SUMMARY
Background
We investigated the determinants of sexual identity in the budding yeast, Saccharomyces cerevisiae. The higher fungi are divided into the Ascomycetes and the Basidiomycetes. Most Ascomycetes have two mating types: one (called α in yeasts and MAT1-1 in filamentous fungi) produces a small, unmodified, peptide pheromone, and the other (a in yeasts and MAT1-2 in filamentous fungi) produces a peptide pheromone conjugated to a C terminal farnesyl group that makes it very hydrophobic. In the Basidiomycetes, all pheromones are lipid-modified, and this difference is a distinguishing feature between the phyla. We asked whether the asymmetry in pheromone modification is required for successful mating in Ascomycetes.
Results
We cloned receptor and pheromone genes from a filamentous Ascomycete and a Basidiomycete and expressed these in the budding yeast, Saccharomyces cerevisiae, to generate novel, alternative mating pairs. We find that two yeast cells can mate even when both cells secrete a-like or α-like peptides. Importantly, this is true regardless of whether the cells express the a- or α-mating type loci, which control the expression of other, sex-specific genes, in addition to the pheromones and pheromone receptors.
Conclusions
We demonstrate that the asymmetric pheromone modification is not required for successful mating of ascomycete fungi and confirm that, in budding yeast, the primary determinants of mating are the specificity of the receptors and their corresponding pheromones.
INTRODUCTION
Sex costs time and resources and represents a critical moment in an organism’s life cycle. Most eukaryotes are sexual, and all those that have been intensively investigated have sexual forms (recently reviewed in [1]). The ability of fungi to mate with themselves is determined by the inheritance of mating potential during mitotic divisions [2]: homothallic strains generate progeny that can mate with each other, whereas heterothallic strains generate progeny that cannot mate with each other, for example Neurospora crassa and most lab strains of the budding yeast, S. cerevisiae. There are two forms of homothallism. In one, genetically identical cells can mate with each other (e.g. Sordaria macrospora), in the other, the mitotic divisions of cells of one mating type can give rise to another mating type, allowing mating between two strains that differ only at their mating type loci (e.g. most wild S. cerevisiae isolates). Budding yeast has a single mating type locus, MAT, and encodes silent copies of both types of mating information (HMLα and HMRa). Wild cells can undergo mating type switching, a gene conversion event that copies information from the silent cassette to the MAT locus, but most lab strains are unable to switch and maintain their mating type stably. A haploid budding yeast cell can only express a single mating locus at a time and mating occurs only between two haploid cells, one expressing MATα (defining the α-cells) and the other expressing MATa (defining the a-cells).
The genotype at MAT distinguishes three cell types (reviewed in [3]): diploids (MATa/MATα cells), which cannot undergo sexual fusion, but are capable of meiosis and sporulation, and two haploid cell types, a (MATa) and α (MATα), which can fuse with each other to form the diploid, a/α cells (Figure 1A). Three regulatory proteins, Matα1, Matα2 and Mata1, control the expression of cell type specific genes. The presence of Matα1 induces the expression of genes that are only expressed in α cells (α specific genes), while Matα2 blocks the expression of genes that are only expressed in a cells (a specific genes). These regulators are only present in α cells. The a mating type is the “default”, and it is the one expressed in the absence of Matα1 and Matα2. In diploid a/α cells, Matα2 represses the a specific genes and binds to Mata1 to block the expression of haploid specific genes (genes expressed in both a and α haploids, but not in a/α diploids). Cells that express neither a nor α specific genes (α cells that lack Matα1), and cells that express both a and α specific genes (a cells that lack Matα2) show significant mating difficulties [4–5] The two haploid mating types sense each other’s presence by reciprocal sets of pheromones and pheromone receptors, with a cells secreting a-factor and expressing the α-factor receptor, Ste2 (which allows them to respond to α-factor), and α cells secreting α-factor and expressing the a-factor receptor, Ste3 (which allows them to respond to a-factor) (Figure 1A). Beyond the receptors, the signaling pathways are identical in both mating types.
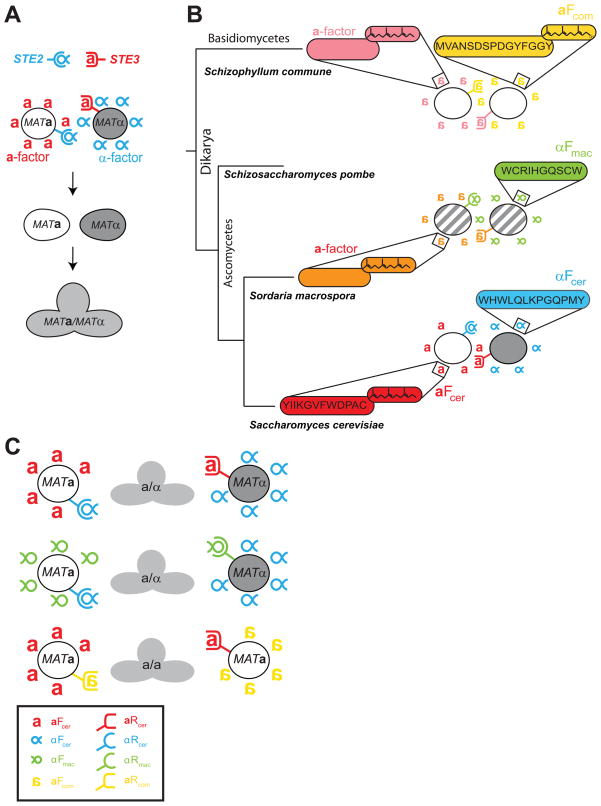
Figure 1. Sexual communication via pheromones and their corresponding receptors.
