Using valproic acid as an example, the authors demonstrate that drug response signatures derived from genome-wide expression data can identify individuals likely to respond to a drug, and propose that this method could select optimal populations for clinical trials of new therapies.
Keywords: biomarkers, cancer, pharmacogenomics
Abstract
Identifying the best drug for each cancer patient requires an efficient individualized strategy. We present MATCH (Merging genomic and pharmacologic Analyses for Therapy CHoice), an approach using public genomic resources and drug testing of fresh tumor samples to link drugs to patients. Valproic acid (VPA) is highlighted as a proof-of-principle. In order to predict specific tumor types with high probability of drug sensitivity, we create drug response signatures using publically available gene expression data and assess sensitivity in a data set of >40 cancer types. Next, we evaluate drug sensitivity in matched tumor and normal tissue and exclude cancer types that are no more sensitive than normal tissue. From these analyses, breast tumors are predicted to be sensitive to VPA. A meta-analysis across breast cancer data sets shows that aggressive subtypes are most likely to be sensitive to VPA, but all subtypes have sensitive tumors. MATCH predictions correlate significantly with growth inhibition in cancer cell lines and three-dimensional cultures of fresh tumor samples. MATCH accurately predicts reduction in tumor growth rate following VPA treatment in patient tumor xenografts. MATCH uses genomic analysis with in vitro testing of patient tumors to select optimal drug regimens before clinical trial initiation.
Introduction
Matching treatments to patients who will most benefit using streamlined computational and biochemical analyses will enable effective clinical trial design for the many drugs currently available (Adjei et al, 2009; Dancey et al, 2010; Freedman et al, 2010). An efficient approach for accurate identification of cancer patients who will benefit from a specific therapy prior to clinical trial initiation will enable optimal selection of patients for clinical trials. The benefits of identifying the right target population for a drug include (1) improved patient response, (2) minimization of time lost treating patients non-responsive to therapy, (3) avoiding side effects in people who will not respond, and (4) smaller sample sizes needed for clinical trials because of larger expected effect sizes. As the pharmaceutical industry estimates that there are over 800 agents and biologics in use or under development for treatment of human malignancies, knowing best how to target these drugs to the cancer patients who will benefit most is critical (PhRMA, 2009).
Historically, most clinical trials evaluate a therapeutic regimen on an unselected patient population, measuring efficacy based on the response in the entire group. Therefore, these studies may fail to identify a significant response rate when a drug is effective in only a subset of patients (Bast and Hortobagyi, 2004). By incorporating prior knowledge about drug sensitivity or the underlying signaling pathways driving cancer progression, relevant subpopulations can be defined prior to a clinical study, improving the likelihood of obtaining a significant drug response (Bild et al, 2006; Downward, 2006; Lamb et al, 2006; Dutta and Maity, 2007; Huang et al, 2007; Rhodes et al, 2007; Du et al, 2009). Given the heterogeneity of cancer, the future of personalized medicine is to have a large number of drugs to choose from, each associated with a biomarker that predicts cancer responsiveness. For example, ∼15–20% of breast cancers have HER2 amplification, which corresponds to high levels of responsiveness to trastuzumab (Barron et al, 2009; Coulson et al, 2010). Without knowing the relevant population of breast cancer patients to treat with this inhibitor, the drug would not have had significant improvement in the overall population. However, even when we use available tools to predict patient sensitivity, such as FISH for HER2 amplification, we still are not able to perfectly predict who will respond. As an example, approximately one third of breast cancer patients with amplified HER2 do not respond to trastuzumab, in part due to deregulation of downstream or parallel pathway components such as PTEN or PI3K (Marty et al, 2008; Stemke-Hale et al, 2008).
With the increasing use of high-throughput technologies in the study of cancer, genomic approaches are finding their way into the oncologists' toolkit (Neve et al, 2006; Tinker et al, 2006; Massague, 2007; Foekens et al, 2008; Gusterson, 2009; Sotiriou and Pusztai, 2009). The majority of commercially available genomic assays focus on prognosis but not prediction (Desmedt et al, 2008; Haibe-Kains et al, 2008; Sotiriou and Pusztai, 2009). Therefore, these tests provide risk of recurrence but do not predict response to particular chemotherapy or non-chemotherapy drugs. For most drugs newly entering the clinical trial pipeline, we still lack the means to rationally and efficiently choose an optimal drug regimen targeted toward the characteristics of a specific tumor, thus impeding the promise of personalized medicine (Desmedt et al, 2008). While creating a predictive drug response signature using actual patient response data is ideal, this approach cannot be used before a drug is tested in a large number of people with different kinds of cancers. Recent studies have identified subsets of patients characterized by gene expression profiles indicating drug sensitivity, and some have translated these findings into clinical trials (Chang et al, 2003; Lamb et al, 2006; Huang et al, 2007; Lamb, 2007; Rhodes et al, 2007; Haibe-Kains et al, 2008). One genomic approach is the ‘Connectivity Map,’ which includes a compendium of gene expression profiles based on chemical and genetic perturbations (Lamb et al, 2006). However, individual assessments of optimal drug sensitivity in cancer patients using high-throughput profiling and the relationship of these profiles to drug response in actual human tumors have not been thoroughly evaluated. Selecting effective drugs by leveraging both genomic and focused preclinical testing with actual patient tumors streamlines the identification of optimal therapeutic regimens.
In this study, we make progress toward this goal by using genomic methods to identify drugs that will be effective for individual tumors. We provide a method, which we call MATCH (Merging genomic and pharmacologic Analyses for Therapy CHoice), to determine the individuals most likely to benefit from a drug prior to a clinical trial. We use the histone deacetylase (HDAC) inhibitor valproic acid (VPA) as our proof-of-principle. We focus on VPA because its optimal target population is not well defined, it is readily available, oral, inexpensive, and has relatively low toxicity. VPA also happens to be attractive from an informatics standpoint because it can be tested in vitro without needing metabolic activation and there are sufficient samples in the Connectivity Map to achieve a robust signature on leave-one-out cross-validation (LOOCV) analysis. MATCH leverages extensive gene expression data and biochemical validation to rationally match drugs to patients. We first use a genomic approach to predict drug sensitivity across cancer types and between cancer and normal tissue to identify those cancers most likely to respond to a drug. We then focus on breast cancer and examine VPA sensitivity for general breast cancer subtypes and for individual tumors within these subtypes. Importantly, this approach allows for drug response characterization in actual human tumors and may represent a more accurate assessment of drug sensitivity than the use of cell lines alone. In silico validation of MATCH's ability to link drugs to optimal target cancer phenotypes is provided by the characterization of a B-RAF inhibitor that has been proven in vitro and in the clinic to effectively treat melanoma: predictions by MATCH correspond to the observed clinical activity of this B-RAF inhibitor. As the ideal target population for VPA is unknown, the accuracy of genomic drug sensitivity predictions for VPA is verified in vitro with breast cancer cell lines grown in two-dimensional (2D) culture. We then use patient tumors, grown in both three-dimensional (3D) cultures and mouse xenografts, to further confirm accurate drug sensitivity predictions for individual samples. From these analyses, VPA is identified as a potential therapy in the treatment of aggressive breast cancers, and our drug response signature is shown to predict in vivo response to VPA in individual patient tumors. More generally, MATCH provides an efficient methodology for determining the cancer patients most likely to respond to a particular drug without any prior knowledge of drug mechanism or target population.
