Abstract
A missense polymorphism in the NRG1 gene, Val > Leu in exon 11, was reported to increase the risk of schizophrenia in selected families from the Central Valley region of Costa Rica (CVCR). The present study investigated the relationship between three NRG1 genetic variants, rs6994992, rs3924999, and Val > Leu missense polymorphism in exon 11, in cases and selected controls from an isolated population from the CVCR. Isolated populations can have less genetic heterogeneity and increase power to detect risk variants in candidate genes. Subjects with bipolar disorder (BD, n = 358), schizophrenia (SZ, n = 273), or unrelated controls (CO, n = 479) were genotyped for three NRG1 variants. The NRG1 promoter polymorphism (rs6994992) was related to altered expression of NRG1 Type IV in other studies. The expression of NRG1 type IV in the dorsolateral prefrontal cortex (DLPFC) and the effect of the rs6994992 genotype on expression were explored in a postmortem cohort of BD, SZ, major depressive disorder (MDD) cases, and controls. The missense polymorphism Val > Leu in exon 11 was not significantly associated with schizophrenia as previously reported in a family sample from this population, the minor allele frequency is 4%, thus our sample size is not large enough to detect an association. We observed however an association of rs6994992 with NRG1 type IV expression in DLPFC and a significantly decreased expression in MDD compared to controls. The present results while negative do not rule out a genetic association of these SNPs with BD and SZ in CVCR, perhaps due to small risk effects that we were unable to detect and potential intergenic epistasis. The previous genetic relationship between expression of a putative brain specific isoform of NRG1 type IV and SNP variation was replicated in postmortem samples in our preliminary study.
Keywords: neuregulin 1 isoform expression, schizophrenia, isolated population, Costa Rica, bipolar disorder, major depressive disorder, hippocampus, dorsolateral prefrontal cortex
1. Introduction
Schizophrenia (SZ) is a debilitating brain disease characterized by delusions, hallucinations, decreased emotional affect, paranoia, and motor deficiencies (Liddle et al. 1994). Though the exact neurobiology of schizophrenia is not wholly understood, family, twin, and adoption studies have demonstrated that schizophrenia is a complex disease with a significant genetic component. First-degree biological relatives of patients with schizophrenia have an estimated 10% risk of developing the disease, compared to 1% for the general population (Gottesman and Erlenmeyer-Kimling 2001). In twin studies, concordance rates of 41% – 65% have been seen in monozygotic twins compared to 0% – 28% in dizygotic schizophrenic twins, suggesting heritability as high as 85% (Tsuang et al. 2001). In light of this evidence, much effort has been directed toward the discovery of genes that increase the risk of schizophrenia.
Schizophrenia linkage to chromosome 8p has been identified in multiple studies (DeLisi et al. 2002; Kaufmann et al. 1998; Levinson et al. 1996; Pulver et al. 1995; Shaw et al. 1998). Neuregulin 1 (NRG1) is located at 8p12 and is involved in neurodevelopment, regulation of glutamate, and synaptic plasticity (Tosato et al. 2005). Stefansson et al (Stefansson et al. 2002) first reported an association between NRG1 and schizophrenia in an Icelandic population via a haplotype (HAPICE) consisting of five SNPs (SNP8NRG221132, SNP8NRG221533, SNP8NRG241930, SNP8NRG243177, SNP8NRG433E1006) and two microsatellite markers (478B14-848, 420M91395) located at the 5′ end of the gene that doubled the risk for the disorder. Since this initial report, confirmatory studies of NRG1 association have been reported in different populations in Scotland (Stefansson et al. 2003), China (Li et al. 2004), Hungary (Keri et al. 2009), Japan (Fukui et al. 2006), Sweden (Alaerts et al. 2009), and a second Scottish cohort (Thomson et al. 2007), though the associated haplotype varies between studies. Negative studies of association have also been reported in Japan (Ikeda et al. 2008; Iwata et al. 2004), Ireland (Thiselton et al. 2004), Denmark (Ingason et al. 2006), Spain (Rosa et al. 2007), and the United States (Crowley et al. 2008).
Variation in NRG1 has been found to be associated with various biological indices that are known to underlie schizophrenia. For example, brain imaging measurements of the anterior cingulate (Wang et al. 2009), prefrontal functioning (Mechelli et al. 2008; Mechelli et al. 2009), overall white matter integrity (Zuliani et al. 2011), excitatory synapse development (Ting et al. 2011), GABA interneuron dysfunction (Ting et al. 2011), and immune system dysregulation (Marballi et al. 2010; Shibuya et al. 2010)have been linked to genetic variation in NRG1. These observations contribute to the validity of the association of NRG1 with schizophrenia and its likeliness to contribute to the underlining basis for the development of this illness.
