The developed, directed mass spectrometry workflow allows to generate consistent and system-wide quantitative maps of microbial proteomes in a single analysis. Application to the human pathogen L. interrogans revealed mechanistic proteome changes over time involved in pathogenic progression and antibiotic defense, and new insights about the regulation of absolute protein abundances within operons.
Keywords: absolute quantification, directed mass spectrometry, Leptospira interrogans , microbiology, proteomics
Abstract
Over the past decade, liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS) has evolved into the main proteome discovery technology. Up to several thousand proteins can now be reliably identified from a sample and the relative abundance of the identified proteins can be determined across samples. However, the remeasurement of substantially similar proteomes, for example those generated by perturbation experiments in systems biology, at high reproducibility and throughput remains challenging. Here, we apply a directed MS strategy to detect and quantify sets of pre-determined peptides in tryptic digests of cells of the human pathogen Leptospira interrogans at 25 different states. We show that in a single LC–MS/MS experiment around 5000 peptides, covering 1680 L. interrogans proteins, can be consistently detected and their absolute expression levels estimated, revealing new insights about the proteome changes involved in pathogenic progression and antibiotic defense of L. interrogans. This is the first study that describes the absolute quantitative behavior of any proteome over multiple states, and represents the most comprehensive proteome abundance pattern comparison for any organism to date.
Introduction
System-wide investigation of gene expression at the mRNA transcript level has become routine and is widely used in systems biology and clinical studies to identify sets of genes that show distinct transcript profiles for a specific cellular state and to classify samples according to their respective molecular patterns (van ’t Veer et al, 2002; Gilchrist et al, 2006; Ishii et al, 2007). It has also been shown that neither the concentration of transcripts (Gygi et al, 1999; Griffin et al, 2002) nor their quantitative change in response to perturbations (MacKay et al, 2004; Kislinger et al, 2006; de Godoy et al, 2008) strongly correlate with the quantitative change of their corresponding proteins, the main functional products of gene expression. Therefore, quantitative proteomics holds great promise to enhance or complement the picture of gene expression in cells, and thus to contribute to the understanding of most molecular mechanisms in a cell. However, owing to the large heterogeneity in the amount and the physico-chemical properties of proteins, along with the lack of protein amplification methods, system-wide quantitative proteome analysis has been more technically challenging than transcriptome analysis.
Recent advances in liquid chromatography–tandem mass spectrometry (LC–MS/MS), currently the method of choice for large-scale protein studies, have made the reliable identification and quantification of thousands of proteins in a single study a reality (Brunner et al, 2007; de Godoy et al, 2008; Ahrens et al, 2010). However, particularly due to the selection of precursor ions using a simple intensity driven heuristics (data-dependent analysis, DDA), results from such studies still show a bias against the detection of low abundant protein species and a decreasing level of reproducibility of identified peptides with decreasing abundance. Comprehensive and more highly reproducible proteome coverage can be achieved by extensive sample pre-fractionation and the mass spectrometric analysis of each fraction, albeit at a cost that multiplies analysis time and limits throughput. Additionally, the detection of different proteome subsets in repetitive LC–MS analyses of similar samples impairs the generation of consistent, reproducible quantitative data sets across multiple samples, a crucial prerequisite in systems biology studies (Ideker et al, 2001; Rifai et al, 2006; Schiess et al, 2009).
Therefore, several alternative or complementary MS strategies have been developed to overcome some of the limitations of current LC–MS/MS workflows (Schmidt et al, 2008; Picotti et al, 2009; Domon and Aebersold, 2010). They make use of a priori information gathered from previous MS studies to increase the reliability, reproducibility and/or throughput of subsequent measurements. Specifically, in each of these strategies, MS analysis is focused on a few proteotypic peptides (PTPs) per protein, thereby minimizing instrument time without compromising analytical sensitivity. Two specific implementations of such strategies have been proposed (Pan et al, 2009; Schmidt et al, 2009; Domon and Aebersold, 2010), which we have termed targeted and directed MS, respectively. Targeted MS is based on selected reaction monitoring (SRM also known as multiple reaction monitoring) and is typically carried out on triple quadrupole mass spectrometers. Because of very high selectivity and sensitivity, it is capable of covering the full dynamic range of proteomes in moderately complex organisms such as yeast (Picotti et al, 2009). However, since each LC–MS/MS run is limited to a few hundred targeted peptides (Stahl-Zeng et al, 2007), the throughput required for proteome-wide measurements is currently difficult to achieve. Directed MS makes use of inclusion mass lists in order to guide the MS sequencing to a desired, pre-determined subset of peptides (Jaffe et al, 2008; Schmidt et al, 2008, 2009). Directed sequencing is carried out on the same types of instruments as discovery measurements by DDA. In contrast to the SRM methodology, directed MS monitors far larger sets of peptides per analysis. However, because the precursor ion signal of the peptide of interest has to be explicitly detected to trigger its identification, the overall dynamic range and sensitivity of directed sequencing is lower than that of SRM and more dependent on the sample matrix (Domon and Aebersold, 2010).
Here, we have studied global and time-resolved changes in the proteome of cells of the human pathogen Leptospira interrogans that were perturbed by antibiotic stress and serum stimulation. Overall, in 31 samples, representing 25 cellular states, 1669 proteins, representing 75% of the Leptospira proteome discovered by saturation sequencing using DDA MS, were consistently detected and their cellular concentrations determined (Supplementary Table SV). This unique data set was generated via an integrated inclusion list driven MS strategy that maximizes protein coverage in individual samples by focusing precious MS-sequencing time on the best flying, PTPs of each protein (Mallick and Kuster, 2010). The cellular concentrations of the detected proteins were estimated in each sample by correlating the average of the signal intensities of the three most highly responding peptides per protein with a calibration curve generated with a set of isotopically labeled reference (Malmström et al, 2009). We show that the protein components of entire pathways can be quantified across several time points and, for the first time, large-scale, consistent proteome data sets can be subjected to cluster analysis, a tool that was previously limited to the transcript level due to incomplete sampling on protein level. We show that the proteomic changes measured differ from the available transcriptomics data. We demonstrate that Leptospira cells adjust the cellular abundance of a certain subset of proteins as a general response to stress while other parts of the proteome respond highly specific. They furthermore react to individual treatments by ‘fine tuning’ the abundance of certain proteins and pathways in order to cope with the specific cause of stress. Using serum treatment we simulated the host environment and elucidate which proteomic adjustments underlie virulence. The method can be implemented with standard high-resolution mass spectrometers and software tools that are readily available in the majority of proteomics laboratories. It is scalable to any proteome of low-to-medium complexity and can be extended to post-translational modifications or peptide-labeling strategies for quantification. We therefore expect the approach outlined here to become a cornerstone for microbial systems biology.
Results
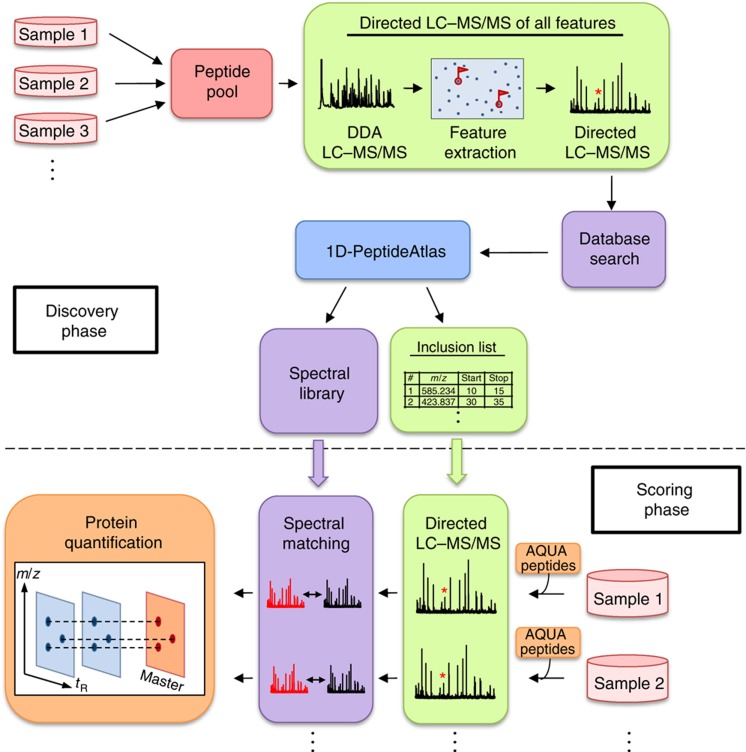
To consistently detect and absolutely quantify the same, extensive subset of the L. interrogans proteome in multiple samples, we developed and deployed the general workflow displayed in Figure 1. It consists of two main phases, proteome discovery and scoring. During the initial discovery phase, a comprehensive atlas of peptides and proteins identified by LC–MS/MS was generated by saturation sequencing of the L. interrogans proteome. To maximize proteome coverage, a pooled sample was generated and analyzed that consisted of aliquots from cells at different states. Subsequently, during the scoring phase, selected PTPs were detected in individual samples via inclusion list driven sequencing and quantified based on the ion current of the selected peptides, to generate quantitative proteome maps for each cellular state. Using this technique, comprehensive LC–MS/MS maps could be generated without the need for sample and time-consuming pre-fractionation steps, which significantly increases sample throughput.
Figure 1.
