Over 700 direct transcriptional targets of the dFOXO transcription factor are identified in the adult fruit fly. dFOXO-bound genes are conserved between worm and fly, but dFOXO is not the sole mediator of the transcriptional response to changes in insulin signalling in the fly.
Keywords: dFOXO, Drosophila , insulin/insulin-like growth factor signalling, transcription
Abstract
FoxO transcription factors, inhibited by insulin/insulin-like growth factor signalling (IIS), are crucial players in numerous organismal processes including lifespan. Using genomic tools, we uncover over 700 direct dFOXO targets in adult female Drosophila. dFOXO is directly required for transcription of several IIS components and interacting pathways, such as TOR, in the wild-type fly. The genomic locations occupied by dFOXO in adults are different from those observed in larvae or cultured cells. These locations remain unchanged upon activation by stresses or reduced IIS, but the binding is increased and additional targets activated upon genetic reduction in IIS. We identify the part of the IIS transcriptional response directly controlled by dFOXO and the indirect effects and show that parts of the transcriptional response to IIS reduction do not require dfoxo. Promoter analyses revealed GATA and other forkhead factors as candidate mediators of the indirect and dfoxo-independent effects. We demonstrate genome-wide evolutionary conservation of dFOXO targets between the fly and the worm Caenorhabditis elegans, enriched for a second tier of regulators including the dHR96/daf-12 nuclear hormone receptor.
Introduction
The insulin/insulin-like growth factor (IGF) signalling (IIS) pathway, conserved throughout the animal kingdom, affects a variety of traits, including growth and development, metabolic homoeostasis, stress resistance, fecundity and adult lifespan (for review see Russell and Kahn, 2007; Piper et al, 2008). Forkhead Box-O (FoxO; note that we use FoxO to refer to all the members of the group) transcription factors (TFs) are regulated by IIS. Stimulation of IIS activates the Akt kinase, which in turn inactivates Foxo3A through phosphorylation resulting in nuclear exclusion (Brunet et al, 1999). Conversely, inactivation of IIS results in activation of FoxOs. FoxOs are also controlled by other signalling pathways, and have complex and important roles during animal development and adulthood. They are involved in metabolism, stress protection, cellular differentiation, cell-cycle arrest and apoptosis (for review see Greer and Brunet, 2008; Partridge and Bruning, 2008; Salih and Brunet, 2008). Recently, FoxOs have been shown to act as lineage-restricted tumour suppressors and to be important in stem cell maintenance in mice (Paik et al, 2007, 2009; Tothova et al, 2007).
Reduced IIS activity extends lifespan in distantly related model organisms such as the nematode worm Caenorhabditis elegans, the mouse Mus musculus and the fruit fly Drosophila melanogaster, at the same time delaying or diminishing age-associated functional decline (Kenyon et al, 1993; Clancy et al, 2001; Tatar et al, 2001; Wessells et al, 2004; Martin and Grotewiel, 2006; Selman et al, 2008). The molecular basis of this lifespan extension is currently under intense investigation. Work on C. elegans has established the critical role of FoxOs in lifespan. The single worm FoxO orthologue (daf-16) is essential for prolonged lifespan and other traits upon reduction in IIS (Kenyon et al, 1993), indicating that transcriptional reprogramming effected by DAF-16 is the basis of this enhanced longevity. Indeed, daf-16 is crucial for the transcriptional response to reduced IIS (Murphy et al, 2003). However, the requirement for dfoxo in the transcriptional or lifespan response to reduced IIS in Drosophila or other organisms has not been defined.
FoxOs have a role in lifespan beyond the IIS pathway: they are also required for lifespan extension achieved by manipulations of the Jun N-terminal kinase (JNK) pathway in flies (Wang et al, 2005) and of the Ste20-like kinase (MST) and AMP-activated protein kinase in worms (Lehtinen et al, 2006; Greer et al, 2007), and also for some forms of dietary restriction in the worm (Greer et al, 2007; Honjoh et al, 2009; Zhang et al, 2009). Furthermore, adult-onset and tissue-restricted over-expression of the single Drosophila FoxO orthologue (dfoxo) is sufficient to enhance longevity in the fly (Giannakou et al, 2004; Hwangbo et al, 2004). Further emphasising the pivotal and evolutionarily conserved role that FoxOs have in lifespan, genetic variation in the Foxo3A gene in humans is strongly associated with longevity (Kuningas et al, 2007; Willcox et al, 2008; Flachsbart et al, 2009). Thus, FoxOs are emerging as potentially important targets for intervention into ageing and ageing-related diseases of humans.
A crucial part of understanding the functioning of TFs, such as dFOXO, is determining their in vivo genome-wide binding locations and the specific transcriptional programmes they orchestrate from these locations. In the case of FoxOs, such information is only emerging. A number of genes are bound by DAF-16 in the worm, but <100 transcriptionally regulated direct targets are known (Oh et al, 2006; Schuster et al, 2010). In Drosophila, genome-wide dFOXO targets have been only examined in larvae during starvation (Teleman et al, 2008) and these may have only limited relevance to adult-specific traits such as ageing.
In this study, we use genomic approaches to discover >700 direct dFOXO targets in the adult female fly. We show that the dFOXO genomic binding locations do not change during stress or downregulation of IIS, but that different target genes are regulated in wild-type and IIS mutant flies. We define the part of the IIS response that requires the action of dFOXO directly as well the indirect effects. Surprisingly, we uncover a substantial portion of the IIS response that does not require dfoxo. In parallel to this study and corroborating our findings, Slack et al (2011) have shown that dfoxo is only required for a subset of physiological changes brought on by reduced IIS in the fly, unlike the situation in C. elegans where all known phenotypic outputs of reduced IIS require daf-16. Despite this difference in the architecture of the IIS response between the worm and the fly, we find conservation of FoxO-dependent transcriptional effects, and a significant genome-wide conservation of genes bound by dFOXO and DAF-16.
Results
dFOXO binds ∼1400 genomic locations in the adult female fly that are distinct from those bound in larvae or cultured cells
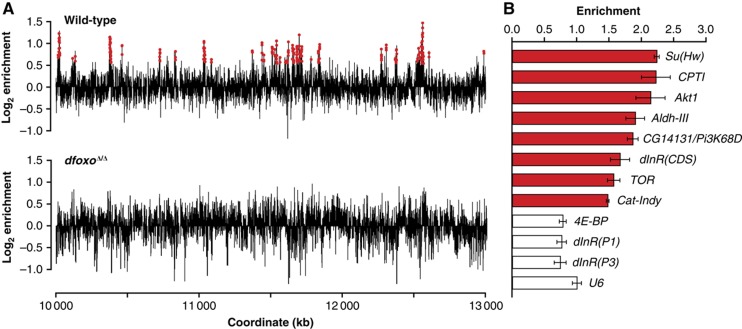
dfoxo has an important role in adult fly physiology, as evidenced by a substantial reduction in lifespan upon removal of dfoxo function (Giannakou et al, 2008; Min et al, 2008; Slack et al, 2011), a reduction that is also observed in loss-of-function mutants for the worm orthologue daf-16 (Larsen et al, 1995; Garigan et al, 2002). This prompted us to capture a snapshot of genomic locations bound by dFOXO in adult flies kept under normal conditions. We prepared chromatin from 7-day-old females and pulled-down dFOXO-associated DNA with an affinity-purified anti-dFOXO antibody (Giannakou et al, 2007). As a control, we performed a mock immunoprecipitation (IP) using the pre-immune serum. By hybridisation of the pulled-down DNA to genome-wide tiling arrays and determination of binding peaks (see Materials and methods), we identified 1423 dFOXO-bound genomic regions, averaging 908 bp in length. The sites bound by dFOXO tended to cluster together in a non-random manner: 78% of the peaks were within 10 kb of another, whereas one peak per 99 kb would be expected by chance. An example of the peaks identified is given in Figure 1A. The locations of the bound regions, as well as all other lists mentioned in the paper are given as Supplementary information. The binding was reproducible, as demonstrated by high concordance of the three biological replicates (Supplementary Figures 1 and 2; Supplementary Figure 2 shows Parson correlations of all ChIP-chip experiments performed). To validate the array data, we tested for enrichment of the bound regions by qPCR. Eight out of eight dFOXO-bound and three out of three non-bound regions were verified by qPCR (Figure 1B), indicating high reliability of the data set. To further establish the specificity of the antibody used, we performed ChIP-chip on dfoxoΔ/dfoxoΔ (dfoxoΔ/Δ) flies that completely lacked the dFOXO protein (Slack et al, 2011). None of the peaks identified in the wild type were present in the dfoxoΔ/Δ (for an example see Figure 1A), confirming that these genomic regions were specifically bound by dFOXO.
Figure 1.
Genome-wide dFOXO binding in whole adult flies. (A) ChIP-chip assays were carried out on 7-day-old females using anti-dFOXO antibody. ChIP-chip traces, showing the enrichment (log2-transformed) of the dFOXO-immunoprecipitated DNA over total chromatin, are averages of three biological repeats after subtraction of the mock (pre-immune serum) control and are shown over a 3-Mbp region of chromosome 2R in wild-type flies (top) or dfoxoΔ/Δ flies (bottom). Red dots denote the peaks identified in the ChIP-chip signal. Note that no peaks were identified in this region in the dfoxoΔ/Δ flies. (B) qPCR was used to confirm the enrichment observed in dFOXO ChIP-chip in the three biological repeats of the wild-type chromatin. Relative enrichment was calculated as proportion of chromatin recovered in the IP for a single region divided by average recovered for all regions for that chromatin, with U6 enrichment set to one. The data are presented as means with standard errors. Red indicates regions that were expected to be enriched, white indicates those that were not. Significant difference was detected by ANOVA (P<10−4, n=3), and t-test revealed that the regions indicated in red were significantly different from the others (P<0.05).