A novel missense mutation present in NRG1 (Val > Leu in exon 11) that increased the risk of schizophrenia in individuals from Costa Rica was reported in a family based association analysis (Walss-Bass et al. 2006a). Our research group has been independently studying the genetics of schizophrenia in families and unrelated individuals from the same region of the Central Valley of Costa Rica (CVCR) (Bertisch et al. 2009; Cooper-Casey et al. 2005; DeLisi et al. 2002); therefore we attempted to replicate the missense association with the Val > Leu polymorphism in exon 11 in this study. We do not believe that we have any overlap in samples with those reported in the Walss-Bass et al. (2006) paper (Walss-Bass et al. 2006a), although we have not made a formal comparison due to potential IRB ramifications. We also selected rs3924999 for analysis as it has been linked to lower prepulse inhibition, an endophenotype of schizophrenia (Hong et al. 2008), and for its association with schizophrenia in a Chinese Han cohort (Zhang et al. 2009). In addition rs6994992 was selected based on its inclusion in the HAPICE risk haplotype (Stefansson et al. 2002), and based on evidence that the T/T genotype is associated with decreased activation of frontal and temporal lobe regions and increased risk of psychosis (Hall et al. 2006). This SNP was also selected for investigation for its function in promoting expression of the NRG1 type IV transcript in postmortem tissue (Law et al. 2006);(Shamir and Buonanno 2010). Therefore, we studied the expression of NRG1 type IV in postmortem brain sample and association with genotypes of rs6994992. Though there is NRG1 intragenic epistasis between 5′ and 3′ markers (Nicodemus et al. 2010) in functional imaging studies, our group selected the present markers to attempt replication of implicated SNPs that were also functionally relevant to the CVCR collection. The SNPs chosen for this study were selected primarily based on conclusions published in the literature when designing the study. As associations between schizophrenia and the HAPICE haplotype have been both supported and refuted across varying populations, we were interested in adding the CVCR results to the pool of association data. rs6994992 was also selected based on reports of its function in promoting expression of the NRG1 type IV transcript in postmortem tissue. Finally, cost of the materials needed to test a wider range of markers was also a factor in determining which SNPs to investigate. As this was a preliminary association study to test our samples with these three implicated SNPs, the present data can be added to meta-analysis using SNPs in NRG1.
In addition to schizophrenia and control subjects, we have included bipolar subjects in the genotyping and brain gene expression as well. Bipolar disorder (BD) illness affects approximately 0.8 – 1.6% of the population (Kessler et al. 1997) and is characterized by cyclical episodes of mania and depression, with a return to normal state between episodes (Berns and Nemeroff 2003). There is a significant genetic component to BD based upon twin studies; BD has an estimated heritability as high as 93% (Kieseppa et al. 2004). Though schizophrenia and bipolar disorder have been historically categorized as divergent psychopathologies, there is a growing body of evidence suggesting that causative commonalities exist between the two disorders (Badner and Gershon 2002; Berrettini 2003). To date, multiple studies have shown schizophrenia implicated loci and genes having a positive association to BD (Craddock et al. 2006; Harrison and Weinberger 2005), including 8p12 (Park et al. 2004) and NRG1 (Green et al. 2005; Prata et al. 2009). NRG1 in particular has also been shown to possibly play a role in bipolar psychopathology, pointing to a common involvement of this cell-cell interaction and growth involved protein in both bipolar disorder and schizophrenia (Georgieva et al. 2008; Thomson et al. 2007).
A population sample from the central valley of Costa Rica was chosen for this study due to its geographical isolation and genetic homogeneity. The Costa Rican genome is comprised of largely European and Amerindian ancestry (Morera et al. 2003), as a result of six waves of Spanish colonization and admixture with the indigenous Amerindians (Carlos, 1997). It is estimated that fewer than 1000 families gave rise to the three million residents of the CVCR (Mathews et al. 2004), and due to mountainous boundaries and dense lowland jungle, the CVCR population has remained isolated from immigration and emigration for 500 years. Consequently, the CVCR region presents a homogenous population perfect for studying genes underlying complex genetic disorders. Chromosomal areas of interest have already been identified in this cohort for both schizophrenia (Cooper-Casey et al. 2005; DeLisi et al. 2002; Walss-Bass et al. 2006b) and bipolar disorder (Freimer et al. 1996).
While large association studies have shown the relative value of identifying common variants that contribute statistically significant associations, there are usually small relative risks for disease attributable to an individual SNP (e.g. (Purcell et al. 2009; Ruderfer et al. 2011). However, family based studies in relatively isolated populations can offer knowledge about regions of interest that might contain rare variants in pathways that have etiological relevance to schizophrenia. Thus, the CVCR collection offers a resource for exploring the effects of genes that have been largely implicated in multiple studies, and perhaps can increase the association signal by increased genetic homogeneity that is lacking in larger association studies. Thus, as sample sizes for schizophrenia and bipolar disorder are projected to be larger in the future, the relative risks attributable to a single variant will most likely be decreasing due to genetic heterogeneity. With a modest sample size pursued in the present study, there is adequate power in pursuing biologically based genes on an a priori basis. The purpose of this study is to examine NRG1 SNPs previously associated with SZ in a geographically isolated, relatively homogenous population from the CVCR.
2. Materials and Methods
Sample Collection
Subjects were recruited with the approval of the Ministry of Health of Costa Rica and the ethics committee for the Hospital Nacional Psiquiatrico and by the Institutional Review Board at the University of California at Irvine. Unrelated individuals with diagnoses of schizophrenia (n = 273) and bipolar disorder (n = 358) whose four grandparents were of Spanish descent were obtained by screening patients admitted to the National Psychiatric Hospital of Costa Rica as previously described(DeLisi et al. 2001; DeLisi et al. 2002). Control subjects (n = 479) with the same ancestral criteria were recruited from large companies via questionnaires administered by the companies’ health services department. Controls were selected if no family history of schizophrenia, bipolar disorder, suicide or hospitalization for psychiatric reasons were present, and if self-reports of psychosis, diagnosis of schizophrenia, bipolar disorder, use of medications for depression or psychiatric conditions, and suicide attempts were negative. Interviews of affected subjects were conducted with the Diagnostic Interview for Genetic Studies (Nurnberger et al. 1994) using a translated Spanish DIGS 2 (DeLisi et al. 2001) and diagnoses were based upon DSM-IV criteria (Association 1994) by a consensus of two independent local psychiatrists. A third independent psychiatrist made a final diagnosis, based on family history, medical records, and a summary of personal interviews. All participants gave written informed consent for participation.
