Abstract
Diffusion-weighted images of the human brain are acquired more and more routinely in clinical research settings, yet segmenting and labeling white matter tracts in these images is still challenging. We present in this paper a fully automated method to extract many anatomical tracts at once on diffusion tensor images, based on a Markov random field model and anatomical priors. The approach provides a direct voxel labeling, models explicitly fiber crossings and can handle white matter lesions. Experiments on simulations and repeatability studies show robustness to noise and reproducibility of the algorithm, which has been made publicly available.
Keywords: White matter tracts, DTI Segmentation, Markov random field modeling
1. Introduction
Diffusion-weighted imaging (DWI) has become a major tool for the in vivo study of connectivity in the human human brain in health and disease (Bihan et al., 2001; Basser and Jones, 2002). Clinical researchers have been increasingly interested in using DWI in their studies, and have widely adopted diffusion tensor derived indices of white matter (WM) integrity such as fractional anisotropy (FA) and mean, parallel and perpendicular diffusivity (Horsfield and Jones, 2002). Furthermore, recent works have shown that a tract-based analysis of the white matter has the promise to offer a deeper insight into its characteristics in health and disease (Reich et al., 2006; Yushkevich et al., 2008; O’Donnell et al., 2009).
Obtaining an anatomical segmentation of white matter tracts, however, has proven to be difficult for several reasons. DWI tractography, which provides information about the connections between remote regions of the brain (Mori and van Zijl, 2002), does not directly lead to the characterization of fiber bundles (tracts) corresponding to known anatomy, and requires grouping, trimming, and labeling of the numerically computed fibers (Mori et al., 2005; Kouby et al., 2005; O’Donnell and Westin, 2007; Maddah et al., 2007; Lawes et al., 2008; Ziyan et al., 2009; Mayer et al., 2011). Fiber tracking reliability can further vary with imaging resolution, noise and patient orientation (Wakana et al., 2007) as well as decreased anisotropy or lesions that occur with disease, and more advanced methods (Tuch, 2004; Behrens et al., 2007; Jones, 2008; Descoteaux et al., 2009; Reisert et al., 2011) require longer imaging times, often impractical for clinical imaging. Finally, the presence of pathology influences many of these methods and may confound the results in studies of diseases involving WM lesions (Reich et al., 2010).
Alternative approaches have been proposed to represent and study the white matter, most prominently a skeleton representation built on the FA map (Smith et al., 2006) and a region-based parcellation (Mori et al., 2008). Both methods rely on deformable registration strategies to align the diffusion images to labeled templates or group averages (Yang et al., 2008; Ceritoglu et al., 2009). These methods provide indirect information about the tracts and may be misled by WM lesions, which disrupt the FA map and provide undesirable features for registration algorithms to match between subjects.
In this work, we present a new approach to fiber tract segmentation capable of accurate performance on routine clinical acquisitions. With an atlas-based Markov random field (MRF) representation, we derive a fast, scalable estimation algorithm to reliably segment most of the major anatomical fiber tracts within the human brain. The approach bypasses tractography and associated issues and is therefore considered a “direct” tract segmentation approach. Our approach models overlapping and crossing fibers and robustly handles the presence of noise and white matter lesions without manual initialization or pre-filtering. The method was tested and validated on simulations and repeatability studies, and has been successfully applied to the analysis of WM tracts in multiple sclerosis (MS). The algorithm (coded in the cross-platform Java programming language) together with its statistical tract atlas are available for free on the NITRC.org website.
1.1. Related Works
Previous methods for direct tract segmentation include a method based on level sets (Lenglet et al., 2006), one using non-parametric fuzzy classification (Awate et al., 2007), and tensor-space clustering approaches (Goh and Vidal, 2008; Rodrigues et al., 2009). These methods required the specification of initial regions of interest (ROIs) corresponding to specific tracts of interest. Some probabilistic tractography methods evaluate the connection strength between regions or voxels (Parker and Alexander, 2003; Fletcher et al., 2007; Yendiki et al., 2008); however, the connection pathways they can define are not necessarily identifiable as known anatomical tracts.
Recent studies indicate that multi-atlas registration combined with image-based segmentation approaches have improved accuracy (Heckemann et al., 2010). Indeed, statistical atlas-based approaches incorporated in voxel classification techniques (Maddah et al., 2008; Hagler et al., 2009) or in parametric deformable models (Eckstein et al., 2009) have shown some promising results for segmenting anatomical tracts. In particular, (Hagler et al., 2009) proposed to use a statistical atlas essentially equivalent to the one presented here. However, these methods have so far considered each fiber tract as a separate structure without explicitly addressing either joint segmentation or crossing tracts. MRFs have been previously considered for regularizing diffusion tensors (Poupon et al., 1998), but have not been used for segmentation. Our work brings together MRF modeling and atlas-based segmentation in a global and computationally efficient algorithm that explicitly handles fiber crossings and lesions in clinical-quality images.
2. Methods
Our segmentation method combines the global information of location and direction contained in a probabilistic atlas of known white matter tracts with the local diffusion information obtained from the image within a MRF model (Li, 2009). In a MRF, the conditional probability distribution for the labels l at x is entirely determined by the conditional probability distribution in their neighborhood. Due to the Hammersley-Clifford theorem, the joint distribution follows a Gibbs distribution:
| (1) |
for a MRF including a unary term V1(x, l) and a pairwise term V2(x, y, l, m), with y and m the location and label of a voxel in the neighborhood of x, l, T a scaling parameter and Z a normalization factor. The terms V1 and V2 are often referred to as energy terms, and we will also use U(x, l) = V1(x, l) + ∑y,m V2(x, y, l, m) to denote the overall energy at voxel x. In our model, the unary term V1(x, l) combines the tensor direction and diffusion characteristics of the data with the tract atlas to encode the likelihood of each label l at x, and the pairwise term V2(x, y, l, m) encodes the local connections between neighboring diffusion tensors.
As it is well known that there is extensive overlap and crossing between multiple tracts at the current voxel resolution in diffusion MRI, it would be unrealistic to expect a given voxel to contain a single tract everywhere, and so our set of labels {l} must not only include labels for each separate tract but also overlapping tracts within a single voxel. To avoid combinatorial explosion, we limit ourselves to a maximum of two tracts per voxel and exclude unlikely pairs (see Section 2.4). The approach can in principle be extended to multiple overlaps, provided that the underlying diffusion information is sufficient to differentiate multiple crossings from isotropic regions.
To obtain a tract segmentation, we need to first model V1 and V2 as a function of the diffusion data for the possible labels and then to maximize P({l}) with respect to the label field {l}. The resulting segmented tracts are represented by membership functions (probabilistic assignments at each voxel) and can be analyzed in a variety of ways including computation of tract volume and determining average diffusion properties (e.g., FA, MD).
Our segmentation algorithm, referred to as Diffusion-Oriented Tract Segmentation (DOTS), is organized as follows. First, the diffusion weighted images are combined and pre-processed to obtain tensor images (Section 2.1). DOTS starts by registering our probabilistic atlas (described in Section 2.2) to the DTI data, and then using the MRF model to estimate the posterior probability of the voxel label given the data (Section 2.3). The model incorporates information about the local anisotropy (Section 2.3.1), the connectivity between neighboring voxels (Section 2.3.2) and the statistics of the atlas (Section 2.3.3). Section 2.3.4 brings all the different components of the MRF into the probability function P({l}) to be maximized by the algorithm. Additional optional modeling is provided to exclude anatomically irrelevant crossing configurations (Section 2.4) and to handle white matter lesions (Section 2.5). Additional implementation details are given in Section 2.6.