Global protein profiling workflow. In the first phase of the study (discovery phase), the peptide samples representing different cell states were mixed and analyzed by data-dependent acquisition (DDA) followed by directed 1D-LC–MS/MS. To achieve comprehensive proteome coverage, all detectable precursor ions, referred to as features, were extracted, sequenced in sequential directed LC–MS/MS analyses and identified by database searching. All identified peptide sequences were stored in a 1D-PeptideAtlas together with their precursor ion signal intensity, elution times and mass-to-charge ratio. For each protein, mass and time coordinates from the five most suitable peptides (PTPs) for quantification were extracted from the PeptideAtlas and stored in an inclusion list. Additionally, a spectral library was generated from the identified spectra to improve both, the sensitivity and speed of spectral matching in the quantification phase. In this phase (scoring phase), LC–MS/MS analysis was focused on the pre-selected PTPs as well as a set of heavy labeled reference peptides that were added to each sample. This determined the concentrations of the corresponding proteins in the sample, which could be used as anchor points to translate the MS response of each identified protein into its concentration (Malmström et al, 2009). After spectral matching, label-free quantification was employed to extract and align identified features and monitor their corresponding protein abundances redundantly over all samples.
Generation of a L. interrogans PeptideAtlas
To build a PeptideAtlas (Desiere et al, 2006; Deutsch et al, 2008) with maximal coverage of the L. interrogans proteome, we generated a pooled sample in which aliquots of extracts from different cell states were combined. Specifically, one aliquot of an untreated control sample and four aliquots of the individual perturbated cells (24 h treatments only, see Figure 3) were pooled. We used a single dimension high-performance LC–MS/MS platform in combination with the recently introduced directed MS technique (Schmidt et al, 2008) to maximize proteome coverage. In such measurements, precursor ion chromatograms are first extracted from two initial data-dependent (DDA) LC–MS/MS runs and the precursor ion maps (retention time versus mass over charge) that are also generated by these measurements are subjected to a peak extraction algorithm (Mueller et al, 2007) to detect precursor ions not identified by DDA MS. In subsequent injections of the same sample, the mass spectrometer was then directed to acquire product ion spectra of previously non-selected precursor ions, to incrementally increase proteome coverage to saturation. We have shown earlier that this procedure maximizes the coverage of moderately complex proteomes at the peptide level while minimizing measurement and computational time (Schmidt et al, 2008).
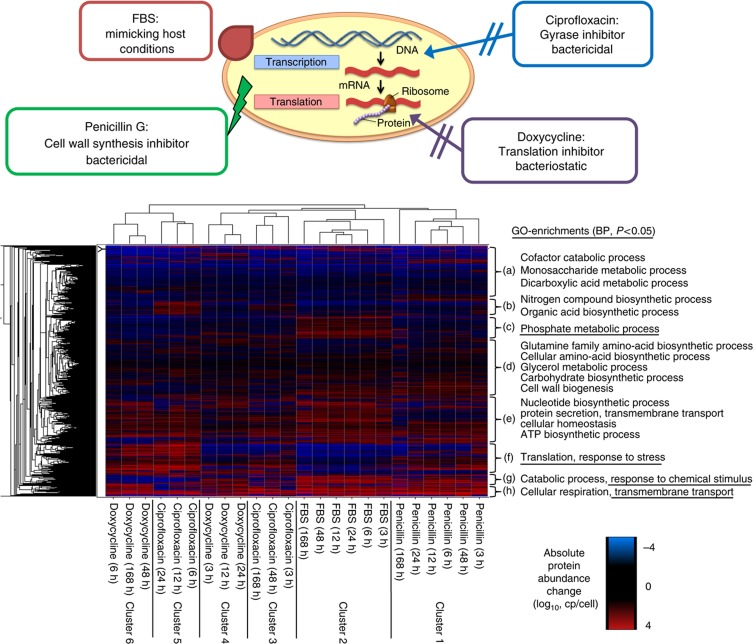
Figure 3.
Hierarchical clustering of protein concentration changes. Hierarchical clustering of absolute protein abundance changes to the corresponding untreated control samples in copies/cell (log10) for all 24 treatments. The column dendrogram representing the clustering of the differentially perturbed samples is displayed and the clusters (1–6) obtained are indicated. Significantly enriched (P<0.05) biological processes (BP) based on GO are indicated for all eight protein clusters obtained (a–h).
Specifically, the following sequence of analyses was carried out to collect the data for the L. interrogans PeptideAtlas. LC–MS/MS runs #1 and #2 were conventional DDA runs where precursor ions of different charge states (2 and >2, respectively) were selected. In subsequent LC–MS/MS runs #3–#20, precursor ions selected by the following criteria were added to inclusion lists and identified by directed precursor ion selection: (i) all features detected by a feature detection algorithm (Mueller et al, 2007) in the initial DDA runs; (ii) precursor ions corresponding to all PTPs extracted from a recently published large-scale proteome analysis on the same species (Beck et al, 2009); and (iii) predicted precursor ion signals for all PTPs that were computed but not observed from the L. interrogans genomic sequence. PTP predictions were carried out by the algorithm PeptideSieve (Mallick et al, 2007). The L. interrogans proteome is highly accessible for the LC–MS analysis employed here since for the majority of gene products (3402/3658) five or more PTPs could be predicted (Supplementary Figure S1). The fragment ion spectra generated from each of these analyses were database searched and the resulting data were filtered to a peptide and protein level false discovery rate (FDR) of 1% (Reiter et al, 2009). At each stage, already identified features as well as proteins identified with more than five PTPs were excluded from further analysis in the subsequent stages.
In the two initial DDA LC–MS/MS runs, we detected 37 833 unique features of which 7776 could be assigned to a peptide sequence, resulting in 6861 peptide identifications corresponding to 1223 proteins (Table I). The remaining features (27 968) for which no MS/MS spectra were acquired were split into four inclusion lists, each comprising around 7000 features. These were then specifically sequenced by directed LC–MS/MS analyses. Thereby, the PeptideAtlas could be extended by 2356 (228) additional peptides (proteins). Finally, 12 and 10 additional directed LC–MS/MS-sequencing runs for the identification of missing proteins using PTPs from a recently published PeptideAtlas or predicted PTPs, respectively, increased the overall number of identifications to a total of 13 113 features, corresponding to 11 611 peptides and 1680 proteins. To reach this coverage, 28 LC–MS/MS runs were required (Table I). As is evident from Figure 2A, the number of protein identifications reaches saturation toward completion of each experimental phase, after rising at the beginning of the phase, indicating that different peptide subsets are identified at each of the analytical stages. The final feature map generated in this discovery phase contains the exact mass and time coordinates of each identified feature and represents a rich resource for the directed sequencing of all detected proteins in the scoring phase. Importantly, the identified features are well distributed by time and mass (Figure 2C), which allowed their specific sequencing in a high number of samples by directed LC–MS/MS.
Table 1. Number of unique features and peptides identified in the discovery phase.
| Data filtering | Number of entries for all 1680 proteins identified in the discovery phase | Number of entries for GroEL |
|---|---|---|
| aDetected using the SuperHirn algorithm (Mueller et al, 2007). | ||
| bIdentified by database searching. FDR was set to 1%. | ||
| cPTPs are defined as features that assigned peptide sequences show full tryptic cleavage, contain no modification and only match to one protein sequence in the database used. | ||
| dUp to the five most intense PTPs per protein were selected for screening phase. | ||
| Detected unique featuresa | 37 833 | Not available |
| Identified unique featuresb | 13 113 | 86 |
| Identified unique peptidesb | 11 611 | 76 |
| Identified unique PTPsb,c | 6889 | 23 |
| Selected unique PTPsd | 4953 | 5 |
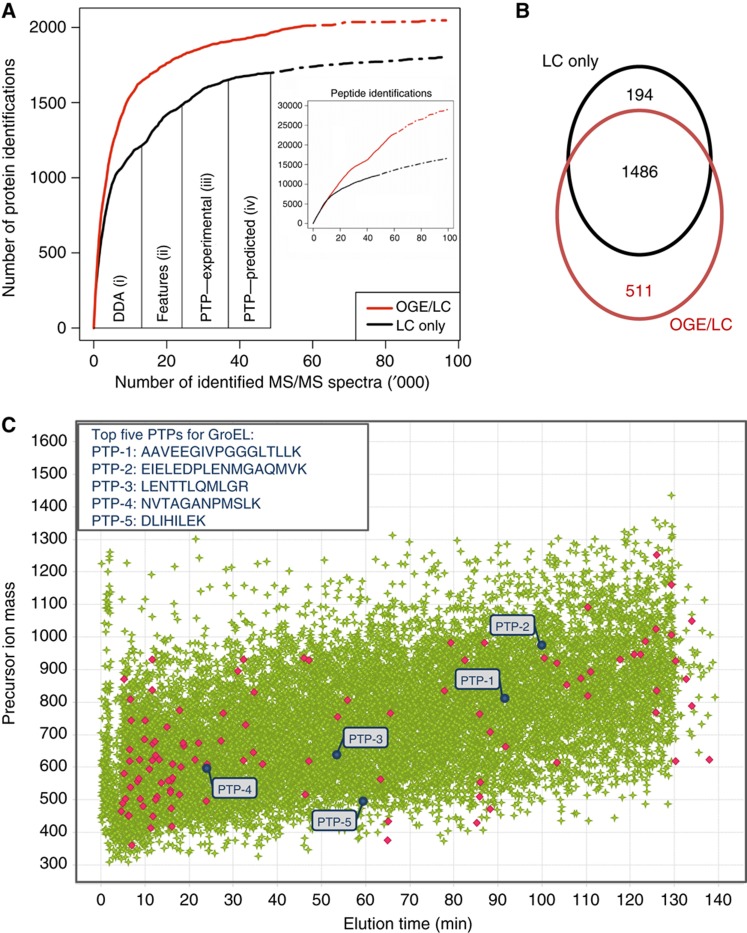
Figure 2.