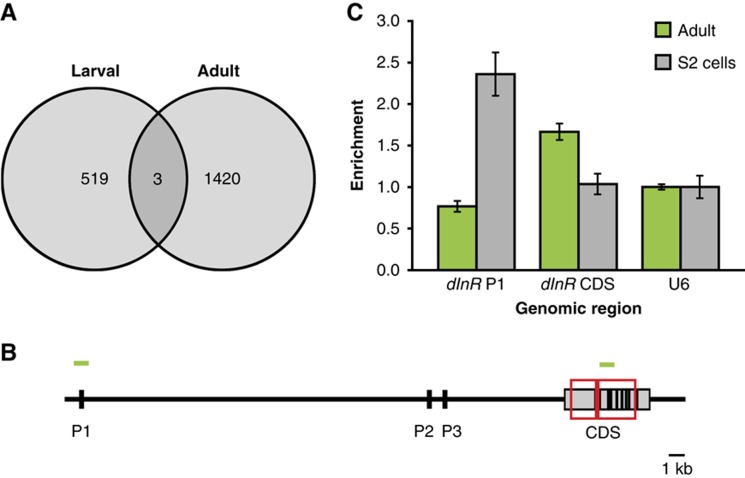
The sites bound by dFOXO in the adult fly were distinct from those previously described as occupied in larvae (Figure 2A) (Teleman et al, 2008), and the overlap was slightly less than expected by chance (overlap of nine peaks expected by chance, three observed, P=0.02). This revealed that dFOXO binding may be influenced by developmental stage and/or tissue composition of the animal. The sites bound were also distinct from those previously observed in cell culture. For example, ectopically expressed dFOXO was bound to the promoter of the Drosophila insulin receptor (dInR) gene in cultured cells (Puig et al, 2003; Puig and Tjian, 2005), whereas we found it bound to the coding region of the gene in adult females. To confirm that this difference was not due to different antibodies or different ChIP protocols used in our and previous studies, we examined the binding of endogenous dFOXO to DNA in S2 cells after 2 h serum starvation. We found that dFOXO was bound to the P1 promoter of the dInR gene in S2 cells, while it bound the coding region of the same gene in adult females (Figure 2B and C). Since the same antibody and the same IP conditions were used, this difference reflects a true difference in dFOXO binding in S2 cells and adults. Hence, the sites of dFOXO binding are dependent on cell type. Note that the binding within coding/transcribed regions was a general feature of dFOXO binding in adult female flies (Supplementary Figure 3).
Figure 2.
dFOXO-binding sites in adults are distinct from those in larvae or S2 cells. (A) Overlap between the genomic sites bound by dFOXO in larvae and adults. The data for larvae were generated by Teleman et al (2008). The observed overlap was slightly smaller than expected by chance (P=0.02). Expected overlap of nine peaks was determined from simulation of 103 random peak sets, of identical size, length and chromosomal distribution. (B) A schematic of the dInR locus is given with grey boxes representing exons, black marks the P1, P2 and P3 promoters (Casas-Tinto et al, 2007), red boxes the sites bound by dFOXO in adult flies (observed in ChIP-chip data) and green bars the location of amplicons (left—P1, right—coding region) used for ChIP-qPCR shown in (C). (C) dFOXO binding within the dInR locus in adults and S2 cells. The qPCR results show relative enrichment of the P1 promoter and the coding region (CDS) of dInR, or the U6 control, in three biological repeats of adult chromatin, or three IPs from a single chromatin sample from 2 h serum-starved S2 cells. The data are presented as in Figure 1B. ANOVA detected significant differences in enrichment (n=3, P<10−4), with P1 promoter being enriched in S2 cells and the coding region in adults (t-test, P<0.05).
To gain an insight into the DNA sequence recognised by dFOXO in adult females, we looked for statistical over-representation of known binding motifs in the DNA recovered from ChIP using Clover analysis (Frith et al, 2004). Several forkhead-like motifs containing the core FoxO-recognition sequence WWAACA (Biggs et al, 2001) were enriched, such as WWWRTAAASAWAA and WNTATAAACAWNNR (Table I), indicating that these are a good match to the motif recognised by dFOXO. We attempted to generate de novo the dFOXO motif present in the genomic DNA bound by dFOXO using MEME analysis. Unfortunately, MEME failed to identify a forkead-like motif but isolated variants of a CTGCTG sequence (Supplementary Table 1). This sequence is similar to the motif bound by ADF1 (England et al, 1990), the motif that was also identified as highly enriched in our ChIP-recovered sequences by Clover (Table I), indicating that ADF1, a Drosophila Myb-like transcriptional activator (Cutler et al, 1998), may share genomic sites with dFOXO.
Table 1. Representative enriched motifs identified by Clover.
| All sequences bound by dFOXO | |||
|---|---|---|---|
| Motif | TF | Raw score | P * |
| For the comprehensive lists please refer to Supplementary information. | |||
| aRelative to wild type. | |||
| bRelative to daGAL4>UAS-dInRDN. | |||
| *Relative to whole chromosome 2L. | |||
| **Relative to all sequences bound by dFOXO. | |||
| ***Relative to all promoter sequences of the genes present on the expression arrays. | |||
| VCGCYGCMGYCGCTGMCNGCG | ADF1 | 665 | <10−3 |
| WWWRTAAASAWAA | BRCZ4 | 659 | <10−3 |
| WNTATAAACAWNNR | XFD2 | 222 | <10−3 |
| dFOXO-bound and gene(s) downregulated in dfoxoΔ/Δ a | |||
| Motif | TF | Raw score | P ** |
| NNNGCCASCAGRKGGCRSNN | CTCF | 117 | <10−3 |
| TRTAAACAANWN | FOXO3A | 104 | 0.003 |
| dFOXO-bound and gene(s) upregulated in dfoxoΔ/Δ a | |||
| Motif | TF | Raw score | P ** |
| WWWRTAAASAWAA | BRCZ4 | 75.2 | <10−3 |
| TRTAAACAANWN | FOXO3A | 56.7 | <10−3 |
| dFOXO-bound and gene(s) downregulated in dfoxoΔ/Δ daGAL4>UAS-dInRDN b | |||
| Motif | TF | Raw score | P ** |
| NNNGCCASCAGRKGGCRSNN | CTCF | 271 | <10−3 |
| TRTAAACAANWN | FOXO3A | 208 | <10−3 |
| dFOXO-bound and gene(s) upregulated in dfoxoΔ/Δ daGAL4>UAS-dInRDN b | |||
| Motif | TF | Raw score | P ** |
| NWAAACAAN | FOXO1 | 63.5 | <10−3 |
| TRTAAACAANWN | FOXO3A | 57.8 | 0.002 |
| Promoters of genes downregulated in daGAL4>UAS-dInRDN indirectly dependent on dfoxo | |||
| Motif | TF | Raw score | P *** |
| NNWGATAASA | GATA2 | 80 | <10−3 |
| MNAGATAANR | GATA1 | 39.9 | <10−3 |
| Promoters of genes upregulated in daGAL4>UAS-dInRDN independent of dfoxo | |||
| Motif | TF | Raw score | P *** |
| NNNWAAAYAAAYANNNNN | FOXJ2 | 115 | 0.004 |
| WWWRTAAASAWAA | BRCZ4 | 110 | <10−3 |
| Promoters of genes downregulated in daGAL4>UAS-dInRDN independent of dfoxo | |||
| Motif | TF | Raw score | P *** |
| NNWGATAASA | GATA2 | 8.11 | <10−3 |
| NCWGATAACA | GATA1 | 4.77 | 0.003 |
dFOXO directly regulates 356 genes in the wild-type adult female
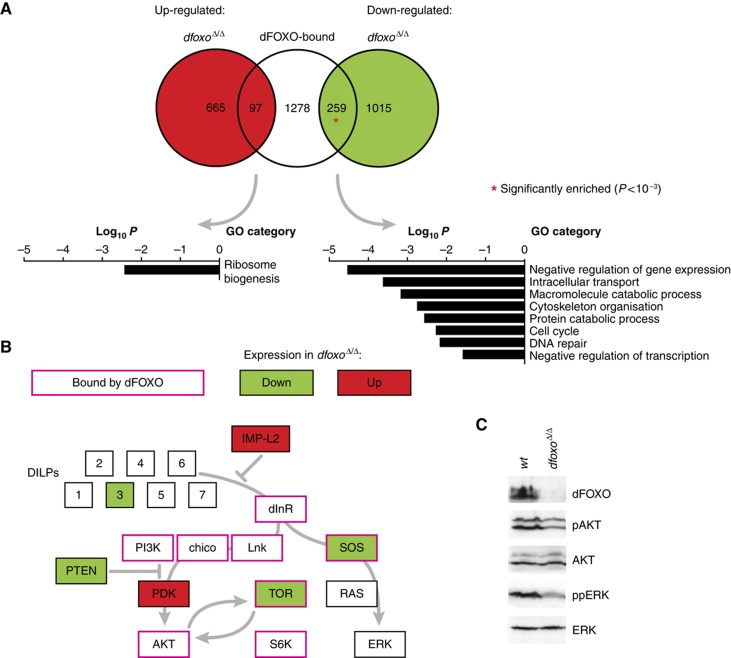
In all, 1755 unique genes were no further than 1 kb away from a dFOXO-bound site, defining a large set of potential dFOXO target genes. To identify which of these genes are direct dFOXO targets in the wild type, we identified the genes that require dfoxo for their normal expression in the adult female under standard conditions. A large portion of the transcriptome (2036 genes) was altered upon dfoxo removal, confirming the importance of this TF to adult physiology. Furthermore, there was a significant overlap (P=10−10) between the genes whose expression changed in dfoxoΔ/Δ and the set of putative dFOXO targets obtained from ChIP-chip, revealing a total of 356 direct dFOXO targets in the adult female (Figure 3A). The enrichment was specific to the subset of genes that were downregulated upon deletion of dfoxo (P=2 × 10−14), indicating that dFOXO tends to act as an activator of transcription, while also directly repressing some genes. The DNA sequences bound by dFOXO and associated with the 356 direct targets that were up/downregulated in dfoxoΔ/Δ flies were further enriched for forkhead-like motifs relative to all dFOXO-bound sites (Table I). Thus, a high density of binding motifs correlates with observable transcriptional control.
