Determination of the average cellular copy number of 400 proteins under different growth conditions and integration with protein turnover and absolute mRNA levels reveals the dynamics of protein expression in the genome-reduced bacterium Mycoplasma pneumoniae.
Keywords: mRNA–protein, Mycoplasma pneumoniae , protein homeostasis, protein turnover, quantitative proteomics
Abstract
Biological function and cellular responses to environmental perturbations are regulated by a complex interplay of DNA, RNA, proteins and metabolites inside cells. To understand these central processes in living systems at the molecular level, we integrated experimentally determined abundance data for mRNA, proteins, as well as individual protein half-lives from the genome-reduced bacterium Mycoplasma pneumoniae. We provide a fine-grained, quantitative analysis of basic intracellular processes under various external conditions. Proteome composition changes in response to cellular perturbations reveal specific stress response strategies. The regulation of gene expression is largely decoupled from protein dynamics and translation efficiency has a higher regulatory impact on protein abundance than protein turnover. Stochastic simulations using in vivo data show how low translation efficiency and long protein half-lives effectively reduce biological noise in gene expression. Protein abundances are regulated in functional units, such as complexes or pathways, and reflect cellular lifestyles. Our study provides a detailed integrative analysis of average cellular protein abundances and the dynamic interplay of mRNA and proteins, the central biomolecules of a cell.
Introduction
Acquiring and integrating large-scale, quantitative biological data is a common feature of Systems Biology studies (Joyce and Palsson, 2006; Sauer et al, 2007). Following enormous technological and methodological advances over the last years, abundance differences of both mRNA (‘t Hoen et al, 2008) and proteins (Ong and Mann, 2005) can be reproducibly measured for complex biological samples. High-throughput approaches determining unbiased average protein copy numbers on a large scale (Jaffe et al, 2004; Lu et al, 2007; Ishihama et al, 2008; Malmström et al, 2009; Tolonen et al, 2011) as well as individual protein turnover rates (Beynon and Pratt, 2005) have been reported recently. However, integrating these diverse data and providing additional functional understanding of cells remain an important challenge for the field of Systems Biology (Joyce and Palsson, 2006). A plausible approach to gaining novel biological insights from large-scale data sets lies in the combined application of these independently developed methodologies in a suitable model organism to the same biological sample, but under different growth and stress conditions.
We report a detailed, integrative analysis of genome-wide experimental data of mRNA levels, average cellular protein abundances and half-lives generated under various relevant perturbation conditions (Box 1). We use Mycoplasma pneumoniae, a human pathogenic bacterium causing atypical pneumonia as model system for our study. Containing a reduced genome with only 690 ORFs, this bacterium is an ideal organism for exhaustive quantitative and systems-wide studies, avoiding technical limitations due to exceeding sample complexity, constrained by limitations in dynamic range and resolution of current generation mass spectrometers. Available data on the transcriptome (Güell et al, 2009), on protein complexes (Kühner et al, 2009), as well as on metabolic pathways (Yus et al, 2009) facilitate the integration of the data generated for this study into an organism-wide context. Additionally, M. pneumoniae represents a relevant organism to study stochastic noise in living systems. The cells are significantly smaller than other bacteria, such as Escherichia coli (0.05 and 1 μm3, respectively), resulting in principle in an increased susceptibility to abundance fluctuations of cellular molecules.
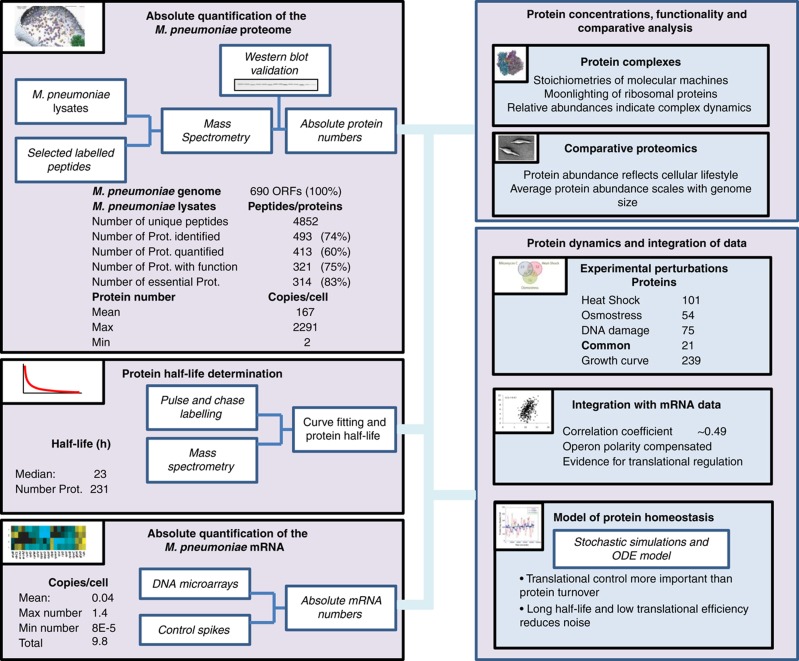
Overview over the generated and analysed data sets and summary of the main findings.
 |
Results
Average cellular protein abundances and dynamics
We determined average cellular protein abundances for 413 different proteins in M. pneumoniae, covering 60% of all predicted open reading frames, 83% of the proteome observable by extensive mass spectrometric mapping (Jaffe et al, 2004; Kühner et al, 2009), 75% of all proteins with annotated function and 83% of all proteins predicted as essential (Glass et al, 2006), respectively (Box 1; Supplementary Table S1). We measured individual protein levels in average copies per cell under control conditions (growth for 96 h), along a 4-day time course, in response to heat shock, DNA damage and osmotic stress (Supplementary Table S2). The reported numbers are averages from cells grown in batch culture. Cellular protein abundances span three orders of magnitude ranging from about 2300 copies (Ef-Tu) to two copies (uncharacterized protein MPN554; Supplementary Figure S1; Supplementary Table S2) with an average abundance of 167 copies per cell. The 20 most prominent proteins in M. pneumoniae account for nearly 44% of the total protein mass. Highly abundant proteins are involved in glucose metabolism (24% of total protein mass), compensating by enzyme abundance for the inefficient generation of two to four ATP molecules per consumed glucose molecule (Yus et al, 2009). Proteins involved in cell adherence used for attachment to lung cells of the host in situ and to the culture dish in vitro account for 8% of the total protein mass. Cellular chaperones GroEL/ES, DnaK/DnaJ/GrpE and trigger factor make up over 9% of the total cellular protein mass. Ribosomal proteins account for 5.6–12.3% of the total protein mass, depending on stationary or exponential growth.
