In a comprehensive analysis of phosphoregulatory evolution in yeast, the authors observe that phosphorylation sites tend to be lost after gene duplication and protein network turnover reshuffles kinase–substrate relationships over time.
Keywords: evolution, phosphorylation, PTMs, regulatory network
Abstract
Elucidating how complex regulatory networks have assembled during evolution requires a detailed understanding of the evolutionary dynamics that follow gene duplication events, including changes in post-translational modifications. We compared the phosphorylation profiles of paralogous proteins in the budding yeast Saccharomyces cerevisiae to that of a species that diverged from the budding yeast before the duplication of those genes. We found that 100 million years of post-duplication divergence are sufficient for the majority of phosphorylation sites to be lost or gained in one paralog or the other, with a strong bias toward losses. However, some losses may be partly compensated for by the evolution of other phosphosites, as paralogous proteins tend to preserve similar numbers of phosphosites over time. We also found that up to 50% of kinase–substrate relationships may have been rewired during this period. Our results suggest that after gene duplication, proteins tend to subfunctionalize at the level of post-translational regulation and that even when phosphosites are preserved, there is a turnover of the kinases that phosphorylate them.
Introduction
Genomes and organisms gain in complexity during evolution by gene duplication followed by the functional divergence of the duplicates (Hurles, 2004). Signaling and regulatory proteins are thought to have a particularly important role in the evolution of organismal complexity (Gough and Wong, 2010). We know very little about the early evolutionary steps that follow the duplication of regulatory proteins and of the substrates they regulate. Studies on short time scales and on well-characterized organisms are needed in order to estimate the contribution of the different evolutionary forces to the assembly of novel regulatory pathways and networks.
Here, we address the evolution of phosphoregulatory networks by directly studying phosphoproteins and their associated protein kinases. Protein phosphorylation regulates several if not most of protein functions by affecting their stability, localization, activity and ability to interact (Moses and Landry, 2010). When maintained, paralogous proteins may diverge in function following two evolutionary paths, which are not mutually exclusive. First, one paralog may evolve new functions (neofunctionalization) (Conant and Wolfe, 2008). Second, degenerative mutations may accumulate in one or both paralogs leading to the loss of redundant functions (subfunctionalization) (Force et al, 1999; Lynch and Force, 2000). If we assume a model under which each phosphosite in a protein has a function (Holmberg et al, 2002), neofunctionalization would correspond to sites acquired after the duplication event and subfunctionalization to sites lost in one of the two paralogs. In the first case, new connections are created in the kinase–substrate network; in the second case, no new function has evolved and regulatory links are lost rather than created. We (Landry et al, 2009) and others (Lienhard, 2008) have recently suggested that a fraction of phosphorylation sites may have no specific functions and represent the result of kinase–substrate interactions that evolved neutrally or nearly neutrally. Accordingly, a fraction of the links that are created or lost after gene duplication in these networks would represent gains and losses of phosphosites without subfunctionalization or neofunctionalization of the proteins.
In this study, we used the budding yeast Saccharomyces cerevisae phosphorylation network as a model. The lineage leading to the budding yeast underwent a whole-genome duplication (WGD) 100 million years (My) ago (Wolfe and Shields, 1997) that affected its signaling networks significantly: while only 10% of all genes (∼500 pairs) were maintained as duplicates, 30 and 33% of protein kinases and phosphatases have been retained as duplicates, respectively (Seoighe and Wolfe, 1999). Furthermore, phosphoproteins were significantly more likely to be retained as paralogs than nonphosphorylated proteins (Amoutzias et al, 2010). Finally, duplicated kinases and their regulatory proteins differ in sequence and functions (Musso et al, 2008) and many of them show accelerated amino acid changes after the WGD (Kellis et al, 2004). Using computational and experimental analyses, we examined the extent to which phosphosites diverged after gene duplication, we addressed whether there have been accelerated gains and losses of phosphosites among these phosphoproteins and whether kinase–substrate relationships have been modified since the WGD.