Figure 3.
Direct dFOXO targets in wild-type adult flies. (A) Overlap between the genes that neighbour a dFOXO-bound site and those with transcript levels altered in dfoxoΔ/Δ flies relative to wild type. For consistency with later experiments, both dfoxoΔ/Δ and wild-type flies also carried the daGAL4 driver. The probability of overlap was calculated based on hypergeometric distribution and an overlap significantly larger than expected by chance (P<10−3) is indicated with a red asterisk. Note that only the dFOXO-bound genes that were present on the expression arrays were taken into account. Representative biological functions enriched within the overlaps are shown. (B) dFOXO binding and regulation of IIS components. dFOXO binding and altered transcript levels in dfoxoΔ/Δ flies were mapped onto a schematic of IIS. Note that PI3K denotes the p110 subunit. (C) The levels of Serine 505-phosphorylated AKT (pAKT) and the dually phosphorylated ERK (ppERK) were measured in wild-type and dfoxoΔ/Δ females, as well as the levels of total AKT, ERK and dFOXO. dfoxoΔ/Δ females had 70% (±10%) of the wild-type pAKT/AKT ratio, and 40% (±3%) of wild-type ppERK/ERK. In both cases, the difference to wild type was significant (P<0.05, n=3, t-test).
Functional analysis of direct dFOXO targets (Figure 3A) revealed dFOXO to be an activator of genes involved in cell cycle, DNA repair, cytoskeletal organisation, intracellular transport and protein catabolism. dFOXO also directly repressed certain ribosome biogenesis genes. Interestingly, a significant number of genes involved in repression of gene expression, particularly at the level of transcription, were downregulated in the absence of dfoxo, including the insulator proteins su(Hw), CTCF (Bushey et al, 2008) and a member of a polycomb group protein complex—dSfmbt (Muller and Verrijzer, 2009), revealing that dFOXO might be important for establishment, demarcation and maintenance of repressive chromatin states. Interestingly, CTCF-recognised DNA motifs were enriched in the sequences bound by dFOXO and associated with loss of transcription in dfoxo nulls (Table I), indicating that CTCF may be important at sites of dFOXO-driven transcriptional activation.
Furthermore, we found that dFOXO directly regulated the expression of several important sequence-specific TFs, including Bigmax, Mio and dHR96, thus uncovering a substantial second tier of regulators. Bigmax and Mio are a pair of basic helix-loop-helix leucine zipper TFs that are the fly orthologues of the MondoA and Mlx TFs involved in regulating metabolism in mammals (Sans et al, 2006), while dHR96 encodes a nuclear hormone receptor regulating xenobiotic resistance in flies (King-Jones et al, 2006). Metabolic and detoxification genes were not significantly represented within the direct dFOXO targets, even though FoxOs have been implicated in the control of metabolic and detoxification processes (McElwee et al, 2003, 2007; Murphy et al, 2003; Matsumoto et al, 2007). In the fly, substantial control of these processes may be mediated via secondary effectors, such as Bigmax/Mio, which are directly repressed by dFOXO, and dHR96, which is directly activated by dFOXO.
Feedback regulation of dInR by dFOXO through transcriptional upregulation has been previously demonstrated in experiments with cultured Drosophila cells (Puig et al, 2003). This feedback onto the IIS pathway may be more extensive than previously thought, because dFOXO was also bound to the insulin-receptor substrates chico and Lnk, the Akt kinase and the Sos adaptor protein genes, as well as to components of the IIS-interacting TOR signalling pathway, S6K and TOR itself (Figure 3B). Importantly, dFOXO was directly required for the maintenance of TOR and Sos transcription in the adult female, since these genes were both bound by dFOXO and their mRNA decreased in the dfoxoΔ/Δ mutant. Interestingly, the transcriptional changes to IIS pathway components, including the upregulation of Imp-L2, an IGF-binding protein homologue and a negative regulator of IIS (Honegger et al, 2008; Alic et al, 2011), and a downregulation of dilp3, a Drosophila insulin-like peptide gene (Brogiolo et al, 2001) (Figure 3B), imply that dfoxoΔ/Δ flies may behave as mutants with reduced IIS activity with respect to the components upstream and/or parallel to dFOXO itself. At the same time, the observed changes in PTEN and PDK could partially compensate for this loss of IIS (Figure 3B).
To further investigate the effect of the direct regulation by dFOXO of TOR and Sos transcription, we determined the consequences of loss of dfoxo on the relevant signalling pathways. AKT is phosphorylated on S505 by the TOR kinase as part of the TOR complex 2 (Sarbassov et al, 2005), while SOS activity results in phosphorylation and activation of the ERK kinase (Biggs et al, 1994). In dfoxoΔ/Δ flies, levels of both S505-phosphorylated AKT and phosphorylated ERK were significantly reduced (Figure 3C), demonstrating that dFOXO-mediated regulation of signalling components has an effect on downstream signalling events. Note that a reduction in AKT S505 phosphorylation in a dfoxo mutant has also been observed by others (Shen and Tower, 2010).
dFOXO genomic locations are unaltered but binding is increased upon stress or IIS reduction
dfoxo is thought to be an important regulator of stress responses, with a well-documented role in resistance to oxidative stress and starvation (Junger et al, 2003; Puig and Tjian, 2005; Zheng et al, 2007; Teleman et al, 2008; Villa-Cuesta et al, 2010). These two assaults may pose different demands on fly physiology, and it might thus be expected that dFOXO would change its binding locations to regulate different groups of genes during these two different stresses.
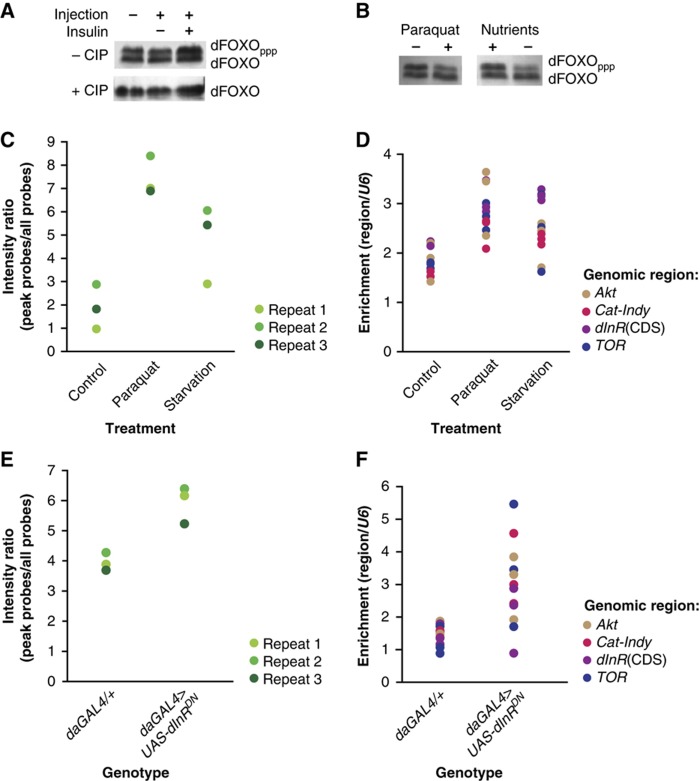
We determined conditions of paraquat (a superoxide generator) or starvation exposure that activated dFOXO, by examining its phosphorylation status. In cell extracts, AKT-phosphorylated, inactive dFOXO is retarded on SDS–PAGE (Puig et al, 2003). Two bands were also present in extracts from 7-day-old female flies, and the proportion of the slower migrating, phosphorylated dFOXO (dFOXOppp) was increased in flies injected with recombinant human insulin compared with mock-injected or uninjected controls (Figure 4A), consistent with AKT phosphorylation. The phosphorylation of the top band was confirmed with calf intestinal phosphatase (CIP) treatment. Treatment of flies with 20 mM paraquat in food (18 h) or starvation (48 h) resulted in an increase in the proportion of unphosphorylated dFOXO (Figure 4B), indicating its activation.
Figure 4.
dFOXO binding under stress conditions or on downregulation of IIS. (A) Phosphorylation of dFOXO upon insulin injection. In all, 7-day-old females were injected with recombinant human insulin, mock-injected or not injected, and frozen after 5 min. The proteins were extracted, some treated with CIP and separated by SDS–PAGE. The phosphorylated (dFOXOppp) and unphosphorylated dFOXO is indicated. (B) dFOXO phosphorylation after 18 h of 20 mM paraquat administration or after 48 h of starvation. (C) Increased genome-wide enrichment of dFOXO-bound regions upon stress. Three biological repeats of the ChIP-chip assay were performed with anti-dFOXO antibody on flies treated with paraquat, starved or untreated controls. The intensity ratios of the peak probes (bound by dFOXO) to all probes, each taken at 0.75 quantile, is shown and was significantly greater for all treatment replicates (Wilcox rank sum test, n=3, P=0.024). (D) Increased region-specific enrichment of dFOXO-bound regions upon stress. The IPs were repeated on the same chromatin samples and the enrichment relative to U6 of Akt, dInR, TOR and the region between the Cat and Indy genes was determined by qPCR. The effect of treatment was found to be significant (two-way ANOVA, n=3, effect of treatment P<10−4, effect of genomic region P=0.02, no significant interaction of the two main effects). The same genome-wide (E) or region-specific (F) analysis was performed on daGAL4>UAS-dInRDN flies or the driver alone control (daGAL4). This resulted in significant increase in the enrichment of dFOXO-bound regions, both on genome-wide scale (Wilcox rank sum test, n=3, P=0.05) and to the four target regions examined (two-way ANOVA on log-transformed data, n=3, effect of treatment P=7 × 10−4, no significant effect of genomic region).