Results
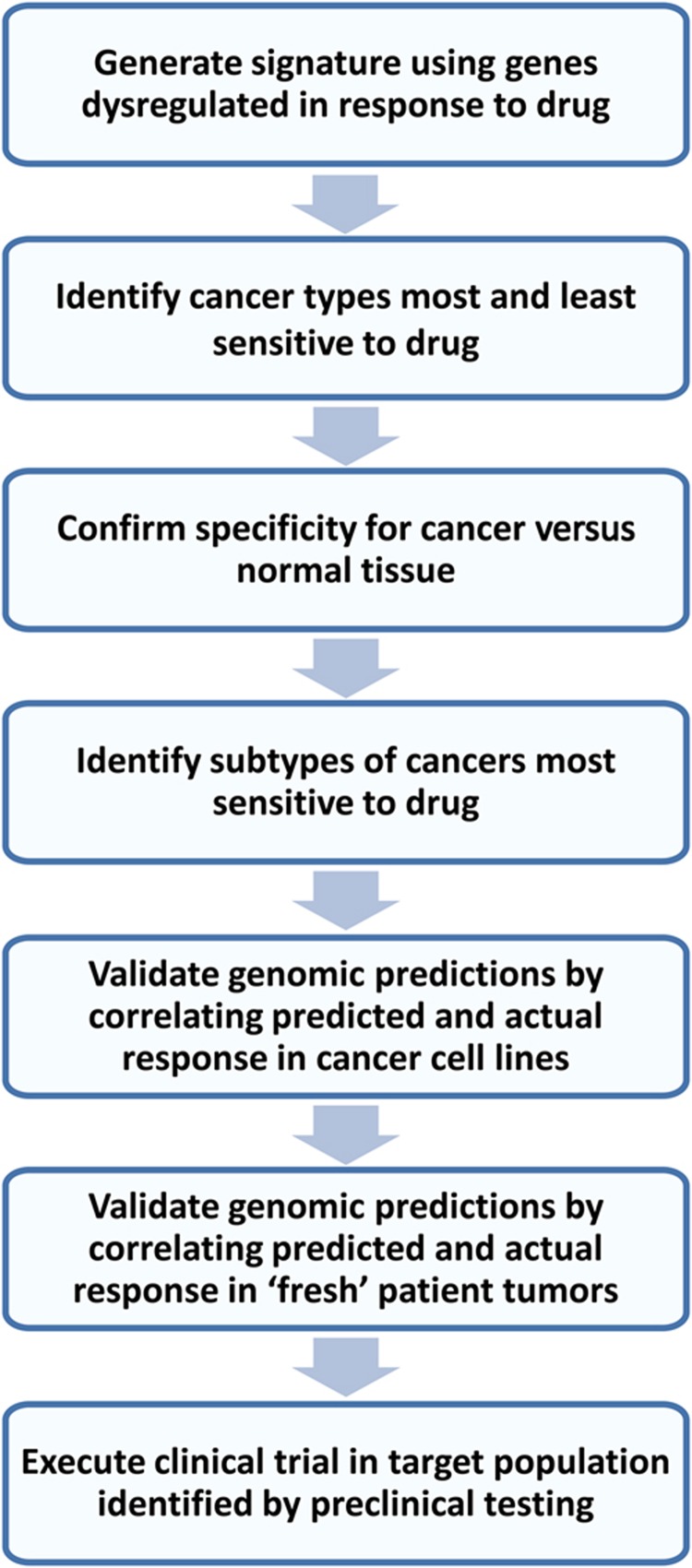
Flowchart of MATCH approach for optimal patient selection
Figure 1 outlines the MATCH approach for choosing an optimal target patient population for a drug. We first generate a genomic signature of cellular response to the drug by using cancer cell lines treated with drug or a placebo vehicle. Gene expression between the untreated and treated cells is then compared to identify genes consistently reflective of drug response, and those genes are used to generate the predictive drug response signature. Following internal and external computational validation, the signature is used to predict drug sensitivity in data sets with multiple tumor types in order to determine the cancer types with little or no sensitivity that should be excluded from further testing. If desired, this analysis can be performed for actual patient tumors, which may provide a more accurate measure of drug sensitivity than cell lines, which often gain additional genetic changes compared with the primary tumor following growth in culture. Next, the signature is used to predict drug sensitivity in data sets with matched tumor and normal tissue in order to exclude those tumor types where there is no significant increase in tumor sensitivity compared with normal tissue. Tumors in such tissues would likely have limited sensitivity to drug given no increase from baseline tissue response levels, and perhaps even show unacceptable toxicity at pharmacologically relevant doses of the drug if high baseline response is seen. Once the optimal target cancer is identified, projection of the signature into data sets of that cancer type allow comparisons of drug sensitivity across subtypes defined by established prognostic or predictive features, such as estrogen receptor (ER) or HER2 status for breast cancer, histologic type for lung cancer, and grade or MSI status for colon cancer. In vitro validation begins with the cancer type(s) predicted to be most sensitive to drug in genomic analyses using standard dose-responsiveness testing of established cell lines in culture, which can be accomplished in a matter of days. If that step is successful, further validation is performed using a panel of fresh patient tumor samples grown in relevant in vitro conditions, such as 3D matrix for breast cancer. If both of these models show a relationship between predicted and actual sensitivity, then the genomic signature can be used to identify an optimal target population for clinical trial initiation.
Figure 1.
Flowchart of streamlined method to identify target population for a drug.