2.1. Diffusion-weighted image pre-processing
To process a series of diffusion-weighted images, we first perform a tensor reconstruction to get a set of diffusion eigenvectors and associated eigenvalues {υ⃗n(x), λn(x)}1, N at each voxel x with the standard linear reconstruction method (Basser and Jones, 2002). The images are first aligned to the b0 image and corrected for distortions against a structural image within the CATNAP software (Landman et al., 2007). Extra-cranial tissues are removed with an automatic skull-stripping method (Carass et al., 2007) applied to a co-registered structural image or using a semi-automatic method (Bazin et al., 2007) applied to the mean diffusivity image if a structural image is not available.
2.2. White Matter Tract Atlas
Our statistical atlas is based on the atlas of Mori and Wakana (Mori et al., 2005). Following the atlas tract definitions and delineation protocols, we obtained a set of semi-manual delineations of major tracts, which we augmented with additional expert delineations of a few other tracts of interest. All tracts were delineated using tractography and multiple regions of interest (ROI) selection in DTI Studio.1 Our standard atlas currently includes the following fiber tracts (separated for left and right when applicable): anterior, superior, and posterior thalamic radiations, corpus callosum and tapetum, inferior and superior longitudinal and fronto-occipital fasciculi, cingulum, fornix, uncinate fasciculus, optic tract, optic radiation, cortico-spinal and cortico-pontine tracts, medial lemniscus, inferior, middle, and superior cerebellar peduncles (see Table 1).
Table 1.
Tract Labels
| Acronym | Name |
|---|---|
| ATRL ATRR |
Anterior thalamic radiation (left, right) |
| CCF CCP CCS |
Corpus callosum (frontal, posterior and superior) |
| CGL CGR |
Cingulum (left, right) |
| CPTL CPTR |
Cortico-pontine tract (left, right) |
| CSTL CSTR |
Cortico-spinal tract (left, right) |
| FNXL FNXR |
Fornix (left, right) |
| ICPL ICPR |
Inferior cerebellar peduncle (left, right) |
| IFOL IFOR |
Inferior fronto-occipital fascicle (left, right) |
| ILFL ILFR |
Inferior longitudinal fascicle (left, right) |
| MCP MLL MLR |
Middle cerebellar peduncle Medial lemniscus (left, right) |
| OPRL OPRR |
Optic radiation (left, right) |
| OPTL OPTR |
Optic tract (left, right) |
| PTRL PTRR |
Posterior thalamic radiation (left, right) |
| SCPL SCPR |
Superior cerebellar peduncle (left, right) |
| SFOL SFOR |
Superior fronto-occipital fascicle (left, right) |
| SLFL SLFR |
Superior longitudinal fascicle (left, right) |
| STRL STRR |
Superior thalamic radiation (left, right) |
| TAP | Tapetum |
| UNCL UNCR |
Uncinate fascicle (left, right) |
Our atlas includes both the spatial probability pl(x) of the existence of label l at voxel x as well as the most probable diffusion direction d̄l(x) along the tract. Deterministic tractography usually provides very conservative delineations, including only the portion of the tracts that can be reconstructed through multiple ROIs. For this reason, we extended the definition of the tracts in the vicinity of the delineated regions with a smoothing function:
| (2) |
where is the delineated tract region, the smoothed delineation, k a smoothing kernel, the principal direction at voxel x in image t for tract l, and N (x) is the neighborhood defined by the smoothing kernel. Note that the directions are meaningful at first only inside the delineated regions , and must be extrapolated from higher probability to lower probability regions. The directions are orientation-independent (i.e. d̄ and −d̄ are the same direction), so the sums of directions are performed as follows:
| (3) |
where d⃗ is the oriented vector corresponding to direction d̄. The averaged direction in Eq. (2) may not have unit norm, unless all the directions are the same. This provides information about the uncertainty in direction at each location in the atlas. In this work we use a linear smoothing kernel of radius 5 mm to construct our atlas. The size of the kernel has some impact on the segmentation, as a larger scale is better at representing tracts with large individual variations but also increases the risk of mislabeling regions with too many overlapping candidates. Our experiments however indicate that a small but reasonable smoothing (5 mm corresponds to about two voxels at the atlas’s original resolution) improves the results.
Examples from the atlas are given in Figure 1. Although this atlas includes most of the major WM tracts in the human brain, the segmentation also includes isotropic regions like the gray matter structures or the ventricles, and some undefined regions of white matter, for instance the “U” fibers that connect neighboring gyri of the cortex. We model the isotropic regions by creating a low anisotropy mask (FA ≤ 0.1) and we capture the undefined white matter regions by subtracting from the opposite mask (FA > 0.1) any region included in one of the tracts defined above.
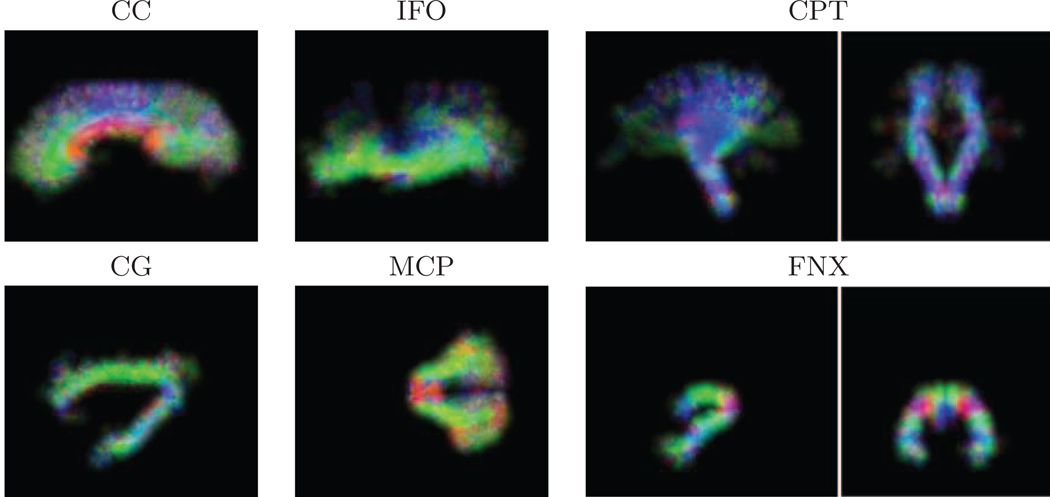
Figure 1.
Examples from the WM tract atlas: maximum intensity projection along the sagittal direction (except for MCP in the axial direction, and FNX, CPT in sagittal and coronal directions) of the shape prior multiplied by the color-coded direction prior.
2.3. Markov Random Modeling of Diffusion
Given the DTI data and the statistical atlas above, we derive below the terms of the MRF model of Eq. (12): unary terms representing local diffusion properties of the tensor and the contributions of the tract atlas in terms of shape and direction, and a binary term representing connectivity between neighboring tensors.
Our set of possible labels includes the individual tracts listed in Table 1, undefined fiber tracts and isotropic regions, as well as overlapping pairs of tracts. In the following, we describe general properties that apply to any individual tract, group of overlapping tracts or isotropic region independently of the specific label. Let us denote by T, O, and I general attributes of the single tract, overlapping tracts and isotropic regions respectively.
2.3.1. Diffusion Type
At every voxel x, we can have one of three types of structures in this model: isotropic diffusion, diffusion along a single tract, or diffusion along multiple tracts overlapping or crossing. To model the type of diffusion, we use indices of diffusion along a single tract (T), overlapping tracts (O), and isotropic regions (I) derived from the linear, planar and spherical indices of (Westin et al., 2002):
| (4) |
Note that water may still diffuse along a single direction for overlapping but aligned tracts, so we cannot differentiate between the two in regions of linear diffusion and dO ≥ dT (see Fig. 2b).
