Highlights
► Transferrin receptor 1 (TfR1) is the cellular receptor for New World hemorrhagic fever arenaviruses. ► Commonalities between human TfR1 and TfR1 of arenaviral host species make zoonoses possible. ► TfR1 upregulation during the acute-phase response may amplify arenaviral replication. ► TfR1 may make a useful target for much needed therapies for arenaviral hemorrhagic fevers.
Abstract
At least five New World arenaviruses cause severe human hemorrhagic fevers. These viruses are transmitted to humans through contact with their respective South American rodent hosts. Each uses human transferrin receptor 1 (TfR1) as its obligate receptor. Accidental similarities between human TfR1 and TfR1 orthologs of arenaviral host species enable zoonoses, whereas mice and rats are not infectable because they lack these TfR1 determinants of infection. All pathogenic New World arenaviruses bind to a common region of the apical domain of TfR1. The ability of a New World arenavirus to use human TfR1 is absolutely predictive of its ability to cause hemorrhagic fevers in humans. Nonpathogenic arenaviruses, closely related to hemorrhagic fever arenaviruses, cannot utilize human TfR1 but efficiently enter cells through TfR1 orthologs of their native rodent hosts. Mutagenesis studies suggest that minor changes in the entry glycoproteins of these nonpathogenic viruses may allow human transmission. TfR1 is upregulated as a result of iron sequestration during the acute-phase response to infection, and the severity of disease may result from amplification of viral replication during this response.
Introduction
Most enveloped viruses infect target cells by associating with a cellular receptor through their entry glycoproteins (GPs). A viral receptor can contribute in a number of ways to viral replication. It can mediate a high affinity attachment to target cells. It can induce conformational changes in the entry protein that prime it for the subsequent steps leading to fusion of the viral and target-cell membranes. It can facilitate internalization to an intracellular compartment hospitable to the fusion process, for example one that is acidic or abundant in activating proteases. It can position the entry protein for a subsequent interaction with an obligate cellular cofactor or coreceptor. Finally, such a receptor can mark target cells advantageous for viral replication, for example those important to the immune control of virus, or those that would enable transmission to the next host. Accordingly, identification of a viral receptor can shed light on a range of questions. Knowledge of the receptor can clarify the tissue tropism of a virus and therefore help describe the course of disease, explain the differential susceptibility of various species to a particular virus, and illuminate distinctive properties of the viral fusion mechanism. In addition, mice transgenic for the receptor can serve as models of infection, and small molecules and antibodies that bind the receptor can make effective therapeutics.
Here we discuss transferrin receptor 1 (TfR1), a receptor for all New World hemorrhagic fever arenaviruses as well as for some of their nonpathogenic cousins. In particular we will describe properties of this receptor that enable efficient arenavirus transmission from the host species to humans. We will also discuss TfR1's potential role in amplifying an initial infection, contributing to a feedback loop that ultimately leads to an often deadly hemorrhagic fever.
New World arenaviruses
The family Arenaviridae consists of a single genus (Arenavirus) with at least 28 recognized viruses [1, 2, 3, 4, 5, 6]. Arenaviruses are enveloped, bisegmented RNA viruses. The L segment includes genes for the viral polymerase and a zinc-binding protein. The S segment encodes the viral nucleocapsid and the viral entry GP precursor. Before assembly in the virus-producing cell, GP is cleaved by a cellular protease into two components, a receptor-binding component GP1, and a transmembrane component GP2. Conformational changes in GP2 promote mixing of viral and target cell lipids necessary for membrane fusion and delivery of the viral RNA to the target cell cytoplasm.