Results and discussion
Paralogous phosphoproteins substantially diverged after WGD
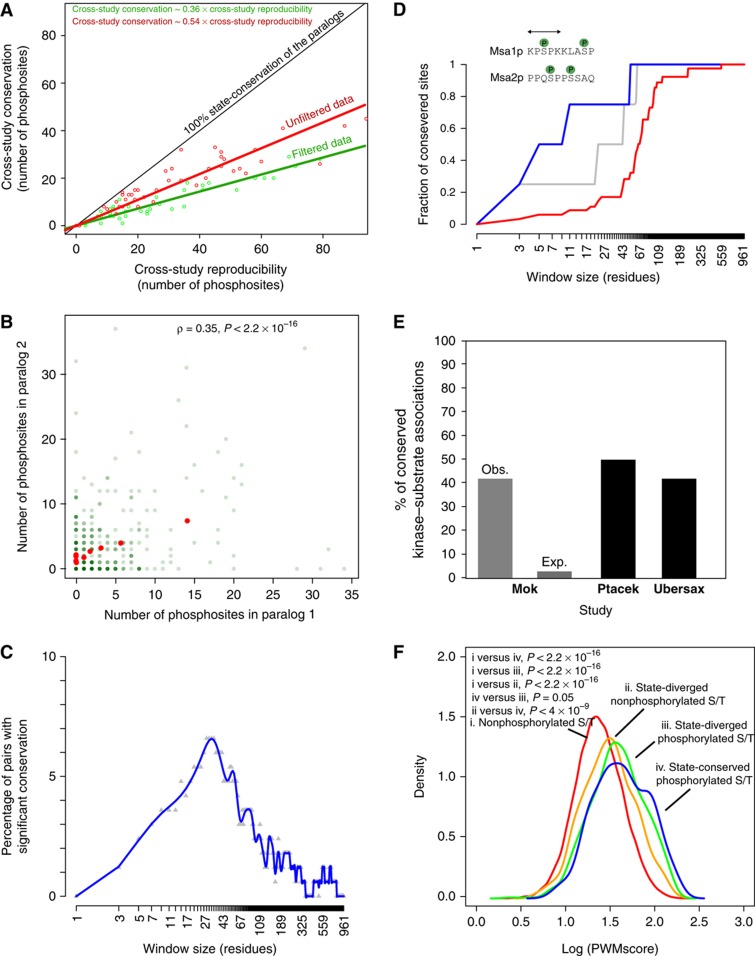
Our data set consists of 2726 phosphosites (serines (S), 82%; threonines (T), 16%; tyrosines (Y), 2%) that belong to one or the other member of the 352 pairs of yeast WGD paralogs for which at least one of the two proteins is a phosphoprotein. In this work, we focused on S/T phosphosites as they make up 98% of all phosphosites. Among these sites, 2445 are unique to one paralog and 118 (that correspond to 236 phosphosites) occur at homologous positions, a number 7.4 times higher than expected by chance (P≪0.001; Supplementary Figure S1). Phosphosites diverge in two ways. First are cases where a S/T residue is phosphorylated in a protein and a residue that cannot be phosphorylated occupies the homologous position in its paralog (site-divergence). Site-divergence accounts for 69% of the sites that are unique to one paralog. Second, a S/T is phosphorylated in one paralog and its homologous position is conserved (S/T) but not observed to be phosphorylated (state-divergence). Eighty-six percent of homologous sites that are phosphorylated are in fact state-diverged. This measure of state-divergence is strongly upwardly biased by false-negative (FN) and false-positive identifications and also by the fact that phosphopeptides that match more than one protein are not included in this data set. We considered these issues by comparing the cross-study conservation with the cross-study reproducibility. We found that state-conservation between paralogs is around 36% for filtered peptides (considering only phosphopeptides that match a single position in the proteome) and 54% for unfiltered peptides (considering all phosphopeptides) (Figure 1A). Protein sequence, function, localization and/or recognition by protein kinases have diverged to such extent in 100 My that only 36–54% of their post-translational regulation by phosphorylation appears to be conserved despite a conservation of the actual residues.
Figure 1.
Conservation and divergence of phosphoregulation among WGD paralogs. (A) The state-conservation of paralogous proteins was estimated as a regression of the cross-study conservation on the cross-study reproducibility. A 1:1 relationship is expected if all phosphosites were state-conserved. Deviation from this 1:1 relationship provides an estimate of state-divergence. Filtered data: phosphopeptides that match a single protein; unfiltered data: all phosphopeptides. (B) Positive correlation in the number of phosphosites of WGD paralogous proteins. Red dots indicate average numbers in binned data and green dots the actual data. Green intensities indicate the number of points at these positions. (C) Proportion of paralogous pairs with significant conservation as a function of the window size considered. A site is considered as being conserved if there is a phosphorylated site in the other paralog within the window (excluding the exact position). (D) Case of putative local compensation. The fraction of conserved sites as a function of window size is shown. Blue: observed value; Grey: 95th quantile (100 permutations); Red: average of the expected distribution. (E) Fraction of paralogous phosphosites or phosphoproteins assigned to the same protein kinase. Assignments are based on PWMs from Mok et al (2010). The observed fraction is calculated using these assignments while the expected fraction is estimated after shuffling the assigned kinases among the pairs of paralogous sites. Ptacek: large-scale in vitro kinase–substrate interactions on microarrays (Ptacek et al, 2005). Ubersax: in vitro Cdc28–substrate interactions (Ubersax et al, 2003). (F) Distributions of the PWM scores for different classes of sites.