Generation of drug response signature
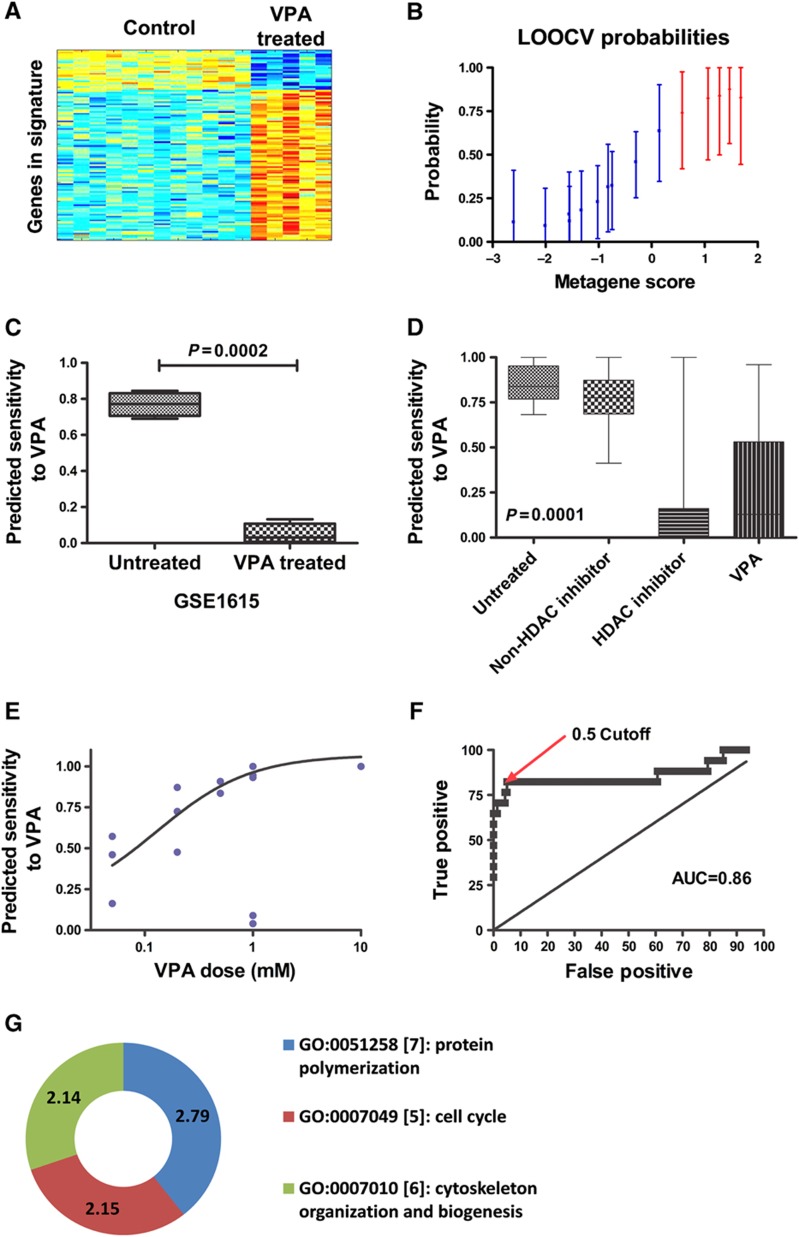
Gene expression profiles of cancer cells following drug treatment are a useful tool to better understand cellular changes reflective of drug treatment. These expression profiles can be used to identify genes reflective of drug response: tumors with genes upregulated that turn off following drug treatment are most likely to be sensitive, and vice versa. One resource that has extensively cataloged genes modulated by specific drug treatments is the Connectivity Map (Lamb et al, 2006). We generated a response signature for VPA using samples from the Connectivity Map build 1 as the training set. Gene expression microarray data from treated and untreated cells were analyzed using a binary regression algorithm to determine the genes that best distinguished treated from untreated samples. Parameters for the binary regression were optimized to maximize the consistency and robustness of predictions within the training set. Internal consistency and robustness of the signature was measured by LOOCV, a process in which each sample is successively left out of the training set and a model is fit to the remaining samples and then applied to the left out sample to obtain a prediction. Prefiltering of gene expression data and number of genes in the signatures were optimized to maximize the accuracy of these predictions. The VPA signature used 200 probes, which represent 188 unique genes, because some genes have more than one probe on the microarray. The heatmap and LOOCV graph for the VPA signature are shown in Figure 2A and B.
Figure 2.
VPA signature. (A) The heatmap columns are the Connectivity Map samples with the 10 controls on the left and 5 treated samples on the right. Each row is a probe in the signature. Red indicates upregulation and blue indicates downregulation of the gene. (B) LOOCV from the Connectivity Map training sample. Blue samples (1–10) are the control samples. Red samples (11–15) are the VPA-treated samples. (C) Bar graph of mean and standard error of predicted VPA sensitivity on ovarian theca cells before and after treatment with VPA. (D) Graph of predicted sensitivity to VPA in Connectivity Map samples from nine independent batches. Samples are grouped as untreated controls, samples treated with a drug other than an HDAC inhibitor, samples treated with an HDAC inhibitor other than VPA, and samples treated with various doses of VPA. (E) Graph of sensitivity predictions versus actual treatment dose for Connectivity Map samples treated with various doses of VPA. The line is a best-fit sigmoidal curve excluding the two outliers. (F) ROC curve based on data from Figure 2D comparing VPA-treated samples with samples that were untreated or treated with a non-HDAC inhibitor. (G) Doughnut plot of the Gene Ontology terms for the genes in the VPA signature with Bayes factor >2. Bayes factor for each term is given on the doughnut.
We validated the accuracy and sensitivity of the signature in two ways. First, an external and independent data set was used in which normal ovarian theca cells were treated with VPA or a control agent, and RNA from these cells were placed onto microarray. Our analyses show a change in gene expression in the VPA-treated cells compared with controls, further validating the accuracy of our signature (Figure 2C). Specifically, the predicted sensitivity dropped from a mean of 0.77 before treatment to 0.05 after treatment (P=0.0002).
Second, we applied the VPA signature to 169 samples from 9 batches of the Connectivity Map build 2 that had at least one sample treated with VPA and one sample treated with another drug. The predictions are shown in Figure 2D. Untreated cells and cells treated with random drugs show significantly more predicted sensitivity to VPA than cells treated with VPA or other HDAC inhibitors (P<0.0001). The variation in prediction in VPA-treated cells is due to different doses, with increasing dose of VPA correlating to a decrease in predicted sensitivity (Figure 2E). Based on the ROC curve, the optimal cutoff for VPA sensitivity is ∼0.5, with an AUC of 0.86 (Figure 2F).
In order to better understand the meaning of the VPA signature, we performed a Gene Ontology analysis of the genes in the signature using http://gather.genome.duke.edu. Gene Ontology categories with a disproportionate number of genes in the signature are shown in Figure 2G. In particular, genes involved in cell growth and communication (FGFR1, IGF2, PDGFRB, TGFB2, and MET) and genes involved in cell-cycle regulation (CDK2, CDK10, and CDKL1) are dysregulated by VPA.