Figure 2.
Example data set depicting diffusion type and connectivity functions for single (top) and overlapping tracts (bottom): a) original image color map, b) dT, dO, c) sT, sO in the X direction, d) sT, sO in the Y direction, e) sT, sO in the Z direction. The diffusion functions are defined in [0, 1], whereas the connectivity functions are defined in [−1, 1]. Although the functions for single and overlapping tracts appear very similar, there are some subtle differences: dT is darker in regions of crossing (e.g. the junction between ATR, IFO and CCF), and the single tract connectivity maps sT appear sharper because they include more negative values (it is more likely to find similar directions when using first and second eigenvectors, as in sO).
2.3.2. Local tensor connectivity
The evidence for fiber tracts comes from the diffusion tensors: if two tensors are aligned they likely correspond to the same tract or overlapping tracts. The connectivity between neighboring tensors is modeled as follows, assuming a single tract:
| (5) |
where υ⃗xy represents the direction vector between voxels x and y, υ⃗1(x) and υ⃗1(y) represent the principal eigenvector directions at x and y, and arccos(|υ⃗1 · υ⃗2|). The function θ(υ⃗1, υ⃗2) gives the angle between directions υ⃗1 and υ⃗2 normalized in [0, 1]. Because it is linear, it is more sensitive to small angular variations than the product υ⃗1 · υ⃗2. In some of the following cases, the vectors in this formula have less than unit norm as a way to encode uncertainty in their direction as a lower bound on angles. In such cases, the function θ(υ⃗1, υ⃗2) returns values in [α, 1], where arccos(|υ1| |υ2|). Note that other measures involving the complete tensor are possible and they may be interesting to explore in the future.
The similarity function sT(x, y) is close to +1 when the diffusion directions at x and y are aligned with each other and with the path from x to y. If both diffusion directions are orthogonal to that path, we cannot assume that they are related even if they are aligned: many fiber tracts have “kissing” fibers that follow the same direction before diverging. In such cases, sT(x, y) goes to zero in order to model the uncertainty. When the diffusion directions are orthogonal and one of them is aligned with the path, then it is clear that both points cannot be part of the same tract, which translates into a negative value up to −1 (see Fig. 2c–e).
For crossing or overlapping fibers, the main diffusion direction can easily switch from υ⃗1 to υ⃗2 if the tensor is “flat”, and so we extend the connectivity to consider the secondary directions:
| (6) |
The first part of this equation means that we select the pair of directions at x and y that are best aligned among the first two eigenvectors {υ⃗1(x), υ⃗2(x), υ⃗1(y), υ⃗2(y)}.
We penalize the υ⃗2 by to account for the uncertainty due to the fact it is required to be orthogonal to υ⃗1. Note also that these directions are not the true fiber directions, which cannot be retrieved with the tensor representation. However, the erroneous tensors will be similar and share directions as long as the two fibers are crossing along similar directions and the diffusion is slightly stronger in one direction. In practice, many regions of overlap between the tracts that we can observe at clinical resolution involve primarily kissing fibers as opposed to crossing ones, for which the principal direction of diffusion as estimated from a tensor reconstruction is still meaningful.
Finally, isotropic regions are assumed to have uniform connectivity sI(x, y) = sI, given as a prior parameter. The value of sI will influence how labels from isotropic regions propagate into anisotropic ones as compared to fibers and overlapping regions. Because all the regions evolve simultaneously, we set sI = 1/Nt in our experiments, where Nt is the total number of regions in the atlas.
2.3.3. Atlas information
The atlas provides a prior on tract location pl and tract direction d̄l, which is integrated in the MRF model through a shape prior term ul and a direction coefficient cl as follows.
Because we allow the presence of multiple tracts at a location, the prior probability for a particular label defined by the atlas cannot be used directly. Instead, we define the shape prior term ul(x) for a single tract l as follows:
| (7) |
giving high values where the prior probability pl(x) is high and likely to be a single tract (∑m pm(x) ≈ pl(x), for all tract labels m).
To compute cl, we compare the direction prior d⃗l(x) with the principal direction υ⃗1(x) of the tensor in the image to segment:
| (8) |
The coefficient cl is positive when the directions coincide, and negative when they are orthogonal. The uncertainty on the direction prior, represented by its norm, lowers both positive and negative values of cl. For two crossing tracts l and m, the shape prior term combines the priors of both objects with the likelihood for two overlapping tracts:
| (9) |
If the tracts have different orientations, the reconstructed tensor will represent a mixture pointing to a different direction than the individual diffusion tensors for each tract. However, if we assume the true tensors with principal directions vectors d⃗1,l(x) and d⃗1,m(x) to be prolate tensors with high anisotropy, we can derive from the Stejskal-Tanner equation that the estimated tensor for the mixture will have for its principal direction the vector d⃗l,m(x) = d⃗1,l(x)±d⃗1,m(x) with largest norm.
The composite tensor direction d⃗l,m(x) is then renormalized so that its norm is . From this composite vector, we can define the following direction coefficient as we did before in the single tract case:
| (10) |
Finally, isotropic regions use the same shape prior as individual tracts and assume no preferred direction, setting .
2.3.4. Propagation of the tract probabilities
Once we have the various elements of diffusion and atlas information above, we can derive the terms of the energy function. From the diffusion and prior-based terms, we derive the following unary term:
| (11) |
for single tracts, overlapping tracts and isotropic regions respectively. We combine all the terms as a product since we require all three conditions to be met jointly in order to attribute a label l to a given voxel (unlike with more classical probabilistic models, we cannot separate the terms of V1 into a sum of conditionally independent elements).
The neighborhood term propagates the unary energy values along the most likely fiber directions or isotropically depending on the type of label. Let x+ = argmaxy∈N+(x)sT(x, y), where N+(x) is the half of the 26-neighborhood N(x) of x such that υ⃗xy · υ⃗1(x) > 0, and similarly x− = argmaxy∈N−(x)sT(x, y), with N−(x) such that υ⃗xy · υ⃗1(x) ≤ 0. The same definition holds for overlapping tracts, substituting sO(x, y) to sT(x, y). The binary term V2(x, y, l, m) is only non-zero if y = x+ or y = x− and l, m include one same label, leading to the following formula for the energy term U(x, l) = V1(x, l) + ∑y,m V2(x, y, l, m) at x:
| (12) |
These three equations list all the possible cases: single tract labels, pairs of overlapping labels, and isotropic regions. Because regions of overlap interact with single tract regions, the energy can propagate between different labels as long as they share a common tract.
With this model, only the neighboring voxels most likely to be along the same tract are considered for single or overlapping tracts, whereas the contribution of all neighbors is averaged for isotropic regions. By using such a subset of the neighborhood, we greatly simplify the structure of the MRF and improve computational stability, as the field is locally oriented along the tracts, removing many loops in the neighborhood graph. The probability function can be efficiently maximized through an iterated conditional modes algorithm which converges quickly in practice and requires little computational overhead (Besag, 1986). This simplified neighborhood structure enable us to estimate the MRF efficiently without the help of a more elaborate MRF solver (Bazin et al., 2009a; Kolmogorov and Zabih, 2004).
2.4. Anatomical constraints
Even with a limit of two overlapping tracts per voxel, the number of possible pairs grows quickly when including more white matter tracts. However, it is well known from anatomy that certain pairs of tracts will not overlap, even in the presence of deforming pathology or trauma. For instance, it is clear that the superior longitudinal fasciculus (SLF) should not interact with the uncinate tract (UNC), or the cortico-spinal tract (CST) with the frontal forceps (CCF). From this observation, we restrict the possible label pairs to a list set a priori from anatomical information. The list may depend on image resolution, as more tracts will overlap in a voxel of coarser resolution.