A) The mating system of S. cerevisiae. The two haploid S. cerevisiae cell types, a (white) and α (dark gray) express seven-transmembrane, G-protein coupled receptors that detect the presence of small peptides secreted by the opposite mating type. MATa cells secrete a-factor (red a) and express the α-factor receptor (Ste2, blue semi-circular receptor) at their surface; MATα cells secrete α-factor (blue α) and express the a-factor receptor Ste3 (red, U-shaped receptor) at their surface,. When the two cells find each other, they can fuse to form a diploid (light gray) cell that can divide mitotically. B) Pheromone asymmetry between the Ascomycetes and the Basidiomycetes: the Ascomycetes, which include Saccharomyces cerevisiae and Sordaria macrospora, communicate via two different types of pheromones, unmodified, α-factor like peptides and lipid-modified, a-factor-like peptides. The Basidiomycetes, like Schizophyllum commune, only use lipid-modified a-factor-like pheromones. The letter a represents lipid-modified pheromones and the letter α represents α-factor-like peptides. Letters are reversed for pheromones derived from species other than S. cerevisiae. The sequences for the α-factor like peptides from S. cerevisiae and S. macrospora are shown in blue and green, respectively. The sequences for the a-factor like pheromones for S. cerevisiae and S. commune are shown in red and yellow, respectively. This color code is common to all figures. C) Examples of artificial mating types. In S. cerevisiae, two cells, expressing different mating loci (MATa or MATα) communicate using asymmetric pheromones (a- and α-factor-like peptides) and their corresponding receptors (top line). We disrupted these asymmetries by generating artificial mating types that communicate via two different α-factor-like (middle line) or two different a-factor-like peptides, and also produced cells that expressed complementary pairs of receptors and pheromones but expressed the same mating type locus (bottom line). Legend: Red: a-factor (aFcer) and a-factor (aRcer) receptor from S. cerevisiae. Blue: α-factor (αFcer) and α-factor receptor (αRcer) from S. cerevisiae. Green: α-factor (αFmac) and α-factor receptor (αRmac) from S. macrospora. Yellow: a-factor (aFcom) and a-factor receptor (aRcom) from S. commune.
The pheromones for the two mating types are asymmetric with respect to size and hydrophobicity. While both peptide sequences are amphipathic, α-factor is an unmodified peptide, but a-factor is farnesylated and carboxymethylated at a C-terminal CAAX box [6]. As a result, a-factor is very hydrophobic, and is secreted from the cytoplasm by a specific transporter, Ste6, a homolog of multidrug transporters, whereas α-factor is secreted by the standard protein secretion machinery [7]. This asymmetry is conserved across the Ascomycetes, but the Basidiomycetes only express lipid-modified, a-factor-like pheromones (Figure 1B), and this is a distinguishing feature between the phyla [8]. Mutations of the CAAX box result in non-farnesylated (or non-carboxymethylated) peptides and lead to significant reductions in mating efficiency, suggesting that the lipid tail is required for recognition and activation of the corresponding a-factor receptors, in both the budding yeast [9] and a common smut [10]. The high hydrophobicity of the a-factor pheromone makes it difficult to work with, and most studies looking at the yeast pheromone signaling pathway are done with a cells being stimulated with α-factor. Therefore, little is known about the extracellular behavior of a-factor and how this behavior might influence mating efficiency.
In S. cerevisiae, mating requires that the two cells express complementary pheromones and receptors, raising the question of whether the pheromone-receptor pairs determine whether cells mate as a or α. Previous approaches to this question involved genetic manipulations to the mating locus to alter the pheromones that each cell produces and force expression of Ste2 receptor in MATα cells [4–5, 11]. Such manipulations led to the conclusion that the receptors and pheromones are the major determinants of mating specificity [5]. Receptors and pheromones have also been swapped within species in Ustilago maydis [12], a heterothallic Basidiomycete and in Cryptococcus neoformans [13], a Basidiomycete with an unusual mating type locus. Both studies conclude that mating specificity is determined by the set of pheromones and receptors that a cell expresses.
This earlier work did not address the question of whether the asymmetry between a and α factors was essential for mating. Because all Ascomycete matings involve one cell that expresses an unmodified α-factor-like pheromone, and one cell that expresses a lipid-modified, a-factor-like pheromone, we hypothesized that this asymmetry is essential for mating.
By using pheromones and receptors from distantly related fungi, we made cognate pairs of pheromones and receptors (an a-like pheromone interacting with an a-like receptor or an α-like pheromone interacting with an α-like receptor) that would not cross-react with the budding yeast pheromones and receptors (Figure 1B and 1C). These combinations make it possible to ask what role the asymmetry between the chemical nature of a-like and α-like receptors plays in mating. Using heterologous pheromone-receptor pairs allowed us to ask if mating type identity is defined by mating-type specific proteins beyond the pheromones and receptors. If it were, a MATa cell should mate significantly worse with another MATa cell than with a MATα cell (Figure 1C), regardless of which pheromones and receptors it expresses.
We show that yeast cells can mate with each other as long as they express complementary sets of receptors and pheromones, suggesting that the identities of these molecules is necessary and sufficient to determine which cell types mate with each other.
RESULTS
S. cerevisiae can mate using heterologous receptor and pheromone pairs
To generate multiple mating types that could communicate via different receptor/pheromone pairs, we chose two fungal species whose receptors had been successfully expressed in S. cerevisiae [14–15]. Schizophyllum commune is a heterothallic Basidiomycete that is predicted to encode at least 18 different receptors and more than 75 pheromones, all of which display the C-terminal farnesylation motif, CAAX, (Figure 1B). Expressing different combinations of the pheromones and receptors defines more than 15000 possible mating types (for a review on S. commune mating see [16] and [2]). To generate artificial “a-mating types”, we cloned one of S. commune’s a-factor-like receptors (Bbr1) and one of the a-like pheromones that bind this receptor (Bbp2(4)) and expressed them in S. cerevisiae (Figure 1B). We refer to this receptor as aRcom (for a-factor receptor from S. commune), to the pheromone as aFcom (for a-factor pheromone from S. commune), and the pair has been color-coded in yellow in all of the figures and tables (Table 1).
Table 1.
| Symbol | Protein | Organism | Nomenclature |
|---|---|---|---|

|
a-factor | S. cerevisiae | aFcer |

|
STE3 | S. cerevisiae | aRcer |

|
αfactor | S. cerevisiae | αFcer |

|
STE2 | S. cerevisiae | αRcer |

|
PPG1 | S. macrospora | αFmac |

|
PRE2 | S. macrospora | αRmac |

|
BBP2(4) | S. commune | aFcom |

|
BBR1 | S. commune | aRcom |
Sordaria macrospora is a homothallic filamentous fungus closely related to Neurospora crassa (for information on S. macrospora mating see [17] and [2], especially chapter 10). To generate artificial α mating types, we cloned its α-factor-like receptor (Pre2) and its corresponding α-like pheromone (Ppg1) (Figure1B). We refer to this receptor as αRmac (for α-factor receptor from S. macrospora), to the pheromone as αFmac (for α-factor pheromone from S. macrospora) and the pair has been color coded in green in all of the figures and tables (see Table 1 for a summary of the terminology and Supplementary Table 1 for the genotype of strains).
The receptors and pheromone pairs from S. cerevisiae are represented by the letters cer. We refer to S. cerevisiae’s α-factor receptor, Ste2, as αRcer and to S. cerevisiae’s α-factor as αFcer. Both have been color coded in blue. S. cerevisiae’s a-factor receptor, Ste3, is denoted aRcer and the pheromone it binds, a-factor, as aFcer. Both have been colored in red.
