse (Mus musculus) T cell receptor α (TRA) and δ (TRD) variable genes. Dev Comp Immunol. 2003;27:465–497. doi: 10.1016/s0145-305x(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 15.Hawwari A, Krangel MS. Regulation of TCR δ and α repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqual N, Gallagher M, Aude-Garcia C, Loiodice M, Thuderoz F, Demongeot J, Ceredig R, Marche PN, Jouvin-Marche E. Quantitative and qualitative changes in V-Jα rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J Exp Med. 2002;196:1163–1173. doi: 10.1084/jem.20021074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YN, Alt FW, Reyes J, Gleason M, Zarrin AA, Jung D. Differential utilization of T cell receptor TCRα/TCRδ locus variable region gene segments is mediated by accessibility. Proc Natl Acad Sci U S A. 2009;106:17487–17492. doi: 10.1073/pnas.0909723106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CY, Sleckman BP. Developmental stage-specific regulation of TCRα-chain gene assembly by intrinsic features of the TEA promoter. J Immunol. 2007;179:449–454. doi: 10.4049/jimmunol.179.1.449. [DOI] [PubMed] [Google Scholar]
- 19.Bassing CH, Tillman RE, Woodman BB, Canty D, Monroe RJ, Sleckman BP, Alt FW. T cell receptor (TCR) α/δ locus enhancer identity and position are critical for the assembly of TCR δ and α variable region genes. Proc Natl Acad Sci U S A. 2003;100:2598–2603. doi: 10.1073/pnas.0437943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 21.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 22.Cuddapah S, Barski A, Cui K, Schones DE, Wang Z, Wei G, Zhao K. Native chromatin preparation and Illumina/Solexa library construction. Cold Spring Harb Protoc. 2009:pdb prot5237. doi: 10.1101/pdb.prot5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 24.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker JE, Cado D, Raulet DH. Developmentally programmed rearrangement of T cell receptor Vγ genes is controlled by sequences immediately upstream of the Vγ genes. Immunity. 1998;9:159–168. doi: 10.1016/s1074-7613(00)80598-1. [DOI] [PubMed] [Google Scholar]
- 29.Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agata Y, Katakai T, Ye SK, Sugai M, Gonda H, Honjo T, Ikuta K, Shimizu A. Histone acetylation determines the developmentally regulated accessibility for T cell receptor γ gene recombination. J Exp Med. 2001;193:873–880. doi: 10.1084/jem.193.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong N, Baker JE, Kang C, Raulet DH. The genomic arrangement of T cell receptor variable genes is a determinant of the developmental rearrangement pattern. Proc Natl Acad Sci U S A. 2004;101:260–265. doi: 10.1073/pnas.0303738101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chien YH, Iwashima M, Wettstein DA, Kaplan KB, Elliott JF, Born W, Davis MM. T-cell receptor δ gene rearrangements in early thymocytes. Nature. 1987;330:722–727. doi: 10.1038/330722a0. [DOI] [PubMed] [Google Scholar]
- 33.Elliott JF, Rock EP, Patten PA, Davis MM, Chien YH. The adult T-cell receptor δ-chain is diverse and distinct from that of fetal thymocytes. Nature. 1988;331:627–631. doi: 10.1038/331627a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu CR, Schaffer L, Head SR, Feeney AJ. Reciprocal patterns of methylation of H3K36 and H3K27 on proximal vs. distal IgVH genes are modulated by IL-7 and Pax5. Proc Natl Acad Sci U S A. 2008;105:8685–8690. doi: 10.1073/pnas.0711758105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 36.Kondilis-Mangum HD, Cobb RM, Osipovich O, Srivatsan S, Oltz EM, Krangel MS. Transcription-dependent mobilization of nucleosomes at accessible TCR gene segments in vivo. J Immunol. 2010;184:6970–6977. doi: 10.4049/jimmunol.0903923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kienker LJ, Ghosh MR, Tucker PW. Regulatory elements in the promoter of a murine TCRD V gene segment. J Immunol. 1998;161:791–804. [PubMed] [Google Scholar]
- 39.Carabana J, Watanabe A, Hao B, Krangel MS. A barrier-type insulator forms a boundary between active and inactive chromatin at the murine TCRβ locus. J Immunol. 2011;186:3556–3562. doi: 10.4049/jimmunol.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.