DNA Extraction
Blood samples were collected from participants in Costa Rica and sent to the laboratory at UC Irvine. DNA was isolated from these samples via 10% SDS and Proteinase K digestion, phenol-chloroform extraction, followed by a sodium acetate precipitation (Bell et al. 1981). Purity and concentration were assessed by 260 nm and 280 nm absorbances on the SpectraMax Plus spectrophotometer (Molecular Devices, Sunnyvale, CA) and aliquots were diluted to a working concentration of 2.5ng/μl.
Genotyping
DNA samples were genotyped for NRG1 SNPs rs3994992 and rs6994992 using pre-validated TaqMan 5′-allele discrimination assays (Applied Biosystems, Foster City, CA). The third polymorphism G/T in exon 11 (Val > Leu) was genotyped with a custom TaqMan assay using the following reference sequence in the Walss-Bass study (Walss-Bass et al. 2006a): GAACATGGACAATGTCATGCAGCATGCCCACTGTTTGGTTGTAGTCAGTCCTGGCAAGTGGAAGTGACCTGTGATGACATCTGCTCTCATCCCTTTCCAGAGGCGGAGGAGCTGTACCAGAAGAAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGCATCATGTGT[G/T]TGGTGGCCTACTGCAAAACCAAGTAAACCTTCTTTCTCCATGCCTTTCTCTCTCCTTCATGCAGAGACAGCTTAGATGGCCAGGGCTTTGCAGAATCTGAGCTCCACAGCCTAGTCTTGGGG. The Walss-Bass assay was performed on an ABI Prism 7000 Sequence Detection system, in a total reaction of 25 μl (4 μl DNA, 12.5 μl TaqMan Universal PCR Master Mix, No AmpErase UNG, 1.25μl 40× TaqMan assay, 7.25 μl H2O) using the following amplification conditions: denaturation at 95 °C for 10 minutes, followed by 40 cycles at 95 °C for 15 seconds and at 55 °C for 1 minute. Individual PCR reactions for rs3924999 and rs6994992 were carried out on an ABI Prism 7900 Sequence Detection system in a total reaction of 12 μl (4 μl DNA, 5 μl TaqMan Universal PCR Master Mix, No AmpErase UNG, 0.5μl 40× TaqMan assay, 2.5 μl H2O) using the following amplification protocol: denaturation at 95 °C for 10 minutes, followed by 50 cycles at 92 °C for 15 seconds and at 58 °C for 1.5 minutes. The genotype of each sample was determined by measuring allelic-specific fluorescence using SDS 2.3 software for allelic discrimination (Applied Biosystems).
Genotype and allelic statistical analyses were performed using Yates corrected χ2 for continuity and Fisher’s Exact Test for analyses that contained low cell numbers. Deviation from Hardy–Weinberg equilibrium (HWE) was tested using on line calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl).
Brain gene expression
Brain samples were obtained at the UC Irvine/UC Davis Brain Repository through a uniform process approved by the Institutional Review Board. Postmortem diagnoses were made through an extensive review of multiple sources of information including the medical examiner’s conclusions, coroner’s investigation, medical and psychiatric records, toxicology results, interviews of the decedents’ next-of-kin and a neuropathological examination as previously described (Tomita et al. 2004; Vawter et al. 2006). The human brain dissection and freezing protocol were performed as previously described (Jones et al. 1992; Vawter et al. 2006) and brains were stored in −80°C freezers until further dissected. RNA was extracted from 100 mg samples of dissected brain using a standard Trizol (Life Technologies, Carlsbad, California) procedure. Integrity of total RNA was evaluated via 18S and 28S ratios and RNA integrity numbers (RIN) using the 2100 Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). Only samples with pH values higher than 6.5 were included in the analysis. cDNA was synthesized with oligo dT primers using the Superscript procedure (Invitrogen, Carlsbad CA). Expression levels of NRG1 IV were interrogated by TaqMan expression assay (ABI) using the same primer and probe sequence provided in the Law et al. study in which differences in gene expression for this subtype were observed (Law et al. 2006): forward 5′GCTCCGGCAGCAGCAT3′; reverse 5′GAACCTGCAGCCGATTCCT3′; internal Probe 5′ FAM ACCACAGCCTTGCCT-MGB-3′. Since the NRG1 E187 exon has high homology with other genes (Steinthorsdottir et al. 2004), a TaqMan probe provided specificity for amplification of the NRG1 gene as the probe spanned the junction of the E187 - Ig1 exons (Law et al. 2006).
After amplification, the PCR product was run on a 2% agarose gel, the amplicon’s specificity was confirmed by sequencing using the same primers used in the TaqMan assay. We compared the NRG1 type IV TaqMan expression assay results for diagnoses groups (Table 1) to controls by an analysis of covariance taking into account age, sex, and RNA quality. The TaqMan expression was normalized with GAPDH. Brain gene expression levels were compared in bipolar disorder (n = 10), control (n = 22), MDD (n = 13) for a total of 45 postmortem subjects that had genotypes for NRG1 rs69949992.
Table 1.
The number of subjects for expression of NRG1 type IV isoform by genotype shown in Figure 1. There were no subjects with SZ that are homozygous for the T allele, so those subjects were not included in the analysis
| Diagnosis * rs69949992 genotype | C/C | C/T | T/T | Total |
|---|---|---|---|---|
| BP | 2 | 5 | 3 | 10 |
| Control | 8 | 11 | 3 | 22 |
| MD | 4 | 6 | 3 | 13 |
| Total | 14 | 22 | 9 | 45 |
Results
Association testing of polymorphisms
The genotype counts for each SNP are shown in Table 2. The recessive and dominant association tests for three NRG1 SNPs genotyped were not significant with either BD or SZ (Table 3). There was a trend for SNP (Val > Leu exon 11) in BD cases only to not be in Hardy Weinberg equilibrium (nominal p = 0.043).