To obtain such information from atlases and anatomy experts is challenging when the number of tracts to be estimated grows. We define an overlap probability for each pair of tracts in the atlas as follows:
| (13) |
where pm(x), pl(x) are the spatial probabilities for tracts m, l. We then restrict the possible label pairs to those such that PO > 1/2.
Similarly, we make the assumption that our blurred atlas priors should fully include the corresponding tracts, thus estimating only probabilities for tracts with non-zero prior at each voxel. These two modeling constraints greatly reduce the combinatorial increase and computational burden when additional tracts and potential crossings are added to the atlas, which allowed the method to scale well from an initial atlas of 10 tracts to our current atlas of 39 tracts.
2.5. Handling WM pathology
Despite the development of many diffusion MRI processing algorithms in recent years, studies of the WM structures have been difficult when pathology is present. In diseases like multiple sclerosis, Alzheimer’s disease, or even in normal aging, lesions appear inside the WM, changing the diffusion properties of the tissue (Reich et al., 2010; Wheeler-Kingshott and Cercignani, 2009).
Our approach is already robust to most lesions as it combines information coming from multiple directions around the area of lesion, however lesions with a sharp decrease in anisotropy will likely be segmented as isotropic regions rather than tracts. If we have an estimate of the lesion location from co-registered structural MRI (using (Shiee et al., 2010) in our case), we can use that information as an additional prior: to compensate for the low anisotropy inside lesions, we simply update the indices in all lesion voxels i by:
| (14) |
This update enforces that DOTS segments lesions as part of at least one tract, provided that the local diffusion direction matches the atlas. Note that DOTS includes a tract label for unidentified WM regions and allows isotropic regions to spread to their neighborhood, so the lesions are not forced to merge with a nearby tract if their tensor directions are not compatible.
2.6. Algorithmic Details
The complete segmentation process is performed as follows. First we register the shape and direction atlas to the skull-stripped tensor image to be segmented with a multi-scale gradient descent method that maximizes ER = ∑x ∑l ‖a(x)pl(T(x))‖2, where a(x) is the fractional anisotropy and T a rigid transform. The direction atlas is rotated accordingly. Next, the estimation algorithm is initialized with U(x, l) = V1(x, l), and then refined using iterated conditional modes and registration until convergence. Convergence is measured by the proportion of changed labels for each iteration.
To reduce computational overhead, we only record the energy function for the NB labels with highest energy at a given voxel, and approximate the others to zero. In our experiments, no significant differences could be observed in the results for NB ≥ 8. Also note that the connectivities sT(x, x+), sT(x, x−), sO(x, x+), sO(x, x−) are functions of the data alone, and can be precomputed to improve computation speed. A 181×217×181 voxel image (1mm cubic resolution) is processed with this method in less than 30 minutes, and a more typical 256×256×60 voxel image takes about 10 minutes to converge on a modern workstation with 8GB of available memory.
Once the algorithm has converged, we obtain a hard segmentation by selecting the labeling of highest energy, and attributing the voxel to the underlying tract or tracts in case of overlap; see Fig. 3. The corresponding membership function is given by:
| (15) |
where g0 is a parameter controlling the sharpness of the membership.
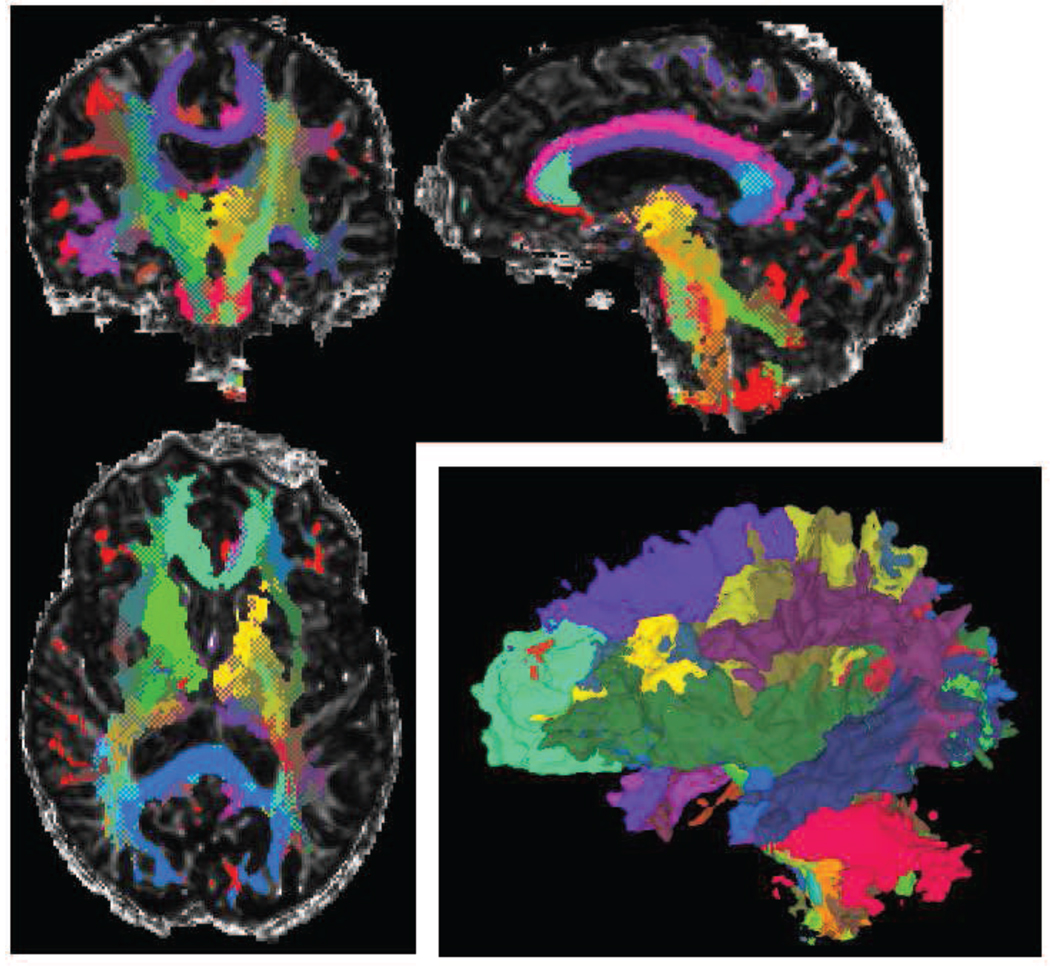
Figure 3.
DOTS segmentation example for one subject of our reproducibility study in axial, coronal, sagittal views and 3D rendering. Regions of overlapping tracts are displayed as a checkerboard pattern of the tracts’ colors in the 2D views. It is notable that many regions of overlap are found in practice. The recovered tracts boundaries are also somewhat irregular, as such boundaries are poorly defined on the tensor images.
The DOTS algorithm has been implemented as a plug-in for the MIPAV software package (McAuliffe et al., 2001) and the JIST pipeline environment (Lucas et al., 2010). The software is freely available on the NITRC repository2.
3. Experiments
We present here a simple synthetic experiment as well as real data experiments on clinical-quality data sets using our 39 tract labels atlas. Additional experiments performed with preliminary atlases have been reported in (Bazin et al., 2009b).
3.1. Synthetic Crossing Experiments
We first investigate the ability to recover crossing fibers with a simulated image depicting crossing fiber tracts with various levels of noise; see Fig. 4. The noise increases the number of voxels treated as isotropic as they diverge from the directions learned in the atlas. The crossing is correctly estimated as belonging to both fiber tracts, although with lower certainty. Note that the tensors from the crossing region were not included in the atlas, as would be expected from a simple tractography-based delineation.