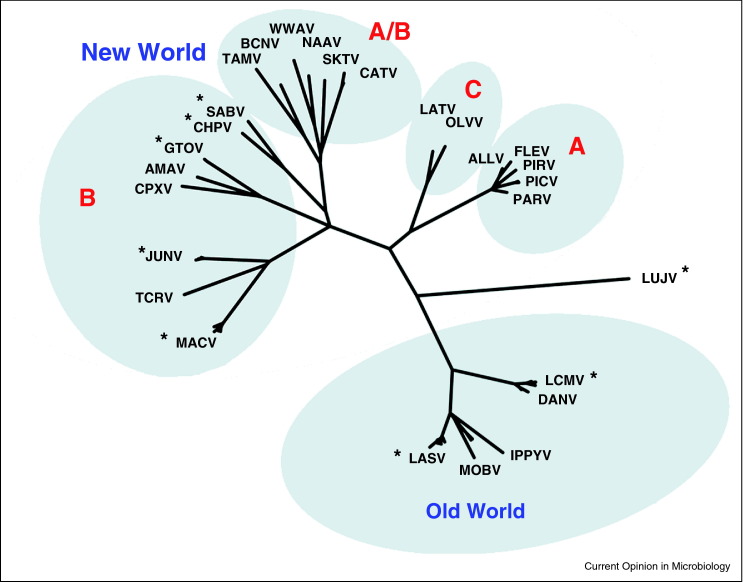
Arenaviruses are classified into two serologically distinct groups, the Lassa–lymphocytic choriomeningitis serocomplex (‘Old World arenaviruses’) and the Tacaribe serocomplex (‘New World arenaviruses’) [6]. Well-known Old World arenaviruses include Lassa fever virus (LASV) and lymphocytic choriomeningitis virus (LCMV). Most Old World viruses including LASV and LCMV utilize cellular α-dystroglycan as their cellular receptors. New World arenaviruses have been divided into three clades (A, B, and C). Clade C viruses also utilize α-dystroglycan, whereas the receptors for clade A viruses remain undefined. Clade A, B, and C viruses are found in South America. In addition, several North American viruses are recombinants of clade A and B viruses (clade A/B). All five known New World hemorrhagic fever viruses — Machupo virus (MACV), Junin virus (JUNV), Guanarito virus (GTOV), Sabia virus (SABV) and Chapare virus (CHPV) — are clade B (see Table 1 for abbreviations, host species, and receptors). Three of these viruses (MACV, JUNV, and GTOV) have been classified as NIAID Category A priority pathogens, because of the severity of disease they cause, their high potential for misuse, and the frequency with which new members of this family emerge, for example SABV in 1990 and CHPV in 2003. In addition to the five known hemorrhagic fever viruses, three additional nonpathogenic clade B viruses have been identified: Tacaribe virus (TCRV), Amapari virus (AMAV), and Cupixi virus (CPXV). Figure 1 shows a phylogenetic tree based on the sequences of their entry GPs [7, 8]. Note that the clade A/B viruses group with clade B viruses in this analysis, because their GP derives from a clade B progenitor. Note also that nonpathogenic viruses such as AMAV and TCRV are more closely related to pathogenic viruses in their respective sublineages than the latter are related to each other [5, 9, 10]. For example the hemorrhagic fever virus MACV is more closely related to TCRV than to other hemorrhagic fever viruses; similarly GTOV (pathogenic) is closer to AMAV (nonpathogenic) than to other hemorrhagic fever viruses. Thus although New World hemorrhagic fever viruses belong to a common clade, their relatedness within that clade is not predictive of their abilities to cause human disease. Another intriguing aspect of New World arenaviruses is that each is harbored in one or two distinct reservoir species, typically a South American rodent (listed in Table 1).
Table 1.
Old and New World arenaviruses. Representatives of the 28 known arenaviruses together with their groups and clades are listed. Eight of these cause human diseases. Each virus has a distinct reservoir species, listed with the country or region where it can be found. The cellular receptor for Old World (OW) viruses and NW clade C viruses is α-dystroglycan (αDG) [45, 47]. All five NW hemorrhagic fever viruses — all clade B — utilized human TfR1 as well as their host-species TfR1 orthologs. Two other nonpathogenic clade B viruses, AMAV and TCRV, use their host-species TfR1 orthologs, but not human TfR1 [17]. All known clade B viruses are represented. Clade A/B viruses have GP proteins that derive from a clade B progenitor virus.