Directed LC–MS/MS analysis of the L. interrogans proteome. A pool of peptide samples generated from different perturbations was LC–MS analyzed to generate a comprehensive protein/peptide atlas of L. interrogans. (A) This was achieved by accumulating the MS data obtained from (i) two non-directed (DDA) LC–MS runs followed by (ii) directed LC–MS analysis of all detected features, (iii) previously detected PTPs (Beck et al, 2009) and (iv) predicted PTPs (Mallick et al, 2007). Proteins detected with five or more PTPs were excluded in the following analysis. The numbers of identified proteins (y axis) and peptides (inset) versus the number of identified tandem mass spectra are displayed. For comparison, the protein discoveries obtained from a non-directed LC–MS analysis of 24 OGE fractions (OGE/LC) are shown in red. A recently developed algorithm was deployed to estimate the increase in protein/peptide discoveries with additional LC–MS experiments (dashed lines) (Claassen et al, 2009). (B) Venn diagram showing the overlap of proteins identified with the LC-only and the OGE/LC–MS approach. (C) LC–MS map of all identified features (green). Precursor ions identified as tryptic peptides of the GroEL protein are shown in red. The sequences as well as the coordinates of the five PTPs selected for GroEL monitoring in the scoring phase are indicated in blue.
We next evaluated the extent of proteome coverage achieved by this iterative directed sequencing strategy with that achieved by more conventional proteome analyses via extensive sample fractionation and DDA analysis of each fraction. For the latter strategy, the same peptide sample used for inclusion list sequencing was fractionated by isoelectric focusing using off-gel electrophoresis (OGE) (Heller et al, 2005) and each of the 24 fractions was analyzed once by DDA LC–MS/MS analysis. Intriguingly, this data set contained 60% more peptide identifications, but only 19% additional protein hits (number versus number, Figure 2A), indicating a higher peptide per protein ratio of 12 (OGE) over 7 (LC only). We thus conclude that 81% of the proteins detected by the OGE–LC–MS/MS approach were also detected by the directed LC–MS/MS method, most of them with a sufficient number of peptides for accurate quantification in the scoring phase. Notably, only a slight increase in protein identifications is expected by additional LC–MS/MS analyses (Claassen et al, 2009), demonstrating that we have detected most of the proteins identifiable by the two LC–MS/MS strategies employed (Figure 2A, dashed lines). As expected, the majority of proteins (67.9%) were identified with both approaches. However, 23.3/8.9% of identified peptides were exclusively detected by the OGE–LC/LC-only approach, respectively (Figure 2B). Functional annotation revealed that many of the 194 protein hits exclusively identified by the directed (LC only) LC–MS/MS approach and missed by the OGE–LC–MS/MS approach are membrane proteins (Supplementary Figure S2), suggesting a decreased recovery of hydrophobic peptides after OGE. Conversely, the OGE–LC–MS/MS strategy showed an increased coverage, particularly of low abundant proteins, like transcription factors and regulators, confirming the higher protein concentration range accessible after extensive sample fractionation. In general, extensive proteome coverage was achieved with both strategies, which is supported by the lack of biases against any functional groups (Supplementary Figure S2).
Overall, of the 13 113 different features identified by directed LC–MS/MS (Supplementary Table SII), 6889 represented suitable PTPs for protein quantification (Supplementary Table SIII). For each protein, the five most suitable PTPs for protein quantification, referred to as top five PTPs, were extracted from the feature list considering the following attributes; (i) specificity to a single database entry, (ii) true tryptic cleavage termini, (iii) lack of modifications and (iv) high MS-signal response determined by the SuperHirn algorithm (Mueller et al, 2007). The selected 4953 PTPs (Table I) covered the whole feature intensity range (Supplementary Figure S3) and all 1680 identified proteins (Table I). The feature intensity range for the PTP precursor ions on the inclusion list spanned more than three orders of magnitude, a dynamic range that is expected to capture most of the L. interrogans proteome (Malmström et al, 2009). The benefits of focusing on the most suitable PTPs for monitoring each protein can be demonstrated in the case of the chaperone GroEL. For this abundant protein, 86 different features could be identified (Table I) of which the five most intense fulfill all PTP selection criteria (Figure 2C, blue), supporting the observation that unspecifically proteolyzed or modified peptides constitute a minor but detectable fraction of the total ion current generated by the peptides from a protein (Picotti et al, 2007). By focusing on these PTPs, >90% of the MS-sequencing cycles required to detect and monitor GroEL levels in the following scoring phase could be saved and thus used for measuring different proteins of interest. It is important to note that this effect is more pronounced for highly abundant and larger proteins for which high numbers of peptides are identified.
Finally, 38 heavy labeled reference peptides from 19 proteins were added to estimate absolute protein concentration on a system-wide scale in each sample following a recently described protocol (Malmström et al, 2009) (Figure 1; Supplementary Table SI). Thus, the final inclusion mass list was distributed over two LC–MS/MS runs and the coordinates of the heavy reference peptides and their endogenous counterparts were included in both runs. Therefore, the data generated in the discovery phase of the project allowed us to establish a method in which 1680 proteins per sample could be detected and absolutely quantified in two inclusion list LC–MS/MS runs with a total analysis time per sample of 4 h.
To increase the speed and identification yield of the selected PTPs in the scoring phase, we computed a spectral library from the acquired MS-sequencing data in the discovery phase using SpectraST (Lam et al, 2009). We included additional MS data from a recent large-scale LC–MS/MS study on the same species (Beck et al, 2009) to further enhance the quality of the consensus spectra in the spectral library and applied very stringent filtering criteria to keep the overall FDR <0.2%. Overall, 321 498 identified MS2 spectra were merged to 33 766 distinct consensus spectra covering >2300 proteins. The library was added to the current L. interrogans PeptideAtlas and can be downloaded from http://www.peptideatlas.org.
Next, we assessed the performance of the described approach by analyzing a single control sample and comparing the number of identified peptides/proteins to the conventional shotgun LC–MS/MS methodology using the same number of runs. While the non-directed DDA LC–MS/MS analysis (Supplementary Figure S4A, blue) identified a larger number of peptides, 404 (40%) additional proteins could be detected by the directed strategy (1593) (Supplementary Figure S4A, red). The coverage was particularly enhanced for proteins of mid-to-low abundance, indicating an increased identification efficiency for these proteins by the directed MS approach compared with DDA LC–MS/MS-based strategies (Supplementary Figure S4B).
Finally, we assessed the utility of the generated inclusion list/spectral library on a different LC–MS platform in a different proteomics laboratory. After adjusting the retention times of the PTPs to the new LC system, the identified proteins could be detected with the same high consistency (Supplementary Figure S5A and B) and coverage (Supplementary Figure S5C) as on the LC–MS platform that was used to build the inclusion list and spectral library. This demonstrates the value of the generated data for the application in other laboratories and the usefulness of the generated, global PeptideAtlas and inclusion mass list for the proteomics community.
Quantitative time course measurements of perturbed L. interrogans cells
We next used the method established above to acquire quantitative proteome profiles of Leptospira cells grown under different conditions. Specifically, cells were cultured in EMJH supplement (control samples) and in the presence of fetal bovine serum (FBS; 10% v/v) and antibiotics (5 μg/ml ciprofloxacin, 10 μg/ml penicillin G, 15 μg/ml doxycycline, respectively) in EMJH supplement. The underlying molecular mechanisms of the individual treatments are displayed in Figure 3. Samples were taken after 3, 6, 12, 24, 48 and 168 h of treatment. Thus, overall 31 protein samples were generated, including 7 controls. We used label-free quantification to generate proteome maps of all detected PTPs and employed them for absolute protein quantification within each sample as well as relative protein quantification across all samples. Two technical replicates were acquired and averaged for all samples, to improve quantification accuracy.
We first evaluated the combined technical and biological reproducibility of the relative protein quantification by comparing the proteome maps of three different control samples (Supplementary Figure S6). The high squared Pearson correlation R2 (0.945–0.965) and the near straight lines indicated the nearly optimal linear relationship between the replicates. Specifically, minimal abundance variations between the replicate samples were observed by the inclusion list driven LC–MS/label-free quantification approach even for proteins of low abundance (Supplementary Figure S6A–C). Consequently, with the measured coefficient of variances of the protein ratios being <26% between all controls, 1.5-fold changes (2 × σ) with a P-value <0.05 (ANOVA) can be confidently detected for most proteins by the described approach (Supplementary Figure S6D–F).