Table 2.
Genotype frequencies for three SNPs in association study of CVCR subjects with bipolar disorder (BD) and schizophrenia (SZ)
| NRG1 Exon 11 (Val>Leu) | BD | SZ | Control | Total |
|---|---|---|---|---|
| G/G | 327 | 256 | 446 | 1029 |
| G/T | 28 | 17 | 33 | 78 |
| T/T | 3 | 0 | 0 | 3 |
| Total | 358 | 273 | 479 | 1110 |
| rs6994992 | BD | SZ | Control | Total |
| C/C | 149 | 116 | 201 | 466 |
| C/T | 159 | 119 | 218 | 496 |
| T/T | 50 | 38 | 60 | 148 |
| Total | 358 | 273 | 479 | 1110 |
| rs3924999 | BD | SZ | Control | Total |
| A/A | 40 | 27 | 62 | 129 |
| A/G | 159 | 132 | 209 | 500 |
| G/G | 159 | 114 | 208 | 481 |
| Total | 358 | 273 | 479 | 1110 |
Table 3.
The NRG1 association results for BD and SZ were not significant for the study of CVCR subjects
| Model | SNP | Disorder | |||
|---|---|---|---|---|---|
| NRG1 Exon 11 | BD | SZ | |||
| Recessive | Yates Chi-Square/p | Pearson Chi-Square/p | Yates Chi-Square/p | Pearson Chi-Square/p | |
| 0.68/0.40 | 0.91/0.34 | 0.04/0.84 | 0.12/0.72 | ||
| Dominant | Fisher exact p | Fisher exact p | |||
| 0.077 | 1 | ||||
| rs6994992 | |||||
| Recessive | Yates Chi-Square/p | Pearson Chi-Square/p | Yates Chi-Square/p | Pearson Chi-Square/p | |
| 0/1 | 0.01/0.92 | 0/1 | 0.02/0.88 | ||
| Dominant | Yates Chi-Square/p | Pearson Chi-Square/p | Yates Chi-Square/p | Pearson Chi-Square/p | |
| .26/.61 | .37/.54 | .19/.66 | .3/.58 | ||
| rs3924999 | |||||
| Recessive | Yates Chi-Square/p | Pearson Chi-Square/p | Yates Chi-Square/p | Pearson Chi-Square/p | |
| .45/.50 | .6/.43 | 1.28/0.25 | 1.55/0.21 | ||
| Dominant | Yates Chi-Square/p | Pearson Chi-Square/p | Yates Chi-Square/p | Pearson Chi-Square/p | |
| 0.05/.82 | .08/.77 | .13/.71 | .2/.65 |
Expression of NRG1 Type IV in brain
Two brain regions (hippocampus and DLPFC) were analyzed for expression differences of NRG1 Type IV. For the hippocampus results, there were no statistically significant differences in expression for SZ, BD, or MDD cases compared to controls; however, in the DLPFC, the MDD cases (n = 12) showed decreased expression (p = 0.004) compared to controls (n = 22) when pH, age, RIN, and gender are included in the ANCOVA model. The RIN factor was significant (p = 0.05). The hippocampus showed very low levels of amplification indicating low expression of intact poly-adenylated mRNA.
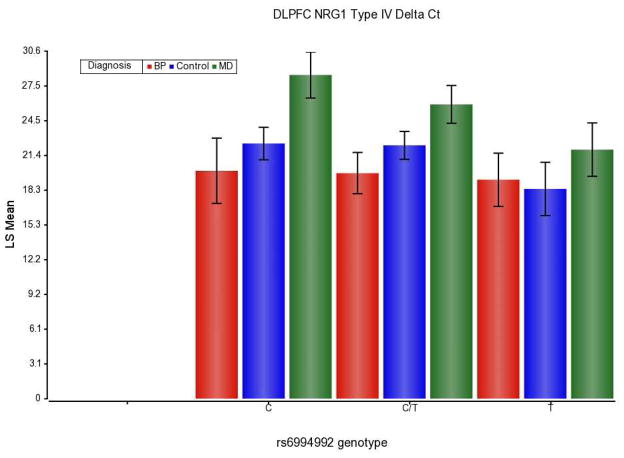
We next investigated the rs6994992 genotype effect on NRG1 type IV expression in hippocampus and DLPFC. The genotype effect was significant (p = 0.040) only in DLPFC; however there were no homozygous T carriers in the SZ group. We did confirm that NRG1 type IV expression was increased in the TT compared to the CC group in the DLPFC and that the effect was significant in the direction previously reported as the TT genotype showed a significantly increased expression (p = 0.024) of 15.3 fold compared to CC genotype (Figure 1). The CT group also showed a significant difference compared to the TT group (p = 0.023); again, the direction supported the dominant effect of the T allele increasing expression of the NRG1 type IV expression. The CT group did not show a significant difference from the CC group (p = 0.83). Interestingly, the MDD subjects showed a decreased expression compared to controls for both the CC and CT genotype group comparisons (p = 0.009, p = 0.018), the TT genotypes were not different comparing expression of NRG1 type IV between MDD and controls (p = 0.21).
Figure 1.
The delta Ct values for NRG1 type IV expression in DLPFC by genotype and diagnosis were analyzed with an ANCOVA. The Diagnosis main effect was significant (p = 0.0015), Diagnosis x Genotype was not significant (p-value = 0.68). The Diagnosis main effect was largely due to a decrease in MDD DLPFC NRG1Type IV expression reduction of 33.5 fold compared to controls (p = 0.002) while BD was not changed significantly in DLPFC (p = 0.51). The Genotype effect was significant (p = 0.040). The TT genotype showed a significantly increased expression (p = 0.024) of 15.3 fold compared to CC genotype. Since schizophrenia subjects did not have any TT genotypes, they were omitted from this analysis. The LS Mean (y-axis) is the delta Ct values adjusted for age, RIN, and sex. The bar graph is an inverse to the relative amount of the NRG1 type IV expression, higher bar indicates lower expression since the figure uses Ct values. The levels have been normalized to the endogenous reference gene GAPDH.