ChIP-chip performed on paraquat-treated or starvation-exposed flies revealed that the substantial majority of binding locations remained the same as those in the untreated controls (Supplementary Table 2; Supplementary Figure 2), and visual inspection of the remaining sites indicated that they were actually present in untreated controls but below the peak-calling threshold. While there appeared to be essentially no change in the location of dFOXO, the ChIP-chip data indicated a general increase in the intensity of the dFOXO-bound peaks. Comparison of non-normalised array replicate data showed that the peak height (the ratio of the height of peak probes to background probes) was significantly higher in the treated samples than in the untreated controls (Figure 4C). This general trend was confirmed for four target regions by qPCR (Figure 4D). Hence, upon stress, more dFOXO localises to the same sites already occupied in the absence of stress.
We also determined whether dFOXO binds to different target sites when it is activated by a reduction in IIS, by performing ChIP-chip on flies with dampened IIS through ubiquitous expression of a dominant-negative form of dInR using the daugtherlessGAL4 driver (daGAL4). Importantly, daGAL4>UAS-dInRDN flies have an extended lifespan (Ikeya et al, 2009). This genetic intervention also resulted in increased binding to pre-existing sites on a genome-wide scale (Figure 4E), and this was confirmed for four specific regions by qPCR (Figure 4F). Thus, upon activation, dFOXO increases its occupancy on pre-existing sites.
dFOXO binding is enriched within the genes upregulated upon IIS reduction
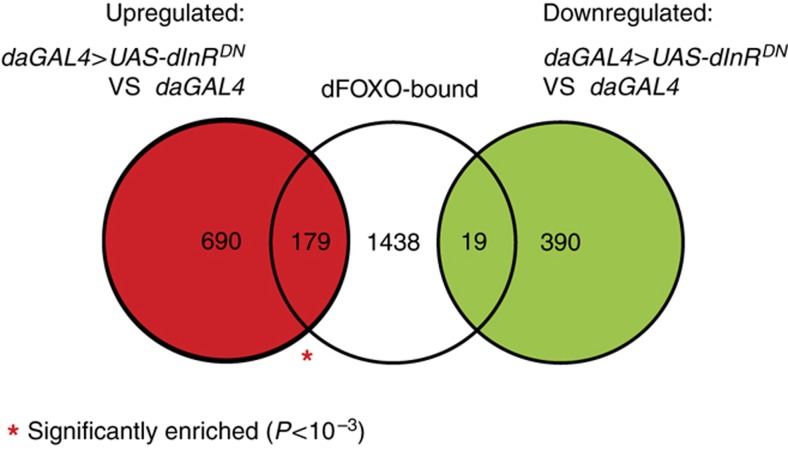
To further examine the relationship between the dFOXO regulon and IIS, we looked at the proportion of dFOXO-bound genes among the genes that are regulated by IIS (Figure 5). As a canonical model of IIS reduction, we generated and compared the expression profiles of daGAL4>UAS-dInRDN flies to their controls, the same genotypes that were used for ChIP-chip above. dFOXO-bound genes were enriched within the genes upregulated in the whole daGAL4>UAS-dInRDN flies (178 genes, P=3 × 10−11), confirming that dFOXO functions as an important transcriptional activator within the IIS response. Interestingly, out of the 198 genes bound by dFOXO and regulated in daGAL4>UAS-dInRDN flies, only 38 had mRNA levels detectably altered in dfoxoΔ/Δ. This overlap was significant (P<10−15 with respect to all genes on expression arrays) but was not complete. Hence, dFOXO may regulate different genes under different conditions even though its binding locations stay the same.
Figure 5.
Enrichment of dFOXO-bound genes within IIS transcriptional response. Overlaps between the genes regulated in whole daGAL4>UAS-dInRDN flies relative to driver only controls (daGAL4) and genes bound by dFOXO. A red asterisk denotes an overlap significantly larger than expected by chance (P<10−3), as computed from a hypergeometric distribution.
dFOXO is directly required for correct expression of 645 genes upon IIS reduction
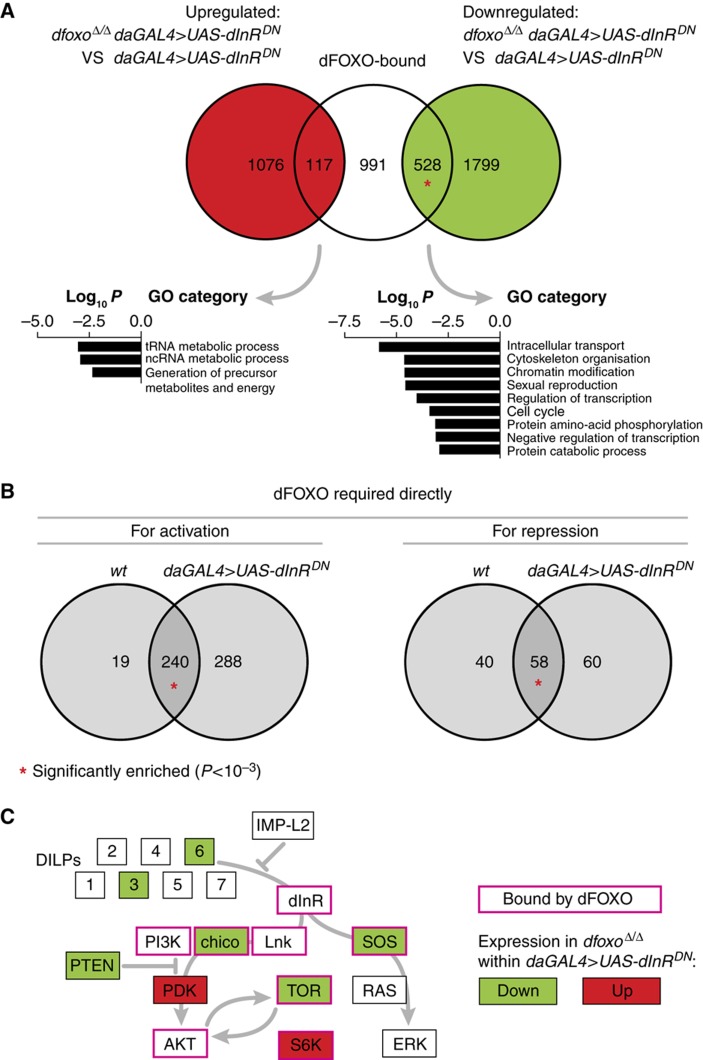
While dFOXO locations on the genome were unaltered upon reduction in IIS, it was possible that different dFOXO-bound genes required dfoxo for correct expression in an IIS mutant and in the wild-type fly. To determine direct targets of dFOXO in daGAL4>UAS-dInRDN flies, we compared the mRNA expression profiles of these flies in the presence or absence of dfoxo. A very large set of genes (3520) was misregulated upon deletion of dfoxo in this genetic context. There was a significant overlap between this set of genes and dFOXO-bound genes (P<10−15), revealing 645 direct targets of dFOXO (Figure 6A). Again this overlap was specifically significant for genes downregulated upon dfoxo deletion (P<10−15), confirming that dFOXO predominantly acts as a transcriptional activator within the IIS response. The dFOXO-bound sequences from which it was active in daGAL4>UAS-dInRDN flies were again enriched for forkhead-recognition motifs relative to all dFOXO-bound sequences (Table I), indicating that transcriptional activity correlates with a high density of binding motifs.
Figure 6.
Direct dFOXO targets in an IIS mutant. (A) Overlap between the genes that neighbour a dFOXO-bound site and those with transcript levels altered in dfoxoΔ/Δ daGAL4>UAS-dInRDN flies relative to daGAL4>UAS-dInRDN. Representative biological functions enriched within the overlaps are shown. (B) Comparison of direct dFOXO targets in wild-type and in daGAL4>UAS-dInRDN flies. The Venn diagram on the left compares genes that directly require dFOXO for activation of transcription in the two genetic contexts (i.e. genes bound by dFOXO and downregulated upon deletion of dfoxo), while the one on the right compares genes that directly require dFOXO for repression in the two genetic contexts (i.e. genes bound by dFOXO and upregulated upon deletion of dfoxo). In both (A, B), the probability of overlap was calculated based on hypergeometric distribution and an overlap significantly larger than expected by chance (P<10−3) is indicated with a red asterisk. Note that only the dFOXO-bound genes that were present on the expression arrays were taken into account. (C) dFOXO binding and regulation of IIS components. dFOXO binding and altered transcript levels in dfoxoΔ/Δ daGAL4>UAS-dInRDN flies were mapped onto a schematic of IIS. Note that PI3K denotes the p110 subunit.