| Name | Clade | Abbr. | Human disease | Host species and region | Receptor |
|---|---|---|---|---|---|
| Lassa | OW | LASV | Lassa fever | Mastomys natalensis (rat); West Africa | αDG [45] |
| Lymphocytic choriomeningitis | OW | LCMV | Lymphocytic choriomeningitis | Mus musculus (house mouse); ubiquitous | αDG [45, 46] |
| Lujo | OW | LUJV | HF | Unknown, South Africa | αDG |
| Pichinde | A | PICV | – | Oryzomys albigulans; Columbia | ? |
| Parana | A | PARV | – | Oryzomys buccinatus; Paraguay | ? |
| Pirital | A | PIRV | – | Sigmodon alstoni; Venezuela | ? |
| Tamiami | A/B | TAMV | – | Sigmodon hispidus (cotton rat); South Florida, USA | ? |
| Bear Canyon | A/B | BCNV | – | Peromyscus californicus; Southern California, USA | ? |
| Whitewater Arroyo | A/B | WWAV | – | Neotoma micropus (woodrat); Texas, USA | ? |
| Junin | B | JUNV | Argentine HF | Calomys musculinus (corn mouse); Argentina | TfR1 [11] |
| Machupo | B | MACV | Bolivian HF | Calomys callosus (vesper mouse); Bolivia | TfR1 [11] |
| Tacaribe | B | TCRV | – | Artibeus jamaicensis (bat), Trinidad | Host TfR1 [17] |
| Guanarito | B | GTOV | Venezuelan HF | Zygodontomys brevicauda (cane mouse); Venezuela | TfR1 [11] |
| Amapari | B | AMAV | – | Neacomys spinosus (bristly mouse); Brazil | Host TfR1 [17] |
| Cupixi | B | CPXV | – | Oryzomys goeldii (rice rat); Brazil | ? |
| Sabia | B | SABV | Brazilian HF | Unknown; Brazil | TfR1 [11] |
| Chapare | B | CHPV | HF | Unknown; Bolivia | TfR1 |
| Oliveros | C | OLVV | – | Necromys benefactus; Argentina | αDG [47] |
| Latino | C | LATV | – | Calomys callosus (vesper mouse); Bolivia | αDG [47] |
Figure 1.
Phylogenetic relationships among representative arenaviruses. Analysis is based on GP regions alone. Asterisks indicate viruses that cause human diseases. Only clade B viruses, which include all New World hemorrhagic fever arenaviruses, are fully represented.
Adapted from Briese et al. [8].
Transferrin receptor 1 is the New World hemorrhagic fever arenavirus receptor
The New World arenaviral receptor was identified as TfR1 using an approach previously successful in the identification of viral receptors for the SARS coronavirus (SARS-CoV) and Nipah and Hendra paramyxoviruses [11, 12, 13]. Specifically, the receptor-binding component, GP1, of the viral entry protein was fused to the Fc-region of human IgG1, and this immunoadhesin was used to precipitate TfR1 from lysates of cells susceptible to infection. Several insights from the SARS-CoV studies in particular expedited identification of TfR1. First, as the study of various SARS-CoV isolates made clear, there tends to be a correlation between receptor affinity and severity of disease, at least in the case of acute infections [12, 14]. Thus, the entry protein of the New World arenavirus reported as most deadly, MACV, was used to precipitate TfR1. Subsequent studies indeed confirmed that the MACV GP1 has the highest affinity among arenaviral GP1 proteins for human TfR1. Second, in several cases including SARS-CoV and HIV-1, smaller fragments of the receptor-binding component (S1 and gp120, respectively) were shown to bind receptor with higher affinity than the full protein [15]. These previous observations motivated the generation of truncation variants of MACV GP1 to identify fragments that bound target cells with higher affinity. One such fragment, lacking 20 amino-acids at the mature GP1 amino-terminus, bound target cells and TfR1 with markedly higher affinity than full-length MACV GP1 [16]. This fragment was subsequently used to solve the GP1/TfR1 cocrystal complex shown in Figure 2 . Following identification of TfR1 through mass spectrometry, a series of studies were undertaken to demonstrate that it was indeed an obligate receptor for MACV, GTOV, JUNV, and SABV. Later studies demonstrated that CHPV also used human TfR1 as its receptors (SJ, MF, and HC, in preparation), and that in every case where the arenaviral host species was known, the TfR1 ortholog of that species served as an efficient receptor for hosted arenavirus [17, 18]. The ability of each virus to infect cells using the specific TfR1 ortholog of its respective host highlights the coevolution of virus and host species, and confirms the role of TfR1 in host species as well as in humans.