Rational determination of relevant tumor type for drug treatment
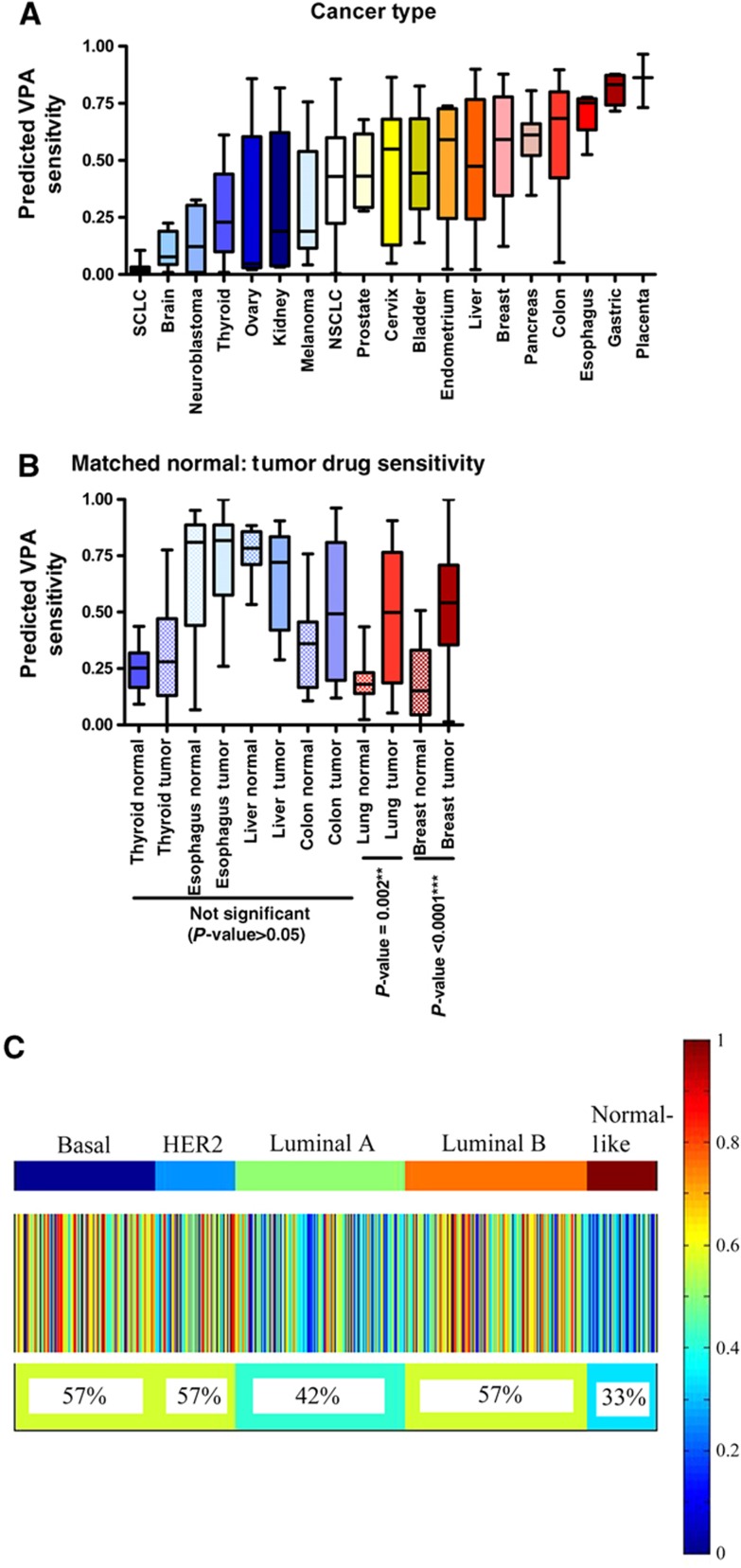
We examined the pattern of predicted sensitivity to VPA across different types of cancer in order to identify the tumors most likely to respond to drug treatment (Figure 3A). Specifically, using the VPA signature defined above (Figure 2), we predicted drug responsiveness in a panel of 310 diverse cancer cell lines (https://array.nci.nih.gov/caarray/project/woost-00041). These cancer cells were placed onto microarray as a group and can be used to compare profiles across over 40 different tumor types. The results highlight the broad variation in responsiveness of different tumor types and show that specific tumor types such as breast, lung, and pancreas have high average predicted response and others, such as brain, ovarian, and melanoma, have low predicted response. Therefore, this approach provides a mechanism to rapidly assess predicted drug sensitivity between different tumor types in order to exclude from further study those with gene expression patterns reflective of little drug sensitivity and to focus on those with subpopulations predicted to be sensitive to the drug.
Figure 3.
Predictions across cancer types and subtypes. (A) Box-whisker plot for predicted VPA sensitivity across GSK cell lines for epithelial cancers. Median is indicated by a horizontal line. The box gives the interquartile range, and the error bars indicate the total range. (B) Box-whisker plot for VPA sensitivity across cancer types in GSE5364. Boxes for normal adjacent tissue are checkered. Median is indicated by a horizontal line. The box gives the interquartile range, and the error bars indicate the total range. (C) Heatmap of samples from 11 breast cancer data sets, divided by intrinsic subtype, with the predicted response to VPA displayed as a color, with red representing a high predicted response, and blue a low predicted response. Each column is an individual sample, and the heterogeneity of predicted response to VPA within and between subtypes is clearly visible. The percent of samples with predicted sensitivity >0.5 is given below the heatmap.
While assessing predicted drug sensitivity between different tumor types is an important first step in determining the optimal tumor types for a specific therapy, these results must be taken in the context of tissue type. The goal is to identify tumor-specific response to drug and minimize tissue-specific effects. To this end, we used a gene expression data set comprised of matched normal and tumor tissue profiles. In order to minimize artifact such as batch effect, we looked within a single previously published data set containing several different types of cancer (Yu et al, 2008), although other data sets were compared and similar results were found (data not shown). The average sensitivity within each type of cancer and normal is shown in Figure 3B. Significantly, there is high correlation between the cancer cell line (Figure 3A) and patient tumor gene expression predictions (Figure 3B), thereby confirming the drug response patterns identified in the cancer cell lines. Further, breast and lung cancers were significantly more sensitive to VPA than the corresponding normal tissues, P=6 × 10−6 and P=0.005, respectively. Liver (P=0.14), esophageal (P=0.3), colon (P=0.22), and thyroid (P=0.36) cancers did not differ in sensitivity from the corresponding normal tissue. These results are consistent with prior research; for example, VPA is known to be toxic to normal liver cells in vitro and in vivo, to cause gastrointestinal-related side effects, and can cause hypothyroidism with long-term use (Fisher et al, 1991; Gau et al, 2010). We chose to focus on breast cancer for further study due to its high predicted sensitivity to VPA compared with other cancers and its highly significant sensitivity in tumor tissue compared with normal breast tissue.
Meta-analysis of drug sensitivity patterns in breast cancer phenotypes
We then examined patterns of sensitivity across different phenotypes of breast cancer. We applied the VPA response signature to each of the 1803 human breast cancer samples in 11 large independent data sets of microarray data from snap-frozen tumors (Supplementary Table S1). A cutoff of 0.5 was then used to predict sensitivity or resistance in all cases. Homogeneity of the data sets was tested by ANOVA limited to each breast cancer subtype across data sets for the VPA response signature (Supplementary Figure S1). Overall, the data sets were homogeneous in the relative predicted sensitivity of each subtype to VPA, the differences in mean VPA sensitivity between data sets within each subtype were <0.2, medians were generally on the same size of 0.5, and removal of individual data sets did not improve homogeneity significantly or affect conclusions. Therefore, all 11 data sets were used.