Conservation and compensation of phosphosite loss by site-position turnover
Surprisingly, despite the low level of site-conservation between paralogous proteins, there is a highly significant correlation in the number of phosphorylation sites between paralogs (ρ=0.35, P-value<2.2 × 10−16; Figure 1B). This correlation remains significant when the number of phosphosites is normalized by protein length (ρ=0.32 P-value <6.9 × 10−14) or the length of disordered regions (ρ=0.27 P-value<3.8 × 10−10), which both tend to be preserved between paralogs. The correlation is also significant when only site-diverged phosphosites are considered (ρ=0.28, P-value=2.0 × 10−11). This correlation suggests that stabilizing selection is acting to maintain the overall number of phosphosites. This result is in agreement with a recent study (Beltrao et al, 2009) reporting that the phosphorylation levels of orthologous protein complexes or pathways between Candida albicans and Saccharomyces cerevisiae tend to be conserved. The turnover of phosphosite position over time could be made possible by the fact that sites that appear at a position nearby a site that is lost can compensate for the loss (Serber and Ferrell, 2007), particularly when the charge of a region rather than that of a specific residue is important. The redundancy in the position of phosphosites has been previously proposed to explain the weak site-conservation among species (Landry et al, 2009), but so far there has been limited evidence for this (Ba and Moses, 2010; Moses and Landry, 2010).
If this local turnover model is responsible for the overall conservation of the number of phosphosites, the proportion of conservation between paralogs should increase significantly if we consider regions of proteins rather than actual positions. We found that to be the case for a significant but limited number of paralogous pairs. We reconsidered the proportion of state-conserved sites as the proportion of sites in a protein that have a phosphosite in the homologous region of a given window size in its paralog. We first found that the window size that maximizes the signal is about 33 amino acids in length (Figure 1C). Then, we found that among the 167 pairs of paralogous proteins where both paralogs have at least one phosphosite, 11 of them (6.6%) showed a significant level of conservation at that window length (an example is shown in Figure 1D). This result may suggest either that compensation by nearby sites is relatively uncommon and is specific to some types of proteins, or that the relatively limited coverage of the yeast phosphoproteome leaves us with limited power to detect significant compensation. Another possibility is that such compensation takes place only in highly phosphorylated proteins. Indeed, we found that paralogous pairs for which there is significant functional compensation have significantly more phosphosites (mean: 9.28 versus 3.87; Wilcoxon test: P-value<9.5 × 10−11) and also tend to contain a larger proportion of disordered residues (mean: 53 versus 42%, P=0.01) when compared with all pairs.
Life after WGD: rewiring the cellular regulatory networks
Phosphosites are phosphorylated by a variety of kinases that recognize specific motifs surrounding the phosphorylatable residue. As for many eukaryotes, around 2% (120 total) of yeast protein-coding genes code for protein kinases (Zhu et al, 2000). We examined the conservation of the relationships between our set of phosphosites and yeast kinases by assigning each phosphosite to a kinase using empirically derived position weight matrices (PWM) for 61 yeast kinases (Supplementary Data set S1 from Mok et al, 2010). We first found that WGD paralogs are generally not biased in terms of the protein kinases that regulate them (ρ=0.99, P-value<2 × 10−16; Supplementary Figure S2). Second, we found that state-conserved sites are assigned to the same kinase 44% of the time, a 20-fold increase over what is expected if phosphosites were randomly matched between paralogs (P-value <0.0001; Figure 1E). This number drops to 23% for state-diverged sites, again supporting the fact that state-divergence does not entirely result from FN identifications. These sites are either not being phosphorylated or being phosphorylated by a different kinase in a different condition not addressed so far in phosphoproteomics studies. This first hypothesis is supported by the fact that, for state-diverged sites, the assigned scores are significantly higher for the phosphorylated sites than the nonphosphorylated ones (Figure 1F). We estimated that the state-diverged nonphosphorylated S/T sites in reality comprise 50% of nonphosphorylated sites (Supplementary Figure S3).