We next used the proteome maps to estimate the absolute quantities of the proteins in each perturbed sample and thus, in conjunction with the number of cells used to generate the samples, the cellular concentrations of the detected proteins. This was accomplished by translating the signal intensities of the high responder peptides from each detected protein into absolute protein quantities, using a recently published approach with some modifications (Malmström et al, 2009). First, the absolute protein quantity of a consistent set of proteins was accurately determined in each sample by comparing the signal intensities of the sample intrinsic peptides with the corresponding signals generated from known amounts by isotopically labeled reference peptides of identical sequence that were added to each sample. Since these peptides were included in the directed LC–MS analysis, no additional SRM LC–MS analyses were required for their quantification. In this way, the precise concentrations of 29 peptides corresponding to 19 proteins could be calculated (Supplementary Table SI). The concentrations of these proteins spanned almost three orders of magnitude, from 68 copies/cell for the flagellar M-ring protein (YP_001355.1) to 13 649 copies/cell for the GroEL protein (YP_001299.1, Supplementary Table SI), confirming the high dynamic abundance range covered by the method (Supplementary Figure S3). In general, the protein abundances determined by multiple heavy reference peptides per protein showed good agreement, even for low abundance proteins (Supplementary Table SI). Moreover, the values determined here matched very well with those published in a recent study and the structural benchmarks employed therein (Malmström et al, 2009) (Supplementary Figure S7). In a second step, these abundance values were aligned with the average intensities of the three PTPs of each protein with the highest MS response, the same peptides that were in the focus of the directed LC–MS analysis for peptide identification. In the same operation, we therefore consistently estimated the absolute abundances of all identified proteins in each of the samples. On average, a high squared Pearson correlation (R2=0.805) of the absolute abundances accurately determined by heavy peptide references and their average feature intensities could be observed (Supplementary Figure S8A). As a result, the error model, calculated using a bootstrapping approach, indicated a mean error of only 1.84-fold with a maximum of 2.8-fold difference (Supplementary Figure S8B).
High-level classification of induced proteome changes
As described above, the quantitative proteomic method used in this study generated highly reproducible data sets over all conditions tested, that is, for the most part, the same proteins were detected and quantified under each condition. To take advantage of this unique property of the data set, in combination with the availability of protein concentration levels, we applied classification methods originally developed for transcript array data to detect systemic responses of the proteome under the given perturbations. A total of 4525 significant protein changes (ANOVA, P<0.05, ratio>1.5) were determined across all samples. These changes revealed that the majority of the detected proteins (944) show a significant change in at least one of the various treatments and time points analyzed. The most intense protein expression changes were observed after long treatments, reaching changes as high as 100-fold. Protein abundance changes detected in the absence of any external factors or stimuli were negligible (Supplementary Figure S9).
Using this data set we asked if the absolute concentration of proteins in the cell correlates with the magnitude of regulation (Supplementary Figure S10A). Interestingly, highly abundant proteins turned out to be regulated to a lesser extent than their lower expressed counterparts. The most highly abundant proteins were, on average, about 1.5-fold up- or 2-fold down-regulated while the least abundant were 2.5-fold up-regulated or 3-fold down-regulated. The observed increase in stability of highly abundant proteins points to an energy saving strategy the L. interrogans cells have developed (Akashi and Gojobori, 2002). Conversely, the impact of the low abundance proteins on the total proteome composition is only marginal and the combined cost for their synthesis and degradation is low (Supplementary Figure S10B).
Therefore, we next investigated whether for the measured proteins, the difference in copies/cell between perturbations represents a better measure for protein clustering than relative abundance changes, since they reflect the actual magnitude of proteome changes in the cell. We first used hierarchical clustering to group the samples (x axis) and the proteins (y axis) according to their changes in absolute level of abundance (in copies/cell) (Figure 3) and relative fold (Supplementary Figure S10C). We observed an improved clustering efficiency, that is samples that are expected to generate the most closely related proteome patterns clustered most closely, when absolute protein changes were compared with fold changes. Specifically, all FBS (cluster 2) and penicillin G (cluster 1) treated samples grouped together and fewer but more distinct clusters were obtained when applying the same thresholds. In addition, proteins belonging to the same complex or sharing similar functions, which are expected to be co-regulated over the various treatments, showed more similar patterns when using absolute expression changes over protein ratios. Therefore, absolute protein changes were employed in all subsequent clustering analyses.
It is apparent from Figure 3 that the patterns at the early time points of doxycycline treatment (cluster 4) strongly resemble the patterns representing very early and very late treatments with ciprofloxacin (cluster 3), while the observed proteome changes in cells treated for 6, 12 and 24 h with ciprofloxacin (cluster 5) more strongly resembled those of late doxycycline treatments (cluster 6). To interpret the observed sample clusters on a functional level, the hierarchically clustered proteins were associated with eight distinct groups (clusters a–h) and subjected to functional annotation and overrepresentation analysis using gene ontology (GO)–Functional groups as the basis of the association (Huang et al, 2007). We found four such clusters (a, d, e, h) that showed a similar response to all perturbations. Cluster ‘d’ essentially consisted of proteins that were unchanged under the applied conditions and these proteins were functionally associated with the general metabolic processes of amino acid, glycerol and carbohydrate metabolism, as well as cell wall synthesis. Proteins involved in cofactor catabolism, monosaccharide and dicarboxylic acid metabolism were preferentially contained in cluster ‘a’. These proteins were commonly down-regulated under perturbed conditions. Proteins involved in ATP synthesis, protein secretion and transport as well as cellular homeostasis were contained in clusters ‘e’ and ‘h’. These proteins were generally up-regulated under perturbed conditions. These findings indicate that L. interrogans cells commonly react to changing environmental conditions by actively rearranging the proteome on the account of specific biosynthesis pathways, while the central amino acid and carbohydrate metabolism remains untouched.
Beyond such ‘default behavior’, response patterns specific to individual perturbations were detected. Cluster ‘f’ consisted of proteins that are involved in translation and response to stress and were down-regulated upon serum and early doxycycline treatments. This pattern likely reflects a redirection of energy from the protein translation and folding systems toward other cellular processes resulting in a reduced growth rate. The same proteins were mostly up-regulated in response to all other treatments, particular in cells treated with antibiotics, indicating induced stress response. The proteins contained in cluster ‘g’ were mostly associated with catabolic processes and response to chemical stimuli and were strongly up-regulated upon serum and penicillin G treatment but down-regulated after ciprofloxacin and doxycycline treatment. Taken together, these data suggest that L. interrogans cells react with more active protein synthesis of stress and elongation factors, like dnaK and tuf, on the account of other cellular systems when coping with DNA-gyrase (ciprofloxacin) or ribosomal (doxycycline) inhibition. In contrast, the inhibition of cell wall synthesis (penicillin G) and stimulation with serum causes an inverse reaction and reduced growth. Besides these clusters that overlap between treatments, highly specific proteome pattern could be detected for serum (cluster ‘c’) and ciprofloxacin (cluster ‘b’) stimulation. In conjunction with the individual clustering of most treatments, this suggests that the proteome regulation follows characteristic patterns corresponding to the different treatments, indicating that specific regulatory mechanisms are activated upon the individual perturbations that are further investigated below.
Pathway classification of individual treatments
To further analyze the detected treatment-specific proteome response patterns, time-resolved protein expression profiles of the individual treatments were grouped according to their changes in copies/cell using K-means clustering (Figure 4A–D). The generated cluster profiles were subjected to an enrichment analysis of pathways (as present in the KEGG database; Kanehisa et al, 2010) using the DAVID algorithm (Huang et al, 2007) to generate a detailed picture of the pathways significantly (P<0.05) enriched in response to the individual treatments (Figure 4E). To better visualize the general regulation of the individual protein clusters, protein profiles showing up- (down-) regulation after 24 h of treatment are indicated in red (blue). Compared with the detection of global changes described above, this analysis reveals the details of response patterns specific to individual stimuli. On average, 4 to 5 meaningful clusters could be identified for each treatment. Intriguingly, the protein profiles obtained clearly indicated a compensatory behavior. An increase in the abundance of some proteins is always compensated by an equivalent down-regulation of other proteins, giving further support to the notion that the total protein mass in a cell stays constant, even under the various and harsh stress conditions applied (Figure 4A–D). This was already observed recently for a limited number of perturbations (Beck et al, 2009) and is now confirmed here with a much larger set of conditions.
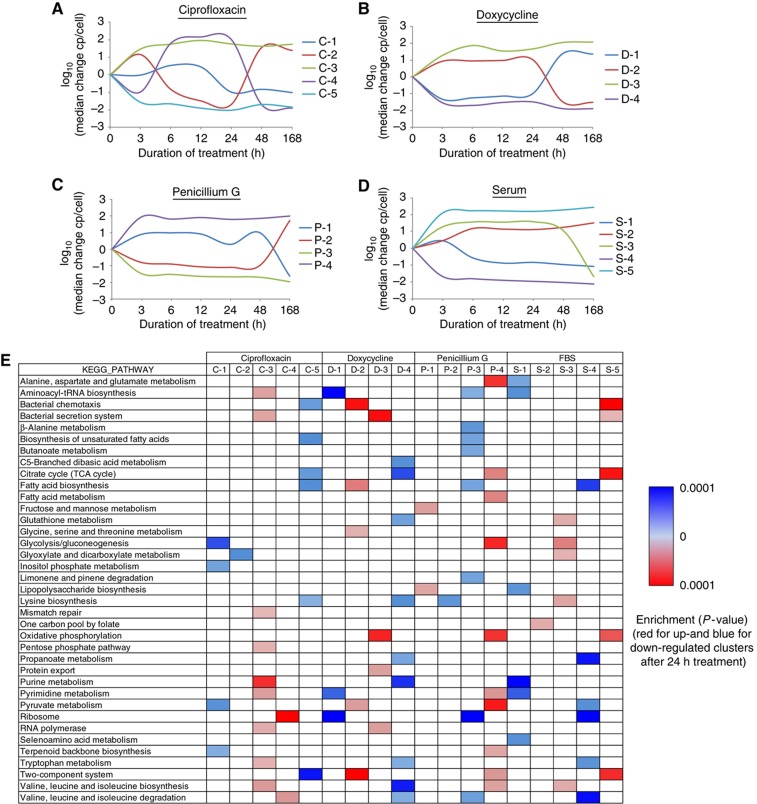
Figure 4.