4. Discussion
Within the isolated CVCR population, this study failed to find an association with schizophrenia or bipolar disorder testing three NRG1 SNPs. Thus while our study had 80% – 86% power for SZ and BD, respectively to find association for two of the more common SNPs (Skol et al. 2007) with risk ratio of ≥ 1.37, it was not adequately powered for the rarer exon 11 missense SNP. This minor allele frequency requires a larger sample to definitively test for association. Though there are positive reports for NRG1 as both a schizophrenia and bipolar susceptibility gene for one or more of these SNPs, this hypothesis has not been consistently proven in the literature as shown in the introduction. Although we tested the same missense mutation previously associated in a family sample in CVCR (Walls-Bass, 2006), we did not replicate these findings in a larger case - control analysis from the same CVCR population, perhaps due to this rare SNP frequency of 4%.
Additionally, we confirmed a prior report of an association between higher expression of NRG1 type IV and the rs6994992 T/T genotype in the DLPFC as previously reported (Law et al. 2006) (Shamir and Buonanno 2010). We also report the preliminary finding of decreased NRG1 type IV expression in MDD in the DLPFC. Although previous studies have not found an association between NRG1 SNPs and MDD in a large sample of European ancestry (Schosser et al. 2010), there was decreased NRG1-alpha protein in MDD and SZ in the prefrontal cortex (Bertram et al. 2007). Since NRG1 has strong pleiotropic effects related to growth factor signaling in the brain, the findings of decreased NRG1 expression in prefrontal cortex could lend support to the overall growth factor hypothesis of depression (Evans et al. 2004).
Two caveats to the postmortem findings are that we cannot rule out the effect of antidepressant medications without animal studies as a potential cause of this decrease in MDD. The decreased expression in DLPFC of NRG1 type IV in MDD requires additional postmortem studies in part based upon small sample size, for validation, but is consistent with a reported decrease of NRG1 protein in MDD frontal cortex. In conclusion, while the present findings do not support association of the NRG1 variants in the CVCR population with schizophrenia or bipolar disorder, the tested SNPs might have some intergenic epistasis or be of a smaller effect size that we do not have power to detect. The present findings continue to show that there is a robust effect of genetic variation on NRG1 type IV expression which is thought to be brain-specific and with multiple impacts on brain function.
Acknowledgments
Support was received from the William Lion Penzner Foundation, Pritzker Neuropsychiatric Disorders Research Consortium for support of the collection and genotyping of the Costa Rica samples and the NIMH Conte Center funding P50 MH60398 for brain sample collection. We appreciate the UC Irvine Davis staff for contributions to brain tissue acquisition, and the Costa Rica staff for subject ascertainment.
Role of the funding source
The authors are members of the Pritzker Neuropsychiatric Disorders Research Consortium, which is supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. A shared intellectual property agreement exists between this philanthropic fund and the University of Michigan, Stanford University, the Weill Medical College of Cornell University, HudsonAlpha Institute of Biotechnology, the Universities of California at Davis, and at Irvine, to encourage the development of appropriate findings for research and clinical applications.
Footnotes
Conflict of interest
All authors have no conflicts of interest to declare.
Contributors
Authors EM, BR, WEB, LED, WB, and MPV conceived and designed the study. EM, BR, LED, WB, and MPV carried out the computational analyses and candidate gene selection. EM, BR, AM, LED conducted subject screening. EM, BR, carried out the genotyping. EM and MPV performed the statistical analysis of the genotyping and expression data. RMM, HA, SJW, JB, EGJ, AS, WEB provided the guidance and additional support on this project. EM, BR, WEB, LED, WB, and MPV wrote the first draft of the paper, all authors revised the current paper. LED, AM, and WB recruited, diagnosed, and gathered patients and controls. BR, EM, LED, AM, WB, MPV contributed to the collection and preparation of control DNA samples.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alaerts M, Ceulemans S, Forero D, Moens LN, De Zutter S, Heyrman L, Lenaerts AS, Norrback KF, De Rijk P, Nilsson LG, Goossens D, Adolfsson R, Del-Favero J. Support for NRG1 as a susceptibility factor for schizophrenia in a northern Swedish isolated population. Arch Gen Psychiatry. 2009;66(8):828–37. doi: 10.1001/archgenpsychiatry.2009.82. [DOI] [PubMed] [Google Scholar]
- Association, AP . Diagnostic and Statistical Manual of Mental Disorders: DSM-IV. Washington DC: American Psychiatric Association; 1994. [Google Scholar]
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7(4):405–11. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- Bell GI, Karam JH, Rutter WJ. Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc Natl Acad Sci U S A. 1981;78(9):5759–63. doi: 10.1073/pnas.78.9.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns GS, Nemeroff CB. The neurobiology of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C(1):76–84. doi: 10.1002/ajmg.c.20016. [DOI] [PubMed] [Google Scholar]