The low percentage of assignment (44%) of the same kinase to state-conserved sites suggests that the kinases that phosphorylate paralogous sites have changed since the WGD (Moses and Landry, 2010). We found independent support for this from large-scale and small-scale kinase–substrate interaction experiments (Ubersax et al, 2003; Ptacek et al, 2005) in which kinase–substrate relationships are also conserved in similar proportions (Figure 1E). Overall, these analyses suggest that while a significant fraction of sites is conserved and phosphorylated in both paralogs, the flanking sequences and/or protein structure and/or localization have diverged enough for the substrate to be regulated by a different protein kinase, a regulatory network turnover that is similar to what is observed for transcriptional networks (Gasch et al, 2004; Moses and Landry, 2010). After 100 My of evolution, up to 50% of kinase–substrate relationships may have been rewired, while preserving the phosphorylation status of the substrates.
Phosphosite loss dominates site-divergence
A recent study on the budding yeast reported putative cases of neofunctionalization and subfunctionalization of phosphosites (Amoutzias et al, 2010), but did not compare the extent of those changes to a null model. We therefore sought to quantify whether site-divergence resulted from losses or gains of phosphosites by reconstructing the ancestral sequences of the paralogous proteins and comparing the observed proportions to the neutral expectations (Figure 2A–C). We found that 25% of sites correspond to gains and 31% of sites correspond to losses. These proportions are, respectively, significantly less and more than expected by chance alone, based on the resampling of phosphorylatable sites in the same set of phosphoproteins (Figure 2C). This remains true for ordered and disordered regions of proteins, which have been shown to evolve at different rates. We consider that these losses represent several subfunctionalization events as nonfunctional phosphosites (Landry et al, 2009) are expected to evolve as randomly selected S/T. These results are also unlikely to result from false positives, as we performed the same analyses on a smaller number of manually curated phosphosites (Ba and Moses, 2010; Supplementary Figure S4) and we observed similar results. Our results are also robust to data filtering (Supplementary Figure S5) and variation in ancestral sequence reconstruction (Supplementary Figure S6).
Figure 2.
Gains and losses of phosphosites after gene duplication. (A) Inference of gains and losses of phosphosites. Serines (S) and threonines (T) are considered equivalent with respect to phosphorylation. !S/!T indicates residues that are not a S nor a T, and pS/pT indicates phosphorylated S/T. (B) Examples of lost (S72), gained (T121) and conserved (S103) sites from the curated data set (Dataset 2). (C) The number of observed losses is greater than expected by chance alone and the number of gains shows the opposite result. Results in ordered and disordered regions agree with each other.
A limitation of this analysis is that we have to assume that the phosphorylatable sites (S/T) of the ancestral sequence that correspond to phosphorylation sites in S. cerevisiae were phosphorylated in the ancestor. Only a direct observation of the phosphorylation state of the ancestral proteins would alleviate this problem. We therefore performed a phosphoproteomics experiment on Lachancea kluyveri (Souciet et al, 2009), a species that diverged from S. cerevisiae before the WGD event and that can be used as a proxy for ancestral functions (van Hoof, 2005). We identified 855 phosphosites on 429 proteins (Supplementary information S1) that we mapped on our alignments. We found that a smaller proportion of phosphosites identified in L. kluyveri are also phosphorylated in the S. cerevisiae WGD paralogs (1:2) compared with the 1:1 S. cerevisiae orthologs (Figure 3A). Assuming that the rate of phosphosite gain in the L. kluyveri lineage was similar in these two categories of genes (1:1 and 1:2 L. kluyveri–S. cerevisiae orthologs), this result confirms that phosphosites were more likely to be lost in the S. cerevisiae WGD paralogs and thus that gene duplication has significantly accelerated the rate of phosphosite divergence. We also found that the proportion of sites that are uniquely phosphorylated in S. cerevisiae (not found to be phosphorylated in L. kluyveri) in the WGD paralogs is actually comparable to the one for the 1:1 orthologs (Figure 3B). Under a scenario where phosphosite gains accelerated the divergence of the WGD paralogs, we would have expected to see a significantly higher fraction of gains for the 2:1 orthologs compared with the 1:1 ones. Our phosphoproteomics results therefore support our bioinformatics analyses based solely on ancestral sequence reconstruction and confirm the prevalence of phosphosite losses in the divergence of paralogous phosphoproteins.