Results from predictions of all data sets were merged and sorted by molecular subtype (upper color bar on Figure 3C), providing an overview of the predicted responsiveness of each subtype to each drug. In all, 57% of basal tumors, 57% of HER2-like tumors, 42% of luminal A tumors, 57% of luminal B tumors, and 33% of normal-like tumors are predicted to be sensitive to VPA. Overall, 51% of breast tumors are predicted to be sensitive to VPA, with significant heterogeneity within subtypes. Overall, more aggressive breast cancer subtypes such as basal, HER2-like, and luminal B have higher levels of predicted sensitivity. This finding is in contrast to other drugs such as fulvestrant, an ER pathway antagonist, which shows subtype response specificity for ER-positive subtypes (luminal A, luminal B, and normal-like) (Supplementary Figure S2). Therefore, even within a particular breast cancer subtype, there remains varied predicted sensitivity, highlighting the importance of individualized predictions.
In silico validation of the MATCH approach using a drug with a known target population
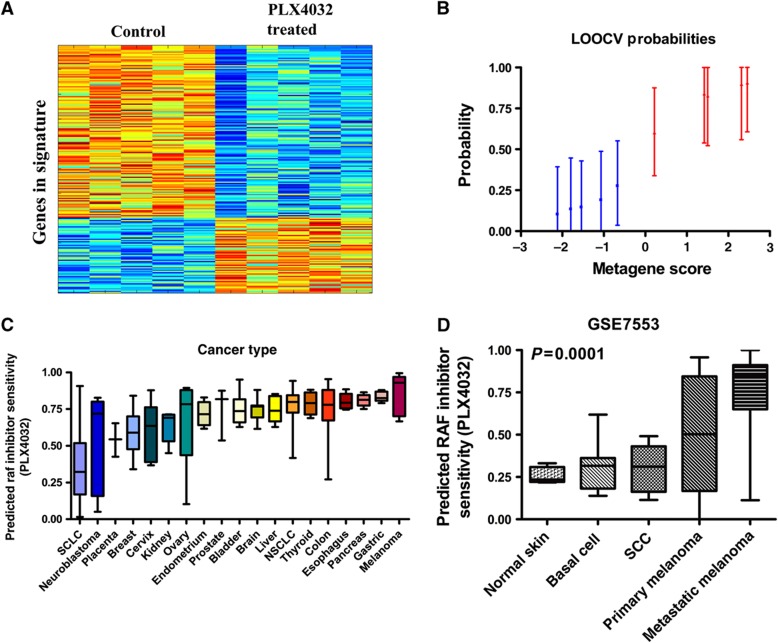
While VPA is attractive to study because of the need for identification of a target population for ongoing clinical trials, we also wanted to validate MATCH using a drug with a well-defined target population. Gene expression analysis on a data set of sensitive melanoma cells treated with a B-RAF inhibitor, PLX4032 (GSE20051) enabled generation of a genomic signature (Joseph et al, 2010). Importantly, we use PLX4032 to validate the ability of our genomic approach to identify responsive cancers, as we know from clinical trials that PLX4032 has an 80% response rate in melanoma with mutant B-RAF (Flaherty et al, 2010). A drug response signature was developed from treated and untreated cells using the approach detailed above. The signature for PLX4032 yielded a signature with 200 probes representing 157 unique genes. The heatmap and LOOCV graph for the B-RAF inhibitor signature are shown in Figure 4A and B. We then validated the ability of the B-RAF inhibitor signature to predict sensitivity in cancer cell lines, and show significant response in tumor cells with activating B-RAF mutations versus those without mutation (Supplementary Figure S3A).
Figure 4.
PLX4032 signature and validation. (A) The heatmap columns are the GSE20051 samples with the five controls on the left and five treated samples on the right. Each row is a probe in the signature. Red indicates upregulation and blue indicates downregulation of the gene. (B) LOOCV from the GSE20051 training set. Blue samples (1–6) are the control samples. Red samples (7–12) are the PLX4032-treated samples. (C) Box-whisker plot for predicted PLX4032 sensitivity across GSK cell lines for epithelial cancers stratified by cancer type. (D) Box-whisker plot for PLX4032 sensitivity across skin cancer types and normal skin in GSE7553. (Cancer and normal types with two or fewer samples were excluded.) Median is indicated by a horizontal line.
We projected this signature into the panel of tumor cell lines and identified high predicted sensitivity to the B-RAF inhibitor PLX4032 in cell lines from melanoma, thyroid cancer, and GI cancers and low predicted sensitivity in cell lines from small cell lung cancer and neuroblastoma (Figure 4C). This inhibitor is known to be active in tumors with the B-RAFV600E mutation, and our findings are consistent with published data on B-RAF mutations in human cancer types. Specifically, neuroblastoma does not have activating mutations in B-RAF or in other members of the PI3K/RAS/RAF/MAPK cascade; therefore, targeting B-RAF in these tumors may not be effective (Dam et al, 2006). Also, RAF activation in small cell lung cancer, in contrast to other cancers, has been shown to be detrimental to cancer cell survival (Ravi et al, 1998). On the other hand, PLX4032 is active in melanoma in clinical trials and thyroid cancer in vitro, supporting the validity of our in silico results (Flaherty et al, 2010; Kopetz et al, 2010; Xing et al, 2011).
Next, we predict sensitivity of patient tumors compared with normal tissue to assess drug specificity. Again, we see that tumor types identified in our initial analysis across a panel of cancer cell lines show high tissue specificity in our analysis of matched normal and tumor samples (Supplementary Figure S3B). Further, as matched normal skin and melanoma cells are not present in the previous data set, we compared normal skin cells versus melanoma using an independent data set and see significant predicted sensitivity to the B-RAF inhibitor in melanoma with little predicted sensitivity in squamous cell cancer of the skin, basal cell cancer of the skin, or normal skin (P<0.0001) (Figure 4D). Therefore, our predictions of the optimal target population for the B-RAF inhibitor correspond to the published and confirmed target populations. Together, these findings highlight the ability of our in silico genomic approach to identify relevant tumor types for specific drugs.
Finally, in order to better understand the meaning of the PLX4032 signature, we performed a Gene Ontology analysis of the genes in the signature using http://gather.genome.duke.edu. Gene Ontology categories with a disproportionate number of genes in the signature are shown in Supplementary Figure S3C. In particular, genes involved in ribosome biogenesis and rRNA metabolism (BRIX1, EXOSC2, EXOSC4), protein amino-acid dephosphorylation (DUSP4, DUSP6, DUSP14, and CDC25A), and regulation of the cell cycle (CCND1, CCNE1, E2F4, and MYC) are dysregulated by PLX4032.