- Berrettini W. Evidence for shared susceptibility in bipolar disorder and schizophrenia. Am J Med Genet C Semin Med Genet. 2003;123C(1):59–64. doi: 10.1002/ajmg.c.20014. [DOI] [PubMed] [Google Scholar]
- Bertisch H, Mesen-Fainardi A, Martin MV, Perez-Vargas V, Vargas-Rodriguez T, Delgado G, Delgado C, Llach M, LaPrade B, Byerley W, Bunney WE, Vawter MP, DeLisi LE. Neuropsychological performance as endophenotypes in extended schizophrenia families from the Central Valley of Costa Rica. Psychiatr Genet. 2009;19(1):45–52. doi: 10.1097/YPG.0b013e3283202816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram I, Bernstein HG, Lendeckel U, Bukowska A, Dobrowolny H, Keilhoff G, Kanakis D, Mawrin C, Bielau H, Falkai P, Bogerts B. Immunohistochemical evidence for impaired neuregulin-1 signaling in the prefrontal cortex in schizophrenia and in unipolar depression. Ann N Y Acad Sci. 2007;1096:147–56. doi: 10.1196/annals.1397.080. [DOI] [PubMed] [Google Scholar]
- Cooper-Casey K, Mesen-Fainardi A, Galke-Rollins B, Llach M, Laprade B, Rodriguez C, Riondet S, Bertheau A, Byerley W. Suggestive linkage of schizophrenia to 5p13 in Costa Rica. Mol Psychiatry. 2005;10(7):651–6. doi: 10.1038/sj.mp.4001640. [DOI] [PubMed] [Google Scholar]
- Craddock N, O’Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32(1):9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Keefe RS, Perkins DO, Stroup TS, Lieberman JA, Sullivan PF. The neuregulin 1 promoter polymorphism rs6994992 is not associated with chronic schizophrenia or neurocognition. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1298–300. doi: 10.1002/ajmg.b.30727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K. Clinical characteristics of schizophrenia in multiply affected Spanish origin families from Costa Rica. Psychiatr Genet. 2001;11(3):145–52. doi: 10.1097/00041444-200109000-00006. [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K, Relja M, Byerley W, Sherrington R. Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet. 2002;114(5):497–508. doi: 10.1002/ajmg.10538. [DOI] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, Lopez JF, Thompson RC, Meng F, Stead JD, Walsh DM, Myers RM, Bunney WE, Watson SJ, Jones EG, Akil H. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci U S A. 2004;101(43):15506–11. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer NB, Reus VI, Escamilla M, Spesny M, Smith L, Service S, Gallegos A, Meza L, Batki S, Vinogradov S, Leon P, Sandkuijl LA. An approach to investigating linkage for bipolar disorder using large Costa Rican pedigrees. Am J Med Genet. 1996;67(3):254–63. doi: 10.1002/(SICI)1096-8628(19960531)67:3<254::AID-AJMG3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Fukui N, Muratake T, Kaneko N, Amagane H, Someya T. Supportive evidence for neuregulin 1 as a susceptibility gene for schizophrenia in a Japanese population. Neurosci Lett. 2006;396(2):117–20. doi: 10.1016/j.neulet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Georgieva L, Dimitrova A, Ivanov D, Nikolov I, Williams NM, Grozeva D, Zaharieva I, Toncheva D, Owen MJ, Kirov G, O’Donovan MC. Support for neuregulin 1 as a susceptibility gene for bipolar disorder and schizophrenia. Biol Psychiatry. 2008;64(5):419–27. doi: 10.1016/j.biopsych.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Erlenmeyer-Kimling L. Family and twin strategies as a head start in defining prodromes and endophenotypes for hypothetical early-interventions in schizophrenia. Schizophr Res. 2001;51(1):93–102. doi: 10.1016/s0920-9964(01)00245-6. [DOI] [PubMed] [Google Scholar]
- Green EK, Raybould R, Macgregor S, Gordon-Smith K, Heron J, Hyde S, Grozeva D, Hamshere M, Williams N, Owen MJ, O’Donovan MC, Jones L, Jones I, Kirov G, Craddock N. Operation of the schizophrenia susceptibility gene, neuregulin 1, across traditional diagnostic boundaries to increase risk for bipolar disorder. Arch Gen Psychiatry. 2005;62(6):642–8. doi: 10.1001/archpsyc.62.6.642. [DOI] [PubMed] [Google Scholar]
- Hall J, Whalley HC, Job DE, Baig BJ, McIntosh AM, Evans KL, Thomson PA, Porteous DJ, Cunningham-Owens DG, Johnstone EC, Lawrie SM. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9(12):1477–8. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- Hong LE, Wonodi I, Stine OC, Mitchell BD, Thaker GK. Evidence of missense mutations on the neuregulin 1 gene affecting function of prepulse inhibition. Biol Psychiatry. 2008;63(1):17–23. doi: 10.1016/j.biopsych.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Takahashi N, Saito S, Aleksic B, Watanabe Y, Nunokawa A, Yamanouchi Y, Kitajima T, Kinoshita Y, Kishi T, Kawashima K, Hashimoto R, Ujike H, Inada T, Someya T, Takeda M, Ozaki N, Iwata N. Failure to replicate the association between NRG1 and schizophrenia using Japanese large sample. Schizophr Res. 2008;101(1–3):1–8. doi: 10.1016/j.schres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Ingason A, Soeby K, Timm S, Wang AG, Jakobsen KD, Fink-Jensen A, Hemmingsen R, Berg Rasmussen H, Werge T. No significant association of the 5′ end of neuregulin 1 and schizophrenia in a large Danish sample. Schizophr Res. 2006;83(1):1–5. doi: 10.1016/j.schres.2005.12.850. [DOI] [PubMed] [Google Scholar]
- Iwata N, Suzuki T, Ikeda M, Kitajima T, Yamanouchi Y, Inada T, Ozaki N. No association with the neuregulin 1 haplotype to Japanese schizophrenia. Mol Psychiatry. 2004;9(2):126–7. doi: 10.1038/sj.mp.4001456. [DOI] [PubMed] [Google Scholar]