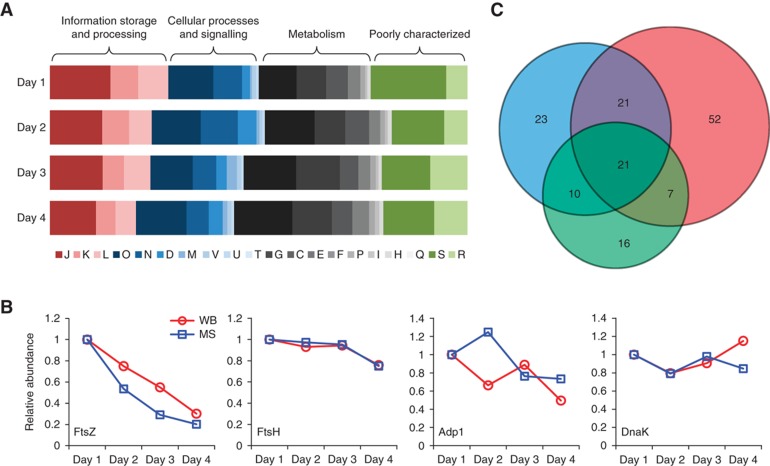
Grouping all quantified proteins in COG functional classes (Supplementary Table S1) revealed a specific increase in cellular proteome mass attributed to metabolic functions (classes G, C, E, F, I, P and H) concomitant with an increase in cellular doubling time during the late stages of 4 days batch culture growth (Figure 1A). We additionally observed a decrease in abundance of proteins involved in information storage and processing (classes J, K and L; Figure 1A), and more specifically a decrease in ribosomal proteins and in FtsZ, a bacterial cell division protein (from 77 to 16 copies per cell, Figure 1B). These data agree well with the slowing down of cell growth and division rate at later stages of the growth curve as previously reported (Yus et al, 2009) and reflects an increased energy requirement for intracellular pH maintenance at later growth stages due to the acidification of the growth medium. Furthermore, the determined protein abundances mirror the described growth stage-related partitioning between acetate and lactate production (Yus et al, 2009): lactate dehydrogenase is upregulated 500% to over 1000 molecules per cell, while acetate kinase shows a 50% reduction in abundance. Additional protein abundance profiles along the growth curve were confirmed by western blotting (Figure 1B). In total, <40% of all quantified proteins show a variation coefficient <33% along the growth curve, indicating global reorganization. However, summing up protein copy numbers and considering their respective molecular weights, the total protein mass per cell stayed constant (3.2 gigadalton, 2.9% standard deviation), indicating a tightly controlled global cellular protein concentration (Supplementary information).
Figure 1.
Proteome composition changes in response to cellular perturbations. (A) COG class dynamics over the course of 4 days growth in batch culture. Functional classes are colour coded. See Supplementary information for COG class nomenclature. (B) Abundance changes of proteins along the growth curve. Blue (MS): mass spectrometry data; red (WB): quantified western blots. (C) Venn diagram for proteins with significantly changed abundance in response to cellular perturbations. Red: heat shock; blue: DNA damage; green: osmotic stress.
We quantitatively analysed the change in proteome composition in response to osmotic stress, mitomycin-induced DNA damage and heat shock (Supplementary Figure S2). Applying stringent cutoff criteria (the observed fold change must be at least 0.5 and larger than the standard deviation of all conditions analysed), we find 54, 75 and 101 proteins with significantly changed abundances following these perturbations, respectively (Figure 1C; Supplementary Table S3). Proteins upregulated in response to heat shock include the chaperons DnaK (+18%), GroES (+29%) and the proteases ClpB (+65%) and Lon (+28%), indicating a concerted response involving re-folding and degradation of heat-damaged proteins. Following mitomycin-induced DNA damage; we observed a doubling of Hit1, an important signalling molecule involved in regulation of DNA replication and repair (Szurmak et al, 2008). Osmotic stress led to only moderate abundance changes in the proteome, including 16 proteins with abundance changes unique to this stress (Figure 1C; Supplementary Table S3). We find a set of 21 proteins with changed abundances in response to all tested perturbations. Some proteins previously unknown for their involvement in general stress response, such as the octameric cell division protein MraZ (Chen et al, 2004) (804 copies per cell), protein p200 (156 copies per cell) involved in cytadherence and gliding motility (Jordan et al, 2007), as well as initiation factor IF1 (25 copies per cell), previously associated with changes in translational control in cold stress (Giuliodori et al, 2007) were among the most upregulated general stress proteins (Supplementary Table S3). Several E. coli stress proteins (Han and Lee, 2006) with orthologues in M. pneumoniae were identified (Supplementary Table S3).
We additionally quantified the M. pneumoniae proteome from cells grown in minimal medium (Yus et al, 2009). Determined protein abundances correlated well with those from cells grown in standard Hayflick medium (rp=0.78). We observed growth rate and nutrient availability related changes in protein abundances, such as a downregulation of ribosomal proteins and oligopeptide transporters in minimal medium (Supplementary information).
In connection with its reduced genome, M. pneumoniae cells contain only a very limited set of proteins involved in transcriptional control. We quantified six out of eight proteins proposed to be transcription factors (Yus et al, 2009) lacking the two proposed sigma-like factors MPN626 (SigD) and MPN424 (XylM), possibly due to their presumed low cellular abundance. Determined abundances for transcription factors range from 4 copies per cell for MPN241 (WhiA) and MPN329 (Fur) to over 300 copies per cell for MPN239 (GntR), which was found to be specifically induced more than four-fold at early stages of the growth curve (Supplementary Table S2). Additionally, the DNA-binding proteins IHF-HU (MPN529, possibly affecting DNA topology; Mouw and Rice, 2007), MraZ (MPN314, octameric cell division protein; Chen et al, 2004) and the transcriptional repressor HrcA are following the same trend (decrease from exponential to stationary phase), making them candidates for global gene expression regulation during M. pneumoniae growth. Aside from these changes along the growth curve, we found no clear induction of either proposed transcription factor in response to the cellular stresses tested for this study.
mRNA–protein integration and dynamics
We used mRNA data from tiling array and deep sequencing experiments (Güell et al, 2009) to analyse the organism-wide correlation between cellular mRNA levels and protein abundances in M. pneumoniae under steady-state and perturbed conditions. In agreement with the published literature on mRNA–protein correlations for large samples (de Sousa Abreu et al, 2009; Maier et al, 2009), we found a modest correlation between quantified mRNA and protein abundances with Pearson's correlation coefficients between 0.41 and 0.51 for different available data sets (average value for all condition=0.52; Supplementary Figure S3). Diverse post-transcriptional factors and individual differences in translation efficiency and protein turnover could contribute to the observed variability of mRNA–protein ratios (Vogel et al, 2010). Certain functional classes (transcription and energy production) appear to be mildly enriched in proteins with biased protein/mRNA ratios under steady-state conditions (Supplementary Figure S4).