To generate artificial mating types, we constructed strains expressing heterologous pheromone receptors and mated them to strains carrying the matching heterologous pheromone genes (Table 1). Since the heterologous receptors and pheromones are being expressed in an organism that is evolutionarily distant, there may be difficulties in the processing and secretion of the pheromones and in the transport of the receptors to the plasma membrane and their communication with the remainder of the pheromone signaling pathway. As a result, strains that have replaced a budding yeast pheromone-receptor pair with the pheromone-receptor pair from S. commune or S. macrospora may mate worse than wild type S. cerevisiae a and α strains, but any mating indicates successful expression and function of the heterologous genes.
In S. cerevisiae, the mating pheromones are each encoded by two genes: α-factor is encoded by MFα1 and MFα2 and a-factor is encoded by MFA1 and MFA2. To replace the endogenous pheromones with the S. macrospora α-like peptide, the αFmac gene was cloned into the two α-factor loci in α cells and into the two a-factor loci in a-cells, replacing the coding sequences for the endogenous, S. cerevisiae, peptides. Both a and α-cells were found to express and secrete mature αFmac pheromone, albeit with significantly lower efficiency than α cells secrete their own endogenous α-factor (Supplementary Figure 1A and B). To replace the endogenous a-factor pheromone with the S. commune a-like peptide, the aFcom gene was cloned into both a-factor loci in a-cells. We could not make the same type of quantitative measurements for aFcom because the farnesyl group of this pheromone leads to non-specific binding to most labware surfaces.
We then replaced the budding yeast pheromone receptors with their homologues from S. macrospora and S. commune, expressing the heterologous receptors from the normal, budding yeast receptor loci. The αRcer receptor was replaced by the αRmac gene in an a-cell, and the aFcer gene was replaced by the aFcom gene in an α-cell (Figure 2A). The heterologous α-factor-like receptor, αRmac, showed difficulties communicating with the downstream MAP kinase signaling components as assayed by measuring the response of cells to their cognate α-like factor (P. Marcenac and J. Gonçalves-Sá, unpublished data). To overcome this problem, we deleted the SST2 gene, which expresses a negative regulator of pheromone signaling. Receptor expression was tested via receptor-green fluorescent protein (GFP) fusions, and we compared localization to that of the endogenous receptors in both a and α cells (Supplementary Figure 2). In all strains, we see a strong signal in cellular compartments (most likely the vacuole, given the high turnover of the receptors) and plasma membrane localization upon pheromone induction. We saw no signal in the strains expressing the aRcom, and this is most likely because we cannot induce the receptor.
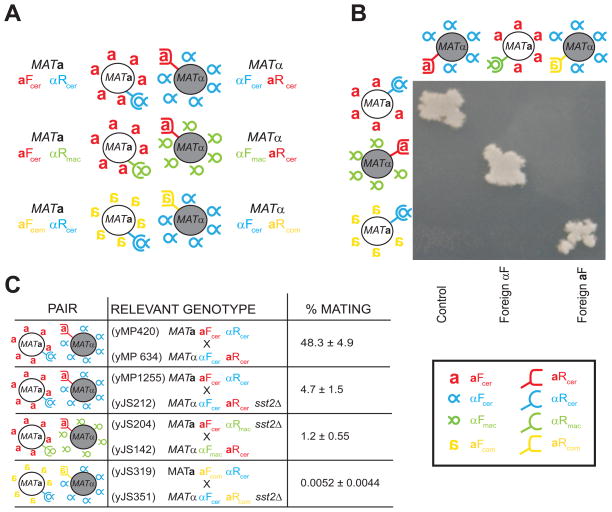
Figure 2. Cells expressing heterologous pheromone-receptor pairs can mate.
A) Mating pairs that conserve the asymmetry between a-factor-like and α-factor-like pheromones. First line: Control mating relying on S. cerevisiae pheromones (aFcer (red a) αRcer (blue α)) and receptors (aRcer (red U-shaped receptor) αFcer (blue semi-circular receptor)). Second line. MATa cells expressing the α-like receptor from S. macrospora (αRmac, green semi-circular receptor) were mated to MATα cells expressing the S. macrospora α-factor-like peptide (αFmac, inverted green α). This pair communicates via the α-factor from S. macrospora and the a-factor from S. cerevisiae (aFcer, red a). Third line: MATa cells expressing an a-factor-like pheromone from S. commune (aFcom, yellow inverted a) were mated with MATα cells expressing the corresponding S. commune receptor (aRcom yellow U-shaped receptor). This pair communicates via the a-factor from S. commune and the α-factor from S. cerevisiae. B) Visualizing mating with heterologous pheromone-receptor pairs. The cells described in A) were streaked, replica plated on top of each other, and allowed to mate over night in complete media. They were then replica plated onto media where only diploids could grow. The absence of off-diagonal mating shows that only strains expressing complementary pheromones and receptor pairs can mate. C) Quantitative mating data. The indicated crosses were allowed to mate on filters for 4 hours (for the cross using two homologous receptor-pheromone pairs) or for 7h (in the case of the sst2Δ and heterologous crosses). Filters were then washed and cells plated on selective media to select for diploids. Mating efficiency is calculated as described in the Materials and Methods. Errors are standard deviations from at least three independent mating trials. Note that the aFcom producing strains (yJS319, last row) were mixed in an excess of 5:1 with the other partner in the mating. All the other crosses were done at a 1:1 ratio. Legend: Red: a-factor (aFcer) and a-factor (aRcer) receptor from S. cerevisiae. Blue: α-factor (αFcer) and α-factor receptor (αRcer) from S. cerevisiae. Green: α-factor (αFmac) and α-factor receptor (αRmac) from S. macrospora. Yellow: a-factor (aFcom) and a-factor receptor (aRcom) from S. commune.
We tested our system by mating a- and α-like cells, which expressed the heterologous receptors and pheromones (Figure 2A). As a control, we mated two wild type S. cerevisiae strains: MATa αRcer aFcer × MATα aRcer αFcer. As expected, these two strains mate with an efficiency of around 50% (Figure 2C and Supplementary Figure 4).
We asked whether the S. macrospora α-like pheromone and its receptor functioned in S. cerevisiae. We mated the following strains, MATa αRmac aFcer and MATα aRcer αFmac, which replace the S. cerevisiae α-factor and α-factor receptor with their homologs from S. macrospora. As expected, this pair can mate, although 40-fold worse than the mating pair that communicates using only S. cerevisiae pheromones (Figure 2B and 2C for quantitation, note that when compared with the sst2Δ pair this difference is only 4-fold).