In vitro validation of drug response signature predictions
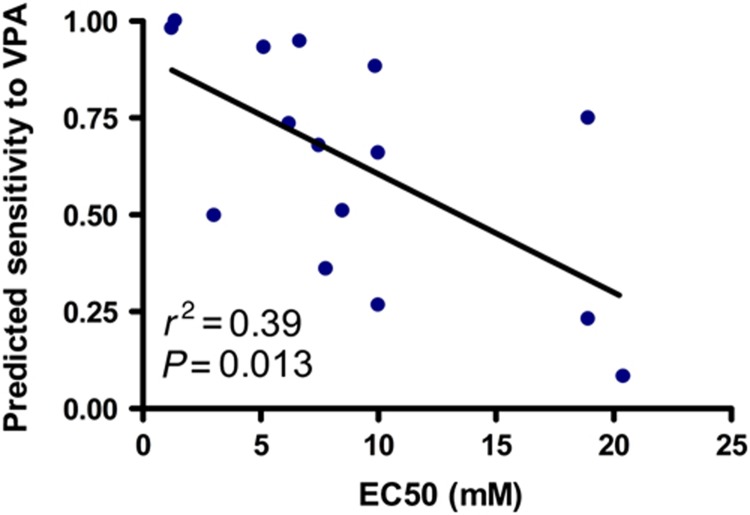
While MATCH leverages genomic resources to guide therapeutic selection for individual tumors in cases where a drug's ideal target population has not previously been defined, a rapid means to validate MATCH predictions is warranted. One method to confirm the accuracy of using drug response signatures is to use in vitro dose-response studies on cancer cell lines in which genomic analysis was used to predict drug sensitivity. After projecting the drug response signatures in a panel of breast cancer cell lines, we correlated predicted and actual drug sensitivity. Following a comprehensive dose-response analysis, we calculated the effective concentration at which the growth rate is inhibited by 50% (EC50) for each cell line and correlated this EC50 to the predicted sensitivity of that cell line to drug treatment. As shown in Figure 5, there is a significant correlation between actual (as determined by a cell line's EC50) and predicted sensitivity for VPA (P=0.013). No cell line predicted to be resistant (predicted sensitivity <0.5) had an EC50 within three-fold of clinically achievable concentration of VPA.
Figure 5.
Correlation of actual response to targeted therapeutics and predicted response from drug response signatures. Breast cancer cell lines were treated with VPA for 96 h and proliferation was assayed using a standard MTT colorimetric method. Scatter plot shows the degree of correlation between proliferation inhibition (EC50) and predicted sensitivity for each cell line. Source data is available for this figure at www.nature.com/msb.
3D validation of drug response signature predictions
Because breast cancer cell lines are known to have ‘drifted’ and, thus, maintain very different expression patterns and phenotypes when compared with actual patient tumors, we validated the genomic predictions for this agent in other model systems: 3D culture of patient tumors and xenografts (Kenny et al, 2007). First, we used short-term, 3D cultures of patient breast tumor samples both from primary tumors and from pleural effusions. The 3D culture microenvironment more closely resembles the in vivo cellular microenvironment and provides a more biologically relevant growth setting than standard 2D microenvironments (Bissell and Labarge, 2005; Griffith and Swartz, 2006). Importantly, this approach allowed for growth of ER-positive tumors, which are difficult to establish in xenograft models.
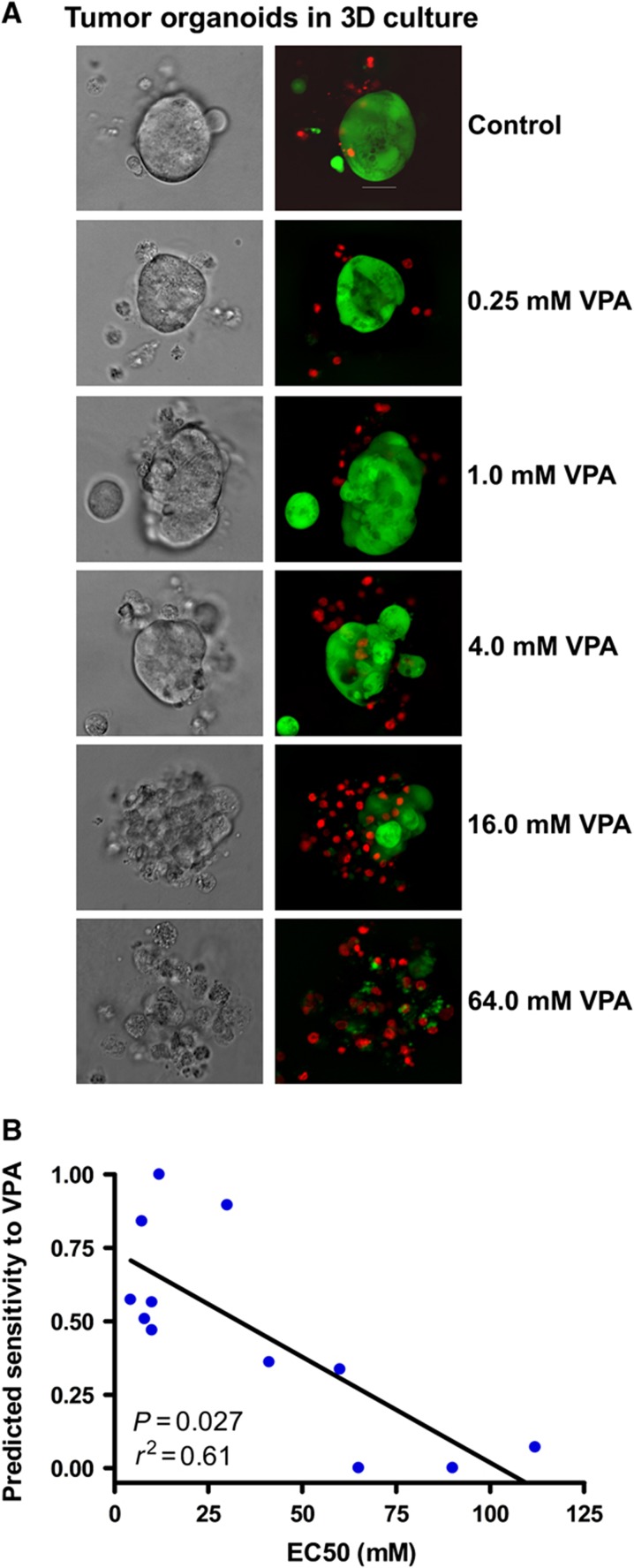
For 3D culture, the cytoxic effect of VPA treatment on the tumor organoids was assessed by light microscopy (a representative example is shown in Figure 6A). As shown in a representative example of a triple negative (basal-like by microarray) tumor predicted to be sensitive to VPA, tumor organoids treated with VPA exhibited reduced structural integrity and increased cellular scatter with increasing concentrations of VPA. A marked increase in ethidium bromide uptake was observed concomitantly with the loss of organoid integrity, demonstrating a cytotoxic effect of VPA on the primary tumor organoids.