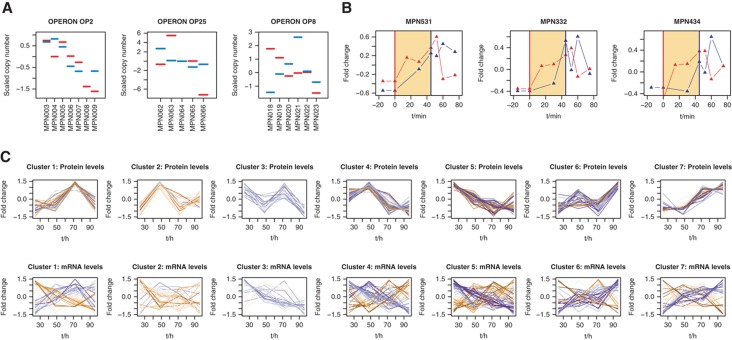
A focused analysis of mRNA–protein abundance correlations on the level of consecutive genes organized in transcriptional units (operons) revealed distinct correlation patterns. We observed similar mRNA–protein profiles in operons, as well as directly anti-correlated patterns (Figure 2A), suggesting operon-specific and selective post-transcriptional regulatory mechanisms. On average, the observed operon polarity of consecutive transcripts (‘staircase-behaviour’) (Güell et al, 2009) tends to be compensated for on the protein level (Supplementary Figure S5).
Figure 2.
mRNA and protein profiles indicate complex post-transcriptional regulatory mechanisms. (A) Examples for abundance dynamics for mRNA and proteins in operons. Red: mRNA; blue: protein. See Supplementary information for additional profiles. (B) Heat-shock response on mRNA and protein levels for ClpB (MPN531), Lon (MPN332) and DnaK (MPN434). Red vertical line: heat shock start; blue vertical line: heat shock end. Red triangles: mRNA; blue triangles: protein. (C) mRNA and protein dynamics along the growth curve. Blue colour: similar patterns of abundance change; orange colour: different patterns of abundance change.
To analyse mRNA–protein abundance dynamics during growth in batch culture over 4 days, we established seven clusters to classify 239 proteins with significant abundance changes (Figure 2C; Supplementary Table S2). Individual mRNA expression patterns correlated moderately with protein abundance profiles; only 24 mRNA and protein profiles fell into identical clusters, suggesting that the regulation of gene expression is largely decoupled from protein dynamics in M. pneumoniae and pointing towards extensive translational regulation. We observed a significant (P<0.05) enrichment of functional classes in some of the clusters and mRNA–protein profiles correlated better for certain metabolic pathways (Supplementary Figure S6). Proteins involved in transcription/translation show a concerted decrease in mRNA–protein abundance when comparing early exponential and late growth (Supplementary Figure S7). Additionally, the mRNA–protein correlation coefficients along 4 days growth are related to gene topology. We observed a higher correlation of mRNA and protein abundances for genes organized in short operons. Additionally, mRNA and protein abundances for genes located at the 3′-end in longer transcriptional units appear to correlate less (Supplementary Figure S7).
Analogously, only part of the proteins significantly changing in response to cellular stress (heat shock, osmotic stress and DNA damage) reflected expression changes on the mRNA level (Supplementary Table S3). However, for classical heat-shock proteins Lon, ClpB and DnaK, we confirm expected mRNA–protein expression dynamics, such as an immediate induction of mRNA and a subsequent increase of corresponding protein abundances (Figure 2B), as well as a consecutive decline of mRNA and protein after the initial heat-shock response. We additionally find corresponding patterns for two proteins lacking a defined heat-shock promotor: the protein translocase subunit SecA and a member of the partitioning protein family, ParA (Supplementary Table S2), suggesting a possible regulatory mechanism on mRNA stability.
Protein turnover, modelling and simulations
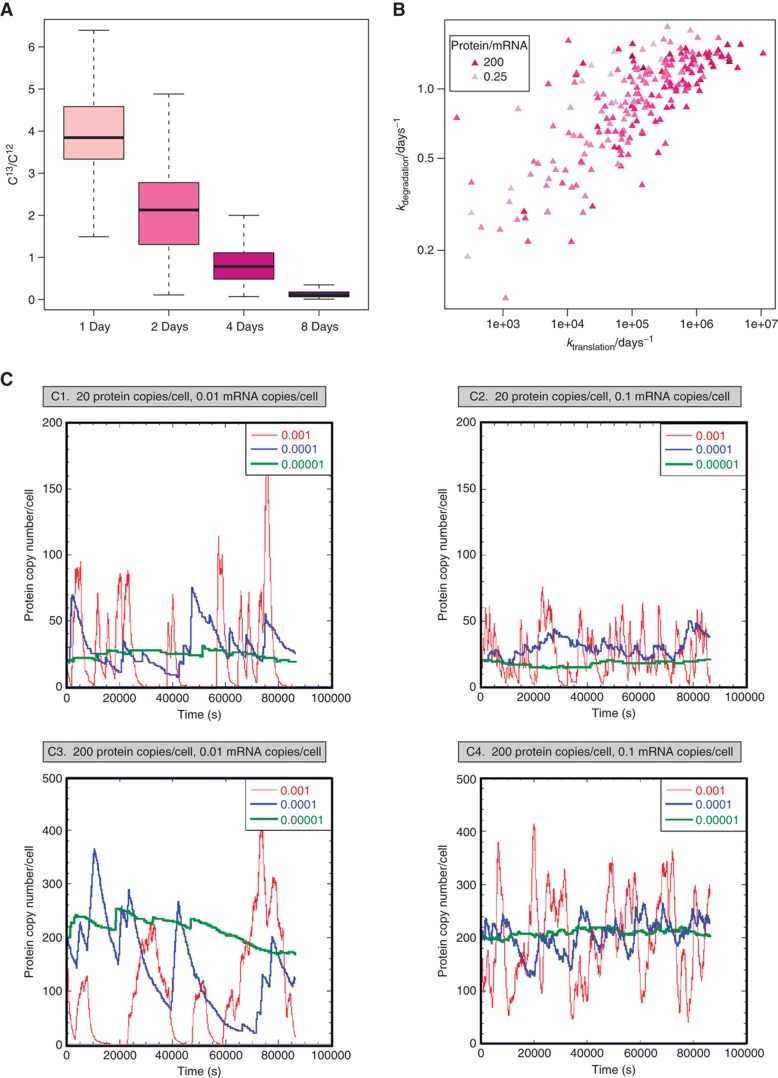
We measured genome-wide individual protein turnover rates using a label-chase approach involving stable isotope-labelled amino acids (Beynon and Pratt, 2005). Compared with other organisms (Belle et al, 2006; Doherty et al, 2009; Jayapal et al, 2010), we obtained longer protein half-lives, averaging 23 h. Most of the determined protein half-lives span from 12 h (10th percentile) to 42 h (90th percentile) (Figure 3A). For a subset of proteins with high degradation rates, only a maximal half-life could be estimated (Supplementary Table S4). We additionally observed very fast degradation of stress-induced proteins during recovery from heat shock, indicating specific proteolytic regulatory mechanisms. For example, for Lon protease, cellular concentration increases by 158% upon shock, but levels turn back to pre-stress values in the time scale of minutes (Figure 2B). The N-end rule (Tobias et al, 1991), predicting protein half-life based on the N-terminal amino-acid context, did not apply in M. pneumoniae (Supplementary Figure S8). We found that proteins involved in transcription, trafficking and secretion are disproportionally more stable under standard growth conditions and proteins involved in energy production and lipid transport have shorter half-lives (Supplementary Figure S9).