Figure 2.
Cocrystal complex of human TfR1 dimer with MACV GP1. The dimer of TfR1 is shown in dark gray and white [16]. TfR1 residues within 5.5 Å of transferrin, as described in Ref. [21], are indicated with magenta. Two MACV GP domains are shown, with key TfR1 residues tyrosine 211 and threonine 348 indicated as red and orange spheres, respectively [18]. The protease-like, helical, and apical domains of TfR1 are indicated on one monomer. Note that transferrin interacts solely with the helical and protease-like domains, whereas MACV GP1 interacts solely with the TfR1 apical domain [16].
TfR1 determinants contributing to zoonoses
TfR1 binds iron-bound transferrin and transports it to an acidic cellular compartment where iron is released and subsequently transported across the vesicle membrane into the cytoplasm [19, 20]. TfR1 is dimeric type II membrane protein with three well-defined domains in its extracellular region (Figure 2). The protease-like and helical domains bind transferrin directly, whereas the prominent and exposed apical domain does not have a known cellular function [21, 22]. Although the GP1 domains of MACV and other hemorrhagic fever arenaviruses bind avidly to human TfR1, they do not associate with murine TfR1, consistent with inability of mice to be infected with these viruses [18]. This difference permitted mapping of the interaction site to the apical domain of TfR1, and in particular, to a region surrounding a prominent tyrosine 211 in human TfR1. Indeed, the presence of a tyrosine at position 211 predicts whether one or more of these arenaviruses can utilize a particular TfR1 ortholog [18]. Thus, the mouse, rat, and guinea pig TfR1 molecules each have an aspartic acid at position 211, and each are refractory to infection by New World clade B arenaviruses. Conversely, each of the rodent hosts of MACV, GTOV, JUNV, AMAV, and TCRV has a tyrosine at 211, as do most Old and New World primates (the host species of remaining clade B arenaviruses are unknown) [17, 18]. Other determinants within the apical domain also affect the efficiency of infection. For example, JUNV, but not MACV or GTOV, tolerate a lysine a position 348. Position 348 is an asparagine in humans and in most clade B arenaviral host species, but it is a lysine in the JUNV host species as well as in mice. Accordingly, MACV and GTOV cannot utilize the JUNV host-species TfR1 ortholog [18]. These studies, as well as antibody neutralization studies make clear that all New World clade B viruses bind a common region of the apical domain of TfR1. Importantly, this region does not overlap with the binding site of transferrin [16, 21, 23], suggesting that targeting the apical domain with small molecules or antibodies could be a safe, effective, and general approach to treating South American hemorrhagic fevers.
Nonpathogenic clade B viruses and their potential for emergence
There are five New World hemorrhagic fever arenaviruses, and three closely related clade B viruses that nonetheless do not cause disease in humans [3, 5, 17]. Given the close relationship between these viruses and hemorrhagic fever viruses, however, it was not surprising that AMAV and TCRV utilized the TfR1 orthologs of their respective host species (the receptor for CPXV is also likely TfR1 from its host species, but this remains undemonstrated) [17]. Interestingly, AMAV and TCRV can infect and replicate in human cells, but they do so independently of human TfR1 [17, 24]. Because hemorrhagic fever arenaviruses are closely related to these viruses — indeed more closely related than individual pathogenic viruses are to each other — we and others [17, 24] have concluded that utilization of human TfR1 determines the ability of a clade B arenavirus to cause human disease, and that therefore that properties of TfR1 contribute to disease pathogenesis. We have also observed that changes in as little as a single human TfR1 residue are sufficient for AMAV and TCRV to gain use of this receptor [16, 17]. This observation raises the unsettling possibility that modest changes in the AMAV and TCRV entry protein will be sufficient for them to become pathogenic in humans. Indeed variants of these viruses may already be circulating in their respective host species that could cause a human hemorrhagic fever.