Figure 6.
Patient tumor cell sensitivity to VPA in 3D culture. Primary tumor and pleural effusion-derived breast cancer cells were embedded in Matrigel and treated with VPA for 96 h. (A) The effect of VPA was assessed by light microscopy (right panel) and fluorescent dye (left panel) to identify live (green) and dead cells (red). (B) Correlation of EC50 of VPA and predicted response from drug response signatures in the fresh tumor samples grown in 3D. Source data is available for this figure at www.nature.com/msb.
A dose-response assay was carried out to determine the relative sensitivity of tumor organoids to VPA treatment. From the dose-response curves for each tumor, we determined the EC50. Individual tumors of the more aggressive subtypes of breast cancer (basal-like, HER2-overexpressing, and luminal B) show higher responsiveness than the less aggressive subtypes (luminal A and normal-like). As shown in Figure 6B, there is a statistically significant correlation between the predicted sensitivity to VPA and EC50 (P=0.006). These studies validate both the genomic predictions and the utility of 3D culture of primary tumor samples for a rapid, efficient, and relevant preclinical analysis of drug sensitivity.
In vivo validation of drug response signature predictions
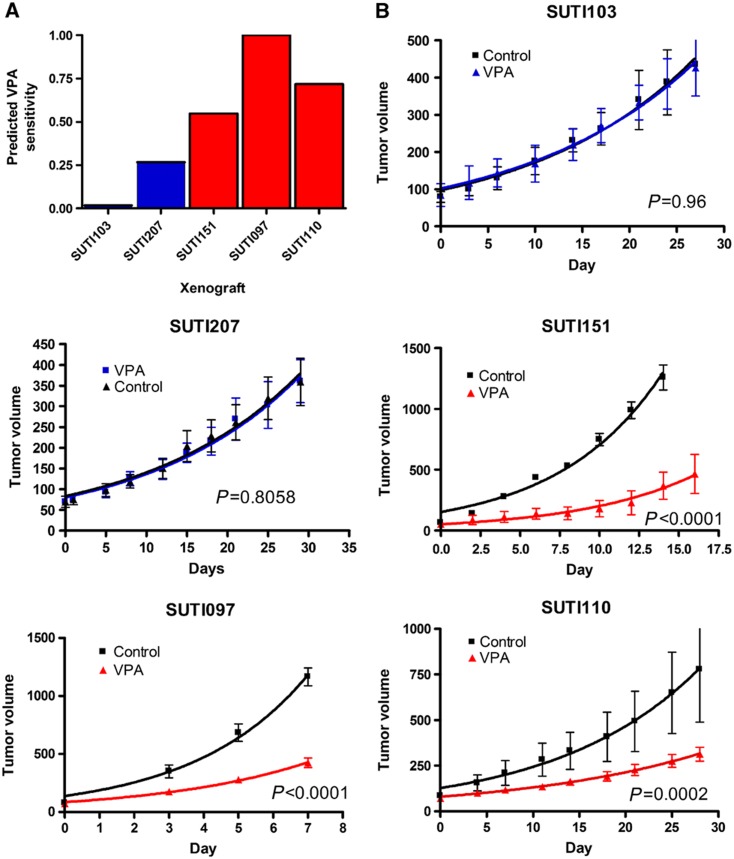
To further assess the accuracy of our predictions, we used mice with tumor-specific xenografts derived from human breast cancers. For this analysis, the gene expression patterns of patient tumors were analyzed using our drug response signature to VPA. We selected different patient tumors predicted to have high or low sensitivity to VPA (Figure 7A) to test our prediction in the corresponding mouse xenograft models generated from these same tumors.
Figure 7.
In vivo validation of computationally predicted responsiveness to VPA using human breast cancer xenografts. (A) Predicted sensitivity of five breast tumors (four basal and one luminal) to VPA. The gene expression patterns of the patient tumors were analyzed using in vitro drug response signatures to VPA. (B) In vivo response to VPA treatment on xenografts generated from the primary tumors in (A). Blue and red lines: VPA group; black line: saline control group. Each group had five mice. Tumor growth rates were plotted as the mean tumor volumes of each group±s.e.m. Source data is available for this figure at www.nature.com/msb.
As shown in Figure 7B, the three tumors predicted to be sensitive to VPA show a highly significant smaller size when treated with VPA compared with placebo (Figure 7B, P<0.0002 for each tumor). No change in tumor size after treatment with VPA was seen in the tumors predicted to be resistant (Figure 6B, P=0.96 and P=0.81). This is an important finding, as it suggests that this genomic approach can accurately predict the response of an individual patient's tumor to a targeted therapy. We next compared conventional chemotherapy with VPA treatment in one tumor predicted to be sensitive to VPA. Each group included 10 mice. In this experiment, VPA decreased tumor growth significantly more than doxorubicin (P<0.0001) (Supplementary Figure S4), further highlighting the potential effectiveness of VPA to inhibit tumor growth in vivo in a sensitive tumor.
Discussion
Clinical trial designs using unselected populations often fail to identify significant responses that may be relevant only to a subset of tumors. By using prior information, clinical trials can be rationally designed to test therapeutic regimens in tumor types identified as most likely to be responsive. However, prior information is not always available. Therefore, we developed MATCH as an unbiased genomic approach to predicting drug sensitivity in individual tumors.
MATCH leverages the large amount of publicly available genomic data to investigate optimal targeted therapy for individual cancers. The example of VPA in breast cancer shows the superiority of individualized genomic predictions over simply using traditional cancer subtypes. Current methods to classify breast cancer phenotypes utilize tumor pathology and signaling pathway status to place the tumor into a molecular subtype (Perou et al, 2000; Sorlie et al, 2001, 2003; Hu et al, 2006; Desmedt et al, 2008; Wirapati et al, 2008; Parker et al, 2009). However, the existing molecular subtypes are not ideal as the sole categorization for defining breast cancer therapy, as heterogeneity exists within each subtype (Di Cosimo and Baselga, 2010; Foulkes et al, 2010). In fact, we are learning that there is increased complexity among subtypes in clinical course and response to therapy. For example, HER2/ERBB2-overexpressing tumors differ in their natural history and/or response to therapy based on lymphocyte-associated genes, PTEN status, and PI3K pathway status (Alexe et al, 2007; Rody et al, 2009; Esteva et al, 2010). Thus, significant heterogeneity remains uncharacterized even within breast cancer subtypes, highlighting the need for approaches investigating drug sensitivity or clinical phenotypes at the individual patient level.