Figure 3.
L. kluyveri phosphoproteomics confirms that phosphosites are preferentially lost in paralogous phosphoproteins. (A) L. kluyveri phosphosites are more likely to be phosphorylated in S. cerevisiae if they are in 1:1 orthologs (142/469 sites in 108 proteins) than in 1:2 orthologs (31/181 sites in 45 proteins). (B) Ratios of the number of sites unique to S. cerevisiae to the number of shared ones with L. kluyveri for 1:1 orthologs (142/6644 sites in 108 proteins) and 2:1 orthologs (62/2681 sites in 45 proteins).
Concluding remarks
A previous study considering the ancestral function of duplicated WGD proteins has shown the importance of subfunctionalization in shaping the function of WGD paralogs acting at the level of protein functions (van Hoof, 2005), whereas investigations of transcriptional regulation have also found a significant contribution of neofunctionalization in the divergence of paralogs (Papp et al, 2003; Tirosh and Barkai, 2007). Our results suggest that at the level of post-translational regulation, subfunctionalization may have been the most important driving force in shaping the yeast regulatory network. One limitation of our analysis is that we consider that, when functional, each phosphosite has an independent function, which may not be necessarily the case, as several cooperative effects among phosphosites have been reported (Kapoor et al, 2000). The combined and individual effects of the subfunctionalized and neofunctionalized sites will need to be addressed experimentally to estimate the functional effects of these divergences. Further integrative analyses will also be required to elucidate the importance of neofunctionalization and subfunctionalization that take place at multiple levels (transcription, protein function, post-translational modifications), as these may be largely dependent on each other (Jensen et al, 2006). Another key finding of our study is that 100 My may be sufficient to rewire half of the kinase–substrate relationships in a cell. This result is in agreement with previous studies showing that protein–protein interaction networks evolve rapidly (Wagner, 2001).
Materials and methods
We compiled a set of 20 342 phosphorylation sites on 2688 proteins from eight large-scale studies. We used 21 068 phosphopeptides from six studies (Gruhler et al, 2005; Bodenmiller et al, 2007; Chi et al, 2007; Li et al, 2007; Reinders et al, 2007; Albuquerque et al, 2008), as compiled by Amoutzias et al (2010) to which we added 3616 phosphopeptides from Beltrao et al (2009) and 3620 phosphopeptides from Gnad et al (2009). Raw phosphopeptides from these studies were filtered according to the following criteria: for the Gnad data set, we considered peptides with a probability score above 0.95; for the Beltrao data set we selected the peptides with score >0.02 and not being acetylated at the amino or carboxy terminus; then for all data sets we selected all the peptides that matched one exact hit on S. cerevisiae proteins using Blat searches (Kent, 2002). Peptides that matched more than one protein were eliminated because they could not be assigned unambiguously to a single protein. We used this data to assemble a first data set (Dataset 1). Thus, we compiled another data set using the same data about the phosphosites, but this time we did not apply the filtering step with Blat (Dataset 2). Finally, we compiled a third data set of manually curated phosphosites that have been shown to be phosphorylated in small-scale experiments and whose function has been determined (Ba and Moses, 2010) (Dataset 3). The compiled data and all the other data described below are available at: http://www.bio.ulaval.ca/landrylab/download/.
We estimated the state-divergence of phosphosites between paralogous proteins by comparing cross-study conservation and reproducibility. Our data set comes from eight distinct studies, so there are 28 possible pairwise comparisons. We only considered sites that were S/T in both paralogs. For each pair of studies we considered two sets of concatenated paralogous proteins, para.1 and para.2. We counted the number of sites found in para.1 in study 1 and examined how many were also found in para.1 in study 2 (cross-study reproducibility) and para.2 in study 2 (cross-study conservation) (Supplementary Figure S7). We did the same comparison for these two studies between sites identified in para.2 of study 1 and also in para.2 of study 2 (cross-study reproducibility) and of para.1 of study 2 (cross-study conservation). Each pair of studies therefore yields two ratios of cross-study conservation/cross-study reproducibility and this ratio gives a measure of the extent of conservation between paralogs while taking into account the reproducibility of the two studies.
 |
A regression of the cross-study conservation on the cross-study reproducibility provides a rough estimate of the state-conservation between paralogs while taking reproducibility into account (Figure 1A).