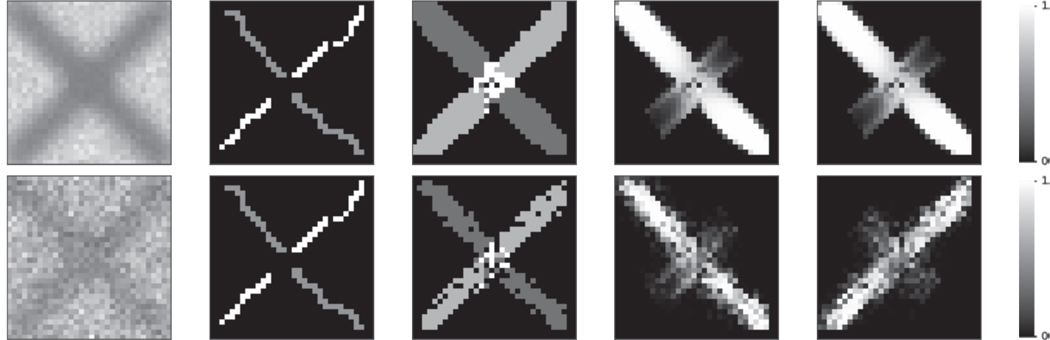
Figure 4.
Simulated crossing experiment at SNR 25 (top) and 5 (bottom). From left to right: a simulated image, the manual “fiber” delineation used in the atlas, the hard segmentation, showing the intersection in white, and the two membership functions for the tracts. Note that the memberships decrease smoothly inside the crossing, as both tracts can have membership values above 0.5 with our model.
3.2. Reproducibility study
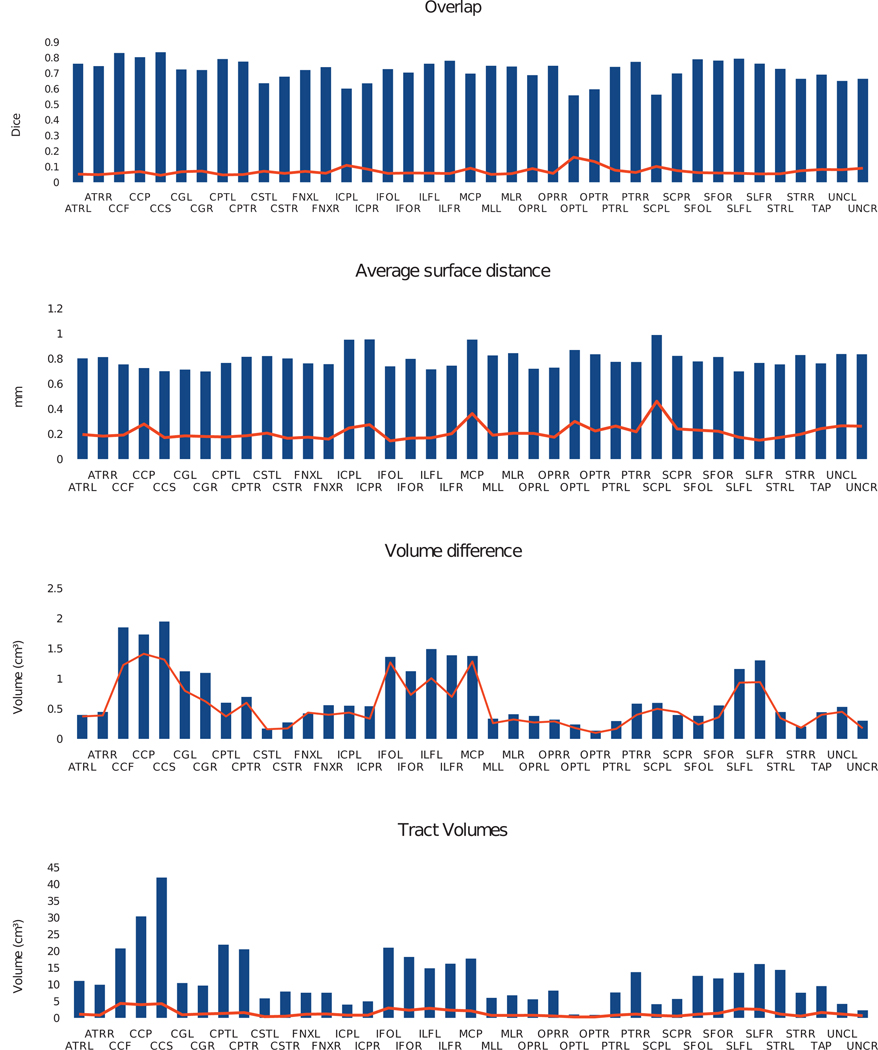
Segmenting tracts in a way that is consistent for repeated scans of a given subject is a requirement for clinical studies. We tested the reproducibility of our segmentation on the publicly available Kirby21 dataset (Landman et al., 2010b), which consists of two sets of MR scans taken on the same day for 21 healthy subjects, following a “Jones 30” protocol (Jones et al., 1999). The scans for each subject were processed separately with DOTS before co-registering their FA maps for comparisons. Figure 5 gives the average and standard deviation of the Dice overlap, average boundary surface distance and volume difference ratio. We also report the fiber tract volumes, as the volume difference ratio and Dice coefficient become more variable for smaller structures (in particular, WM tracts are much smaller than GM structures for which volume measurements and Dice coefficients are usually reported).
Figure 5.
DOTS segmentation results for two successive acquisitions. Mean (bar) and standard deviation (line) for 21 subjects of the following measures (from top to bottom): Dice overlap, average boundary surface distance, volume difference (as a percentage of the average volume), and average volume.
This experiment indicates that DOTS is robust to changes in patient position and scanning noise. Most notably, the tract overlap is above 0.7 for the larger tracts, and stays above 0.6 even for the optic tract, the cerebellar peduncles and the uncinate fasciculus, despite their small sizes. The distance between estimated tract boundaries is about half the imaging voxel size of 2.2 mm.
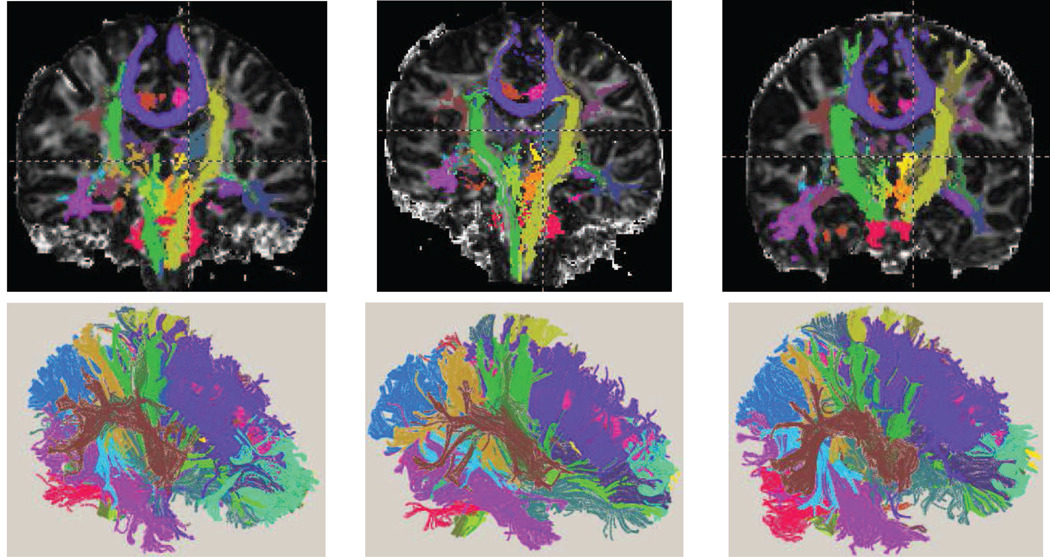
The segmentations from the two separate scans are very similar for each subject, while the labels adapt well to different anatomies; see Figure 6. It is also noticeable that there is a large amount of overlap between estimated labels, confirming the need for modeling overlap at the current resolution of clinical DTI.