Figure 3.
Protein turnover measurements and stochastic simulations. (A) Boxplots showing the variance of measured protein turnover profiles. 13C/12C ratios of metabolically labelled cells were determined by mass spectrometry. (B) Influence of k1 (ktranslation) and k2 (kdegradation) on the ratio protein/mRNA. Colour gradient according to the ratio protein/mRNA. (C) Stochastic simulation of transcription–translation in M. pneumoniae using the range of experimental values for mRNA and protein turnover rates found in this study. We considered 200 ribosomes, 130 RNA polymerase complexes and 400 promoters, RNA lifetime of around 3 min (seen after heat-shock induction; Figure 2B) and a rate for protein production of 0.1 molecules of protein per second (see Supplementary information). Red: protein turnover rate of 0.001/s; blue: protein degradation rate of 0.0001/s; green: protein turnover rate of 0.00001/s. Panels show different mRNA–protein combinations.
We quantified individual average mRNA amounts per cell by spiking known amounts of reference RNAs into mRNA samples analysed by tiling arrays (Supplementary Figure S10). In agreement with findings in E. coli (Taniguchi et al, 2010) and previous estimates for M. pneumoniae (Weiner, 2003) measured mRNA abundances were on average below one copy per cell (mean abundance: 0.04). We determined a cellular average of 9.8 mRNA molecules at any given time. Based on these data, we established an ordinary differential equations model for the estimation of individual in vivo protein degradation (k2) and translation efficiency rates (k1) (Supplementary Table S4). Correlating protein abundance with logk1 (rs=0.5) and logk2 (rs=0.3), respectively, allowed quantifying the relative contribution of k1 and k2 to protein homeostasis: the influence of translation efficiency on protein abundance is 40% higher than the influence of protein turnover (Figure 3B). Interestingly, a subset of previously identified cellular phosphoproteins (Su et al, 2007; Schmidl et al, 2010; Supplementary Table S4) shows significantly higher than average turnover rates under steady-state conditions (k2all=0.94 k2phosphoproteins=1.20, P=0.008).
Stochasticity in gene expression has been studied theoretically, as well as experimentally with model proteins (Ozbudak et al, 2002; Kaern et al, 2005). These studies describe the propagation of transcription bursts and the importance of small molecule numbers as well as high translation efficiency in biological noise. To evaluate the physiological importance of stochastic noise, we performed simulations of transcription–translation with the software SmartCell (Dublanche et al, 2006; Figure 3C). We observed robust gene expression when simulating with representative mRNA and protein amounts as well as average translation efficiencies and experimentally determined turnover rates in M. pneumoniae. As previously suggested, key parameters for compensating noise in gene expression are low translation efficiencies in conjunction with long protein half-lives (Ozbudak et al, 2002; Pedraza and Paulsson, 2008; Figure 3C). Reducing the protein half-life artificially to 2.5 h resulted in a significant increase of gene expression noise, amplified by low mRNA numbers (Figure 3C). Our simulations additionally suggest that high cellular protein amounts represent an effective buffer against spikes in gene expression. In agreement with this finding, essential proteins are on average more abundant in M. pneumoniae (top quartile: 18% non-essential, bottom quartile: 37% non-essential; Supplementary Figure S11), also confirming findings in E. coli (Taniguchi et al, 2010) and S. cerevisiae (Ghaemmaghami et al, 2003). Simulating a reduction of ribosome number as seen for cells grown in minimal medium does not significantly change those results (Supplementary Figure S12).
Protein complex abundances and stoichiometries
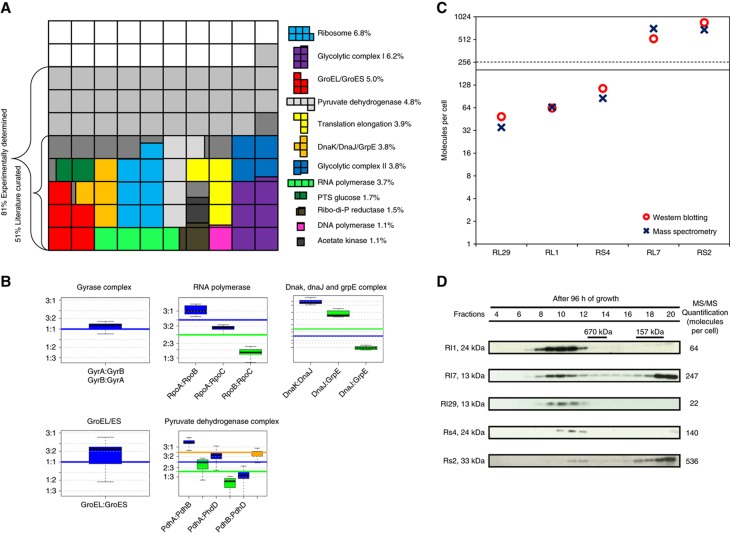
The organizational principle of proteins in macromolecular assemblies is conserved in eukaryotic cells (Gavin et al, 2002; Ho et al, 2002) as well as in bacteria (Kühner et al, 2009). Often, protein complexes, such as the ribosome, RNA polymerase or the GroEL/ES chaperonin system carry out essential biological functions. We used our quantitative data sets to assign cellular abundances and stoichiometries to known protein complexes (Figure 4; Supplementary Figure S13). In total, 51% of all cellular proteins by mass in M. pneumoniae have interaction partners, considering only the literature-curated homomultimeric and heteromultimeric protein complexes (Figure 4A). Extending this analysis to a proteome-wide screen by tandem affinity purification coupled with mass spectrometry (TAP-MS; Kühner et al, 2009) revealed that up to 81% of the cellular proteome by mass may be following this organizational principle.
Figure 4.
Protein complex stoichiometries and dynamics reflect cellular functions. (A) Mapping protein abundances on the literature-curated complexes reveal the importance of this organizational principle. Coloured boxes represent complexes with >1% total abundance by mass. (B) Examples for protein complexes and the stoichiometric abundance of the subunits in the proteome. Boxplots represent measured pairwise protein copy number ratios distributions within protein complexes. All different experimental conditions were used to estimate the distributions. Horizontal continuous lines represent expected stoichiometries. Identical colours associate the measured ratios (boxplots) with expected ratios (horizontal lines) from the literature. (C) Comparison of mass spectrometry and western blotting for the quantification of ribosomal proteins. Red circles: cellular abundances determined by quantitative western blotting; blue crosses: cellular abundances determined by quantitative mass spectrometry. Black line: median (190); dashed line: average (255). (D) Western blots against ribosomal proteins on size exclusion fractionated M. pneumoniae lysates (Superose6 PC 3.2/30 column). High abundant proteins L7 and S2 appear also in the low molecular weight fraction, suggesting secondary functions.