- Jones EG, Hendry SH, Liu XB, Hodgins S, Potkin SG, Tourtellotte WW. A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J Neurosci Methods. 1992;44(2–3):133–44. doi: 10.1016/0165-0270(92)90006-y. [DOI] [PubMed] [Google Scholar]
- Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, Zambuto CT, Schmitt K, Matise TC, Harkavy Friedman JM, Hampe C, Lee H, Shore D, Wynne D, Faraone SV, Tsuang MT, Cloninger CR. NIMH Genetics Initiative Millenium Schizophrenia Consortium: linkage analysis of African-American pedigrees. Am J Med Genet. 1998;81(4):282–9. [PubMed] [Google Scholar]
- Keri S, Kiss I, Kelemen O. Effects of a neuregulin 1 variant on conversion to schizophrenia and schizophreniform disorder in people at high risk for psychosis. Mol Psychiatry. 2009;14(2):118–9. doi: 10.1038/mp.2008.1. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Rubinow DR, Holmes C, Abelson JM, Zhao S. The epidemiology of DSM-III-R bipolar I disorder in a general population survey. Psychol Med. 1997;27(5):1079–89. doi: 10.1017/s0033291797005333. [DOI] [PubMed] [Google Scholar]
- Kieseppa T, Partonen T, Haukka J, Kaprio J, Lonnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry. 2004;161(10):1814–21. doi: 10.1176/ajp.161.10.1814. [DOI] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103(17):6747–52. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mowry BJ, Sharpe L, Endicott J. Penetrance of schizophrenia-related disorders in multiplex families after correction for ascertainment. Genet Epidemiol. 1996;13(1):11–21. doi: 10.1002/(SICI)1098-2272(1996)13:1<11::AID-GEPI2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Li T, Stefansson H, Gudfinnsson E, Cai G, Liu X, Murray RM, Steinthorsdottir V, Januel D, Gudnadottir VG, Petursson H, Ingason A, Gulcher JR, Stefansson K, Collier DA. Identification of a novel neuregulin 1 at-risk haplotype in Han schizophrenia Chinese patients, but no association with the Icelandic/Scottish risk haplotype. Mol Psychiatry. 2004;9(7):698–704. doi: 10.1038/sj.mp.4001485. [DOI] [PubMed] [Google Scholar]
- Liddle P, Carpenter WT, Crow T. Syndromes of schizophrenia. Classic literature. Br J Psychiatry. 1994;165(6):721–7. doi: 10.1192/bjp.165.6.721. [DOI] [PubMed] [Google Scholar]
- Marballi K, Quinones MP, Jimenez F, Escamilla MA, Raventos H, Soto-Bernardini MC, Ahuja SS, Walss-Bass C. In vivo and in vitro genetic evidence of involvement of neuregulin 1 in immune system dysregulation. J Mol Med. 2010;88(11):1133–41. doi: 10.1007/s00109-010-0653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews CA, Reus VI, Bejarano J, Escamilla MA, Fournier E, Herrera LD, Lowe TL, McInnes LA, Molina J, Ophoff RA, Raventos H, Sandkuijl LA, Service SK, Spesny M, Leon PE, Freimer NB. Genetic studies of neuropsychiatric disorders in Costa Rica: a model for the use of isolated populations. Psychiatr Genet. 2004;14(1):13–23. doi: 10.1097/00041444-200403000-00003. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Prata DP, Fu CH, Picchioni M, Kane F, Kalidindi S, McDonald C, Demjaha A, Kravariti E, Toulopoulou T, Murray R, Collier DA, McGuire PK. The effects of neuregulin1 on brain function in controls and patients with schizophrenia and bipolar disorder. Neuroimage. 2008;42(2):817–26. doi: 10.1016/j.neuroimage.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Viding E, Pettersson-Yeo W, Tognin S, McGuire PK. Genetic variation in neuregulin1 is associated with differences in prefrontal engagement in children. Hum Brain Mapp. 2009;30(12):3934–43. doi: 10.1002/hbm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera B, Barrantes R, Marin-Rojas R. Gene admixture in the Costa Rican population. Ann Hum Genet. 2003;67(Pt 1):71–80. doi: 10.1046/j.1469-1809.2003.00010.x. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Law AJ, Radulescu E, Luna A, Kolachana B, Vakkalanka R, Rujescu D, Giegling I, Straub RE, McGee K, Gold B, Dean M, Muglia P, Callicott JH, Tan HY, Weinberger DR. Biological validation of increased schizophrenia risk with NRG1, ERBB4, and AKT1 epistasis via functional neuroimaging in healthy controls. Arch Gen Psychiatry. 2010;67(10):991–1001. doi: 10.1001/archgenpsychiatry.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51(11):849–59. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863–4. [DOI] [PubMed] [Google Scholar]
- Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry. 2004;9(12):1091–9. doi: 10.1038/sj.mp.4001541. [DOI] [PubMed] [Google Scholar]
- Prata DP, Breen G, Osborne S, Munro J, St Clair D, Collier DA. An association study of the neuregulin 1 gene, bipolar affective disorder and psychosis. Psychiatr Genet. 2009;19(3):113–6. doi: 10.1097/YPG.0b013e32832a4f69. [DOI] [PubMed] [Google Scholar]
- Pulver AE, Lasseter VK, Kasch L, Wolyniec P, Nestadt G, Blouin JL, Kimberland M, Babb R, Vourlis S, Chen H, et al. Schizophrenia: a genome scan targets chromosomes 3p and 8p as potential sites of susceptibility genes. Am J Med Genet. 1995;60(3):252–60. doi: 10.1002/ajmg.1320600316. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Gardner M, Cuesta MJ, Peralta V, Fatjo-Vilas M, Miret S, Navarro ME, Comas D, Fananas L. Family-based association study of neuregulin-1 gene and psychosis in a Spanish sample. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(7):954–7. doi: 10.1002/ajmg.b.30511. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Kirov G, Chambert K, Moran JL, Owen MJ, O’Donovan MC, Sklar P, Purcell SM. A family-based study of common polygenic variation and risk of schizophrenia. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosser A, Cohen-Woods S, Gaysina D, Chow PC, Martucci L, Farmer A, Korszun A, Gunashinghe C, Gray J, Jones L, Craddock N, Owen MJ, Craig IW, McGuffin P. NRG1 gene in recurrent major depression: no association in a large-scale case-control association study. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):141–7. doi: 10.1002/ajmg.b.30965. [DOI] [PubMed] [Google Scholar]