The set of direct dFOXO targets in daGAL4>UAS-dInRDN flies contained the majority of the direct targets observed in the wild type (298 out of 357; Figure 6B) but was larger. This increase in the number of direct targets may have resulted from an increase in dFOXO binding leading to more sites passing a threshold of bound dFOXO required for transcriptional regulation. Alternatively, the regulation of dFOXO function in daGAL4>UAS-dInRDN flies might have occurred through processes independent of dFOXO binding, such as activation of cofactors. This increased number of direct dFOXO targets was also reflected in an increased feedback to IIS, including the TOR signalling pathway. For example, the chico and S6 kinase genes were revealed as direct targets of dFOXO in daGAL4>UAS-dInRDN flies (Figure 6C). Functions enriched within these direct dFOXO targets included cell cycle, catabolism, intracellular transport, cytoskeleton organisation, sexual reproduction and tRNA metabolism (Figure 6A). Interestingly, genes involved in protein phosphorylation as well as regulation of transcription were also over-represented, revealing again a potentially important second tier of regulators.
dFOXO is required for only a part of the transcriptional response to reduced IIS
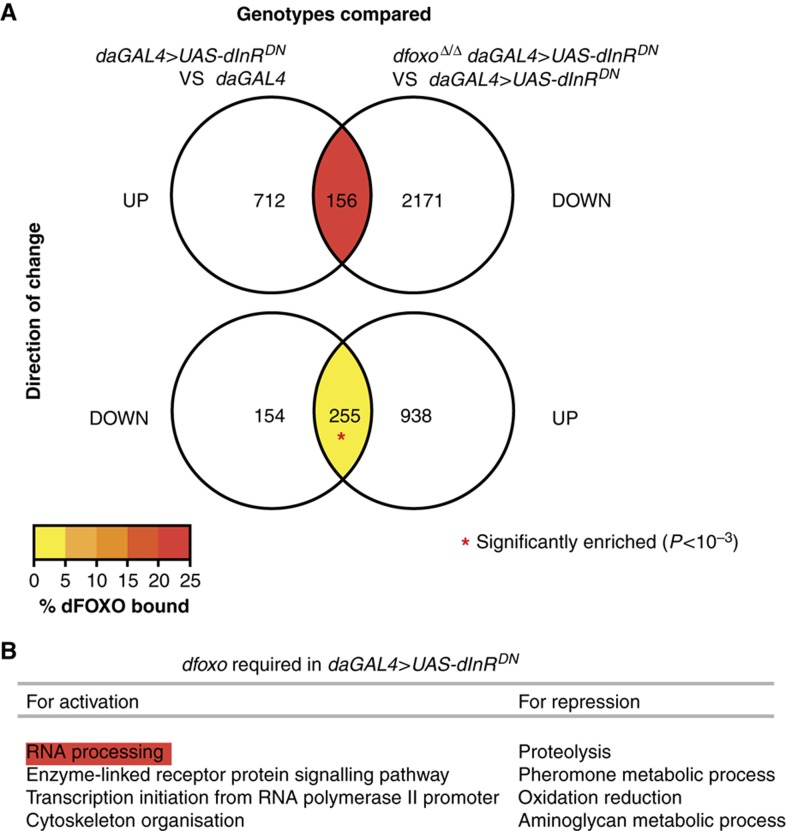
The direct targets of dFOXO in daGAL4>UAS-dInRDN flies identified above could require dFOXO for activation or repression in response to a reduction in IIS, or may require dFOXO for sustained basal level transcription during IIS reduction. To discern between these two possibilities, and uncover genes that require dfoxo for active transcriptional regulation upon reduction in IIS, we compared the genes whose transcripts were altered in daGAL4>UAS-dInRDN flies to those that were changed in the opposite direction upon removal of dfoxo in the background of daGAL4>UAS-dInRDN (Figure 7A). Interestingly, we found a highly significant overlap between genes repressed in daGAL4>UAS-dInRDN and those activated upon removal of dfoxo in that genetic context (P<10−15), while the overlap between genes activated in daGAL4>UAS-dInRDN and repressed upon dfoxo deletion was marginally significant (P=0.01). Hence, dfoxo is mainly required for repression of genes during IIS reduction. On the other hand, the comparison of dFOXO ChIP-chip data with expression in dfoxoΔ/Δ and dfoxoΔ/Δ daGAL4>UAS-dInRDN flies (Figures 3 and 6) showed that dFOXO tends to act as an activator of transcription. Mapping the ChIP-chip data onto the expression data overlaps (Figure 7A) revealed that the genes requiring dfoxo for activation during IIS reduction were enriched for direct dFOXO targets, while those requiring dfoxo for repression tended to be indirect targets.
Figure 7.
dfoxo-dependent part of the transcriptional response to altered IIS. (A) Sets of genes that are transcriptionally upregulated in response to over-expression of dInRDN (daGAL4>UAS-dInRDN vs daGAL4; left) or downregulated upon deletion of dfoxo in a fly over-expressing dInRDN (dfoxoΔ/Δ daGAL4>UAS-dInRDN vs daGAL4>UAS-dInRDN; right) were compared in the Venn diagram above. The overlap is composed of the genes that require dfoxo for upregulation in daGAL4>UAS-dInRDN flies. The Venn diagram below shows the reciprocal situation, and the overlap is composed of the genes that require dfoxo for downregulation in daGAL4>UAS-dInRDN flies. The probability of overlap was calculated based on hypergeometric distribution and an overlap significantly larger than expected by chance (P<10−3) is indicated with a red asterisk. The proportion of the genes in the overlaps that are direct dFOXO targets (bound by dFOXO) is indicated with colour. (B) Non-redundant functional categories that are enriched (P<0.05) in the genes within the overlaps in (A). Those that are enriched within the direct dFOXO targets within the same overlaps are highlighted in red. Note that ‘proteolysis’ included predominantly extracellular proteases.
The functional categories enriched within the set of genes that require dfoxo for upregulation in daGAL4>UAS-dInRDN flies included RNA processing, signal transduction, transcription and cytoskeleton organisation (Figure 7B). Genes involved in RNA processing were directly upregulated by dFOXO in daGAL4>UAS-dInRDN flies. On the other hand, none of the predominantly metabolic functions downregulated in daGAL4>UAS-dInRDN flies in a dfoxo-dependent manner were directly regulated by dFOXO.
To uncover a potential mechanism whereby dFOXO indirectly regulates gene repression in an IIS mutant, we searched for TF-binding motifs over-represented in the promoters of genes that require dfoxo for repression in daGAL4>UAS-dInRDN flies, but are not directly bound by dFOXO. Clover analysis identified several GATA-like motifs (Table I). Indeed, dFOXO directly activates transcription of GATAd in both the wild-type and daGAL4>UAS-dInRDN flies, and this TF may in turn be required for gene repression in daGAL4>UAS-dInRDN flies. Rigorous demonstration of GATAd as a mediator of dFOXO actions awaits further study.
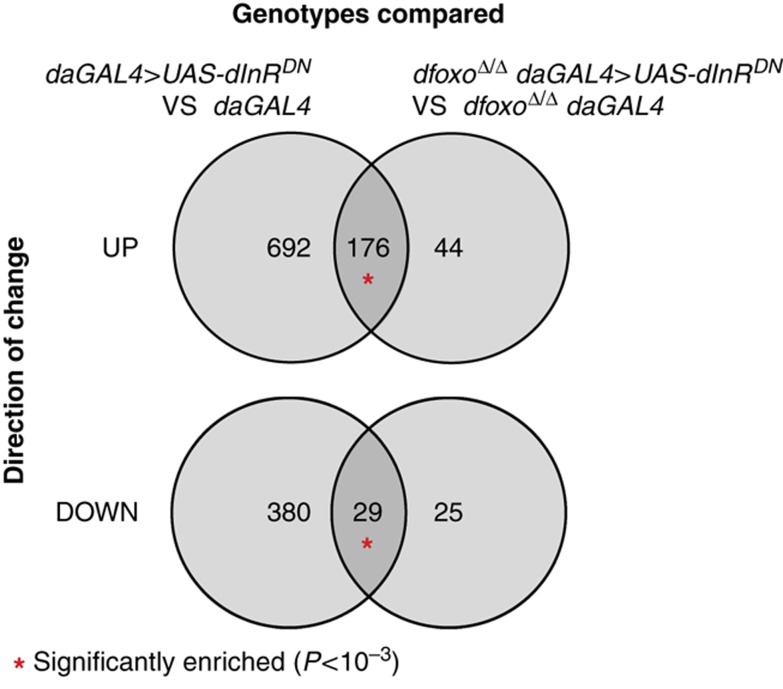
Interestingly, a substantial number of genes changed in daGAL4>UAS-dInRDN flies were not altered upon deletion of dfoxo (Figure 8). Hence, dfoxo appears to be required only for a part of the IIS response in flies. To confirm this surprising finding, we looked at what happens when we induce dInRDN in a dfoxoΔ/Δ background. We compared the transcriptome response to the induction of dInRDN in a dfoxoΔ/Δ background to the response observed in the presence of dfoxo. We found that 176 genes upregulated in daGAL4>UAS-dInRDN were still upregulated in dfoxoΔ/Δ daGAL4>UAS-dInRDN, while 29 genes downregulated in daGAL4>UAS-dInRDN were still downregulated in the absence of dfoxo. This directly demonstrates that a substantial portion of the IIS response, at least 16% of the detectable changes, is independent of dfoxo in adult Drosophila.
Figure 8.
dfoxo-independent part of the transcriptional response to altered IIS. Sets of genes that are transcriptionally upregulated in response to over-expression of dInRDN in the wild-type fly (daGAL4>UAS-dInRDN vs daGAL4; left) or upregulated upon over-expression of dInRDN in an dfoxo null fly (dfoxoΔ/Δ daGAL4>UAS-dInRDN vs dfoxoΔ/Δ daGAL4; right) were compared in the Venn diagram above. The overlap is composed of the genes that do not require dfoxo for upregulation in daGAL4>UAS-dInRDN flies. The Venn diagram below shows the reciprocal situation, and the overlap is comprised of genes that do not require dfoxo for downregulation in daGAL4>UAS-dInRDN flies. The probability of overlap was calculated based on hypergeometric distribution and an overlap significantly larger than expected by chance (P<10−3) is indicated with a red asterisk.
To identify the TFs that may mediate this dfoxo-independent aspect of the IIS response, we looked at over-representation of known TF-binding motifs in the promoters of the 176 genes upregulated and the 29 downregulated upon induction of dInRDN irrespective of the absence of dfoxo. Numerous forkhead-like motifs were associated with the upregulated genes (Table I), indicating that another forkhead factor mediates the dfoxo-independent transcriptional activation in an IIS mutant. The promoters of the downregulated genes were enriched for GATA-like motifs, indicating a GATA factor, but probably not GATAd, is required for gene repression in an IIS mutant, but further studies will be needed to directly demonstrate this mechanism.