Local phosphosite turnover was tested as follows. We took all the pairs of WGD phosphoproteins where both paralogs had one or more phosphosites. For each phosphosite present in the first paralog, we examined a window of length l centered on the site, thus defining a range of positions along the sequence. Excluding all state-conserved sites (at the exact same position), we counted all the phosphosites present in the aligned second paralog inside the corresponding range of positions within the window. A site was conserved if for a given phosphosite in the first paralog there was at least one phosphosite in the second paralog inside the range of positions. We then determined the ratio of conserved sites over all sites for each window size. The random expectation was estimated using 100 randomizations of phosphosites as described below.
The PWM used for the prediction of the protein kinases associated with each of the phosphosites were derived empirically by Mok et al (2010) through in vitro peptide screening using 61 of the 122 kinases from S. cerevisiae. While these data are incomplete, it is the best currently available as it relies on empirically derived consensus motifs rather than completely in silico predictions. In order to assign all of the phosphosites to their most likely corresponding kinases, we extracted all of the 15-mers of the yeast proteome that correspond to the phosphosite and their 14 flanking (±7) residues. All phosphosites were then scored by summing the logarithm of the values present in each kinase PWM matrix corresponding to each of the amino acids of the 15-mer. We then assigned a protein kinase to a particular site based on the highest score for that site (Supplementary Figure S8). Data on kinase–substrate interactions were obtained from Ptacek et al (2005) and Ubersax et al (2003). In the first case, the data represent microarray interactions between 87 different kinases and >4000 potential substrates. We estimated the fraction of paralogs that were phosphorylated by the same kinase, considering only paralogs that were both phosphorylated by at least one kinase. The second data comes from an in vitro experiment testing for interactions between Cdc28 and the yeast proteome. We calculated the number of times both paralogs were phosphorylated by the kinase among all cases where at least one of the two was phosphorylated.
Gains and losses of phosphosites were inferred as described in Figure 2A. We estimated the expected numbers of gains and losses by randomly sampling S/T sites. We divided the phosphosites in the four classes according to the type of the residue (S or T) and the type of region where the residue was located (ordered or disordered), and the representation of each class was respected in the resampling. Disordered regions of proteins were predicted using DISOPRED (Ward et al, 2004), using all the fungal protein sequences as a reference database. We performed a random sampling of S/T positions 1000 times, calculating the number of gains and losses after each resampling. The ancestral residues occupying the phosphosite position were determined as follows. We aligned all of S. cerevisiae proteins to the L. kluyveri and Zygosaccharomyces rouxii orthologs, these two species having diverged from the S. cerevisiae lineage before the WGD (see Supplementary information S2). All the sequences and the orthology relationships were obtained from YGOB (Gordon et al, 2009) and alignments were performed with MUSCLE (Edgar, 2004) using default parameters. Orthology relationships were found for 4401 genes (among which 516 out of 553 of S. cerevisiae paralogous genes). For each quartet of sequences, we inferred the ancestral sequence at the first node joining the two paralogs (Figure 2A). The ancestral protein sequences were inferred using the Codeml method implemented in PAML (Yang, 2007) using the following parameters: fix_alpha=0, α=0.04, fix_blength=2. We reconstructed ancestral sequences using two different substitution matrices (wag and dayhoff) and both gave similar results so we are presenting only results derived from the wag matrix. We examined the robustness of the reconstruction by performing the same analyses including an additional pre-WGD species (Kluyveromyces thermotolerans) to our set. In this case, we were able to reconstruct the orthology relationships and the ancestral sequence for 4388 genes (among which 516 out of 551 of S. cerevisiae paralogous genes) (Dataset 4). All analyses were performed using Perl (http://www.perl.org) and R (http://www.r-project.org/) scripts.
The L. kluyveri phosphosites were identified as follows. L. kluyveri (formerly known as Saccharomyces kluyveri) strain FM628 (MATa ura3) was obtained from Marc Johnston (Washington University). Precultures of 75 ml were grown to OD600∼3 overnight in standard yeast YPD medium at 30°C, agitated at 600 r.p.m. and diluted to OD600=0.1 in the morning in 1 L of YPD. Cells were harvested at OD600∼0.6–0.8 by centrifugation at 4000 r.p.m. for 20 min. The pellets (about 2–3 g) were suspended in 20 ml of lysis buffer following (Albuquerque et al, 2008) with slight modifications: 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.2% NonidetP-50, 1.5 mM MgCl2, 0.2 mM EDTA. The lysis buffer also contained phosphatase inhibitors phosSTOP (Roche), protease inhibitors, complete protease cocktail (Roche) and 1 mM PMSF. Samples were quickly frozen directly in liquid nitrogen drop-by-drop to make 1 cm3 frozen pellets and conserved at −80°C. Yeast powder extracts were then produced using a Freezer-Mill (Spex SamplePrep), which pulverizes cryogenically small pellets with a magnetically driven impactor submerged in liquid nitrogen. The fine powder was then centrifuged at 14 500 r.p.m. (rotor SA600) for 30 min at 4°C. The clear supernatant was treated with Benzonase (Novagen) to eliminate nucleic acids overnight at 4°C and then cold acetone precipitated.