For several well-characterized protein complexes, such as the GroEL/ES chaperonin (160 multimeric complexes per cell), DNA gyrase (50 A2B2 tetramers per cell) or ribonucleoside-diphosphate reductase (300 copies per cell), cellular abundances of the subunits reflect the expected complex stoichiometries closely (Figure 4B; Supplementary Figure S13). As expected, for dynamic protein complexes characterized by the transient interaction of specific subunits, such as the sigma factor RpoD with RNA polymerase or the nucleotide exchange factor GrpE with the chaperone DnaK, cellular protein abundances did not mirror their functional stoichiometries (Figure 4B; Supplementary information). For pyruvate dehydrogenase, the expected overall complex composition is reflected in the respective protein abundances, but the stoichiometries of the heteromultimeric E1 subunits are altered (Figure 4B), suggesting intra-complex subunit rearrangements.
Strikingly, the variance of measured half-lives for proteins involved in complexes with stable subunit stoichiometries, such as GroEL/ES (9.9 × 10−5), pyruvate dehydrogenase (9.8 × 10−5) or phenylalanine-tRNA synthase (0.02), was significantly lower than the total variance for all proteins with determined half-life (0.44, Supplementary Table S4). For several protein complexes, the observed subunit stoichiometries are conserved in the bacterium Leptospira interrogans (see below and Supplementary Table S5), additionally confirming the mapped abundances for M. pneumoniae.
The principle of protein abundances closely following the stoichiometries of stable molecular machines is not maintained for the largest protein complex in the cell, the ribosome. We identified 46 of 51 annotated ribosomal proteins (Supplementary Table S1) and 43 were directly quantified with a corresponding labelled peptide (Supplementary Table S6). Their cellular abundances span two orders of magnitude and range from 24 (RL22) to over 1000 (RS3) copies per cell (median 190 and standard deviation: 238; Supplementary Figure S14). This number agrees well with the 140 ribosomes per cell previously determined for M. pneumoniae by electron tomography (Yus et al, 2009) and is reflected in the determined cellular rRNA abundance (Supplementary Figure S10). A similar abundance range has been reported for L. interrogans (Malmström et al, 2009). We excluded that protein extraction introduced a bias in protein resolubilization of ribosomal proteins (Supplementary Figure S15) and validated the measured abundances by quantitative western blotting for proteins RL1, RL7, RL29, RS2 and RS4 (Figure 4C; Supplementary Figure S16). Size exclusion chromatography experiments revealed that high abundant ribosomal proteins (L7 and S2) are not exclusively associated with the ribosome, but are also found in fractions corresponding to the size of free monomers (Figure 4D; Supplementary Figure S17). This, together with the finding that several ribosomal proteins of M. pneumoniae are found associated with different protein complexes (Kühner et al, 2009), suggests their multi-functionality. We additionally showed by western blotting that ribosomal proteins in high molecular weight fractions, corresponding to intact ribosomes and separate 30S and 50S subunits, fall into a closer abundance range (Supplementary Figure S17). We find several ribosomal proteins with abundances significantly below the median value (190), both by mass spectrometry and by quantitative western blotting. We speculate that those proteins might be dispensable for ribosome function, indicating a degree of plasticity in ribosome composition. A detailed analysis of mRNA–protein ratios in the main ribosomal operons (MPN164–MPN183; Supplementary Figure S18) indicated that a relative increase in ribosomal protein abundance is related to the degree of overlap of the ribosomal binding site of those genes with the consensus Shine-Dalgarno sequence, indicating post-transcriptional regulation of protein abundance.
Comparative analysis with L. interrogans
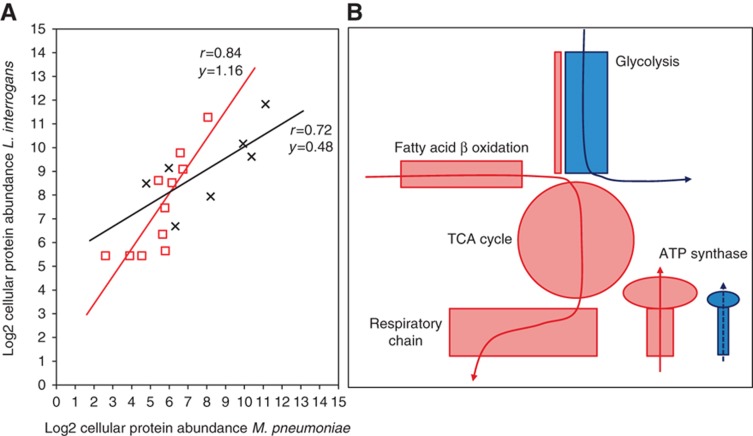
We investigated how genome reduction, cell size and the specific growth environment of M. pneumoniae are reflected in the proteome composition by interspecies comparison with the spirochaete bacterium and human pathogen L. interrogans, the only other organism to date where average cellular protein quantities have been measured on a large scale following a similar methodology (Malmström et al, 2009). L. interrogans cells are considerably larger than M. pneumoniae (0.22 and 0.05 μm3, respectively) (Beck et al, 2009) and have a more complex genome containing 3658 annotated ORFs in the analysed serotype. This is reflected by a 14.5 times higher absolute protein number in L. interrogans while the average protein abundance is only 3.2 times higher. A reciprocal protein BLAST search and a gene name comparison of M. pneumoniae and L. interrogans identified 443 orthologous protein pairs (Supplementary Table S5). For matched pairs under both criteria, determined protein abundances correlated with a Pearson's coefficient of rp=0.67 (Supplementary Table S5). Subdividing this set of proteins into functional categories revealed distinct groups of high correlation, however, with very different abundance ratios. For example, proteins involved in replication, recombination and repair as well as proteins involved in carbohydrate transport and metabolism correlate highly, but show very different relative cellular expression levels (Figure 5A).
Figure 5.
Protein abundances reflect cellular lifestyles. (A) Protein abundances for orthologous proteins within same functional classes correlate highly. Relative enrichment of proteins (slope differences of the trend lines) reflect specific cellular lifestyles of M. pneumoniae and L. interrogans. Red squares: proteins in COG class L (replication, recombination and repair); black crosses: proteins in COG class G (carbohydrate transport and metabolism). (B) Differences in metabolism between L. interrogans and M. pneumoniae are mirrored in protein abundance. Red blocks and line: carbon metabolic route of L. interrogans; blue blocks and line: carbon metabolic routes of M. pneumoniae. Size of coloured shapes represents relative abundances.