Iron sequestration, TfR1 expression, and arenaviral hemorrhagic fevers
The close relationship between human TfR1 use and severe human disease suggests that TfR1 regulation plays a pivotal role in disease severity. We suggest that the early response to viral infection drives TfR1 expression, which in turn accelerates viral replication. This feedback mechanism, not observable in vitro, could distinguishes hemorrhagic fever clade B arenaviruses from their nonpathogenic cousins. There are two general ways in which TfR1 might be upregulated. First, rapid cell division, in response to infection, increases TfR1 expression. Second, low serum iron, as a result of iron sequestration during infection or tissue damage (‘hypoferremia of inflammation’), can drive TfR1 expression higher [25, 26]. Iron sequestration is a consequence of an acute-phase response to infection, and is specifically driven by IL-6 expression. IL-6, observed at high levels in New World hemorrhagic fevers [27, 28], is in turn a major contributor to production of the key iron regulatory hormone, hepcidin [25, 29, 30]. Hepcidin, a 25-amino-acid peptide produced in the liver, binds and inhibits the iron-export protein ferroportin present at the duodenum, where dietary iron is absorbed, and in iron-storing hepatocytes and macrophages [30]. Continued absorption of iron by extra-hepatic tissues depletes the pool of extracellular iron, leading to upregulation of TfR1 in most tissues. Regulation of TfR1 expression is mediated through iron-responsive elements (IRE) at 3′ untranslated (UTR) regions of the TfR1 mRNA [31, 32]. When cellular iron level is low, iron-responsive protein (IRP) binds to the TfR1 mRNA IRE, stabilizing it, and increasing cell-surface expression of TfR1. This in turn promotes more vigorous replication of the infecting virus.
Although the role of such a feedback mechanism remains speculative, there is clear evidence for each step in this process. Significantly elevated IL-6 levels have been measured in South American hemorrhagic fevers [27], and increased IL-6 and hepcidin is observed with several other viral infections, for example in individuals acutely infected with common cold viruses or chronically infected with hepatitis C virus [9, 33]. The links between IL-6 levels, hepcidin concentration, hypoferremia, and TfR1 upregulation have also been established [25]. IL-6 has been demonstrated to be necessary and sufficient for hepcidin expression and hypoferremia in mice and human volunteers [26, 30]. During acute-phase responses in the rats induced by turpentine oil injection or radiation, hepcidin concentration increased, followed by enhanced TfR1 expression in the extra-hepatic tissues [34, 35]. Similarly, during cardiac surgeries in humans, increased hepcidin levels were accompanied by enhanced TfR1 expression [36, 37]. IL-6 was induced within three hours after injection of mice with LPS, and urinary hepcidin peaked at six hours, followed by a significant decrease in serum iron [38]. There is also clear evidence that natural and induced iron deficiency in animals or humans strongly correlates with increased tissue TfR1 expression [39, 40, 41, 42, 43, 44]. Finally, published data demonstrate that iron supplementation inhibits and iron chelation enhances MACV or JUNV GP-mediated entry in two cell lines [11]. Thus each step in this feedback mechanism is established, but its final demonstration awaits interruption of this process in vivo, perhaps with anti-IL-6 or anti-hepcidin antibodies.