121 dFOXO-bound genes are also bound by DAF-16 in the worm
Several physiological roles of FoxOs, as well as of IIS, are conserved across distantly related animals. However, examination of transcriptional changes in worm, fly and mouse IIS mutants failed to identify any significant co-regulation of orthologous genes in the three organisms (McElwee et al, 2007). We realised that the regulatory architecture of the transcriptional response in the worm and the fly is different since in the fly the IIS response only partially requires dfoxo. This prompted us to re-examine the conservation of the transcriptional response between the worm and the fly.
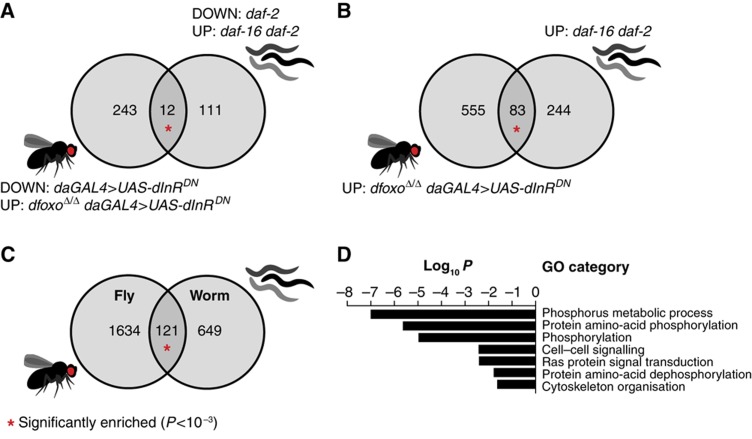
Transcriptional response to IIS changes in the worm has been examined from the perspective of daf-2/daf-16 epistasis. Since we have now performed the equivalent dInR/dfoxo epistasis experiments for the fly, we compared our data set to the ones already published for the worm (McElwee et al, 2003, 2007; Murphy et al, 2003), making sure that equivalent gene sets were being compared. We identified significant conservation of the genes that require dfoxo for downregulation between the worm and the fly. The comparisons that produced statistically significant overlaps are shown in Figure 9A and B. Hence, there is actually evolutionary conservation of dfoxo-dependent aspects of the IIS transcriptional response.
Figure 9.
dFOXO-bound genes are conserved between the fly and the worm. (A) Overlap between the fly orthologues of the genes that are downregulated by reduced function of daf-2 and upregulated by reduced function of daf-16 in the worm (Murphy et al, 2003), on the one hand, and those downregulated in daGAL4>UAS-dInRDN and upregulated in dfoxoΔ/Δ daGAL4>UAS-dInRDN in the fly, on the other (hypergeometric distribution, P=7 × 10−6). (B) Overlap between the fly orthologues of the genes that are upregulated by reduced function of daf-16 in a daf-2 background in the worm (McElwee et al, 2007), on the one hand, and upregulated in dfoxoΔ/Δ daGAL4>UAS-dInRDN in the fly, on the other (hypergeometric distribution, P<10−15). (C) Overlap between the genes bound by dFOXO in the fly and the fly orthologues of the genes bound by DAF-16 in the worm (Oh et al, 2006; Schuster et al, 2010). (D) Representative functional categories enriched within the overlap shown in (C).
Even though the set of genes requiring dfoxo for repression in the fly is comprised predominantly of indirect dFOXO targets, the evolutionary conservation within this set strongly suggested that there would be an underlying conservation at the level of direct dFOXO targets. Hence, we examined if evolutionary conservation could be observed at the level of dFOXO binding between the worm and the fly. We compared the set of genes that are bound by dFOXO in the adult fly with the composite set of DAF-16-bound genes identified by ChIP-cloning (Oh et al, 2006) or Dam-ID (Schuster et al, 2010). Strikingly, we found that there was a significant tendency for orthologous genes to be bound by dFOXO in the fly and DAF-16 in the worm (Figure 9C; P<10−15), indicating significant conservation of dFOXO-bound genes between the two animals. Out of the 121 genes present in the overlap, 44 were disregulated upon dfoxo deletion in the wild-type or daGAL4>UAS-dInRDN flies. The overlap was significantly enriched for genes acting in signal transduction (Figure 9D), including Sos, Akt and PP2A-B′. Hence, direct regulation of signalling components is an evolutionarily conserved role of FoxOs. This overlap not only included conservation of signalling feedback loops, but also of control of other signalling pathways (e.g. CaMKII) and extended to several TFs. For example, FoxOs may link steroid hormone signalling to IIS in both flies and worms through regulation of the dHR96/daf-12 TF.
Discussion
Using ChIP-chip we have defined >1400 genomic locations occupied by dFOXO in the adult fly. Interestingly, we find these locations to be distinct from those observed by others in larvae (Teleman et al, 2008) and in cell culture (Puig et al, 2003). It is possible that the differences between our adult data and the published larval data stem from differences in protocols (e.g. the antibody used) or even experimental design (e.g. sex of the flies used). Importantly, however, we show that the observed differences between S2 cells and adults, in the case of the promoter (P1) and the coding region of the Drosophila InR, represent true biological differences. It is not surprising that dFOXO would occupy different locations during development and in the adult fly. A similar observation has been made for a number of transcriptional events, and even the dInR gene alone is transcribed from three promoters under tight spatio-temporal control (Casas-Tinto et al, 2007). Furthermore, some differences will stem from cell- and tissue-specificity of dFOXO action. Indeed, FoxO factors are known to elicit tissue-specific transcriptional changes in the mouse (Paik et al, 2007; Tothova et al, 2007), and the same tissue-restricted action by dFOXO on the transcription of the myc gene has been observed in Drosophila larvae (Teleman et al, 2008). By binding to different locations in a spatially and temporally determined manner, dFOXO would be able to orchestrate different responses to suit its function in different life stages and tissues. Interestingly, we find a substantial portion of dFOXO bound in transcribed regions. In yeast, forkhead factors regulate Pol II elongation (Morillon et al, 2003), and dFOXO may perform a similar function.
We observe dFOXO bound to a number of genes encoding IIS signalling components. Furthermore, dfoxo may also exert feedback onto other pathways that regulate it: dFOXO was bound near the genes encoding PP2A-B′, 14-3-3ε and JNKKKs (slpr and TAK1), among others. PP2A, 14-3-3ε and JNK have all been shown to regulate FoxO activity (Wang et al, 2005; Nielsen et al, 2008; Yan et al, 2008). A number of these dFOXO-activated genes is also activated on over-expression of superoxide dismutase (Curtis et al, 2007), suggesting that dFOXO, like its mammalian counterparts (Nemoto and Finkel, 2002; Dansen et al, 2009), may be redox regulated. Interestingly, as is evident from Figures 3B and 6C, we detect binding to only the intracellular components of IIS such as chico, Lnk and Akt, while the genes with altered expression level in dfoxoΔ/Δ include extracellular cell-to-cell signalling molecules, such as those encoded by dilp3, dilp6 and Imp-L2. The latter genes have a more localised expression pattern, for example dilp3 is expressed in only ∼14 cells in the whole adult fly (Broughton et al, 2005). It is possible that genes such as dilp3 are also bound and directly regulated by dFOXO but that we did not observe this in the whole fly ChIP-chip due to a very small number of cells in which this binding occurs.
4E-BP (a.k.a. Thor) has been shown to be bound by dFOXO in larvae (Teleman et al, 2008) and cell culture (Puig et al, 2003), and its regulation has been reported as consistent with dFOXO acting as a direct activator of its expression (Junger et al, 2003; Puig et al, 2003). On the other hand, we do not observe dFOXO binding in the vicinity of this gene in adults (see Figure 1B), and the 4E-BP transcript is actually elevated in a dfoxo null. It is possible that dFOXO is required for direct activation of this gene in only a limited number of cells/tissues in the adult, thus escaping detection by ChIP-chip on whole animals. Furthermore, the role of dFOXO in 4E-BP regulation may be sexually dimorphic, as has recently been indicated (Shen and Tower, 2010). Alternatively, 4E-BP might be a target of a different forkhead factor in the adult female fly. Indeed, Forkhead (Fkh, the fly FoxA orthologue) is able to activate transcription of 4E-BP in larvae (Bulow et al, 2010). Since dfoxo nulls have reduced levels of TOR, and TOR is an inhibitor of Fkh activity (Bulow et al, 2010), it is likely that Fkh is activated in dfoxo nulls leading to increased levels of the 4E-BP transcript. It remains to be established whether Fkh might indeed be directly binding to the 4E-BP locus in adult flies.