We then looked at the mating efficiency using S. commune proteins by mating MATa αRcer aFcom with MATα aRcom αFcer (Figure 2A). This pair, which replaces the S. cerevisiae a-factor and a -factor receptor with their homologs from S. commune, showed almost no mating. We hypothesized that the low mating efficiency could be explained by low levels of receptor and/or pheromone expression. Over expressing the pheromones and receptors from a multi copy plasmid with a strong promoter showed that pheromone expression was the limiting factor, as increasing pheromone receptor expression did not significantly increase the number of mating events (Supplementary Figure 3 and data not shown). To increase the amount of pheromone experienced by cells expressing aRcom, the experiments involving the S. commune mating genes were done with the aFcom-expressing strains at a 5:1 ratio to the aRcom-expressing strains. While mating efficiency was improved, it remained 200-fold lower compared to crosses using the S. macrospora proteins (Figure 2C). These differences can be rationalized as reflecting the larger phylogenetic distance between S. commune and S. cerevisiae: the Basidiomycete pheromone and receptor might be expressed at lower levels than their Ascomycete homologs and/or the receptor communicates with the MAP kinase signaling components less effectively.
Because we expressed heterologous pheromones and receptors and sensitized the pheromone signaling pathway (by using sst2Δ strains,) we asked if the receptors maintain their specificity for the corresponding pheromones. We used replica plating to mix cells expressing the different receptors with cells expressing the different pheromones. These crosses were allowed to mate, and we selected for the presence of diploids. Figure 2B shows that all three cognate pairs (expressing either S. cerevisiae, S. macrospora or S. commune proteins) can mate and that the receptors are specific for their pheromones, as no off-diagonal mating can be observed.
Recapitulating Basidiomycete matings in an Ascomycete
We then generated a mating pair where both cells express a-like pheromones and a-factor-like receptors. In one a-cell, we replaced the endogenous αRcer receptor with the S. commune a-factor-like receptor (aRcom), to make a MATa aRcom aFcer. In another a-cell, we replaced the endogenous a-factor pheromone genes with the S. commune pheromone, aFcom, and the αRcer receptor with the aRcer receptor, usually expressed in α cells, to make the strain MATa aRcer aFcom, as shown in Figure 3A (this strain also lacks ASG7, an a-specific gene that interacts with the a-factor receptor, Ste3, to interfere with pheromone-signaling by altering the localization of Ste4, the Gβ protein that transmits the pheromone signal [18–19]). These strains (MATa aRcom aFcer and MATa aRcer aFcom) now express complementary pairs of a-factor-like pheromones and receptors: (Figure 3A). While mating efficiency was low, it was higher than that of the cross that used the S. commune pheromone and receptor, but maintained the asymmetry between a- and α-like receptors in both pheromones and cell-type background (Figure 3C).
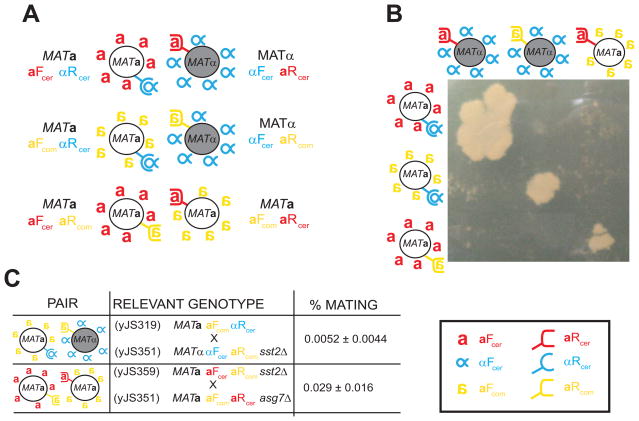
Figure 3. Mating between cells that can only communicate using a-factor-like pheromones.
A) Artificial mating pairs with the S. commune pheromones/receptors. First line: the control mating pair using S. cerevisiae pheromones described in Figure 2A. Second line: the mating pair that expresses heterologous a-factor-like pheromone and the corresponding receptor from S. commune described in Figure 2A. Third line: a mating pair composed of two MATa strains, one expressing the a-factor-like pheromones from S. commune (aFcom, yellow inverted a) and the a-factor receptor from S. cerevisiae, usually expressed in MATα cells (aRcer, red U-shaped receptor), and the other expressing S. cerevisiae a-factor (aFcer, red a) and the corresponding S. commune receptor (aRcom, yellow U-shaped receptor). This pair communicates via the a-factor from S. commune and the a-factor from S. cerevisiae. B) Visualizing mating with heterologous pheromone-receptor pairs. The cells described in A) were streaked, replica plated on top of each other, and allowed to mate over night in complete media. They were then replica plated onto media where only diploids could grow. The absence of off-diagonal mating shows that only strains expressing complementary pheromones and receptor pairs can mate. C) Quantitative mating data. The indicated crosses were allowed to mate on filters for 7 hours. Filters were then washed and cells plated on selective media to select for diploids. Mating efficiency is calculated as described in the Materials and Methods. Errors are standard deviations from at least three independent mating trials. Note that the aFcom producing strains (yJS319, yJS360) were mixed in an excess of 5:1 with the other partner in the mating. All the other crosses were done at a 1:1 ratio. Legend: Red: a-factor (aFcer) and a-factor (aRcer) receptor from S. cerevisiae. Blue: α-factor (αFcer) and α-factor receptor (αRcer) from S. cerevisiae. Yellow: a-factor (aFcom) and a-factor receptor (aRcom) from S. commune.
Observing mating between two strains that both express a-factor-like peptides has two implications. First, there is no requirement for having pheromone dimorphism (farnesylated vs. non-farnesylated pheromones) for Ascomycete mating, raising the question as to what the functional or evolutionary significance of this asymmetry might be. The second is that self-stimulation does not prevent the growth of diploid cells. The result of mating MATa aRcom aFcer with MATa aRcer aFcom cells are MATa/MATa diploids, which fail to express Matα2, which normally represses the expression of a factor and the a factor receptor in diploid cells. As a result, the MATa/MATa cells should express two cognate a-like pheromone/receptor pairs (aFcer binding to aRcer, and aFcom binding to aRcom), leading to self-stimulation of pheromone signaling and G1 arrest. To see if diploids were forming but failing to divide, we mixed MATa aRcom aFcer with MATa aRcer aFcom cells and imaged them. We never observed the formation of a diploid under the microscope (Supplementary Movies 1 and 2), which suggests that these cells arise at a frequency of less than 10−4. This can be because the a-factor pheromones are expressed at lower levels, are less efficient at inducing G1 arrest, because the aRcom signals to the cascade very weakly, or a combination of these. But the fact that we never observed cell fusion microscopically suggests that cells expressing S. commune’s genes mate at very low frequency, and that the low number of colonies that we observe is not due to the failure of self-stimulated diploid cells to give rise to colonies.