Protein abundances reflect the respective lifestyles of L. interrogans and M. pneumoniae. Even though both bacteria have similar doubling times under exponential growth (Saengjaruk et al, 2002; Yus et al, 2009), their catabolic metabolism routes differ fundamentally. L. interrogans utilizes predominantly fatty acid β-oxidation as carbon source and oxidative phosphorylation coupled with an electron transport chain for energy production (Ren et al, 2003; Figure 5B). M. pneumoniae on the other hand relies mainly on glycolysis for ATP generation (Yus et al, 2009). Hence, even though most glycolytic enzymes are present in L. interrogans, their cumulative abundance only accounts for 1.3% of all quantified proteins. Contrarily, 19.7% of the all quantified proteins in M. pneumoniae (24% of the total protein mass) are involved in glucose metabolism. Strikingly, the relative abundance ratios of glycolytic enzymes are conserved in both bacteria, suggesting that the adaption to different carbon and energy sources involves global abundance regulation of metabolic pathways, rather than the alteration of individual enzymatic activities.
The observed 150-fold relative enrichment of thioredoxin in M. pneumoniae (1265 copies per cell) further highlights their distinct metabolic routes. While organisms with an electron transport chain, such as L. interrogans utilizes NADH as electron donors during end oxidation, thioredoxin could have an active role in balancing the cellular redox-state during acetate production in M. pneumoniae by serving as electron acceptor for reduced coenzymes NADH and NADPH (Zeller and Klug, 2006). Owing to its drastic genome reduction, M. pneumoniae relies on the import of precursors for proteins, RNA and DNA rather than synthesizing them. Correspondingly, peptide importers, proteases, as well as RNA degradation enzymes are found to be of higher concentration in M. pneumoniae. Reflecting similar doubling times during exponential growth (Saengjaruk et al, 2002; Yus et al, 2009), we find in both cases a similar proportion of ribosomal mass of the total proteome (8% in L. interrogans and 5.6–12.3% in M. pneumoniae). This contrasts with values up to 21% in the fast dividing bacterium E. coli (Arnold and Reilly, 1999).
Conclusions and novel insights
We integrated large-scale average abundance data for mRNA and proteins with turnover rates in the bacterium M. pneumoniae, an ideal model organism for systems-wide studies. Measured protein abundance changes in response to several perturbation conditions revealed a highly dynamic proteome including specific sets of stress response proteins. In addition to sequence signatures, mRNA abundance (Vogel et al, 2010) and measurement variation (Nie et al, 2006), we found that predominantly post-transcriptional rather than post-translational regulatory mechanisms control cellular mRNA to protein abundance ratios. These findings are confirmed for mammalian cells using a complementary approach (M Selbach, personal communication).
Quantitative simulations of mRNA and protein homeostasis showed how long protein half-life and poor translational efficiency buffers gene expression noise propagating from low cellular mRNA levels in vivo. Integration of our data with previous work (Kühner et al, 2009) revealed that unusual subunit stoichiometries indicate protein complex dynamics and suggested possible moonlighting for several ribosomal proteins. Finally, a quantitative comparison with the pathogenic bacterium L. interrogans revealed metabolic adaption involving regulation of entire pathways and highlighted how protein abundances reflect different cellular lifestyles. We expect our data to serve as a reference point for future integrative large-scale quantitative studies in other organisms, as well as a valuable resource for further functional studies and for refined, organism-wide mathematical models.
Materials and methods
Cell culturing and protein extraction
M. pneumoniae cell cultures were grown in Hayflick rich medium as previously described (Yus et al, 2009) and samples were taken at 24 h intervals. For cellular perturbations, cells grown for 96 h (control conditions) were treated for 20 min with 5 μg/ml mitomycin C (DNA damage) or with 0.5 M NaCl (osmotic stress) before lysis. For heat-shock treatment, cell culture dishes were placed in a 42°C water bath for 45 min and samples were taken in 15 min intervals starting 30 min after heat shock start. Attached cells were washed twice with ice-cold PBS, harvested by scraping and centrifuged at 4000 g for 10 min. Cell pellets were resuspended in lysis buffer (8 M urea, 150 mM ammonium bicarbonate) and lysed by a 5-min treatment in an ice-cold sonification bath. The cell lysate was centrifuged in a cooled desktop centrifuge at 16 000 g for 5 min and the supernatant further processed for mass spectrometry or western blotting. The protein concentration of the supernatant was determined with the Pierce BCA protein assay kit (Thermo Scientific). A comparison with SDS-based cell lysis and extraction of proteins showed no significant differences in lysis and protein resolubilization efficiency (Supplementary Figure S15). In total, 2.4% of all proteins in SDS-treated samples and 1.8% in urea-treated samples remained insoluble after the extraction procedure.
Mass spectrometry
Protein abundances were determined using an LC-MS based approach involving 30 stable isotope-labelled reference peptides spanning the full abundance range of the M. pneumoniae proteome (Supplementary Table S6) and extracting ion currents of the three most dominant precursor ions per protein (Silva et al, 2006; Malmström et al, 2009). We used an additional set of 47 reference peptides to accurately determine the abundances of ribosomal proteins, since they proved intrinsically difficult to quantify (Supplementary Tables S6 and S7). The setup of the μRPLC-MS system was as described previously (Schmidt et al, 2008). Each survey scan acquired in the ICR-cell at 100 000 FWHM was followed by MS/MS scans of the three most intense precursor ions in the linear ion trap with enabled dynamic exclusion for 60 s. After converting the acquired raw files to the centroid mzXML format (readW, http://tools.proteomecenter.org), MS/MS spectra were searched using the SEQUEST algorithm (Yates et al, 1995). The database search results were further validated using the PeptideProphet (Keller et al, 2002) and ProteinProphet (Nesvizhskii et al, 2003) program and the peptide false discovery rate was fixed to 1% in both cases by adjusting the probability and spectrum counts thresholds.
Protein profiling and quantification
A rolling inclusion mass list was generated based on the recently generated PeptideAtlas (Kühner et al, 2009) in combination with the masses of the 30 spiked in reference peptides. The list was imported as global mass lists into the mass spectrometer and the PTPs sequenced in each sample by directed LC-MS/MS analysis (Schmidt et al, 2009). The Progenesis LC-MS software (v2.5, Nonlinear Dynamics Limited) was employed for label-free protein and peptide quantification. Protein MS abundances were calculated for each LC-MS analysis by summing up the MS intensities of its corresponding PTPs, respectively. The average cellular abundances of all identified proteins were determined as recently specified (Malmström et al, 2009).