Figure 6.
DOTS segmentation examples for three different subjects in coronal views for two separate acquisitions (top and middle). The bottom row displays selected regions of overlap, which are indicated with a checkerboard pattern of the tracts’ colors. Regions of unlabeled white matter are indicated in red. Note that the labeling is very consistent even in the regions of overlap where the tensors are more likely to be influenced by noise.
3.3. Integration with fiber tractography
Although DOTS labels give a reliable representation of the white matter tracts, they are quite different from the segmentation one would obtain from manual bundling of tractography results. Because our MRF model does not require long range interactions between voxels, the DOTS labels are more inclusive. On the other hand, one can independently reconstruct individual sample paths through the white matter (generally referred to as “fibers,” although the term is misleading) with any appropriate tractography technique (Mori et al., 1999; Koch et al., 2002; Behrens et al., 2007; Reisert et al., 2011).
Given a set of fibers computed by any tractography method, we can associate a fiber f to a tract l using the following method:
where l0 is a minimum length parameter set to 20 mm, bl a binary function equal to 1 inside tract l and 0 otherwise, and r a minimum inclusion ratio set to 75%.
We performed fiber tractography on the Kirby21 dataset using the FACT algorithm (Mori et al., 1999) with the following parameters: FA start 0.2, FA stop 0.1 and maximum angle 60 degrees, and labeled the generated fibers with the above method. As shown on Fig. 7, the labeled fibers form compact bundles with many fibers. It is notable that the fibers extend further toward the outer boundaries of the WM than the corresponding DOTS labels, as the deterministic tractography procedure extends fibers until they reach a FA of 0.1, whereas the DOTS model assigns lower probability to these regions of lower FA and higher variability. Note that DOTS labels can easily be extended to always include such regions if necessary using a white matter mask to define a priori the boundaries of the isotropic regions.
Figure 7.
Tractography results automatically labeled with DOTS for the three subjects of Fig. 6 in coronal view and 3D rendering. The bundles are overall rather homogeneous, and the included regions are similar in size to the original DOTS labels, indicating that the reconstructed fibers follow quite closely the DOTS segmentation.
3.4. Robustness to pathology
Finally, we assessed the application of DOTS on white matter lesions on a data set of 10 multiple sclerosis subjects. We identified white matter lesions on co-registered structural MRI (MPRAGE and FLAIR) with (lesion)TOADS, an automated brain and lesion segmentation tool (Shiee et al., 2010). DOTS was run with and without the prior lesion map, to measure the impact of lesions on the labeling.
We measured average fractional anisotropy (FA) and volume for the tracts, two measures of interest when assessing the impact of lesions on white matter. When comparing uncorrected and corrected measures with a t-test, we find small but significant differences (generally a decrease) in FA for ATRR, CCS, FNXL, PTRL and more important differences in volume (either increasing or decreasing by up to 5% with the correction) for CSTL, ICPL, MLR, OPTL, PTRL, SCPR, SLFL, SLFR, TAP and UNCL (p < 0.05). These results clearly show that the presence of lesions confounds DTI measurements unless they are properly accounted for in the model.
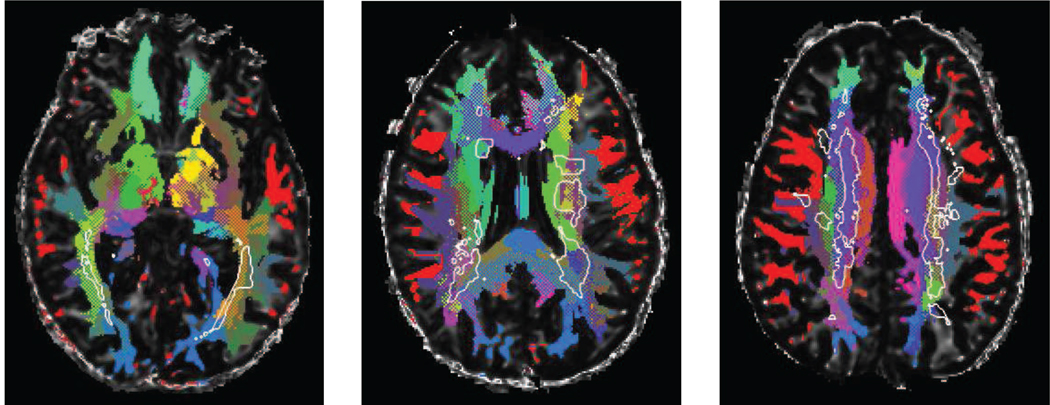
The individual segmentations, as depicted in Fig. 8 highlight the accumulation of lesions along the peri-ventricular tracts (especially the optic radiation) for subjects with large lesion loads, while the interplay of focal lesions with other tracts appears more complex. DOTS handles well the damaged regions on clinical quality DTI, and we hope it will offer a more systematic way to study the relationship of WM lesions with cerebral circuits and their relative impairment.
Figure 8.
DOTS segmentation examples for three different subjects with multiple sclerosis. The white matter lesions segmented from the co-registered structural images (T1 and FLAIR) are indicated with a white outline. Although the lesions do not follow given tracts, it is clear that certain tracts are largely spared while others are strongly affected.
4. Discussion
We presented here an atlas-based segmentation technique to reliably extract known anatomical tracts from clinical quality diffusion tensor MRI. As in the work of (Hagler et al., 2009), the method combines global statistical priors of shape and direction with the local connectivity information extracted from the tensors. The prior direction information in particular is important to disambiguate many possible tracts. However, we also noticed that the atlas alone was only partially successful, and that many parts of the tracts were not directly identified where eigenvector directions were not well aligned with the atlas. The MRF model plays a central role in solving this problem, as it propagates well-defined labels to the neighboring voxels along the tracts of the subject, reinforcing label probabilities.
The explicit modeling of crossings and overlapping regions is also a key element, as many tracts overlap at the current resolution. The algorithm is presently limited to crossings between two tracts, which may be a source of error in regions of triple-perpendicular crossings (Jeurissen et al., 2011). The main limitation resides in the tensor representation: because these crossings would be indistinguishable from isotropic regions, adding the extra degrees of freedom needed for triple intersection may reduce the accuracy of the results. However, the model could be extended easily from pairwise to multiple overlaps, given a more elaborate model of diffusion at higher angular resolution or even using regular DTI sequences (Landman et al., 2010a). Finally, the regions of multiple crossings are not necessarily estimated as isotropic in the current model, because neighboring regions from the converging tracts can propagate their labels into the crossing (sT and SO are usually lower but non-zero in isotropic voxels), and we did not observe any clear mislabeling in regions of multiple overlap.
Our experiments show that the DOTS segmentation is highly reproducible, even for small and variable tracts. The tracts segmented can be used readily as regions of interest for localized analysis of specific white matter pathways. Tract based statistics can be extracted separately for each tract, and effects of lesions or atrophy on anisotropy measures are less likely to confound the results. The segmented regions are somewhat larger than ones obtained through bundling tractography results, inherently because the MRF model does not require each voxel of the tract to be part of a complete path. On the other hand, a larger region of interest can be beneficial when computing statistics of the white matter. The results of DOTS can also be combined with any tractography technique to generate labeled fiber bundles in a fully automated fashion as well.