Protein pellets were resuspended in 1% SDS/50 mM ammonium bicarbonate (AB) and microBCA (Pierce) was used to determine protein concentration. Proteins extracts (1 mg) were reduced for 20 min at 37°C with 0.5 mM Tris (2-carboxyethyl)phosphine, TCEP (Pierce), alkylated with 50 mM iodoacetamide for 20 min at 37°C and quenched by adding 50 mM DTT. Samples were diluted 10 × with 50 mM AB, digested overnight at 37°C with sequencing grade trypsin (enzyme:substrate, 1:100) (Promega). The digestion was stopped by adding trifluoro acetic acid (TFA) and was followed by evaporation on a SpeedVac (Thermo Fisher Scientific, San Jose, CA). Phosphopeptides were enriched on home-made TiO2-affinity columns (1.25 mg Titansphere, 5 μm, GL Sciences), using 250 mM lactic acid (Fluka) and eluted with 30 μl of 1% ammonium hydroxide, as described previously (Thingholm et al, 2006). Samples were acidified with 1 μl of TFA, desalted using 30 mg HLB cartridge (Waters Corporation, Milford, MA), dried and resuspended in 2% acetonitrile, ACN (Thermo Fisher Scientific), 0.2% formic acid, FA (EMD Chemicals Inc., Gibbstown) before analysis.
Triplicate 2D-nanoLC–MS/MS analysis of phosphopeptides was performed on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to an Eksigent LC system. Online SCX separation (Opti-Guard 1 mm cation column, Optimize Technologies) was performed using five different ammonium acetate salt fractions, pH 3.0 (0, 250, 500, 1000 and 2000 mM) in 2% ACN (0.2% FA). Peptides eluted from each salt fraction were transferred to a precolumn reverse phase trap (4 mm length, 360 μm i.d.) and injected on a reverse phase analytical column (10 cm length, 150 μm i.d.) (Jupiter C18, 3 μm, 300 Å, Phenomenex). A linear gradient (2–25% ACN over 63 min followed by 25–40% ACN over the next 15 min) was applied to separate phosphopeptides, which were directly injected into the mass spectrometer at a flow rate of 600 nl/min. Detailed MS operation procedure is described in Marcantonio et al (2008). Mascot Distiller v2.1.1 (Matrix Science, London, UK) was used to extract and preprocess MS/MS spectra from raw data files. Peptide identification was done with Mascot v2.2 using L. (Saccharomyces) kluyveri protein sequence database (http://www.ebi.ac.uk/embl/). The following parameters were used: parent and fragment tolerance of 0.02 and 0.5 Da, respectively, trypsin with two missed cleavages and the following modifications: carbamidomethyl (C), deamidation (NQ), oxidation (M), phosphorylation (STY). ProteoConnections (Courcelles et al, 2011) was used to limit peptide false discovery rate to 1% and evaluate the confidence of phosphorylation site localization. MS/MS of all peptide identifications are available at http://www.thibault.iric.ca/proteoconnections (and were submitted to the PRIDE database (accession numbers: 17339)). Phosphosites with a confidence score above 60% were considered for the evolutionary analyses (711 sites in 396 proteins).
Supplementary Material
Acknowledgments
We thank H Wurtele and A Verreault for the use of their facilities and N Lartillot and A Moses for comments on the manuscript. This work was supported by a Canadian Institute of Health Research (CIHR) Grant GMX-191597 and FRSQ to CRL, and CIHR Grant GMX-152556 to SWM. CRL is a CIHR New Investigator. LF was supported by a Quebec Research Network on Protein Function, Structure and Engineering (PROTEO) fellowship.