In total, 37 quantitative LC-MS maps for the M. pneumoniae proteome were generated. Controls (cells after 4 days of growth) were measured in nine replicates. Samples subjected to perturbations (different time points after heat shock, mitomycin-induced DNA damage, osmotic stress, cells at different days during batch culture growth) were each measured in duplicate. The error rates of the abundances thus determined were assessed by bootstrapping the measured precursor ion intensities against the protein concentrations directly determined using the labelled reference peptides (Malmström et al, 2009; Supplementary Figure S19). The estimated average error rate is 1.77-fold for all quantified proteins and 1.54-fold for proteins quantified by three independent peptides (80% of all proteins). Additionally, error estimation was carried out using a bootstrap analysis (Supplementary Figure S19). The MS/MS data files can be retrieved via the Tranche website (https://proteomecommons.org/tranche/, ‘Mycoplasma_MSB-11-2933’, hashcode dMFS6Of7sYZyKATdLL3nJMYU8uVzpbZIn6IgmwCB4yHsenNoST3j5eUrF8umj7NHcRtap+n5ORQMlKsVLi4sphzLrbwAAAAAAAAXIA==).
Analysis of protein turnover rates
Proteins were isotopically labelled for 14 days using the SILAC approach (Ong et al, 2002) as recently specified (Jayapal et al, 2010) by spiking labelled amino acids to a final concentration of 10 mM into the medium of a growing M. pneumoniae culture and passaging them every 4 days. After full labelling was achieved, cells were harvested and a fully labelled sample was collected. Fresh cultures were inoculated with 10 mM unlabelled arginine and lysine and cells were harvested after 1, 2, 4 and 8 days of growth with intermittent passaging for the latter time point. Absolute protein amount was determined for each time point and set in relation to the starting amount, thereby serving as a correction factor for loss of labelled signal due to cell growth. After protein extraction and digestion, the generated peptide samples were analysed as described above. The Xpress algorithm of the TPP (http://tools.proteomecenter.org/TPP.php) was employed to determine the precise ratios of the individual identified peptides over all samples. The median of the corresponding peptide ratios for each protein was used to calculate the final turnover rates. We identified protein turnover profiles for 231 proteins.
RNA quantification
mRNA copy numbers have been estimated from an Affymetrix tiling array (Supplementary Table S8; Güell et al, 2009) which was deposited here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM722501. Twelve RNA spikes in controls spanning more than the dynamic range of the M. pneumoniae transcriptome were used to estimate the amount of each mRNA in 10 μg of total RNA. Assuming a 90% of rRNA in total RNA, 150 ribosomes per cell (Kühner et al, 2009) and no free rRNA in the cell, we estimated mRNA copy number per cell.
Data integration and analysis
Acquired data were analysed with Microsoft excel and the software R. Dynamic changes in mRNA and protein abundance were considered to be significant if they were higher than a 0.5-fold change and higher than the respective standard deviation over all conditions measured. Only proteins having a coefficient of variation >0.33 were considered for clustering of the growth curve data. Proteins have been scaled to equal median and equal median absolute deviation. Fuzzy c-means algorithm has been used to derive seven clusters from the scaled data. Protein profiles were compared with corresponding mRNA profiles for each member of all clusters. mRNA profiles were considered to be equivalent to the protein profiles only if: (a) standard deviation of mRNA levels along the profile is >0.4; (b) mRNA profile is correlated against all protein cluster medoids. Only if the highest correlation corresponds with the cluster of the protein profile and the Pearson's correlation coefficient is >0.5, the mRNA and protein profiles were considered equivalent.
Stochastic simulations
We used SmartCell, a software designed for modelling biological processes occurring in a cell (Ander et al, 2004; Dublanche et al, 2006). The stochastic simulator uses the Gibson and Bruck (2000) optimization of the Gillespie Algorithm. In the transcriptional–translational simulations we performed, we consider competition of RNA polymerase binding to the promoter of our target protein with the rest of the chromosomal promoters, assuming that all chromosomal promoters have the same properties and are thus represented by a single species (C). The number of C was assumed to be 400 based on the number of monocistronic operons (Güell et al, 2009). Simulations are made in a virtual M. pneumoniae cell represented by a single voxel with a lattice length of 0.6 μm. See Supplementary information for detailed simulation parameters.
Size exclusion chromatography
M. pneumoniae cell cultures after 96 h were washed, pelleted and resuspended in lysis buffer (50 mM Tris pH 7.5, 5% glycerol, 1.5 mM MgCl2, 100 mM NaCl, 0.2% NP40, 1 mM DTT, 1 mM AEBSF, 1 mM PMSF, 1 μg/ml pepstatin A, 1 μg/ml antipain, 2 μg/ml aprotinin, 1 μg/ml leupeptin and 16 μg/ml benzamidin) and lysed mechanically using a douncer. After two steps of centrifugation at 10 000 g and 100 000 g, the supernatant was collected for gel filtration (GF) chromatography. GF chromatography was performed at 10°C on a Pharmacia SMART system at a flow rate of 40 μl/min by using a Superose6 PC 3.2/30 column and a Superdex 200 column, equilibrated with lysis buffer. The chromatographic profile was monitored at 280 nm by using the μPeak monitor (Pharmacia). Volumes of 50 μl of M. pneumoniae lysates were loaded on a column and 60 μl fractions were collected and analysed by SDS–PAGE and western blotting (Figure 4D; Supplementary Figure S17). Polyclonal antibodies produced in rabbits have been used to detect the ribosomal proteins. Quantitative western blotting was carried out as previously described (Kühner et al, 2009).
Supplementary Material
Supplementary Data
Supplementary Figures S1–S19
All tables in Excel format in one .zip file
Acknowledgments
We thank Ben Lehner (CRG Barcelona, Spain) for comments on the manuscript; JA Wodke (CRG Barcelona, Spain) for pBLAST and essentiality analysis; A Leitner (ETH Zurich, Switzerland) for help with protein turnover sample processing; I Vonkova and V Rybin (EMBL Heidelberg, Germany) for help with size exclusion chromatography; H Molina (CRG Barcelona, Spain) for additional mass spectrometry experiments; E Yus (CRG Barcelona) for minimal medium mRNA data. This work was supported by the European Research council (ERC) advanced grant, the Fundacion Marcelino Botin, the Spanish Ministry of Research and Innovation to the ICREA researcher LS; by the EU grants Prospect and Trireme, by the European Research Council (grant #ERC-2008-AdG 233226) to RA and by SystemsX.ch.