Pathway analysis of K-means clustered proteome changes. (A–D) The individual protein profiles (changes in copies/cell, log10) obtained were grouped and numbered by K-means clustering for each treatment, respectively. Each cluster profile is indicated with the first letter of the treatment and its number. (E) All identified protein clusters were searched for significant overrepresentation of proteins belonging to specific pathways according to the KEGG database (Kanehisa et al, 2010) using DAVID (Huang et al, 2007), respectively. Protein groups that are up-regulated after 24 h of treatment are shown in red, while clusters containing mostly proteins with reduced expression after 24 h are indicated in blue.
The treatment with serum is of particular interest because it can, to some extent, replicate conditions under which Leptospira cells adapt to a host environment and become virulent. For this treatment, we obtained five meaningful protein clusters (Figure 4D). Three of them showed an immediate and strong regulation of protein abundance after 3 h of treatment, whereby clusters ‘S-4’ and ‘S-5’ showed a further slight increase upon longer treatments and cluster ‘S-3’ showed a rapid down-regulation after 7 days of treatment. Proteins involved in motility, tissue penetration and virulence (Lux et al, 2000; Ren et al, 2003) showed the highest increase in expression (cluster ‘S-5’) and were also found to be significantly enriched in cluster ‘c’ from our global analysis (Figure 3). Most proteins of the chemotaxis pathway and the two-component system were up-regulated in cluster ‘S-5’ (Supplementary Figure S11), demonstrating a strong co-regulation of the members within this protein group.
Further, strongly enriched pathways after serum treatment include the citrate cycle (TCA cycle, Supplementary Figure S12) and oxidative phosphorylation (Supplementary Figure S13), suggesting that aerobic respiration is the preferred energy source for Leptospira in FBS-containing media. The pathway analysis also confirmed the reduced abundance of ribosomal proteins after serum treatment (cluster S-4). These findings are in agreement with recent transcriptomics (Patarakul et al, 2010) and proteomics (Eshghi et al, 2009) studies that found that several ribosomal and heat shock proteins were regulated after incubation of L. interrogans with serum. However, for most proteins, the correlation between mRNA and protein levels was found to be very poor. For instance, the confirmed virulence surface protein Loa22 (Ristow et al, 2007) and the potential virulence factor OmpL1 (Barnett et al, 1999) with confirmed expression in vivo were clearly up-regulated on the protein level (both in cluster ‘S-5’), but not differentially expressed on the mRNA level (Patarakul et al, 2010), underlining the importance of quantitative proteome studies. In fact, we found the concentration of these proteins Loa22 and OmpL1 to be increased by 14 754 and 11 985 molecules per cell, respectively, after 7 days of serum treatment. This represents the second and third highest increase in abundance of any cellular protein induced by this treatment (Supplementary Table SV), indicating the relevance of these proteins for adaptation of the cell to a host-like environment (Becker et al, 2006). Notably, the list of proteins with a high increase in expression further contains potential virulence factors like catalase (Lo et al, 2010) and chemotaxis proteins, but also several hypothetical and membrane proteins that have not yet been associated with Leptospira virulence or any other function.
In contrast to the perturbation by serum exposure, the ribosomal proteins were found to be strongly up-regulated after 6, 12 and 24 h of antibiotic ciprofloxacin treatment (cluster C-4). This increase was compensated by an equivalent down-regulation of proteins involved in glyoxylate metabolism (cluster ‘C-2’). The regulation of these proteins is inverted after 48 h of treatment, suggesting that the cells have adapted to the treatment or reduced the antibiotic concentration to tolerable levels. Interestingly, immediately after ciprofloxacin exposure, the cells activate a highly specific cascade of pathways to cope with the DNA-topo-isomeric stress (cluster ‘C-3’). The group of proteins that was exclusively up-regulated after 6, 12 and 24 h ciprofloxacin treatment (see also Figure 3 cluster ‘b’), contains mainly proteins involved in transcriptional and translational processes, like DNA mismatch, RNA polymerization, aminoacyl-tRNA synthesis, purine and pyrimidine metabolism, as well as the secretion system and the SOS response (Fonville et al, 2010), like recombinase A and J. These data indicate that the cells are trying to compensate the DNA-topo-isomeric stress induced by the ciprofloxacin treatment (Michel, 2005; Cirz et al, 2007; López et al, 2007; Vlasić et al, 2008). Intriguingly, we also found the protein TetR in this cluster, which was recently found to be specifically mutated in ciprofloxacin-resistant strains of Bacillus anthracis (Serizawa et al, 2010), underlining the relevance of the specific protein changes detected. In parallel, the proteome abundance of the chemotaxis and two-component systems, the TCA cycle and the lysine and fatty acid biosynthesis are reduced (cluster ‘C-5’). These proteins apparently represent pathways that are lesser important for ciprofloxacin defense. Interestingly, with an average increase of >15 000 copies/cell, the chaperone GroEL was the most heavily induced protein across all antibiotic treatments, whereas no significant regulation of this protein could be detected upon serum stimulation (Supplementary Table SV). Apparently, GroEL is a key protein for Leptospira cells to maintain proper assembly of unfolded polypeptides generated under antibiotic stress.
Upon treatment with doxycycline, a tetracycline-class inhibitor of the ribosomal protein biosynthesis, Leptospira cells show, as with ciprofloxacin stimulation, a converse regulation of a specific proteome subset after 48 h of treatment (cluster ‘D-1’). Proteins involved in translation, like ribosomal proteins and aminoacyl-tRNA biosynthesis, are first reduced in concentration. After 48 h of treatment their abundance increases, a regulation pattern that was also observed by transcriptome analysis of Tropheryma whipplei (Van La et al, 2007). An inverted behavior was detected for the chemotaxis, the two-component and several metabolic pathways (cluster ‘D-2’). As with the ciprofloxacin treatment, the proteome levels of the bacterial secretion system are promptly increased (cluster ‘D-3’) to reduce the doxycycline concentration in the cell. These observations indicate that although Leptospira cells are affected by doxycycline, the drug cannot inhibit protein synthesis entirely because large-scale proteomic changes are apparent. Upon treatment with the drug penicillin G a large-scale proteomic adjustment, namely an instantaneous and strong up-regulation (cluster ‘P-4’) or down-regulation (cluster ‘P-3’) regulation of several pathways comprising a large number of proteins is apparent and remains constant throughout all time points.
To conclude, by using a novel proteomic technology for generating consistent quantitative proteome profiles measuring absolute cellular protein concentrations we could, for the first time, survey the behavior of significant fractions of the proteome over time in multiple samples, allocate the generated protein clusters to most biochemical pathways present in L. interrogans and detect biologically informative patterns. This revealed that the cells have successfully generated systematic and highly specific defense and adaption processes over time for survival in rapidly changing environments.
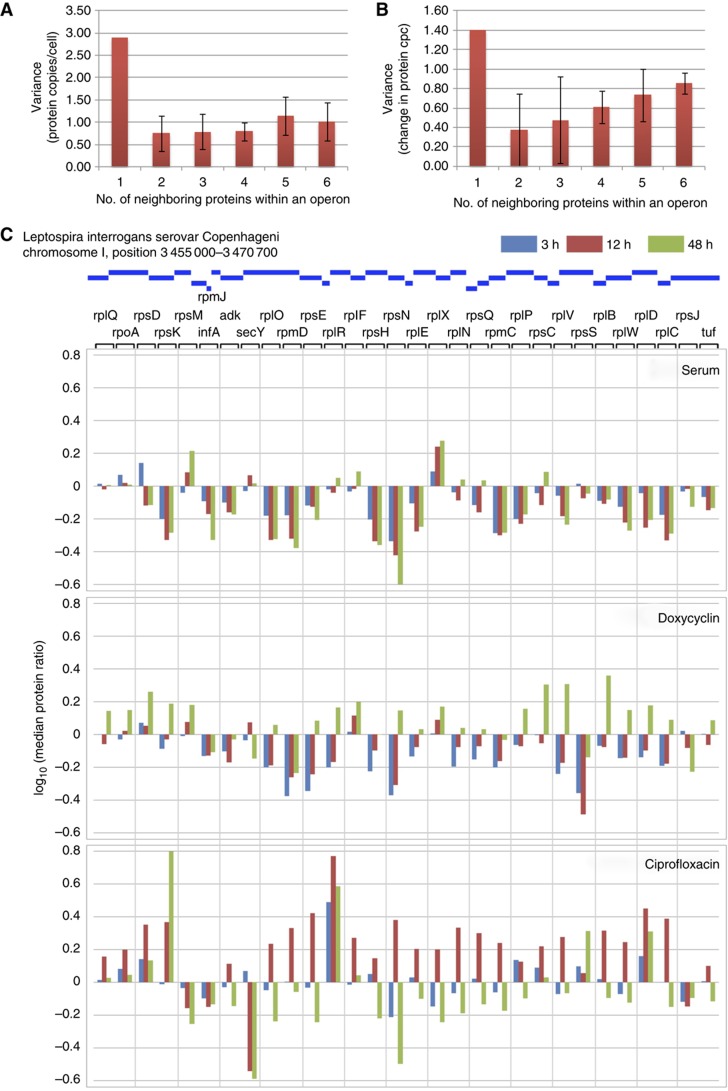
Protein dynamics within operons
Transcriptomics using expression arrays or RNA sequencing can reveal mRNA abundances on a genome-wide scale. The present study contains, to our knowledge for the first time, absolute abundance values on the protein level for an extensive fraction of the proteome. We therefore asked whether the absolute protein quantities could reveal novel properties of the Leptospira proteome. First, we asked if proteins that localize to the same (in silico predicted) operon in the genome (Dehal et al, 2010) have similar absolute abundances, which would be expected because they are being synthesized from the same pool of mRNA species. Indeed, the variance of copy numbers per cell of all proteins was more than three times larger than the variance of copy numbers per cell of proteins within an operon (Figure 5A). Transcriptomics also predicts a higher abundance of proteins at the 5′ end of operons, since the transcription of mRNA is often incomplete, a phenomenon that is also referred to as staircase behavior and has been observed for around half of all operons in other bacteria (Benders et al, 2005; Güell et al, 2009). We investigated this phenomenon on the protein level but could confirm it only for a minority of operons (∼5%). We next asked if proteins organized within operons would respond to the cellular treatments with a similar rate of up- or down-regulation. We observed a general trend that the proteins within an operon responded synchronously, but that the regulation was more pronounced the closer the proteins localized to the 5′ end of an operon (Figure 5B). There were, however, obvious exceptions. To illustrate regulation patterns observed upon serum exposure, doxycycline and ciprofloxacin treatment, we chose a genome region that encodes high abundant ribosomal proteins, translational elongation and initiation factors as well as SecY as an example, specifically position 3 455 000–3 470 700 on chromosome I (Figure 5C). We tracked the abundance of all 32 proteins within this region throughout all time points and stimuli except for the very small protein coded by gene rpmJ that did not generate a sufficient number of MS compatible tryptic peptides to allow conclusive measurement. Upon stimulation with serum, most ribosomal proteins were down-regulated, a few remained constant and two were strongly up-regulated (rpsM and rplX). Almost the same pattern was observed after 3–12 h of treatment with doxycycline, however, in that case after 48 h most ribosomal proteins were strongly up-regulated, indicating that the cell compensates for ribosomal inhibition by synthesizing a higher number of ribosomes. The translocon protein SecY and translational initiation factor infA were down-regulated at the same time. They are likely needed in smaller amounts due to the reduced number of active ribosomes. The regulation pattern observed upon treatment with ciprofloxacin is very different. Most ribosomal proteins go through a maximum and are up-regulated after 12 h but down-regulated after 48 h. There are again a number of proteins that do not follow the general trend but stick out of the overall pattern. RpsK, rplR rpsS and rplD are up-regulated even after 48 h. RpsM, rpsJ, initiation factor infA and SecY are already down-regulated after 12 h. This suggests that although most proteins within an operon respond to regulation synchronously, bacterial cells seem to have subtle means to adjust the levels of individual proteins or protein groups outside of the general trend, a phenomena that was recently also observed on the transcript level of other bacteria (Güell et al, 2009).