Open questions
The identification of TfR1 as the receptor for New World hemorrhagic fever arenaviruses has shed light on a number of issues, but there are still key questions remaining. For example, it remains unclear how nonpathogenic arenaviruses infect human cells. The close relationship between pathogenic and nonpathogenic clade B viruses suggests that there might be an alternative receptor or coreceptor used by both sets of viruses, but this receptor remains unknown. A second question raised is why the arenaviral entry process is so decisive in the ability of these viruses to transmit to humans, whereas other steps in the viral life-cycle do not appear to be species-specific. A central role for the entry process in zoonotic transmission has also been observed with influenza A virus and SARS-CoV. This pattern sharply contrasts with that of retroviruses for example, where species-specific dependencies and restrictions can be observed at many postentry steps. In general, a virus that moves relatively rapidly from species to species will tend to depend on conserved elements of host proteins with which it interacts. However the entry protein becomes more rapidly species-specific than other viral proteins because it is one of the few susceptible to selective pressure by serum antibodies. Accidental or adaptive changes in the entry protein that permit infection of a new species can then be sufficient for transmission to that species. This in turn raises the question of how easy it is for nonpathogenic arenaviruses to gain use of human TfR1. Although a preliminary investigation suggests that modest changes in the arenaviral GP might be sufficient, it is not clear that a pathway for such changes will be available in the host species of TCRV, AMAV, or CPXV. Further laboratory studies and field work clarifying this critical issue are clearly needed. Also, the role of TfR1 in a feedback loop contributing to pathogenesis remains unproven. Such a mechanism would suggest novel therapeutic approaches, for example those that interrupt the process of acute-phase iron sequestration. It also raises the possibility that the receptors of other highly pathogenic viruses are upregulated as a direct consequence of infection. Finally and perhaps most importantly, it is not yet certain that any of these insight can be applied therapeutically. Anti-TfR1 antibodies which blockade GP association but which do not impair iron metabolism have already been described and may effectively limit replication in an infected patient, or protect the uninfected health care worker during an outbreak. It also remains possible that iron supplementation, commonly used to treat anemia, can slow arenaviral replication, although such an approach may have to bypass tight control of iron intake in the duodenum. We suggest that animal and clinical testing of these possible therapeutic approaches is straightforward and warranted.
Acknowledgements
This work was supported by New England Regional Center for Excellence/Biodefense and Emerging Infectious Disease (U54 AI057159; HC, SJ, and MF), the New England Primate Research Center Base Grant, RR000168 (MF) and by the Burroughs Welcome Fund (MF).
References
- 1.Jay M.T., Glaser C., Fulhorst C.F. The arenaviruses. J Am Vet Med Assoc. 2005;227:904–915. doi: 10.2460/javma.2005.227.904. [DOI] [PubMed] [Google Scholar]
- 2.Clegg J.C. Molecular phylogeny of the arenaviruses. Curr Top Microbiol Immunol. 2002;262:1–24. doi: 10.1007/978-3-642-56029-3_1. [DOI] [PubMed] [Google Scholar]
- 3.Bowen M.D., Peters C.J., Nichol S.T. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology. 1996;219:285–290. doi: 10.1006/viro.1996.0248. [DOI] [PubMed] [Google Scholar]