Author contributions: TM designed the study, carried out experiments, analysed the data, prepared figures and wrote the manuscript. AS carried out the mass spectrometry analysis. MG carried out experiments, prepared the model, analysed data and prepared figures. SK carried out the size exclusion chromatography. LS designed the study, performed the simulations, analysed data, discussed results and commented on the manuscript. ACG and RA contributed to the study design, discussed results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ander M, Beltrao P, Di Ventura B, Ferkinghoff-Borg J, Foglierini M, Kaplan A, Lemerle C, Tomás-Oliveira I, Serrano L (2004) SmartCell, a framework to simulate cellular processes that combines stochastic approximation with diffusion and localisation: analysis of simple networks. Syst Biol 1: 129–138 [DOI] [PubMed] [Google Scholar]
- Arnold RJ, Reilly JP (1999) Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal Biochem 269: 105–112 [DOI] [PubMed] [Google Scholar]
- Beck M, Malmström JA, Lange V, Schmidt A, Deutsch EW, Aebersold R (2009) Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods 6: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK (2006) Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA 103: 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon RJ, Pratt JM (2005) Metabolic labeling of proteins for proteomics. Mol Cell Proteomics 4: 857–872 [DOI] [PubMed] [Google Scholar]
- Chen S, Jancrick J, Yokota H, Kim R, Kim S-H (2004) Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif. Proteins 55: 785–791 [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol Biosyst 5: 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J Proteome Res 8: 104–112 [DOI] [PubMed] [Google Scholar]
- Dublanche Y, Michalodimitrakis K, Kümmerer N, Foglierini M, Serrano L (2006) Noise in transcription negative feedback loops: simulation and experimental analysis. Mol Syst Biol 2: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gibson MA, Bruck J (2000) Efficient exact stochastic simulation of chemical systems with many species and many channels. J Phys Chem A 104: 1876–1889 [Google Scholar]
- Giuliodori AM, Brandi A, Giangrossi M, Gualerzi CO, Pon CL (2007) Cold-stress-induced de novo expression of infC and role of IF3 in cold-shock translational bias. RNA (New York, NY) 13: 1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, Smith HO, Venter JC (2006) Essential genes of a minimal bacterium. Proc Natl Acad Sci USA 103: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güell M, van Noort V, Yus E, Chen W-H, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kühner S, Rode M, Suyama M, Schmidt S, Gavin A-C, Bork P, Serrano L (2009) Transcriptome complexity in a genome-reduced bacterium. Science (New York, NY) 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Han M-J, Lee SY (2006) The Escherichia coli proteome: past, present, and future prospects. Microbiol Mol Biol Rev 70: 362–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe JD, Berg HC, Church GM (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4: 59–77 [DOI] [PubMed] [Google Scholar]
- Jayapal KP, Sui S, Philp RJ, Kok Y-J, Yap MGS, Griffin TJ, Hu W-S (2010) Multitagging proteomic strategy to estimate protein turnover rates in dynamic systems. J Proteome Res 9: 2087–2097 [DOI] [PubMed] [Google Scholar]
- Jordan JL, Chang H-Y, Balish MF, Holt LS, Bose SR, Hasselbring BM, Waldo RH, Krunkosky TM, Krause DC (2007) Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect Immun 75: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce AR, Palsson B (2006) The model organism as a system: integrating ‘omics’ data sets. Nat Rev Mol Cell Biol 7: 198–210 [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kühner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castaño-Diez D, Chen W-H, Devos D, Güell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R et al. (2009) Proteome organization in a genome-reduced bacterium. Science (New York, NY) 326: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124 [DOI] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett 583: 3966–3973 [DOI] [PubMed] [Google Scholar]
- Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw KW, Rice PA (2007) Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol Microbiol 63: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W (2006) Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations. Biochem Biophys Res Commun 339: 603–610 [DOI] [PubMed] [Google Scholar]
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Ong S-E, Mann M (2005) Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1: 252–262. Available at: 10.1038/nchembio736 [Accessed 19 July 2010] [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, Paulsson J (2008) Effects of molecular memory and bursting on fluctuations in gene expression. Science (New York, NY) 319: 339–343 [DOI] [PubMed] [Google Scholar]
- Ren S-X, Fu G, Jiang X-G, Zeng R, Miao Y-G, Xu H, Zhang Y-X, Xiong H, Lu G, Lu L-F, Jiang H-Q, Jia J, Tu Y-F, Jiang J-X, Gu W-Y, Zhang Y-Q, Cai Z, Sheng H-H, Yin H-F, Zhang Y et al. (2003) Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422: 888–893 [DOI] [PubMed] [Google Scholar]
- Saengjaruk P, Chaicumpa W, Watt G, Bunyaraksyotin G, Wuthiekanun V, Tapchaisri P, Sittinont C, Panaphut T, Tomanakan K, Sakolvaree Y, Chongsa-Nguan M, Mahakunkijcharoen Y, Kalambaheti T, Naigowit P, Wambangco MAL, Kurazono H, Hayashi H (2002) Diagnosis of human leptospirosis by monoclonal antibody-based antigen detection in urine. J Clin Microbiol 40: 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Heinemann M, Zamboni N (2007) Genetics. Getting closer to the whole picture. Science (New York, NY) 316: 550–551 [DOI] [PubMed] [Google Scholar]
- Schmidl SR, Gronau K, Hames C, Busse J, Becher D, Hecker M, Stülke J (2010) The stability of cytadherence proteins in Mycoplasma pneumoniae requires activity of the protein kinase PrkC. Infect Immun 78: 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Claassen M, Aebersold R (2009) Directed mass spectrometry: towards hypothesis-driven proteomics. Curr Opin Chem Biol 13: 510–517 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Gehlenborg N, Bodenmiller B, Mueller LN, Campbell D, Mueller M, Aebersold R, Domon B (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics 7: 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Su H-C, Hutchison CA, Giddings MC (2007) Mapping phosphoproteins in Mycoplasma genitalium and Mycoplasma pneumoniae. BMC Microbiol 7: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmak B, łouch-Cieszyñska A, Wszelaka-Rylik M, Bal W, rzañska M (2008) A diadenosine 5′,5″-P1P4 tetraphosphate (Ap4A) hydrolase from Arabidopsis thaliana that is activated preferentially by Mn2+ ions]. Acta Biochimica Polonica 55: 151–160 [PubMed] [Google Scholar]
- ‘t Hoen PAC, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, de Menezes RX, Boer JM, van Ommen G-JB, den Dunnen JT (2008) Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, Emili A, Xie XS (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A (1991) The N-end rule in bacteria. Science (New York, NY) 254: 1374–1377 [DOI] [PubMed] [Google Scholar]
- Tolonen AC, Haas W, Chilaka AC, Aach J, Gygi SP, Church GM (2011) Proteome-wide systems analysis of a cellulosic biofuel-producing microbe. Mol Syst Biol 7: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, de Sousa Abreu R, Ko D, Le S-Y, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400 ; DOI: 10.1038/msb.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J III (2003) Transcription profiles of the bacterium Mycoplasma pneumoniae grown at different temperatures. Nucleic Acids Res 31: 6306–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Eng JK, McCormack AL, Schieltz D (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67: 1426–1436 [DOI] [PubMed] [Google Scholar]
- Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen W-H, Wodke JAH, Güell M, Martínez S, Bourgeois R, Kühner S, Raineri E, Letunic I, Kalinina OV, Rode M, Herrmann R, Gutiérrez-Gallego R, Russell RB, Gavin A-C, Bork P et al. (2009) Impact of genome reduction on bacterial metabolism and its regulation. Science (New York, NY) 326: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Zeller T, Klug G (2006) Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes]. Die Naturwissenschaften 93: 259–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Figures S1–S19
All tables in Excel format in one .zip file