Figure 5.
Protein dynamics within operons. (A) Absolute protein concentration variance over all proteins (1) and within operons comprising different number of genes (2–6). (B) Like A, representing the variance in protein copies/cell (cpc) after 168 h of serum treatment over all proteins (1) and within operons covering various numbers of genes (2–6). The standard deviations are indicated as error bars. (C) Median protein ratio (log10) of the proteins present in the largest predicted operon in the L. interrogans genome on chromosome I ranging from position 3 455 000 to 3 470 700. The ratios are displayed for 3 h (blue), 12 h (red) and 48 h (green) of serum, doxycycline and ciprofloxacin treatment, respectively.
Discussion
The two-step quantitative proteomic technique described here comprehensively and reproducibly determines absolute abundance protein abundance patterns at high throughput. As a first step, an atlas of peptides is generated. This 1D-peptide catalog is not a static entity but evolves as data are accumulated and the directed LC–MS/MS workflow and the instrumentation used advance. The subsequent measurements are then focused to a limited number of ‘high-flying’ peptides per protein that are derived from the initially generated atlas. Thereby, thorough coverage of the proteome or selected protein sets in single MS runs is achieved and the peptides are identified quickly and reliably using the previously acquired information. Compared with classical shotgun methods, the throughput is accelerated, efficiency and sensitivity are increased and measurement time and sample amount are minimized. Since the MS data generated by classical and directed LC–MS/MS are very similar, the same well-established and validated data processing tool for protein identification (Yates et al, 1995; Perkins et al, 1999; Keller et al, 2002; Nesvizhskii et al, 2003) and quantification (Mueller et al, 2007) can be employed to mine the large acquired data sets. Because of the low number of MS/MS scans generated, the database searching is accelerated and data storage as well as post-processing is simplified. Additionally, the consistent identification of features across runs improves the alignment of extracted precursor ion chromatograms and enables more reliable label-free protein quantification. Moreover, the method described here could be combined with isotope-labeling approaches (de Godoy et al, 2008) or screenings for post-translational modifications (Huber et al, 2009). Additionally, the determined ‘high-flying’ PTPs in combination with the spectral library serve as an excellent resource for designing SRM assays for the fast analysis of small protein subsets (Jaffe et al, 2008; Picotti et al, 2009). Current bottlenecks include the necessity of cost-intense heavy labeled reference peptides and the dynamic range on the MS1 level that limits the approach to organisms of low-to-intermediate genomic complexity. Nevertheless, new high-throughput methods to generate reference peptides, the combination with sample pre-fractionation strategies (Heller et al, 2005) and further instrumental developments (Makarov et al, 2009) are likely to increase the scope of the approach in the near future.
We applied this method to study the global proteome changes of the human pathogen L. interrogans and could achieve system-wide proteome coverage across 25 differential treated samples that enabled us to perform a detailed investigation of protein subset expression changes of most pathways in this bacterium. Additionally, the determined absolute proteome changes improved the clustering efficiency over usually employed relative fold changes and allowed us to detect common and specific proteome patterns for antibiotic defense and pathogenic adaptation of L. interrogans. In particular, the coherent grouping of all 25 perturbations facilitated the detection of highly specific and information-rich protein clusters for some treatments. These generated fingerprints of cellular states might be compared and deployed to determine these cellular states in future studies. With the possibility to deploy the generated PTP mass lists together with the heavy reference peptides across different high-resolution LC–MS platforms and laboratories, we believe that the method described here will become a corner stone for systems biology of microbes.
Materials and methods
Cell culture and treatment
The Leptospira interrogans serovar Copenhageni of the strain Fiocruz L1–130 were obtained from the American Type Culture Collection (ATCC No. BAA-1198) and cultivated as previously (Haake et al, 1991). In brief, cultures of 10 ml volume were grown in EMJH medium at 30°C to a density of 2 × 107 /ml and then stimulated (or left untreated as a control). The cells were treated for 3, 6, 12, 24, 48 and 168 h with one of the following substances, respectively: 5 μg/ml Ciprofloxacin, 15 μg/ml Penicillin G, 10 μg/ml Doxycycline and 10% FBS in culture medium. Afterwards, the cells were harvested by centrifugation at 3000 g, washed twice in PBS, counted, pelleted again, resuspended in 200 μg lysis buffer (100 mM ammoniumbicarbonate, 8 M urea, 0.1% RapiGest™), sonicated for 5 min and stored at −80°C. Additionally, a small aliquot of the supernatant was taken to determine the protein concentration using a BCA assay (Thermo Fisher Scientific).
Protein cleavage
The proteins obtained from differentially treated cultures were reduced with 5 mM TCEP for 60 min at 37°C and alkylated with 10 mM iodoacetamide for 30 min in the dark at 25°C. After quenching the reaction with 12 mM N-acetyl-cysteine, the samples were diluted with 100 mM ammoniumbicarbonate buffer to a final urea concentration of 1.5 M. Proteins were digested by incubation with sequencing-grade modified trypsin (1/50, w/w; Promega, Madison, WI) overnight at 37°C. Then, the samples were acidified with 2 M HCl to a final concentration of 50 mM, incubated for 15 min at 37°C and the cleaved detergent removed by centrifugation at 10 000 g for 5 min. For absolute quantification, aliquots of a mixture containing 38 heavy labeled reference peptides (10 pmol each, Supplementary Table SI) were added to each sample. Subsequently, all peptides were desalted on C18 reversed-phase spin columns according to the manufacturer's instructions (Macrospin, Harvard Apparatus), dried under vacuum and stored at −80°C until further use.
Off-gel electrophoresis
Aliquots of all samples were pooled, dried and resolubilized to a final concentration of 1 mg/ml in OGE buffer containing 6.25% glycerol and 1.25% IPG buffer (GE Healthcare). The peptides were separated on a 24-cm pH 3–10 IPG strip (GE Healthcare), with a 3100 OFFGEL fractionator (Agilent) as previously described (Heller et al, 2005) using a protocol of 1 h rehydration at maximum 500 V, 50 μA and 200 mW. Peptides were separated at maximum 8000 V, 100 μA and 300 mW until 50 kVh were reached. Subsequently, each of the 24 peptide fractions was desalted using C18 reversed-phase columns according to the manufacturer's instructions (Macrospin, Harvard Apparatus), dried under vacuum and subjected to data-dependent LC–MS/MS analysis.