- 4.Rowe W.P., Pugh W.E., Webb P.A., Peters C.J. Serological relationship of the Tacaribe complex of viruses to lymphocytic choriomeningitis virus. J Virol. 1970;5:289–292. doi: 10.1128/jvi.5.3.289-292.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cajimat M.N., Fulhorst C.F. Phylogeny of the Venezuelan arenaviruses. Virus Res. 2004;102:199–206. doi: 10.1016/j.virusres.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Charrel R.N., de Lamballerie X. Zoonotic aspects of arenavirus infections. Vet Microbiol. 2010;140:213–220. doi: 10.1016/j.vetmic.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 7.Milazzo M.L., Cajimat M.N., Haynie M.L., Abbott K.D., Bradley R.D., Fulhorst C.F. Diversity among tacaribe serocomplex viruses (family Arenaviridae) naturally associated with the white-throated woodrat (Neotoma albigula) in the southwestern United States. Vector Borne Zoonotic Dis. 2008;8:523–540. doi: 10.1089/vbz.2007.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briese T., Paweska J.T., McMullan L.K., Hutchison S.K., Street C., Palacios G., Khristova M.L., Weyer J., Swanepoel R., Egholm M. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cajimat M.N., Milazzo M.L., Rollin P.E., Nichol S.T., Bowen M.D., Ksiazek T.G., Fulhorst C.F. Genetic diversity among Bolivian arenaviruses. Virus Res. 2009;140:24–31. doi: 10.1016/j.virusres.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charrel R.N., Feldmann H., Fulhorst C.F., Khelifa R., de Chesse R., de Lamballerie X. Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem Biophys Res Commun. 2002;296:1118–1124. doi: 10.1016/s0006-291x(02)02053-3. [DOI] [PubMed] [Google Scholar]
- 11.Radoshitzky S.R., Abraham J., Spiropoulou C.F., Kuhn J.H., Nguyen D., Li W., Nagel J., Schmidt P.J., Nunberg J.H., Andrews N.C. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Negrete O.A., Levroney E.L., Aguilar H.C., Bertolotti-Ciarlet A., Nazarian R., Tajyar S., Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 14.Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham J., Corbett K.D., Farzan M., Choe H., Harrison S.C. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat Struct Mol Biol. 2010;17:438–444. doi: 10.1038/nsmb.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham J., Kwong J.A., Albarino C.G., Lu J.G., Radoshitzky S.R., Salazar-Bravo J., Farzan M., Spiropoulou C.F., Choe H. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog. 2009;5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radoshitzky S.R., Kuhn J.H., Spiropoulou C.F., Albarino C.G., Nguyen D.P., Salazar-Bravo J., Dorfman T., Lee A.S., Wang E., Ross S.R. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc Natl Acad Sci U S A. 2008;105:2664–2669. doi: 10.1073/pnas.0709254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews N.C., Fleming M.D., Levy J.E. Molecular insights into mechanisms of iron transport. Curr Opin Hematol. 1999;6:61–64. doi: 10.1097/00062752-199903000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Hentze M.W., Muckenthaler M.U., Andrews N.C. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y., Zak O., Aisen P., Harrison S.C., Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence C.M., Ray S., Babyonyshev M., Galluser R., Borhani D.W., Harrison S.C. Crystal structure of the ectodomain of human transferrin receptor. Science. 1999;286:779–782. doi: 10.1126/science.286.5440.779. [DOI] [PubMed] [Google Scholar]
- 23.Daniels T.R., Delgado T., Rodriguez J.A., Helguera G., Penichet M.L. The transferrin receptor part I: biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Flanagan M.L., Oldenburg J., Reignier T., Holt N., Hamilton G.A., Martin V.K., Cannon P.M. New world clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and -independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J Virol. 2008;82:938–948. doi: 10.1128/JVI.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganz T. Iron in innate immunity: starve the invaders. Curr Opin Immunol. 2009;21:63–67. doi: 10.1016/j.coi.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemeth E., Rivera S., Gabayan V., Keller C., Taudorf S., Pedersen B.K., Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marta R.F., Montero V.S., Hack C.E., Sturk A., Maiztegui J.I., Molinas F.C. Proinflammatory cytokines and elastase-alpha-1-antitrypsin in Argentine hemorrhagic fever. Am J Trop Med Hyg. 1999;60:85–89. doi: 10.4269/ajtmh.1999.60.85. [DOI] [PubMed] [Google Scholar]
- 28.Kunz S. The role of the vascular endothelium in arenavirus haemorrhagic fevers. Thromb Haemost. 2009;102:1024–1029. doi: 10.1160/TH09-06-0357. [DOI] [PubMed] [Google Scholar]
- 29.Ganz T. Hepcidin — a peptide hormone at the interface of innate immunity and iron metabolism. Curr Top Microbiol Immunol. 2006;306:183–198. doi: 10.1007/3-540-29916-5_7. [DOI] [PubMed] [Google Scholar]