The atlas currently includes 39 separate tracts, covering most of the deep white matter pathways. Additional tracts can easily be added provided they can be identified reliably enough on diffusion MRI. Anatomical constraints are however important in order to avoid combinatorial explosion of the number of possible labels to estimate, and the atlas would benefit from incorporating high resolution anatomical delineations (Bürgel et al., 2006). The diffusion properties of a given tract are not fully captured by the principal eigenvector directions, and a more elaborate representation of the expected tensor (or higher order model) shape might further increase accuracy at the cost of higher computational demands and possibly a lowered robustness to noise and lesions.
Even with the few limitations discussed above, essentially inherited from the underlying tensor model of DTI, the DOTS software provides a novel and practical tool for DTI analysis in routinely acquired sequences, and we hope that making this method available to the community will help neuroscientists study in more detail individual white matter tracts in larger cohorts.
Acknowledgments
We thank Dr. Susumu Mori and Mr. Kegang Hua for providing us tensor images and tract delineations for their digital atlas used in preliminary versions of this work (Mori et al., 2005), and Dr. Peter Calabresi for providing the multiple sclerosis data used in the lesion experiments. This work was supported in part by the NIH/NIDA K25DA025356 grant, the NIH/NINDS R01NS056307 and R01NS070906 grants, the National Multiple Sclerosis Society TR3760A3 grant, the NIH K99NS064098 grant, the NIH P41RR015241 grant, the China Scholarship Council and the Intramural Research Program of NINDS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Awate S, Zhang H, Gee J. A fuzzy, nonparametric segmentation framework for DTI and MRI analysis: with applications to DTI-tract extraction. IEEE Trans Med Imaging. 2007 Nov;26(11):1525–1536. doi: 10.1109/TMI.2007.907301. [DOI] [PubMed] [Google Scholar]
- Basser P, Jones D. Diffusion-tensor MRI: theory, experimental design and data analysis - a technical review. NMR Biomed. 2002;15(7–8):456–467. doi: 10.1002/nbm.783. [DOI] [PubMed] [Google Scholar]
- Bazin P-L, Bogovic J, Reich DS, Prince JL, Pham DL. Belief propagation based segmentation of white matter tracts in DTI. Proceedings of the 11th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI’09); London. 2009a. september, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin P-L, Bogovic J, Reich DS, Prince JL, Pham DL. Efficient MRF segmentation of DTI white matter tracts using an overlapping fiber model. Proceedings of the International Workshop on Diffusion Modelling and Fiber Cup (DMFC09); London. 2009b. september, [Google Scholar]
- Bazin P-L, Cuzzocreo J, Yassa MA, Gandler W, McAuliffe M, Bassett S, Pham D. Volumetric neuroimage analysis extensions for the mipav software package. Journal of Neuroscience Methods. 2007;165:111–121. doi: 10.1016/j.jneumeth.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens T, Berg HJ, Jbabdi S, Rushworth M, Woolrich M. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besag JE. On the statistical analysis of dirty pictures. Journal of the Royal Statistical Society, Series B. 1986;48(3):259–302. [Google Scholar]
- Bihan DL, Mangin J, Poupon C, Clark C, Pappata S, Molko N, Chabriat H. Diffusion tensor imaging: concepts and applications. Journal of Magnetic Resonance Imaging. 2001;13(4):534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- Bürgel U, Amunts K, Hoemke L, Mohlberg H, Gilsbach JM, Zilles K. White matter fiber tracts of the human brain: Three-dimensional mappings at microscopic resolution, topography and intersubject variability. NeuroImage. 2006;29:1092–1105. doi: 10.1016/j.neuroimage.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Carass A, Wheeler M, Cuzzocreo J, Bazin P-L, Bassett S, Prince J. A joint registration and segmentation approach to skull stripping. Proceedings of the IEEE International Symposium on Biomedical Imaging; Arlington. 2007. april, [Google Scholar]
- Ceritoglu C, Oishi K, Li X, Chou M-C, Younes L, Albert M, Lyketsos C, van Zijl PC, Miller MI, Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. NeuroImage. 2009;47(2):618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux M, Deriche R, Bihan DL, Mangin J-F, Poupon. C. Diffusion propagator imaging: Using laplace’s equation and multiple shell acquisitions to reconstruct the diffusion propagator; Proceedings of Information Processing in Medical Imaging (IPMI). Vol. 5636 of Lecture Notes in Computer Science (LNCS); 2009. pp. 1–13. [DOI] [PubMed] [Google Scholar]
- Eckstein I, Shattuck DW, Stein JL, McMahon KL, de Zubicaray G, Wright MJ, Thompson PM, Toga AW. Active fibers: Matching deformable tract templates to diffusion tensor images. NeuroImage. 2009;47 Supplement 2:T82–T89. doi: 10.1016/j.neuroimage.2009.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher PT, Tao R, Jeong W-K, Whitaker RT. A volumetric approach to quantifying region-to-region white matter connectivity in diffusion tensor MRI. Proceedings of the International Conference on Information Processing in Medical Imaging 2007 (IPMI’07); Kerkrade; 2007. july, [DOI] [PubMed] [Google Scholar]
- Goh A, Vidal R. Segmenting fiber bundles in diffusion tensor images; Proceedings of the European Conference on Computer Vision 2008 (ECCV’08); 2008. [Google Scholar]
- Hagler DJ, Ahmadi ME, Kuperman J, Holland D, McDonald CR, Halgren E, Dale AM. Automated white-matter tractography using a probabilistic diffusion tensor atlas: Application to temporal lobe epilepsy. Human Brain Mapping. 2009;30(5):1535–1547. doi: 10.1002/hbm.20619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A, Initiative TADN. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. NeuroImage. 2010;51:221–227. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- Horsfield M, Jones D. Applications of diffusion-weighted and diffusion tensor mri to white matter diseases - a review. NMR Biomed. 2002;15(7–8):570–577. doi: 10.1002/nbm.787. [DOI] [PubMed] [Google Scholar]
- Jeurissen B, Leemans A, Jones DK, Tournier J-D, Sijbers J. Probabilistic fiber tracking using the residual bootstrap with constrained spherical deconvolution. Human Brain Mapping. 2011;32(3):461–479. doi: 10.1002/hbm.21032. URL http://dx.doi.org/10.1002/hbm.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Horsfield M, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magnetic Resonance in Medicine. 1999;42:515–525. [PubMed] [Google Scholar]
- Jones DK. Tractography gone wild: probabilistic fibre tracking using the wild bootstrap with diffusion tensor MRI. IEEE transactions on medical imaging In Medical Imaging. 2008;27(9):1268–1274. doi: 10.1109/TMI.2008.922191. [DOI] [PubMed] [Google Scholar]
- Koch MA, Norris DG, Hund-Georgiadis M. An investigation of functional and anatomical connectivity using magnetic resonance imaging. NeuroImage. 2002;16(1):241–250. doi: 10.1006/nimg.2001.1052. [DOI] [PubMed] [Google Scholar]
- Kolmogorov V, Zabih R. What energy functions can be minimized via graph cuts? IEEE Transactions on Pattern Analysis and Machine Intelligence. 2004;26(2):147–159. doi: 10.1109/TPAMI.2004.1262177. [DOI] [PubMed] [Google Scholar]
- Kouby VE, Cointepas Y, Poupon C, Rivire D, Golestani N, Poline J-B, Bihan DL, Mangin J-F. MR diffusion-based inference of a fiber bundle model from a population of subjects. Proceedings of the 8th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI’05); Palm Springs. 2005. october, [DOI] [PubMed] [Google Scholar]