Data-dependent acquisition (DDA) MS
The setup of the μRPLC–MS system was as described previously (Schmidt et al, 2008). The hybrid LTQ-FT-ICR mass spectrometer was interfaced to a nanoelectrospray ion source (both Thermo Electron, Bremen, Germany) coupled online to a Tempo 1D-plus nanoLC (Applied Biosystems/MDS Sciex, Foster City, CA). In all, 1 μg of total peptide mass was separated on a RPLC column (75 μm × 15 cm) packed in-house with C18 resin (Magic C18 AQ 3 μm; Michrom BioResources, Auburn, CA) using a linear gradient from 96% solvent A (98% water, 2% acetonitrile, 0.15% formic acid) and 4% solvent B (98% acetonitrile, 2% water, 0.15% formic acid) to 30% solvent B over 120 min at a flow rate of 0.3 μl/min. Each survey scan acquired in the ICR cell at 100 000 FWHM was followed by MS/MS scans of the three most intense precursor ions in the linear ion trap with enabled dynamic exclusion for 60 s. Charge state screening was employed to select for ions with at least two charges and rejecting ions with undetermined charge state. The normalized collision energy was set to 32% and one microscan was acquired for each spectrum.
Directed mass spectrometry
Generally, similar settings as with the DDA LC–MS/MS analysis were used for directed LC–MS/MS measurements with a few modifications: the resolution of each survey scan acquired in the ICR cell was reduced to 50 000 FWHM and the preview mode option was disabled. Furthermore, the dynamic exclusion was reduced to 15 s to acquire multiple MS/MS spectra for the parent ions of interest to increase both their identification rates and consensus spectra quality in the generated spectral library. Finally, the non-peptide isotopic pattern filter was disabled to allow more precursor ions to trigger MS-sequencing attempts and increase the overall sensitivity of the directed LC–MS/MS approach (Schmidt et al, 2008).
Database searching
After converting the acquired raw files to the centroid mzXML format using ReAdW (http://tools.proteomecenter.org/wiki/index.php?title=Software:ReAdW), MS/MS spectra were searched using the SORCERER-SEQUEST™ v4.0.3 algorithm (Yates et al, 1995) against a decoy database (consisting of forward and reverse protein sequences) of the predicted proteome from Leptospira interrogans serovar Copenhageni str, complete genome NCBI genome number NC_005823 and NC_005824 (http://www.ncbi.nlm.nih.gov/entrez). The database consists of 3658 Leptospira proteins as well as known contaminants such as porcine trypsin, human keratins and high abundant bovine serum proteins (Non-Redundant Protein Database, National Cancer Institute Advanced Biomedical Computing Center, 2004, ftp://ftp.ncifcrf.gov/pub/nonredundant), resulting in a total of 7480 protein sequences. The search criteria were set as follows: full tryptic specificity was required (cleavage after lysine or arginine residues, unless followed by proline); two missed cleavages were allowed; carbamidomethylation (C) was set as fixed modification; oxidation (M), 13C6-15N2 (K) and 13C6-15N4 (R) were applied as variable modifications; mass tolerance of 15 p.p.m. (precursor) and 0.8 Da (fragments). The database search results were further processed using the PeptideProphet (Keller et al, 2002) and ProteinProphet (Nesvizhskii et al, 2003) program and the peptide FDR was set to 1% on the peptide and 2% on the protein level and validated using the number of reverse protein sequence hits in the data sets.
Generation of 1D-PeptideAtlas
Three different strategies were employed in the discovery phase to characterize as many features as possible within the 2-h LC gradient and establish a comprehensive 1D-LC–MS peptide map of the L. interrogans proteome with the goal to identify at least five PTPs for each protein that can be targeted for accurate quantification in the final scoring phase. PTPs are defined as (i) peptides that sequence is unique to one protein in the proteome, (ii) have two tryptic termini and no missed cleavage and (iii) give a high MS response. To achieve maximal protein expression, one aliquot of each perturbation after 24 h of treatment were pooled and the generated peptide mix was extensively mapped using four different MS strategies.
(i) First, two data-dependent acquisition (DDA) LC–MS/MS runs, focusing on doubly charged and three or higher charged precursor ions, were carried out, respectively. (ii) Subsequently, the SuperHirn peak extraction and alignment algorithm (version 3) was used to extract all MS1 features and generate a MasterMap that includes the MS/MS-spectra assignments (Mueller et al, 2007). All features that did not trigger a MS/MS spectrum were specifically MS sequenced using scheduled, directed LC–MS/MS analysis as recently specified (Schmidt et al, 2008). Next, for proteins with less than five PTP identifications, PTP masses were extracted from peptide identifications obtained in the pre-fractionation (OGE) LC–MS experiment (iii) or, if not available, predicted by the PeptideSieve software tool (iv) (Mallick et al, 2007). Retention time prediction (Spicer et al, 2007) allowed timewise segmentation of the mass lists into five segments, which reduced the number of directed LC–MS runs required to sequence all selected PTPs. All MS/MS spectra were database searched as described and the identified peptides sequences assigned to the generated MasterMap (Schmidt et al, 2008). An additional feature was added to the SuperHirn algorithm (version 3) that employs lower intensity thresholds to all identified precursor ions for which no feature was detected in the initial peak extraction step. This allowed us to also determine the MS intensity, charge state and elution time for most peptide ions identified in phases (iii) and (iv). Up to five PTPs were selected for each identified protein using the above filtering criteria, resulting in a final list of 4953 PTPs. All PTPs for which no peak area could be calculated by the SuperHirn software were ranked according to their number of identified MS2 spectra instead.
Spectral library searching
A spectral library consisting of all confidently identified MS/MS spectra obtained above as well as currently present in the L. interrogans PeptideAtlas (Beck et al, 2009) was prepared. Therefore, all spectra assigned to the same peptide sequence were combined to reduce the presence of interfering fragment ions and improve the overall quality of the spectral library. In total, 321 498 identified MS/MS spectra were combined to 33 766 consensus spectra covering 26 029 unique peptides and unique 2370 proteins with a FDR of <0.2%. The library was added to the current L. interrogans PeptideAtlas and is publicly available (see http://www.peptideatlas.org/builds/) or can be downloaded using the following link: http://www.peptideatlas.org/speclib/ISB_Linterrogans_IT_v1.0.tgz.
The software SpectraST was used to match the acquired MS/MS spectra with the consensus spectra in the spectral library and score each match (Lam et al, 2008). In order to statistically determine matching confidence, decoy consensus spectra were added to the spectral library to calculate FDRs (Lam et al, 2009). Non-matching MS/MS spectra subjected to a conventional database search using Sequest as described above and combined with the spectral matching data while keeping the FDR <1% on the peptide and 2% on the protein level, respectively. The peptide and protein prophet probabilities as well as the number of peptide identifications were used as parameters to set the FDR accordingly.
Global protein profiling
After employing the above filtering criteria, 4953 validated PTPs representing 1680 identified proteins (Supplementary Table SIV) could be detected. For directed mass spectrometric analysis, all detected precursor ion masses of the selected PTPs were equally distributed over two mass lists. To each list, the detected precursor ion masses and retention times of 38 heavy labeled reference peptides (Thermo Fisher Scientific, Supplementary Table SI) and their endogenous counterparts were added. These inclusion mass lists were imported as global mass lists into the mass spectrometer and the PTPs sequenced in each sample using two single directed LC–MS/MS runs applying the same parameters as described above. The acquired MS/MS spectra were searched against the spectral library built and protein database as described above and pepxml-files covering all LC–MS runs of the individual time courses, respectively, were generated. These were imported into the Progenesis LC–MS software (v2.5, Nonlinear Dynamics Limited), which was used for label-free protein quantification applying the default parameters. Only unmodified peptides having a PeptideProphet score of 0.85, corresponding to an FDR of <1%, were considered for quantification. The quantitative data obtained were further normalized and statistically analyzed according to Brusniak et al (2008) using the Spotfire Decision Site program (version 9.1.1, TIBCO) and the guides provided for analyzing large transcriptomics data sets. In brief, we set a nominal lower bound value (noise level) as the minimum measured intensity and replaced with it missing values and values below it. We then calculated fold-change ratios (in log-scale) between control and perturbated samples. On the protein level, the ProteinProphet probability were employed to set the FDR to 2% based on the number of reverse protein hits. Only proteins with a 1.5-fold change in abundance and a P-value <0.05 (ANOVA) were considered significant (Supplementary Figure S6). The protein ratios and absolute abundances of all identified proteins across the individual treatments are displayed in Supplementary Table SV. The corresponding primary MS/MS data files can be retrieved via the Tranche website (https://proteomecommons.org/tranche/, ‘Leptospira_Time_Course_MSB-11-2792’, hashcode H4hv0MiRqwiPc0gONayV7oou/d4eRD8VviwIh6ORNP+UK+CR72ZZgKuujLsgCRP6DLRjUOLPZpAIkiFFJRMMRtHg3V8AAAAAAAApWg==).
Absolute protein quantification
The absolute abundances of all identified proteins were determined as recently specified (Malmström et al, 2009). First, the concentrations of the 19 anchor proteins were calculated from the ratio of the signal intensities of the heavy labeled reference peptides (known concentration of 100 fmol) and their endogenous counterparts (Supplementary Table SI). Then, the three most intense PTPs of each protein were selected, their MS-intensity values as determined by the Progenesis software averaged and aligned with the absolute abundances of the 19 reference proteins (Supplementary Figure S8A). After correlating the calculated protein concentrations with the number of cells used for each experiment, the abundance of each protein in copies/cell could be estimated. Additionally, error estimation was carried out using a bootstrap analysis (Supplementary Figure S8B) according to Malmström et al (2009). Absolute protein concentrations were determined for all perturbations (Supplementary Table SV).
Cluster analysis
To cluster temporal or regulatory patterns of protein abundance, we used the Spotfire Decision Site program (version 9.1.1, TIBCO) and the guides implemented in the functional genomics suite for microarray data analysis. We used either protein fold ratios (log-scale) or changes in protein copies/cell (also log-scale) for Hierarchical and K-means clustering employing the following default parameters: for hierarchical clustering, UPGMA was set as clustering methods, Euclidean distance was set as Similarity measure, Average value was set as Ordering function and calculate column dendrogram was enabled. For K-means clustering, data centroid based search was set as cluster initialization and Euclidean distance was set as similarity measure.