- 30.Nemeth E., Valore E.V., Territo M., Schiller G., Lichtenstein A., Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461–2463. doi: 10.1182/blood-2002-10-3235. [DOI] [PubMed] [Google Scholar]
- 31.Fleming R.E., Britton R.S. Iron imports. VI. HFE and regulation of intestinal iron absorption. Am J Physiol Gastrointest Liver Physiol. 2006;290:G590–G594. doi: 10.1152/ajpgi.00486.2005. [DOI] [PubMed] [Google Scholar]
- 32.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann N Y Acad Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 33.Hoppe M., Hulthen L. Capturing the onset of the common cold and its effects on iron absorption. Eur J Clin Nutr. 2007;61:1032–1034. doi: 10.1038/sj.ejcn.1602614. [DOI] [PubMed] [Google Scholar]
- 34.Sheikh N., Dudas J., Ramadori G. Changes of gene expression of iron regulatory proteins during turpentine oil-induced acute-phase response in the rat. Lab Invest. 2007;87:713–725. doi: 10.1038/labinvest.3700553. [DOI] [PubMed] [Google Scholar]
- 35.Christiansen H., Sheikh N., Saile B., Reuter F., Rave-Frank M., Hermann R.M., Dudas J., Hille A., Hess C.F., Ramadori G. x-Irradiation in rat liver: consequent upregulation of hepcidin and downregulation of hemojuvelin and ferroportin-1 gene expression. Radiology. 2007;242:189–197. doi: 10.1148/radiol.2421060083. [DOI] [PubMed] [Google Scholar]
- 36.Vokurka M., Lacinova Z., Kremen J., Kopecky P., Blaha J., Pelinkova K., Haluzik M., Necas E. Hepcidin expression in adipose tissue increases during cardiac surgery. Physiol Res. 2010;59:393–400. doi: 10.33549/physiolres.931759. [DOI] [PubMed] [Google Scholar]
- 37.Maruna P., Lindner J., Kunstyr J., Plocova K., Hubacek J. Plasma prohepcidin as a negative acute phase reactant after large cardiac surgery with a deep hypothermic circulatory arrest. Physiol Res. 2009;58:827–833. doi: 10.33549/physiolres.931678. [DOI] [PubMed] [Google Scholar]
- 38.Kemna E., Pickkers P., Nemeth E., van der Hoeven H., Swinkels D. Time-course analysis of hepcidin, serum iron, and plasma cytokine levels in humans injected with LPS. Blood. 2005;106:1864–1866. doi: 10.1182/blood-2005-03-1159. [DOI] [PubMed] [Google Scholar]
- 39.Fry M.M., Kirk C.A., Liggett J.L., Daniel G.B., Baek S.J., Gouffon J.S., Chimakurthy P.M., Rekapalli B. Changes in hepatic gene expression in dogs with experimentally induced nutritional iron deficiency. Vet Clin Pathol. 2009;38:13–19. doi: 10.1111/j.1939-165X.2008.00081.x. [DOI] [PubMed] [Google Scholar]
- 40.Liu C.Y., Liu Y.F., Zeng L., Zhang S.G., Xu H. The expression of TfR1 mRNA and IRP1 mRNA in the placenta from different maternal iron status. Zhonghua Xue Ye Xue Za Zhi. 2007;28:255–258. [PubMed] [Google Scholar]
- 41.Lu J., Hayashi K., Hu X. Transferrin receptor and iron deposition pattern in the hepatic lobules of the iron-deficient and iron-overloaded rats. Zhonghua Bing Li Xue Za Zhi. 1995;24:75–77. [PubMed] [Google Scholar]
- 42.Collins J.F., Franck C.A., Kowdley K.V., Ghishan F.K. Identification of differentially expressed genes in response to dietary iron deprivation in rat duodenum. Am J Physiol Gastrointest Liver Physiol. 2005;288:G964–G971. doi: 10.1152/ajpgi.00489.2004. [DOI] [PubMed] [Google Scholar]
- 43.Siddappa A.J., Rao R.B., Wobken J.D., Casperson K., Leibold E.A., Connor J.R., Georgieff M.K. Iron deficiency alters iron regulatory protein and iron transport protein expression in the perinatal rat brain. Pediatr Res. 2003;53:800–807. doi: 10.1203/01.PDR.0000058922.67035.D5. [DOI] [PubMed] [Google Scholar]
- 44.Pietrangelo A., Rocchi E., Casalgrandi G., Rigo G., Ferrari A., Perini M., Ventura E., Cairo G. Regulation of transferrin, transferrin receptor, and ferritin genes in human duodenum. Gastroenterology. 1992;102:802–809. doi: 10.1016/0016-5085(92)90161-q. [DOI] [PubMed] [Google Scholar]
- 45.Cao W., Henry M.D., Borrow P., Yamada H., Elder J.H., Ravkov E.V., Nichol S.T., Compans R.W., Campbell K.P., Oldstone M.B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 46.Reignier T., Oldenburg J., Noble B., Lamb E., Romanowski V., Buchmeier M.J., Cannon P.M. Receptor use by pathogenic arenaviruses. Virology. 2006;353:111–120. doi: 10.1016/j.virol.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 47.Spiropoulou C.F., Kunz S., Rollin P.E., Campbell K.P., Oldstone M.B. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J Virol. 2002;76:5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]