From the 1400 dFOXO-bound locations, using transcriptional profiling of dfoxo null flies under normal conditions or with reduced IIS, we define >700 direct transcriptional targets of dFOXO in the adult. Several functions associated with these genes have been linked with FoxO biology previously, such as cell cycle (Medema et al, 2000), DNA repair (Tran et al, 2002), cytoskeleton organisation (Kamei et al, 2004), negative regulation of gene expression such as translation (Puig et al, 2003; Teleman et al, 2008) and regulation of protein catabolism (Stitt et al, 2004). dFOXO is known to be involved in the repression of protein synthetic machinery via myc in larvae (Teleman et al, 2008) but our study also revealed a significant regulation of ribosome biogenesis genes effected directly by dFOXO in the adult female. We also identified other, previously unknown functions, such as control of negative regulators of transcription and chromatin modifiers, hinting at the importance of dFOXO in establishment and maintenance of repressive chromatin states. Yet other functions were completely unexpected. For example, dFOXO appears as a positive regulator of sexual reproduction, including oogenesis, in an IIS mutant (see Figure 6C). This surprising finding is backed up by phenotypic epistasis analysis that shows removal of dfoxo to exacerbate the fecundity defect of several IIS mutants (Slack et al, 2011). Hence, dFOXO actually positively regulates some aspects of IIS. Indeed, one of the most surprising findings of our study is that dFOXO is directly required for expression of several components of IIS and interacting pathways, including TOR and Sos, in the wild-type fly, with consequences for the downstream signalling events. Importantly, this is not just simple feedback in response to alteration in the levels of insulin/IGF-like signal, but rather dFOXO is active in the normal adult and its activity promotes signalling through the IIS pathway. This observation can also explain why dfoxo deletion is lethal in combination with certain IIS mutants (Slack et al, 2011), since the combined reduction in IIS will be too great for the flies to survive. This potentiation of IIS by FoxOs could also explain why mice with reduced IIS through mutation of IRS1 have mild insulin resistance but preserved old-age glucose homoeostasis (Selman et al, 2008). In this case, the mild insulin resistance would be the primary effect of the mutation of IRS1, while the resulting activation of FoxOs would be responsible for sustained IIS in old age and thus for the observed preservation of glucose homoeostasis.
dFOXO directly regulates an extensive second tier of regulators; throughout this study we have repeatedly encountered different transcriptional and post-transcriptional regulators as predominant dFOXO targets. This aspect of dFOXO biology is also conserved in the worm (Schuster et al, 2010). Indeed, some of the potential secondary effectors are directly conserved between the worm and the fly, such as the nuclear hormone receptor dHR96/daf-12, highlighting their importance. Our study also illustrates the role this second tier of regulators may play. dFOXO is directly required for the maintenance of GATAd mRNA levels in both the wild-type and IIS-compromised flies, and this in effect may constitute an IIS feed-forward loop, since GATAd in turn may be an important transcriptional repressor in response to reduced IIS. Hopefully, subsequent studies will demonstrate the existence of such a feed-forward loop.
Since daf-16 is strictly required for all phenotypic outputs of reduced IIS in the worm (Kenyon et al, 1993; Gems et al, 1998), and also appears strictly required for the transcriptional response to reduced IIS (Murphy et al, 2003), it was very surprising to find that dFOXO was only required for part of the transcriptional response to reduced IIS in the fly. On the other hand, this is in accordance with phenotypic epistasis experiments in the fly where lifespan extension and xenobiotic resistance are dependent on dfoxo, while lowered fecundity and body size, delayed development and resistance to paraquat are not (Slack et al, 2011). This implies that phenotypes such as fecundity are negatively regulated via other factors in the fly. Our study indicates that GATA factors are the most likely candidates for mediating transcriptional repression in response to reduced IIS. Studies in the worm have also revealed the presence of a GATA-recognition sequence in the promoters of IIS-regulated genes (Murphy et al, 2003; Budovskaya et al, 2008). Furthermore, at least one of the 14 worm GATA TF (elt-3) is regulated by IIS, and reduced function in any of the three GATA TFs (elt-3, egr-1, egl-27) blocks the lifespan extension by a daf-2 mutant (Budovskaya et al, 2008). The role of GATA factors in lifespan in other organisms awaits examination. At the same time, our study reveals the potential involvement of other forkhead factors, besides dFOXO, in the transcriptional activation response to IIS reduction. Fkh is the prime suspect, since it is regulated by TOR signalling in the fly (Bulow et al, 2010), and Foxa2 is involved in the IIS response in mammals (Wolfrum et al, 2003). Indeed, Foxa2 is directly inactivated by Akt via phosphorylation of a single site that is conserved in the fly Fkh (Wolfrum et al, 2003). While our study provides hints, further work will be needed to determine the identity of other TFs involved in the fly IIS response.
Our study reveals that the transcriptional response to IIS in the fly is clearly more complex than that in the worm. The parallel genetic study performed by Slack et al (2011) shows that the genes directly regulated by dFOXO must still effect the lifespan extension by reduction in IIS. Importantly, we have now identified these genes. Their characterisation is the next step towards understanding the physiological and molecular changes that can extend animal lifespan, keeping in mind that it is now crucial to determine the architecture of the mammalian response to reduced IIS.
Materials and methods
Fly handling
For experiments on wild-type flies, the Dahomey stock (Clancy et al, 2001) was used. daugterlessGAL4 (Bloomington Stock Center), UAS-dInRDN (Wu et al, 2005) and dfoxoΔ94 (Slack et al, 2011) were backcrossed at least six times into Dahomey background carrying w1118 mutation (Giannakou et al, 2004), and which was Wolbachia positive. All experiments were performed at 25°C, 12-h light/dark cycle and controlled humidity. Flies were reared at standard density on SYA food (5% sucrose, 10% yeast, 1.5% agar) and females were sorted on day 3 of adulthood. For chromatin preparation, flies were kept at 200 females per bottle, 10 per vial for all other experiments. For starvation, flies were kept on 1% agar for 48 h, and for paraquat treatment for 18 h on food containing 1% agar, 5% sucrose, 20 mM paraquat, starting on day 5, and immediately frozen in liquid nitrogen. In all other cases, the flies were frozen on day 7. For insulin injections, 20 7-day-old females were gassed with carbon dioxide, injected with 50 nl of PBS with 0.1 μg/ml blue food dye (FD&C Blue Dye no. 1) with or without insulin (10 IU/ml, Actrapid, Novo Nordisk), allowed to wake up for 5 min and frozen.
Chromatin preparation, IP, array hybridisation and qPCR
Biological triplicates were done for all fly chromatin preparations. For each experiment, all the batching was done so that the treatments to be compared were carried out in parallel. The ChIP protocol as described by Kuras and Struhl (1999) was adapted for adult Drosophila. In all, 1000 females were crushed to a fine powder under liquid nitrogen and re-suspended in 6 ml of PBS supplemented with Protease Inhibitors Cocktail (10 μl/ml; Sigma). The flies over-expressing dInRDN were smaller than their controls so that they were re-suspended in 4 ml PBS to maintain the fly weight/buffer volume ratio. Cross-linking was performed with 0.5% formaldehyde for 10 min and quenched with addition of 1.5 ml of 2.5 M glycine. The cross-linked chromatin was recovered by centrifugation and washed twice with FA/SDS buffer (50 mM Hepes-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Na deoxycholate, 0.1% SDS, 1% Triton X-100 and 1 mM PMSF) re-suspended in the same and incubated for 1 h at 4°C. Chromatin was again recovered by centrifugation and sheared to an average size of 400 bp by sonication, giving on average 6 ml of chromatin in FA/SDS. For IPs, 1 μl of affinity-purified anti-dFOXO antibody (Giannakou et al, 2007), or 1 μl of the corresponding pre-immune serum (mock control) were bound to Protein-G Dynabeads (Invitrogen) and incubated for 2 h at room temperature with 450 μl of chromatin. Beads were washed once with FA/SDS, three times with FA/SDS containing 500 mM NaCl, once with TE and once with 10 mM Tris–HCl pH 8, 250 mM LiCl, 1 mM EDTA, 1% NP40 and 0.5% Na deoxycholate. DNA was recovered, treated with protease, de-cross-linked, treated with RNase and purified with Qiagen PCR purification kit (Qiagen, UK).
For array hybridisation, the entire IP after volume reduction, or 50 ng of total chromatin DNA, were amplified two times (Whole Genome Amplification kit, Sigma) as per the manufacturer's instructions. The material from the IP was hybridised against the input material. The labelling and hybridisations were carried out by Nimblegen Systems, using custom Drosophila whole-genome tiling arrays with probes spaced approximately every 300 bp, as described (Choksi et al, 2006).
Chromatin was prepared from S2 cells based on a published method (Andrulis et al, 2000; Puig et al, 2003). In all, 10 ml of 5 × 106 cells/ml were incubated in Schneider's medium without serum for 2 h at 25°C after which formaldehyde was added to 0.1% and quenched 3 min later with 0.5 ml of 2.5 M glycine for 3 min. The cells were collected by centrifugation and taken up in 2 ml of FA/SDS supplemented with PMSF. The chromatin was washed, sheared by sonication and the IPs performed as for whole flies above.
For qPCR, a suitable dilution of total chromatin and IP was used for quantification with primer pairs indicated, using Power SYBR Green PCR Master Mix (ABI) on ABI Prism 7000. Unless otherwise noted, the value reported is the percentage of the total chromatin recovered in the IP for the target sequence divided by the same for the U6 control. The primers used are given in Supplementary information.
Peak identification and analysis
ChIP-chip data were normalised using the LIMMA package (Smyth and Speed, 2003) in Bioconductor (Gentleman et al, 2004), applying loess normalisation within each array and quantile normalisation between arrays. Replicate information was pooled by taking the median probe value for each set of arrays and was smoothed along each chromosome using a running median within a window of three probes. Experimental signal was adjusted by mock control (pre-immune serum) data by direct subtraction of median probe intensity values. Peaks were called using the Ringo package (Toedling et al, 2007) in R, using a y0 threshold of 0.97 and a distance cutoff of 600 bp. Peaks were padded with 1000 bp upstream and downstream of the outermost peak probe position and genes were considered associated with the peak where any part of a gene taken from the FlyBase release 4.3 gene set (Drysdale and Crosby, 2005) overlapped with this region. When required, the observed peak set was compared with simulations of 1000 random peak sets, of identical size, length and chromosomal distribution.
RNA isolation, expression array hybridisation and analysis
RNA was extracted using Trizol (Invitrogen) from four biological repeats of 10 females of the following genotypes: daGAL4, dfoxoΔ/Δ daGAL4, daGAL4>UAS-dInRDN, dfoxoΔ/Δ daGAL4>UAS-dInRDN. All the batching was done so that the treatments to be compared were carried out in parallel. RNA was purified with RNeasy columns (Qiagen) and its quality and concentration were determined using an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA). The RNA was further processed into cRNA using standard Affymetrix protocols and hybridised to the Affymetrix Drosophila Genome 2.0 Genechip. The data were summarised and normalised using RMA (Bolstad et al, 2003; Irizarry et al, 2003a, 2003b) and quantile normalisation as implemented in the LIMMA package. Differential expression between different samples was assessed using linear models and the empirical Bayes moderated t-statistic implemented in LIMMA. Highly differentially expressed genes were selected by applying an adjusted P-value <0.005 cutoff.