Author contributions: CRL and LF planned the analysis, analyzed the data and wrote the paper. CRL, MC and PT performed the phosphoproteomics experiments. SWM contributed reagents, tools and guidance on the phosphoproteomics experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, He Y, Gordon J, Mossialos D, Oliver SG, Van de Peer Y (2010) Posttranslational regulation impacts the fate of duplicated genes. Proc Natl Acad Sci USA 107: 2967–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba ANN, Moses AM (2010) Evolution of characterized phosphorylation sites in budding yeast. Mol Biol Evol 27: 2027–2037 [DOI] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ (2009) Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol 7: e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods 4: 231–237 [DOI] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA 104: 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Courcelles M, Lemieux S, Voisin L, Meloche S, Thibault P (2011) ProteoConnections: a bioinformatics platform to facilitate proteome and phosphoproteome analyses. Proteomics 11: 2654–2671 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y-L, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB (2004) Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol 2: e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, de Godoy LMF, Cox J, Neuhauser N, Ren S, Olsen JV, Mann M (2009) High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 9: 4642–4652 [DOI] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH (2009) Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5: e1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough NR, Wong W (2010) Focus issue: the evolution of complexity. Sci Signal 3: eg5. [DOI] [PubMed] [Google Scholar]
- Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics 4: 310–327 [DOI] [PubMed] [Google Scholar]
- Holmberg CI, Tran SEF, Eriksson JE, Sistonen L (2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci 27: 619–627 [DOI] [PubMed] [Google Scholar]
- Hurles M (2004) Gene duplication: the genomic trade in spare parts. PLoS Biol 2: 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P (2006) Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature 443: 594–597 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Hamm R, Yan W, Taya Y, Lozano G (2000) Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene 19: 358–364 [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Levy ED, Michnick SW (2009) Weak functional constraints on phosphoproteomes. Trends Genet 25: 193–197 [DOI] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villén J, Elias JE, Gygi SP (2007) Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res 6: 1190–1197 [DOI] [PubMed] [Google Scholar]
- Lienhard GE (2008) Non-functional phosphorylations? Trends Biochem Sci 33: 351–352 [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcantonio M, Trost M, Courcelles M, Desjardins M, Thibault P (2008) Combined enzymatic and data mining approaches for comprehensive phosphoproteome analyses: application to cell signaling events of interferon-gamma-stimulated macrophages. Mol Cell Proteomics 7: 645–660 [DOI] [PubMed] [Google Scholar]
- Mok J, Kim PM, Lam HYK, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, Cameroni E, Niu H, Good M, Remenyi A, Ma J-LN, Sheu Y-J, Sassi HE, Sopko R, Chan CSM, De Virgilio C et al. (2010) Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal 3: ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Landry CR (2010) Moving from transcriptional to phospho-evolution: generalizing regulatory evolution? Trends Genet 26: 462–467 [DOI] [PubMed] [Google Scholar]
- Musso G, Costanzo M, Huangfu M, Smith AM, Paw J, San Luis B-J, Boone C, Giaever G, Nislow C, Emili A, Zhang Z (2008) The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res 18: 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD (2003) Evolution of cis-regulatory elements in duplicated genes of yeast. Trends Genet 19: 417–422 [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee S-J, Mah AS, Meng L, Stark MJR, Stern DF, De Virgilio C, Tyers M et al. (2005) Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Reinders Jr, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der Laan M, Rehling P, Sickmann A, Pfanner N, Meisinger C (2007) Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics 6: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH (1999) Yeast genome evolution in the post-genome era. Curr Opin Microbiol 2: 548–554 [DOI] [PubMed] [Google Scholar]
- Serber Z, Ferrell JE Jr (2007) Tuning bulk electrostatics to regulate protein function. Cell 128: 441–444 [DOI] [PubMed] [Google Scholar]
- Souciet J-L, Dujon B, Gaillardin C, Johnston M, Baret PV, Cliften P, Sherman DJ, Weissenbach J, Westhof E, Wincker P, Jubin C, Poulain J, Barbe Vr, Ségurens Ba, Artiguenave Fo, Anthouard Vr, Vacherie B, Val M-E, Fulton RS, Minx P et al. (2009) Comparative genomics of protoploid Saccharomycetaceae. Genome Res 19: 1696–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingholm TE, Jørgensen TJD, Jensen ON, Larsen MR (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc 1: 1929–1935 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2007) Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol 8: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- van Hoof A (2005) Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171: 1455–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A (2001) The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol Biol Evol 18: 1283–1292 [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M (2000) Analysis of yeast protein kinases using protein chips. Nat Genet 26: 283–289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.