Functional annotation
We used the annotation tools DAVID (Huang et al, 2007) for functional annotation and GO and pathway enrichment analysis (using the KEGG database (Kanehisa et al, 2010)) of protein sets. The P-value threshold was set to 0.05. In case of multiple significant term/pathway enrichments for a given perturbation across multiple clusters, only the enrichment for the cluster having the term/pathway with the lowest P-value is displayed.
Protein changes within Operons
A list comprising all predicted operons of the L. interrogans genome were downloaded from http://www.microbesonline.org (Dehal et al, 2010). The list was matched with the quantitative data set generated using the Spotfire Decision Site program (version 9.1.1, TIBCO). Proteins belonging to the same operons were grouped and clustered according to their number of neighboring proteins.
Supplementary Material
Supplementary figures S1–13, Supplementary table SI
Acknowledgments
This project was funded in part by ETH Zurich, the Swiss National Science Foundation (Grant 31000-10767), federal funds from the National Heart, Lung and Blood Institute, the National Institutions of Health (contract no. N01-HV-28179), SystemsX.ch and the Swiss initiative for systems biology. RA is supported by the European Research Council (Grant # ERC-2008-AdG 233226). JM was supported by a fellowship from the Swedish Society of Medical Research (SSMF), MB was supported by a long-term fellowship of the European Molecular Biology Organization and a Marie Curie fellowship of the European Commission, AS were supported by the Competence Center for Systems Physiology and Metabolic Diseases.
Author contributions: AS, MB and JM conceived and designed the research. AS and MB performed the experiments, developed and performed the analysis, analyzed the data and wrote the manuscript. HL generated the spectral library. MC performed data analysis and proteome coverage predictions. DC generated the PeptideAtlas and performed the PTP prediction. RA was the project leader and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahrens CH, Brunner E, Basler K (2010) Quantitative proteomics: a central technology for systems biology. J Proteom 73: 820–827 [DOI] [PubMed] [Google Scholar]
- Akashi H, Gojobori T (2002) Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA 99: 3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JK, Barnett D, Bolin CA, Summers TA, Wagar EA, Cheville NF, Hartskeerl RA, Haake DA (1999) Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect Immun 67: 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Malmström JA, Lange V, Schmidt A, Deutsch EW, Aebersold R (2009) Visual proteomics of the human pathogen Leptospira interrogans. Nat Meth 6: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Selbach M, Rollenhagen C, Ballmaier M, Meyer TF, Mann M, Bumann D (2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440: 303–307 [DOI] [PubMed] [Google Scholar]
- Benders GA, Powell BC, Hutchison CA (2005) Transcriptional analysis of the conserved ftsZ gene cluster in Mycoplasma genitalium and Mycoplasma pneumoniae. J Bacteriol 187: 4542–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, De Lichtenberg U, Rinner O, Lee H, Pedrioli PGA, Malmstrom J, Koehler K, Schrimpf S, Krijgsveld J, Kregenow F, Heck AJR, Hafen E, Schlapbach R et al. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol 25: 576–583 [DOI] [PubMed] [Google Scholar]
- Brusniak M-Y, Bodenmiller B, Campbell D, Cooke K, Eddes J, Garbutt A, Lau H, Letarte S, Mueller LN, Sharma V, Vitek O, Zhang N, Aebersold R, Watts JD (2008) Corra: computational framework and tools for LC-MS discovery and targeted mass spectrometry-based proteomics. BMC Bioinformatics 9: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, Romesberg FE (2007) Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol 189: 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen M, Aebersold R, Buhmann JM (2009) Proteome coverage prediction for integrated proteomics datasets. J Comput Biol 18: 283–293 [DOI] [PubMed] [Google Scholar]
- de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP (2010) MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38: D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R (2006) The PeptideAtlas project. Nucleic Acids Res 34: D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Lam H, Aebersold R (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep 9: 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon B, Aebersold R (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol 28: 710–721 [DOI] [PubMed] [Google Scholar]
- Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE (2009) Global proteome analysis of Leptospira interrogans. J Proteome Res 8: 4564–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville NC, Bates D, Hastings PJ, Hanawalt PC, Rosenberg SM (2010) Role of RecA and the SOS response in thymineless death in Escherichia coli. PLoS Genet 6: e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A (2006) Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441: 173–178 [DOI] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1: 323–333 [DOI] [PubMed] [Google Scholar]
- Güell M, van Noort V, Yus E, Chen W-H, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kühner S, Rode M, Suyama M, Schmidt S, Gavin A-C, Bork P, Serrano L (2009) Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Walker EM, Blanco DR, Bolin CA, Miller MN, Lovett MA (1991) Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect Immun 59: 1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M, Ye M, Michel PE, Morier P, Stalder D, Jünger MA, Aebersold R, Reymond F, Rossier JS (2005) Added value for tandem mass spectrometry shotgun proteomics data validation through isoelectric focusing of peptides. J Proteome Res 4: 2273–2282 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y et al. (2007) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316: 593–597 [DOI] [PubMed] [Google Scholar]
- Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA (2008) Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics 7: 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125: 173–186 [DOI] [PubMed] [Google Scholar]
- Lam H, Deutsch E, Aebersold R (2009) Artificial decoy spectral libraries for false discovery rate estimation in spectral library searching in proteomics. J Proteome Res 9: 605–610 [DOI] [PubMed] [Google Scholar]
- Lam H, Deutsch EW, Eddes JS, Eng JK, Stein SE, Aebersold R (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat Meth 5: 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M, Murray GL, Khoo CA, Haake DA, Zuerner RL, Adler B (2010) Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect Immun 78: 4850–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López E, Elez M, Matic I, Blázquez J (2007) Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol Microbiol 64: 83–93 [DOI] [PubMed] [Google Scholar]
- Lux R, Moter A, Shi W (2000) Chemotaxis in pathogenic spirochetes: directed movement toward targeting tissues? J Mol Microbiol Biotechnol 2: 355–364 [PubMed] [Google Scholar]
- MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR (2004) Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics 3: 478–489 [DOI] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Lange O (2009) Performance evaluation of a high-field Orbitrap Mass Analyzer. J Am Soc Mass Spectrom 20: 1391–1396 [DOI] [PubMed] [Google Scholar]
- Mallick P, Kuster B (2010) Proteomics: a pragmatic perspective. Nat Biotechnol 28: 695–709 [DOI] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125–131 [DOI] [PubMed] [Google Scholar]
- Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B (2005) After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak M-Y, Vitek O, Aebersold R, Müller M (2007) SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7: 3470–3480 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA (2009) Mass spectrometry based targeted protein quantification: methods and applications. J Proteome Res 8: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarakul K, Lo M, Adler B (2010) Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol 10: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R, Domon B (2007) The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics 6: 1589–1598 [DOI] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller L, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics 8: 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S-X, Fu G, Jiang X-G, Zeng R, Miao Y-G, Xu H, Zhang Y-X, Xiong H, Lu G, Lu L-F, Jiang H-Q, Jia J, Tu Y-F, Jiang J-X, Gu W-Y, Zhang Y-Q, Cai Z, Sheng H-H, Yin H-F, Zhang Y et al. (2003) Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422: 888–893 [DOI] [PubMed] [Google Scholar]
- Rifai N, Gillette MA, Carr SA (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol 24: 971–983 [DOI] [PubMed] [Google Scholar]
- Ristow P, Bourhy P, da Cruz McBride FW, Figueira CP, Huerre M, Ave P, Girons IS, Ko AI, Picardeau M (2007) The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog 3: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiess R, Wollscheid B, Aebersold R (2009) Targeted proteomic strategy for clinical biomarker discovery. Mol Oncol 3: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Claassen M, Aebersold R (2009) Directed mass spectrometry: towards hypothesis-driven proteomics. Curr Opin Chem Biol 13: 510–517 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Gehlenborg N, Bodenmiller B, Mueller LN, Campbell D, Mueller M, Aebersold R, Domon B (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics 7: 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa M, Sekizuka T, Okutani A, Banno S, Sata T, Inoue S, Kuroda M (2010) Genomewide screening for novel genetic variations associated with ciprofloxacin resistance in Bacillus anthracis. Antimicrob Agents Chemother 54: 2787–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer V, Yamchuk A, Cortens J, Sousa S, Ens W, Standing KG, Wilkins JA, Krokhin OV (2007) Sequence-specific retention calculator. A family of peptide retention time prediction algorithms in reversed-phase HPLC: applicability to various chromatographic conditions and columns. Anal Chem 79: 8762–8768 [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics 6: 1809–1817 [DOI] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- Van La M, Barbry P, Raoult D, Renesto P (2007) Molecular basis of Tropheryma whipplei doxycycline susceptibility examined by transcriptional profiling. J Antimicrob Chemother 59: 370–377 [DOI] [PubMed] [Google Scholar]
- Vlasić I, Ivancić-Baće I, Imesek M, Mihaljević B, Brcić-Kostić K (2008) RecJ nuclease is required for SOS induction after introduction of a double-strand break in a RecA loading deficient recB mutant of Escherichia coli. Biochimie 90: 1347–1355 [DOI] [PubMed] [Google Scholar]
- Yates JR, Eng JK, McCormack AL, Schieltz D (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67: 1426–1436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1–13, Supplementary table SI