- Landman B, Wan H, Bogovic J, Bazin P-L, Prince J. Resolution of crossing fibers with constrained compressed sensing using traditional diffusion tensor mri; Proc SPIE Medical Imaging; 2010a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman BA, Farrell JA, Jones CK, Smith SA, Prince JL, Mori S. Effects of diffusion weighting schemes on the reproducibility of dti-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5t. NeuroImage. 2007;36(4):1123–1138. doi: 10.1016/j.neuroimage.2007.02.056. [DOI] [PubMed] [Google Scholar]
- Landman BA, Huang AJ, Gifford A, Vikram DS, Lim IAL, Farrell JA, Bogovic JA, Hua J, Chen M, Jarso S, Smith SA, Joel S, Mori S, Pekar JJ, Barker PB, Prince JL, van Zijl PC. Multi-parametric neuroimaging reproducibility: A 3-t resource study. NeuroImage. 2010b doi: 10.1016/j.neuroimage.2010.11.047. In Press, –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawes I, Barrick T, Murugam V, Spierings N, Evans D, Song M, Clark C. Atlas-based segmentation of white matter tracts of the human brain using diffusion tensor tractography and comparison with classical dissection. Neuroimage. 2008;39:62–79. doi: 10.1016/j.neuroimage.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Lenglet C, Rousson M, Deriche R. DTI segmentation by statistical surface evolution. IEEE Transactions on Medical Imaging. 2006;25(6):685–700. doi: 10.1109/tmi.2006.873299. [DOI] [PubMed] [Google Scholar]
- Li SZ. Markov Random Field Modeling in Image Analysis. 3rd Edition. Springer-Verlag; 2009. [Google Scholar]
- Lucas B, Bogovic J, Carass A, Bazin P-L, Prince J, Pham D, Landman B. The java image science toolkit (jist) for rapid prototyping and publishing of neuroimaging software. Neuroinformatics. 2010 March;18:5–17. doi: 10.1007/s12021-009-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddah M, Wells WM, Warfield SK, Westin C-F, Grimson E. Probabilistic clustering and quantitative analysis of white matter fiber tracts. Proceedings of the International Conference on Information Processing in Medical Imaging 2007 (IPMI’07); Kerkrade. 2007. july, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddah M, Zöllei L, Grimson WEL, Westin C-F, Wells WM. A mathematical framework for incorporating anatomical knowledge in DT-MRI analysis. Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008. 5th IEEE International Symposium on Biomedical Imaging; Paris, France. 2008. pp. 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Zimmerman-Moreno G, Shadmi R, Batikoff A, Greenspan H. A supervised framework for the registration and segmentation of white matter fiber tracts. Medical Imaging, IEEE Transactions on. 2011 jan;30(1):131–145. doi: 10.1109/TMI.2010.2067222. [DOI] [PubMed] [Google Scholar]
- McAuliffe M, Lalonde F, McGarry D, Gandler W, Csaky K, Trus B. Medical image processing, analysis and visualization in clinical research; Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems (CBMS 2001); 2001. [Google Scholar]
- Mori S, Crain BJ, Chacko VP, Van Zijl PCM. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 1999;45(2):265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziottad J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40:570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl P. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15(7–8):468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Mori S, Wakana S, Nagae-Poetscher LM, van Zijl PC. MRI Atlas of Human White Matter. Elsevier; 2005. [DOI] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin C-F. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Transactions on Medical Imaging. 2007 November;26(11):1562–1575. doi: 10.1109/TMI.2007.906785. [DOI] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin C-F, Golby AJ. Tract-based morphometry for white matter group analysis. NeuroImage. 2009;45:832–844. doi: 10.1016/j.neuroimage.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G, Alexander D. Probabilistic monte carlo based mapping of cerebral connections utilising whole-brain crossing fibre information; Proceedings of the International Conference on Information Processing in Medical Imaging 2003 (IPMI’03); 2003. [DOI] [PubMed] [Google Scholar]
- Poupon C, Mangin J-F, Frouin V, Rgis J, Poupon F, Pachot-Clouard M, Bihan DL, Bloch I. Regularization of MR diffusion tensor maps for tracking brain white matter bundles; Proceedings of the International Conference on Medical Image Computing and Computer-Aided Intervention (MICCAI); 1998. pp. 489–498. [Google Scholar]
- Reich D, Smith S, Jones C, Zackowski K, van Zijl P, Calabresi P, Mori S. Quantitative characterization of the corticospinal tract at 3T. American Journal of Neuroradiology. 2006 Nov–Dec;27:2168–2178. [PMC free article] [PubMed] [Google Scholar]
- Reich DS, Ozturk A, Calabresi PA, Mori S. Automated vs. conventional tractography in multiple sclerosis: Variability and correlation with disability. NeuroImage. 2010;49(4):3047–3056. doi: 10.1016/j.neuroimage.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert M, Mader I, Anastasopoulos C, Weigel M, Schnell S, Kiselev V. Global fiber reconstruction becomes practical. NeuroImage. 2011;54(2):955–962. doi: 10.1016/j.neuroimage.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Rodrigues P, Jalba A, Fillard P, Vilanova A, ter Haar Romeny B. A multi-resolution watershed-based approach for the segmentation of diffusion tensor images; Proceedings of the MICCAI Workshop on Diffusion Modelling; 2009. pp. 554–565. [Google Scholar]
- Shiee N, Bazin P-L, Ozturk A, Calabresi P, Reich D, Pham D. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols T, Mackay C, Watkins K, Ciccarelli O, Cader M, Matthews P, Behrens T. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006 Jul;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. URL http://dx.doi.org/10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Tuch D. Q-ball imaging. Magnetic Resonance in Medicine. 2004;52(6):1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck M, Fallon J, Perry M, Gollub R, Hua K, Zhang J, Dubey P, Blitz A, van Zijl P, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage. 2007;36:630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin C-F, Maier S, Mamata H, Nabavi A, Jolesz F, Kikinis R. Processing and visualization of diffusion tensor MRI. Medical Image Analysis. 2002;6(2):93–108. doi: 10.1016/s1361-8415(02)00053-1. [DOI] [PubMed] [Google Scholar]
- Wheeler-Kingshott CAM, Cercignani M. About ”axial” and ”radial” diffusivities. Magnetic Resonance in Medicine. 2009;61(5):1255–1260. doi: 10.1002/mrm.21965. [DOI] [PubMed] [Google Scholar]
- Yang J, Shen D, Davatzikos C, Verma R. Diffusion tensor image registration using tensor geometry and orientation features; Proceedings of MICCAI; 2008. pp. 905–913. [DOI] [PubMed] [Google Scholar]
- Yendiki A, Stevens A, Jbabdi S, Augustinack J, Salat D, Zollei L, Behrens T, Fischl B. Probabilistic diffusion tractography with spatial priors. Proceedings of the MICCAI workshop on Computational Diffusion MRI; New York. 2008. [Google Scholar]
- Yushkevich PA, Zhang H, Simon TJ, Gee JC. Structure-specific statistical mapping of white matter tracts. NeuroImage. 2008;41:448–461. doi: 10.1016/j.neuroimage.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyan U, Sabuncu M, Grimson W, Westin C-F. Consistency clustering: A robust algorithm for group-wise registration, segmentation and automatic atlas construction indiffusion mri. International Journal of Computer Vision. 2009;85:279–290. doi: 10.1007/s11263-009-0217-1. URL http://dx.doi.org/10.1007/s11263-009-0217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]