The lipid tail is not-required for partner recognition and fusion in yeast
The mating pathway in S. cerevisiae has been extensively studied and several proteins involved in membrane fusion have identified [20–22], but the signal that triggers cell-cell fusion remains unknown. We have shown that we can make two a cells fuse even in the absence of an α-like pheromone. Because this situation appears to mimic mating in the Basidiomycetes, it is possible that a-factor, or some unknown a-specific protein, might play a fundamental role in cell-cell fusion. If the farnesyl group of the pheromone is required for membrane fusion, at least one partner would have to express a lipid-modified peptide for mating to occur. Thus, two cells of opposite mating types that secrete only α-factor-like peptides should be able to form pre-zygotes, but be unable to fuse.
To test this hypothesis, we constructed a mating pair that communicates using only α-like pheromones. Starting from an α-cell, we replaced the aRcer receptor with αRmac. This strain is now αMATα αRmac αFcer, producing S. cerevisiae’s α-factor and responding to the S. macrospora pheromone, αFmac. Starting from an a-cell, we replaced both a-factor producing genes with αFma to make a cell that is MATa αRcer αFmac (Figure 4A third row). This MATα αRmac αFcer × MATa αRcer αFmac pair can communicate using only α-like pheromones although the two cell backgrounds, determined by the MAT locus, remain different, a and α (Figure 4A). To our surprise, these cells could now mate with efficiencies of around 1%, comparable to that of matings between strains that express S. cerevisiae’s a factor and S. macrospora’s α-like factor and their cognate receptors (the mating of a and α cells in which the α-factor and α-factor receptor come from S. macrospora, and the a-factor and a-factor receptor come from S. cerevisiae, compare the first and second rows in Figure 4C).
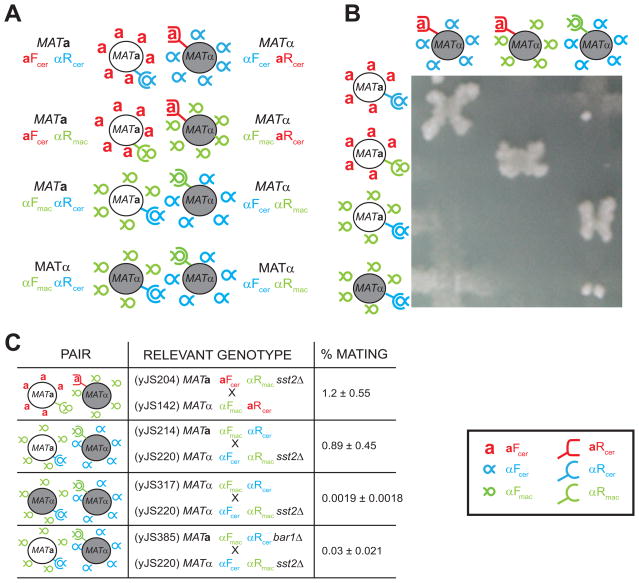
Figure 4. Cells that communicate using only α-factor-like pheromones can mate.
A) Artificial mating pairs using the S. macrospora pheromone and receptor. First line: the mating pair using S. cerevisiae pheromones described in Figure 2A. Second line: the mating pair that expresses heterologous α-factor-like pheromone and the corresponding receptor from S. macrospora described in Figure 2A. Third line: a MATa cell expressing the S. macrospora α-factor pheromone (αFmac, green inverted α) mating with MATα cell expressing the α-factor receptor from S. macrospora (αRmac, green rounded receptor). This pair can communicate via α-factor like peptides only, but maintains the asymmetry at the mating locus (MATa and MATα cells). Fourth line: a MATα strain expressing S. cerevisiae’s α-factor receptor, usually expressed in MATa cells (αFcer, blue rounded receptor), and the pheromone from S. macrospora (αFmac, green inverted α). This cell can now communicate with the MATα αFcer αRmac cell described above, using only α-factor like peptides, in a mating with both cells expressing MATα. B) Visualizing mating with heterologous pheromone-receptor pairs. The cells described in A) were streaked, replica plated on top of each other, and allowed to mate over night in complete media. They were then replica plated onto media where only diploids could grow. The absence of off-diagonal mating shows that only strains expressing complementary pheromones and receptor pairs can mate. C) Quantitative mating data. The indicated crosses were allowed to mate on filters for 7 hours. Filters were then washed and cells plated on selective media to select for diploids. Mating efficiency is calculated as described in the Materials and Methods. Mating pairs where both strains express the MATα locus mate about 450 times worse than mating pairs that express different mating loci. Errors represent standard deviations of at least 3 independent trials. See Materials and Methods and main text for more details. Legend: Red: a-factor (aFcer) and a-factor (aRcer) receptor from S. cerevisiae. Blue: α-factor (αFcer) and α-factor receptor (αRcer) from S. cerevisiae. Green: α-factor (αFmac) and α-factor receptor (αRmac) from S. macrospora.
This result shows that there is no requirement for the lipid-modified pheromone in mating, but it does not rule out the possible contribution of a-specific genes, other than the pheromone, in mating. To address a putative role for other a-specific genes in mating, we started from an α cell and replaced both endogenous pheromone genes with αFmac, and replaced the naturally expressed aRcer receptor with αRcer (Figure 4A, fourth row) allowing us to make a MATα αRmac αFcer × MATα αRcer αFmac pair.
We now have two mating pairs that can communicate via α-factor like peptides only, but in one pair, both cells express the MATα locus, and in the other pair, the cells express different MAT loci. When we compared the mating efficiencies, we found that the MATα/MATα pair mated about 450-fold worse than the MATa/MATα pair (Figure 4C). Several factors could explain this difference: a) the α cells might have problems expressing αRcer (which is usually produced by a cells); b) the α cells, now producing αFmac, might express less pheromone than the a cells used in the earlier cross; c) there is some a-specific protein that is important for efficient mating; d) some feature of the MATα × MATα cross keeps mating partners from finding each other successfully; or e) the α/α diploids have difficulties re-budding after fusion, as they could still self-stimulate and be arrested in G1, since they lack heterozygosity at the MAT locus.