DNA motif identification, EASE analysis and comparison to C. elegans data sets
Identification of known DNA motifs with a statistical over-representation was done using the Clover program (Frith et al, 2004) and the TransFac database (Matys et al, 2003) for input motifs. De novo identification of motifs from peak sequences was conducted with MEME (Bailey et al, 2006) on regions 500 bp padded from the most intense probe in the peak and repeat-masked.
Gene function over-representation analysis within gene sets was conducted using EASE in DAVID v6.7 online (Dennis et al, 2003; Huang da et al, 2009).
For comparison with C. elegans data sets, the two colour array data from McElwee et al (2003) were retrieved from the Stanford Microarray Database (Hubble et al, 2009) and processed using LIMMA in order to define lists of differentially expressed genes. For all other worm data sets, selected gene lists were already provided. The C. elegans data sets were mapped to fly genes using orthology relationships that were retrieved from TreeFam (Li et al, 2006; Ruan et al, 2008) and InParanoid (O'Brien et al, 2005; Berglund et al, 2008) using the FlyMine resource (Lyne et al, 2007).
Western blots
The proteins were obtained by TCA extraction and separated on 8% SDS–PAGE and western blots performed as previously described (Giannakou et al, 2007). Anti-phospho-AKT, phospho-ERK, total AKT and total ERK were obtained from Cell Signaling. Where reported, the blots were quantified as described (Alic et al, 2011).
Statistical analysis
Analyses were performed in R, Excel or JMP. Where required, the data were log-transformed to fit a normal distribution. The details of tests used are given in figure captions.
Note
Array data are available from ArrayExpress under accession numbers E-TABM-751 (ChIP-chip data) and E-TABM-757 (expression data).
Supplementary Material
Peak and gene lists, and their analysis mentioned in the manuscript
Supplementary Information, Supplementary Figures S1–3, Supplementary Tables S1–2
Acknowledgments
We thank AA Teleman and SM Cohen for larval dFOXO ChIP-chip data, and MD Piper for critical reading of the manuscript. We acknowledge funding by the Wellcome Trust (JT and LP), BBSRC (HMC and LP), Max Planck (LP) and EMBO and Marie Curie post-doctoral fellowships (NA).
Author contributions: NA, MEG, CS, HMC and LP designed the experiments. NA, MEG, CS and MPH performed the experiments. TDA, IP, ES and JT designed the data analysis. TDA, IP, NA and ES performed the analysis. NA, HMC and LP wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alic N, Hoddinott MP, Vinti G, Partridge L (2011) Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell 10: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev 14: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL (2008) InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res 36: D263–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs WH III, Cavenee WK, Arden KC (2001) Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome 12: 416–425 [DOI] [PubMed] [Google Scholar]
- Biggs WH III, Zavitz KH, Dickson B, van der Straten A, Brunner D, Hafen E, Zipursky SL (1994) The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J 13: 1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E (2001) An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11: 213–221 [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L (2005) Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA 102: 3105–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow MH, Aebersold R, Pankratz MJ, Junger MA (2010) The Drosophila FoxA ortholog Fork head regulates growth and gene expression downstream of Target of rapamycin. PLoS One 5: e15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG (2008) Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Tinto S, Marr MT II, Andreu P, Puig O (2007) Characterization of the Drosophila insulin receptor promoter. Biochim Biophys Acta 1769: 236–243 [DOI] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH (2006) Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell 11: 775–789 [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L (2001) Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106 [DOI] [PubMed] [Google Scholar]
- Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavare S, Tower J (2007) Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol 8: R262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler G, Perry KM, Tjian R (1998) Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol Cell Biol 18: 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, Szypowska A, Meppelink A, Brenkman AB, Yodoi J, Holstege FC, Burgering BM (2009) Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol 5: 664–672 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- Drysdale RA, Crosby MA (2005) FlyBase: genes and gene models. Nucleic Acids Res 33: D390–D395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- England BP, Heberlein U, Tjian R (1990) Purified Drosophila transcription factor, Adh distal factor-1 (Adf-1), binds to sites in several Drosophila promoters and activates transcription. J Biol Chem 265: 5086–5094 [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A (2009) Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA 106: 2700–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z (2004) Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 32: 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL (1998) Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150: 129–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jacobson J, Vinti G, Leevers SJ, Partridge L (2007) Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell 6: 429–438 [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L (2004) Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305: 361. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Partridge L (2008) Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging Cell 7: 187–198 [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A (2008) FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 192: 19–28 [DOI] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 17: 1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger B, Galic M, Kohler K, Wittwer F, Brogiolo W, Hafen E, Stocker H (2008) Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E (2009) Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457: 726–730 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hubble J, Demeter J, Jin H, Mao M, Nitzberg M, Reddy TB, Wymore F, Zachariah ZK, Sherlock G, Ball CA (2009) Implementation of GenePattern within the Stanford Microarray Database. Nucleic Acids Res 37: D898–D901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566 [DOI] [PubMed] [Google Scholar]
- Ikeya T, Broughton S, Alic N, Grandison R, Partridge L (2009) The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc Biol Sci 276: 3799–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E (2003) The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O (2004) Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 279: 41114–41123 [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464 [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS (2006) The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab 4: 37–48 [DOI] [PubMed] [Google Scholar]
- Kuningas M, Magi R, Westendorp RG, Slagboom PE, Remm M, van Heemst D (2007) Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur J Hum Genet 15: 294–301 [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399: 609–613 [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL (1995) Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139: 1567–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A (2006) A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125: 987–1001 [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, Wong GK, Zheng W, Dehal P, Wang J, Durbin R (2006) TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res 34: D572–D580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, Rana D, Riley T, Sullivan J, Watkins X, Woodbridge M, Lilley K, Russell S, Ashburner M, Mizuguchi K, Micklem G (2007) FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol 8: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Grotewiel MS (2006) Distinct genetic influences on locomotor senescence in Drosophila revealed by a series of metrical analyses. Exp Gerontol 41: 877–881 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D (2007) Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 6: 208–216 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH (2003) Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121 [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D (2007) Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol 8: R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Burgering BM (2000) AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404: 782–787 [DOI] [PubMed] [Google Scholar]
- Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M (2008) Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell 7: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, O'Sullivan J, Azad A, Proudfoot N, Mellor J (2003) Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science 300: 492–495 [DOI] [PubMed] [Google Scholar]
- Muller J, Verrijzer P (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283 [DOI] [PubMed] [Google Scholar]
- Nemoto S, Finkel T (2002) Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 295: 2450–2452 [DOI] [PubMed] [Google Scholar]
- Nielsen MD, Luo X, Biteau B, Syverson K, Jasper H (2008) 14-3-3 Epsilon antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell 7: 688–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer EL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33: D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257 [DOI] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, Hiller D, Jiang S, Protopopov A, Wong WH, Chin L, Ligon KL, DePinho RA (2009) FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5: 540–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Bruning JC (2008) Forkhead transcription factors and ageing. Oncogene 27: 2351–2363 [DOI] [PubMed] [Google Scholar]
- Piper MD, Selman C, McElwee JJ, Partridge L (2008) Separating cause from effect: how does insulin/IGF signalling control lifespan in worms, flies and mice? J Intern Med 263: 179–191 [DOI] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev 17: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Tjian R (2005) Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev 19: 2435–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, Heriche JK, Hu Y, Kristiansen K, Li R, Liu T, Moses A, Qin J, Vang S, Vilella AJ, Ureta-Vidal A, Bolund L, Wang J, Durbin R (2008) TreeFam: 2008 update. Nucleic Acids Res 36: D735–D740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Kahn CR (2007) Endocrine regulation of ageing. Nat Rev Mol Cell Biol 8: 681–691 [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A (2008) FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE (2006) MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol 26: 4863–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, Piper MD, Al-Qassab H, Speakman JR, Carmignac D, Robinson IC, Thornton JM, Gems D, Partridge L, Withers DJ (2008) Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J 22: 807–818 [DOI] [PubMed] [Google Scholar]
- Shen J, Tower J (2010) Drosophila foxo acts in males to cause sexual-dimorphism in tissue-specific p53 life span effects. Exp Gerontol 45: 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C, Giannakou ME, Foley A, Goss M, Partridge L (2011) dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell (advance online publication, doi:10.1111/j.1474-9726.2011.00707.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14: 395–403 [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS (2001) A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W (2007) Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho RA, Gilliland DG (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339 [DOI] [PubMed] [Google Scholar]
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr, DiStefano PS, Chiang LW, Greenberg ME (2002) DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296: 530–534 [DOI] [PubMed] [Google Scholar]
- Villa-Cuesta E, Sage BT, Tatar M (2010) A role for Drosophila dFoxO and dFoxO 5′UTR internal ribosomal entry sites during fasting. PLoS One 5: e11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H (2005) JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121: 115–125 [DOI] [PubMed] [Google Scholar]
- Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R (2004) Insulin regulation of heart function in aging fruit flies. Nat Genet 36: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD (2008) FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA 105: 13987–13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Besser D, Luca E, Stoffel M (2003) Insulin regulates the activity of forkhead transcription factor Hnf-3beta/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc Natl Acad Sci USA 100: 11624–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P (2005) Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci USA 102: 13289–13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Lavin VA, Moser LR, Cui Q, Kanies C, Yang E (2008) PP2A regulates the pro-apoptotic activity of FOXO1. J Biol Chem 283: 7411–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Poplawski M, Yen K, Cheng H, Bloss E, Zhu X, Patel H, Mobbs CV (2009) Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol 7: e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A (2007) FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci USA 104: 15899–15904 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peak and gene lists, and their analysis mentioned in the manuscript
Supplementary Information, Supplementary Figures S1–3, Supplementary Tables S1–2