- Shamir A, Buonanno A. Molecular and cellular characterization of Neuregulin-1 type IV isoforms. J Neurochem. 2010;113(5):1163–76. doi: 10.1111/j.1471-4159.2010.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SH, Kelly M, Smith AB, Shields G, Hopkins PJ, Loftus J, Laval SH, Vita A, De Hert M, Cardon LR, Crow TJ, Sherrington R, DeLisi LE. A genome-wide search for schizophrenia susceptibility genes. Am J Med Genet. 1998;81(5):364–76. doi: 10.1002/(sici)1096-8628(19980907)81:5<364::aid-ajmg4>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Shibuya M, Komi E, Wang R, Kato T, Watanabe Y, Sakai M, Ozaki M, Someya T, Nawa H. Measurement and comparison of serum neuregulin 1 immunoreactivity in control subjects and patients with schizophrenia: an influence of its genetic polymorphism. J Neural Transm. 2010;117(7):887–95. doi: 10.1007/s00702-010-0418-3. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Optimal designs for two-stage genome-wide association studies. Genet Epidemiol. 2007;31(7):776–88. doi: 10.1002/gepi.20240. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, Ingason A, Gulcher JR, Stefansson K, St Clair D. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72(1):83–7. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877–92. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, Olafsson O, Stefansson K, Gulcher JR. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342(1):97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Thiselton DL, Webb BT, Neale BM, Ribble RC, O’Neill FA, Walsh D, Riley BP, Kendler KS. No evidence for linkage or association of neuregulin-1 (NRG1) with disease in the Irish study of highdensity schizophrenia families (ISHDSF) Mol Psychiatry. 2004;9(8):777–83. doi: 10.1038/sj.mp.4001530. image 729. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Christoforou A, Morris SW, Adie E, Pickard BS, Porteous DJ, Muir WJ, Blackwood DH, Evans KL. Association of Neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the Scottish population. Mol Psychiatry. 2007;12(1):94–104. doi: 10.1038/sj.mp.4001889. [DOI] [PubMed] [Google Scholar]
- Ting AK, Chen Y, Wen L, Yin DM, Shen C, Tao Y, Liu X, Xiong WC, Mei L. Neuregulin 1 promotes excitatory synapse development and function in GABAergic interneurons. J Neurosci. 2011;31(1):15–25. doi: 10.1523/JNEUROSCI.2538-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, Overman KM, Atz ME, Myers RM, Jones EG, Watson SJ, Akil H, Bunney WE., Jr Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55(4):346–52. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosato S, Dazzan P, Collier D. Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophr Bull. 2005;31(3):613–7. doi: 10.1093/schbul/sbi043. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Stone WS, Faraone SV. Genes, environment and schizophrenia. Br J Psychiatry Suppl. 2001;40:s18–24. doi: 10.1192/bjp.178.40.s18. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Tomita H, Meng F, Bolstad B, Li J, Evans S, Choudary P, Atz M, Shao L, Neal C, Walsh DM, Burmeister M, Speed T, Myers R, Jones EG, Watson SJ, Akil H, Bunney WE. Mitochondrial-related gene expression changes are sensitive to agonal-pH state: implications for brain disorders. Mol Psychiatry. 2006;11(7):615, 663–79. doi: 10.1038/sj.mp.4001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walss-Bass C, Liu W, Lew DF, Villegas R, Montero P, Dassori A, Leach RJ, Almasy L, Escamilla M, Raventos H. A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biol Psychiatry. 2006a;60(6):548–53. doi: 10.1016/j.biopsych.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Walss-Bass C, Montero AP, Armas R, Dassori A, Contreras SA, Liu W, Medina R, Levinson D, Pereira M, Atmella I, NeSmith L, Leach R, Almasy L, Raventos H, Escamilla MA. Linkage disequilibrium analyses in the Costa Rican population suggests discrete gene loci for schizophrenia at 8p23.1 and 8q13.3. Psychiatr Genet. 2006b;16(4):159–68. doi: 10.1097/01.ypg.0000218616.27515.67. [DOI] [PubMed] [Google Scholar]
- Wang F, Jiang T, Sun Z, Teng SL, Luo X, Zhu Z, Zang Y, Zhang H, Yue W, Qu M, Lu T, Hong N, Huang H, Blumberg HP, Zhang D. Neuregulin 1 genetic variation and anterior cingulum integrity in patients with schizophrenia and healthy controls. J Psychiatry Neurosci. 2009;34(3):181–6. [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Li WQ, Zhang Y, Zhao JP, Lv LX, Yang G. Association analysis of neuregulin 1 gene polymorphism with schizophrenia in Chinese Han population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26(1):16–20. doi: 10.3760/cma.j.issn.1003-9406.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Zuliani R, Moorhead TW, Bastin ME, Johnstone EC, Lawrie SM, Brambilla P, O’Donovan MC, Owen MJ, Hall J, McIntosh AM. Genetic variants in the ErbB4 gene are associated with white matter integrity. Psychiatry Res. 2011;191(2):133–7. doi: 10.1016/j.pscychresns.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]