We investigated the MATα × MATα mating in more detail. The data presented in Supplementary Figure 1 shows that the MATα αRcer αFmac cells actually produce slightly more S. macrospora α-factor than MATa αRcer αFmac a cells (Supplementary Figure 1B), and the αRcer receptor is expressed in both a and α cells (Supplementary Figure 2). To test whether α/α mating pairs had difficulties in the cell-cell fusion step, we followed the formation of mating pairs under the microscope. By mixing the MATα αRmac αFcer with its α mating pair, MATα αRcer αFmac, the cells were found to arrest and induce the mating pathway, but have difficulty polarizing and appear to shmoo in random directions rather than polarizing towards a mating partner. On rare occasions, two cells expressing the matching receptor and pheromone pairs find each other, align their polarities, and the fusion process proceeds normally (Supplementary Movie 3 and data not shown).
These phenotypes mimic those seen in S. cerevisiae MATa bar1Δ × MATα matings (with both partners expressing their normal pheromones and receptors). Bar1 is secreted by a-cells and degrades the α-factor pheromone. Cells that lack Bar1 are supersensitive to α-factor-induced G1 arrest and exhibit reduced mating efficiency [23–24]. Because BAR1 is an a-specific gene, it is not expressed by MATα cells, meaning that the MATα αRcer αFmac should have the same pheromone supersensitivity as MATa αRcer aFcer bar1Δ cells. To test this explanation, we removed Bar1 from the mating between MATa and MATα cells that expressed only α-like pheromones (MATα αRmac αFcer × MATa αRcer αFmac). The frequency of mating in the cross that lacks Bar1 (MATα αRmac αFcer × MATa αRcer αFmac bar1Δ) is 30-fold lower than that of the cross where Bar1 is expressed (Figure 4C, last row). This difference does not result from differences in the level of αFmac secretion: the measured secretion from cells that do or do not produce Bar1 is similar (Supplementary Figure 1B), demonstrating that S. cerevisiae Bar1 does not cleave αFmac. Thus, the absence of Bar1 accounts for much of the difference in the mating efficiency between MATα × MATα and MATα × MATa crosses. The remaining 15-fold difference is most likely due to the reduced plating efficiency of the MATα/MATα pairs (Figure 5 and Supplementary Table 2), which continue to produce pheromone and pheromone receptors after they form diploids because they lack the Matα2/Mata1 heterodimer that is required to repress haploid-specific genes.
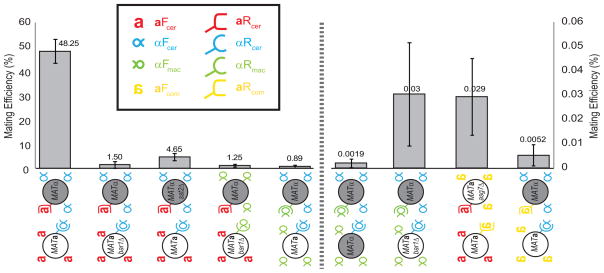
Figure 5. Quantitative mating efficiency of all crosses.
The mating efficiency of all crosses reported in this paper was quantified as described previously. Cells were allowed to mate for 7h (with the exception of the left-most pair, which used only S. cerevisiae pheromones, which was mated for 4h only) and then plated on selective media to isolate diploids. Note that the scale for the mating efficiency on the left side of the graph is 1000 times higher than the one on the right. Error bars represent standard deviations from at least three independent trials. Crosses using the heterologous pheromone/receptor pairs mate worse than the ones expressing the S. cerevisiae genes, but there is no substantial difference between the mating efficiencies of strains expressing only a or only α-like pheromones. SST2 was deleted in all the strains expressing the heterologous receptors and ASG7 was deleted in all MATa strains that expressed an a factor receptor. sst2Δ, asg7Δ and bar1Δ means that the shown strains were deleted for the SST2, ASG7 or BAR1 genes. Legend: Red: a-factor (aFcer) and a-factor (aRcer) receptor from S. cerevisiae. Blue: α-factor (αFcer) and α-factor receptor (αRcer) from S. cerevisiae. Green: α-factor (αFmac) and α-factor receptor (αRmac) from S. macrospora. Yellow: a-factor (aFcom) and a-factor receptor (aRcom) from S. commune.
DISCUSSION
We have shown that fungal cells can mate with each other as long as they express complementary pairs of pheromones and receptors. Thus, budding yeast can mate successfully when both cells express a-like pheromones, when both cells express α-like pheromones, and even when both partners express the same allele (MATα or MATa) at the mating-type locus. These results have four implications. First, the fundamental features of pheromones and their receptors have been sufficiently conserved that they can work in organisms that are as distant as Ascomycetes are from Basidiomycetes, which are estimated to have diverged from each other about 600 million years ago [25]. Second, there is no requirement that one pheromone be α-factor like and the other be a-factor like, even though all Ascomycetes that we investigated show this asymmetry. Third, we can produce colonies of MATa/MATa and MATα/MATα diploids despite the prediction that these cells should express two cognate pairs of pheromones and pheromone receptors, activate the pheromone signaling pathway, and thus arrest in G1. Fourth, there are no undiscovered α- or a-specific genes that are essential for mating.
In matings between wild type MATα and MATa cells, about 50% of the cells form diploids, whereas in our experiments with heterologous pheromones and receptors the efficiency of mating ranges from 5% to 0.002%. We contend that the differences in mating efficiency can be explained by a variety of idiosyncratic problems rather than systematic differences between different types of pheromones. These problems include difficulties in expressing the heterologous receptors and/or pheromones which lead to polarization problems, failure of cells to produce appropriate pheromone-degrading enzymes, and divergence in the response between two strains of a mating pair (with some arresting, others cycling and other shmooing). Mating events between these alternative mating types are rare and hard to track by video microscopy, however, we have never observed the formation of mating pairs which successfully polarize towards each other, but then fail to fuse. This observation argues that there is no α- or a-specific protein that is essential for cell fusion.
The fact that we can isolate a/a and α/α diploids with a certain frequency is not easy to interpret. As discussed above, these cells should self-stimulate, arrest and have difficulties forming colonies. One hypothesis is that these cells might have suffered a mutation in some gene related to the mating pathway or cell cycle progression. This seems unlikely, since the frequency of α-factor resistant mutants is only 5.9 × 10−6 per cell per generation [26]. The diploids would need to grow exponentially for 14 divisions to produce enough cells to ensure that 9% of the resulting colonies had produced at least one pheromone resistant mutation. In addition, we have observed that these diploids have longer cell-cycles and are still inducing the mating pathway, even if only slightly (data not shown). Another explanation could be that the presence of more than one receptor titrates away Gα subunits or other components of the MAP kinase cascade leading to reduced sensitivity to pheromone. This has been observed when both STE2 and STE3 were simultaneously expressed in a cells [27]. We also cannot rule out that an unknown regulator might shut down the pheromone response pathway upon cell-cell fusion, but this is unlikely, as a/a and α/α diploids are known to respond to pheromones and mate as the corresponding haploid cells. It is also possible that the cells have adapted physiologically, rather than genetically, to continual stimulation. At this point, we cannot distinguish between these hypotheses, but our results show that both α cells and a cells can mate with themselves if given the appropriate stimuli (summarized in Figure 5 and Supplementary Table 2).
Even homothallic Ascomycetes, like S. macrospora, express the two types of pheromones (a and α-factor like) whereas Basidiomycetes only express farnesylated peptides. The a-factor-like pheromones seem to be conserved over long evolutionary times, and the budding yeast’s a-factor transporter, Ste6, can substitute for a Drosophila transporter in inducing stem cell migration and is hypothesized to act by exporting a lipid-modified peptide [28]. Given the strong conservation of a-factor, we hypothesized that the farnesyl group could be playing a role in partner discrimination and/or cell-cell fusion. If this were the case, mating pairs that only communicated via two different α-factors like peptides should have significantly impaired mating efficiency. Likewise, if the asymmetry in pheromone hydrophobicity is the fundamental determinant of specificity, mating pairs that only express a-factor-like pheromones should also display reduced mating efficiencies. This is not what we saw: cells that communicate via α-factor-like peptides can mate. The fact that this lipid-modified pheromone is so conserved across phyla raises the question of why mating mechanisms that require α-factor-like pheromones have evolved. We can only speculate on the events that led to the appearance of α-factor like pheromones in the Ascomycetes. The greater solubility of α-factor may have been important for recognition events that required signaling at a distance. Alternatively, it may have allowed sexual selection. In budding yeast, when a cells are given a choice, they prefer to mate with the α cells that secrete more α-factor [29]. As a result, α cells are selected to produce more and more α-factor, which could eventually produce pheromone concentrations that are high enough to overwhelm the ability of a cells to detect the concentration gradients that they use to polarize towards their partners. The evolution of a protease that is induced by α-factor and can destroy α-factor solves this problem. These proteases have been identified in Ascomycetes as distant as S. cerevisiae, C. albicans, and S. pombe. In both S. cerevisiae and C. albicans, these are aspartyl proteases [30], but it is not clear that they are true homologs, since each one is more related to different aspartyl proteases in the genome of its relative. In S. pombe, the protease that degrades the α-like pheromone, Sap30, is a serine carboxypeptidase [31], arguing that pheromone degrading enzymes have arisen independently in different branches of the Ascomycete lineage.
Despite this conservation in pheromone asymmetry and a likely requirement for the presence of an analog Bar1 in all Ascomycete species, the Ascomycetes show a wide variety of mating patterns from strict homothallism to strict heterothallism. In heterothallic and pseudo-homothallic species, the cells that interact with each other possess different mating type loci and express different pheromones and pheromone receptors. As an example, standard laboratory strains of S. cerevisiae are heterothallic and mating occurs only between α and a strains. Many wild isolates are homothallic, allowing them to switch mating types and mate with genetically identical relatives (reviewed in [32]). Population genetic analysis of the sister species, S. paradoxus, shows that 94% of spores mate with their sisters, 5% self-mate as a result of mating type switching, and only 1% outbreed [33].
The clear picture presented by analyzing budding yeast is challenged by three observations. The first is the existence of fully homothallic species, such as S. macrospora, in which genetically identical cells fuse with each other. These species still require pheromone/receptor pairs for successful mating, suggesting that pheromone signaling is needed for cells to communicate with each other [34]. The second is evidence for autocrine stimulation and same sex mating in species previously thought to be exclusively heterothallic, such as Candida albicans, Cryptococcus gatti, and Cryptococcus neoformans [35–37]. In some of these cases, strains of a single mating type produce “inappropriate” pheromones that stimulate their own receptors and lead to mating between genetically identical cells. In budding yeast, a cells transcribe α-factor (MFα1 and MFα2) and a-factor receptor (STE3) genes when they are treated with α-factor [38], even though these genes were previously thought to be expressed in α cells only. This raises the interesting question of why is it that MATa cells don’t mate with each other at a higher frequency, and our results suggest that it may be possible to create truly homothallic budding yeast. Third, many Basidiomycetes require different alleles at each of two different mating loci for mating and normal sexual development, but crosses that only show differences alleles at one locus are capable of some sexual development (reviewed in [39]), and some species have connected the two loci to produce single locus mating systems like those found in Ascomycetes (reviewed in [1]). Cryptococcus neoformans, is a basidiomycete with an unusual and complex mating locus, which appears to represent a transition from having two mating type loci to having only one [40]. But despite the complexity of the mating type locus forced expression of receptors and pheromones demonstrates that pheromones and receptors are sufficient to determine the sexual identity of haploid cells [13].
Our overall conclusion is that two fungal cells can mate as long as each cell can produce a pheromone that stimulates a pheromone receptor on its partner. Thus, it is the ability of pheromones and receptors to interact with each other, more than mating type loci or mating system, that is the primary determinant of sexual identity.
MATERIALS AND METHODS
Strain construction and manipulation
Standard yeast manipulation methods were used. Strains used in this study are listed in Supplementary Table 1. All fluorescent protein cassettes come from plasmids produced by K. Thorn [41]. Strains with fluorescent reporters were constructed by inserting a plasmid containing YFP under the control of the FUS1 promoter at the LEU2 locus. Cassettes were amplified by PCR from plasmids made from the pFA6a backbone with a pair of primers that included 40 to 70 bp upstream and downstream of the targeted genomic region and integrated into the genome by homologous recombination. The heterologous receptor/pheromone strains were cloned into the endogenous genomic locus of the corresponding genes in S. cerevisiae (Supplementary Table 1). SST2 was deleted in all the strains expressing the heterologous receptors. ASG7 was deleted in all the MATa strains expressing the STE3 receptor. sst2Δ, asg7Δ and bar1Δ means that the shown strains were deleted for the SST2, ASG7 or BAR1 genes.
Non-Quantitative Mating assays
Fresh colonies were streaked into selective media and allowed to grow overnight at 30ºC. Mating pairs were replica plated on top of each other into rich media and allowed to mate for approximately 24h. They were then replica plated on diploid selective media and grown for at least 48h before screening.
Quantitative mating assays
Cell cultures were grown, harvested, and mixed 1:1 with the corresponding mating pair (unless otherwise noted). The mixes were then sucked into filters, placed on agar plates and allowed to mate for 4h or 7h at 30ºC. Filters were then washed and approximately 200 cells were plated on media lacking one amino acid (to count haploid cells) and varying numbers (depending on the mating efficiency) were plated on media lacking two amino acids (to select for diploids). Mating efficiency was calculated as the number of zygotes divided by the number of haploid cells (averaged across the two strains) at the end of the mating assay.
Pheromone secretion measurements
Pheromone secretion was measured by harvesting medium that had contained α-factor expressing cells and comparing its activity to synthetic pheromones. Conditioned medium was collected from α-factor producing cells (MP634 for S. cerevisiae pheromone, JS214, JS385 and JS317 for S. macrospora pheromone producing cells). The test, MATa, cells were grown in YPD and were then incubated with different concentrations of synthetic α-factor (from 0 to 500nM) to generate a calibration curve or with different dilutions conditioned media (no dilution, or diluted 2-fold or 10-fold), to estimate the secretion rate, and grown at 30ºC for 2h. The fraction of cells that had formed shmoos was determined by light microscopy at least 4 independent times and at least 200 cells were counted, per condition. S. cerevisiae and S. macrospora’s α-factor like pheromone peptides were synthesized by Biomatik Corporation Wilmington, DE, and HPLC purified to >95% purity. The response curve of MP384 and JS204 to the synthetic α-factors (% of shmoos vs. α-factor concentration) was fit, and this fit was used to estimate the concentration of α-factor the a-cells in conditioned media were sensing. The pheromone secretion rate, η, was estimated using , where r is the average replication time in seconds (5400), N0 is the initial number of cells (200000, 100000 or 20000), T is incubation time in seconds (1800, 3600 or 7200).
Supplementary Material
HIGHLIGHTS.
Asymmetric modification of pheromones is not required for yeast mating
Two yeast strains that express complementary pheromones and receptors mate with each other.
Two yeast strains that express the same mating type allele can mate with each other.
Receptors and the pheromones determine the sexual identity of budding yeast.
Acknowledgments
The authors would like to thank S. Pöggeler for sharing plasmids with Sordaria macrospora’s PRE2 and PPG1 genes and T. Fowler for sharing plasmids with BBR1, BBR2, BBP2(4) and BBP1(1) genes from Schizophyllum commune. We also wish to thank S. Pöggeler, A. Johnson, R. Bennett, J. Heitman, C. Hull and members of the Murray Lab for suggestions and critical reading of the manuscript. This work was supported by grants from NIH (GM 68763 and GM 43987 to A. M), as well a fellowship to J.G-S. from the Fundação para Ciência e Tecnologia (SFRH/BD/15220/2004).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heitman J. Sex in fungi: molecular determination and evolutionary implications. Washington, D.C: ASM Press; 2007. [Google Scholar]
- 3.Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 4.Bender A, Sprague GF., Jr Yeast peptide pheromones, a-factor and alpha-factor, activate a common response mechanism in their target cells. Cell. 1986;47:929–937. doi: 10.1016/0092-8674(86)90808-1. [DOI] [PubMed] [Google Scholar]
- 5.Bender A, Sprague GF., Jr Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics. 1989;121:463–476. doi: 10.1093/genetics/121.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P, Sapperstein SK, Choi JD, Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuchler K, Sterne RE, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus S, Caldwell GA, Miller D, Xue CB, Naider F, Becker JM. Significance of C-terminal cysteine modifications to the biological activity of the Saccharomyces cerevisiae a-factor mating pheromone. Mol Cell Biol. 1991;11:3603–3612. doi: 10.1128/mcb.11.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spellig T, Bolker M, Lottspeich F, Frank RW, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama N, Miyajima A, Arai K. Common signal transduction system shared by STE2 and STE3 in haploid cells of Saccharomyces cerevisiae: autocrine cell-cycle arrest results from forced expression of STE2. EMBO J. 1987;6:249–254. doi: 10.1002/j.1460-2075.1987.tb04746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- 13.Stanton BC, Giles SS, Staudt MW, Kruzel EK, Hull CM. Allelic exchange of pheromones and their receptors reprograms sexual identity in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000860. doi: 10.1371/journal.pgen.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayrhofer S, Poggeler S. Functional characterization of an alpha-factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with its cognate receptor in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:661–672. doi: 10.1128/EC.4.4.661-672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fowler TJ, DeSimone SM, Mitton MF, Kurjan J, Raper CA. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol Biol Cell. 1999;10:2559–2572. doi: 10.1091/mbc.10.8.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raudaskoski M, Kothe E. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell. 2010;9:847–859. doi: 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Poggeler S. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell. 2010;9:894–905. doi: 10.1128/EC.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth AF, Nelson B, Boone C, Davis NG. Asg7p-Ste3p inhibition of pheromone signaling: regulation of the zygotic transition to vegetative growth. Mol Cell Biol. 2000;20:8815–8825. doi: 10.1128/mcb.20.23.8815-8825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Bortz E, Zhong H, Leeuw T, Leberer E, Vershon AK, Hirsch JP. Localization and signaling of G(beta) subunit Ste4p are controlled by a-factor receptor and the a-specific protein Asg7p. Mol Cell Biol. 2000;20:8826–8835. doi: 10.1128/mcb.20.23.8826-8835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKay VL, Welch SK, Insley MY, Manney TR, Holly J, Saari GC, Parker ML. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci U S A. 1988;85:55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289:1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 26.Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci U S A. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivers DM, Sprague GF., Jr Autocrine activation of the pheromone response pathway in matalpha2- cells is attenuated by SST2- and ASG7-dependent mechanisms. Mol Genet Genomics. 2003;270:225–233. doi: 10.1007/s00438-003-0914-3. [DOI] [PubMed] [Google Scholar]
- 28.Ricardo S, Lehmann R. An ABC transporter controls export of a Drosophila germ cell attractant. Science. 2009;323:943–946. doi: 10.1126/science.1166239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson CL, Hartwell LH. Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 1990;63:1039–1051. doi: 10.1016/0092-8674(90)90507-b. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer D, Cote P, Whiteway M, Bennett RJ. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryot Cell. 2007;6:907–918. doi: 10.1128/EC.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladds G, Rasmussen EM, Young T, Nielsen O, Davey J. The sxa2-dependent inactivation of the P-factor mating pheromone in the fission yeast Schizosaccharomyces pombe. Mol Microbiol. 1996;20:35–42. doi: 10.1111/j.1365-2958.1996.tb02486.x. [DOI] [PubMed] [Google Scholar]
- 32.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 33.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci U S A. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayrhofer S, Weber JM, Poggeler S. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics. 2006;172:1521–1533. doi: 10.1534/genetics.105.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 37.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 38.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 39.Kues U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsueh YP, Fraser JA, Heitman J. Transitions in sexuality: recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot Cell. 2008;7:1847–1855. doi: 10.1128/EC.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.