Although VPA had enriched sensitivity in the more aggressive breast cancer subtypes, our studies show that there is great variation in sensitivity of patient tumors to treatment with VPA alone but that there is a definable population of breast tumors that are markedly sensitive to VPA. VPA is known to be a HDAC inhibitor, a class without any well-defined biomarkers of response. VPA is of considerable interest because it is oral, already FDA-approved for treatment of epilepsy, and well tolerated over the long term. The low toxicity of VPA makes this drug an ideal candidate to further assess its potential in the treatment of selected tumors within aggressive breast cancer subtypes, such as triple negative tumors. VPA is currently being studied in clinical trials against breast, lung, ovarian, thyroid, melanoma, prostate, and cervical cancers (Michaelis et al, 2007). The fact that this drug is being tested against so many different cancers without a clear indication of potential efficacy highlights the need for a more targeted approach to finding the populations of patients who will respond. For example, trials of another HDAC inhibitor, SAHA, in lung cancer have shown stable disease but no responses, perhaps due to the small number of subjects receiving therapeutic doses (Vansteenkiste et al, 2008; Traynor et al, 2009). The ability to increase trial efficiency by excluding people who will not respond could improve such trials. By using MATCH to couple personalized computational prediction with in vitro and in vivo validation work, as presented in this study, a systematic methodology for streamlining identification of optimal therapeutic regimens can be leveraged in the clinical setting. We use the genomic predictions to guide our pharmacological studies, followed by experimental validation in actual patient tumor cells to determine clinical significance before initiating a clinical trial. We then rapidly verify the accuracy of these predictions using 2D in vitro cell lines and 3D cell cultures generated from fresh tumors. In the VPA example, we found significant correlation between predicted and actual sensitivity in all experimental conditions.
MATCH has some limitations. First, effective doses of drugs in vitro may not be the same as in vivo due to factors such as increased protein binding in culture media or organ metabolism. Thus, MATCH gives the relative sensitivity of individual tumors but cannot determine absolute doses for use. Second, MATCH cannot predict all off target effects and side effects of drugs. Third, not all tumors predicted to be sensitive by MATCH will be sufficiently sensitive to clinical doses of drugs. MATCH is useful for identifying patient populations most likely to respond (or not) to a drug. Identifying relevant populations for a drug boosts the proportion of possible responders in a trial population, decreasing the chance of falsely negative trials.
MATCH leverages high-throughput pharmacogenomic and individual-based tumor drug response analyses, and serves to identify the appropriate cancer patients for specific drug therapies. This rational approach to drug selection has the potential to streamline preclinical testing, optimize clinical trial design, and identify clinically useful biomarkers early in drug development.
Materials and methods
Calculating drug sensitivity
The VPA response signature was defined using the Connectivity Map build 1 (Lamb et al, 2006). The PLX4032 signature was defined using samples from GSE20051 (Joseph et al, 2010). A detailed description of the signature generation methods can be found at http://io.genetics.utah.edu/files/bildres. In brief, Mas5 normalized gene expression data from the Connectivity Map website were log normalized and then used as the training set in the analysis. Each data set was also Mas5 normalized and then log transformed. Following normalization, signature and data sets were quantile normalized, and 25% of probes on the microarrays with the lowest expression and lowest variability were prefiltered from the analysis. Distance weighted discrimination standardization was used to correct for batch effects between data sets. A Bayesian binary regression algorithm was then used on the training set to generate a list of probes and weights relative to the first principle component for calculating a metagene score that is then converted to a probability. The probability output from the binary regression model was subtracted from one, so that probabilities closer to one indicated higher probability of sensitivity to the drug (Bild et al, 2006). Parameters were optimized to minimize the P-value for a t-test comparing the predictions of the treated and untreated samples in the training set in the LOOCV analysis. To compare independently collected and processed data sets, we log normalized and then linearly transformed the binary regression output for each data set and for each signature to span from 0 to 1. To enable complete reproduction of our results, the input files, output files, and the binary regression program used in this study are available at http://io.genetics.utah.edu/files/bildres. The binary regression program is also available via a web interface through a Genepattern module at http://genepattern.genome.duke.edu/signature/. Genes and probes comprising the VPA response signature are found in Supplementary Table S2. For signature validation, CEL files were downloaded from GEO GSE1615, which contain gene expression data for human ovarian cells before and after treatment with VPA, GEO GSE7553, which contains gene expression data for various skin cancers and normal skin, and from Connectivity Map batches 2, 35, 44, 56, 63, 70, 626, 757, and 767 (Wood et al, 2005).
Study population and analysis of expression data for multiple cancer types
CEL files were downloaded from GEO for data set GSE5364, which contain gene expression data for tumor and adjacent normal tissues for breast, colon, esophagus, lung, liver, and thyroid cancers, and from CaBIG for the woost-00041 data set, which contains gene expression data from over 300 different cell lines provided by Glaxo-Smith-Kline (GSK), https://array.nci.nih.gov/caarray/project/woost-00041 (Yu et al, 2008). All data were mas5 normalized, and samples with a ratio >3 between probes at the 3′ and 5′ end of GAPDH were considered potentially degraded and excluded from further analysis. We excluded sarcomas and hematopoietic malignancies from our analysis as our signatures are generated with epithelial cells. VPA sensitivity predictions were then generated as described above. For duplicate and triplicate samples in the GSK panel, final predictions were averaged.
Study population and analysis of expression data for breast cancer subtypes
Breast cancer samples from 11 microarray studies were employed in this analysis (Supplementary Table S1). Duplicate samples from GEO data sets gse6532, gse7390, and gse3494 and cell line and normal samples from gse7904 were removed. As not all public data sets had clinical annotations, intrinsic breast cancer subtypes were assigned as described previously (Sorlie et al, 2001). Briefly, a training set of 259 samples representing the five subtypes (luminal A, luminal B, HER2-overexpressing, basal-like, and normal-like) and 306 genes were used to build a corresponding set of five centroids (Hu et al, 2006). Features were assigned using Entrez Gene identifiers and duplicate identifiers were collapsed to the mean. Each test case was then compared with the five standardized centroids using Spearman's rank correlation and assigned the subtype of the nearest centroid.
Dose-response assays
Breast cancer cell lines obtained from ATCC (HCC1806, HCC1428, HCC1143, BT549, BT474, MDA-MB-361, MDA-MB-435s, MDA-MB-231, MDA-MB-453, SKBR3, ZR75, CAMA I, MCF7, Hs578t, T47D) were seeded in 384 plates (NUNC) in MEBM media (Lonza) containing 5% fetal bovine serum (GIBCO), at a density to yield 80% confluency in control treated wells at 96 h post-treatment (as determined by growth curves). After 24 h, VPA (Calbiochem) was added at 10 doses ranging from 64 to 0.25 mM. A BIOMEK 3000 (Beckman Coulter) robot was used to seed the cells and dispense the drug. After 96 h, CellTiter-Blue Reagent (Promega) was added to test cell viability. After 2 h of incubation at 37°C, the fluorescence was recorded (560(20)Ex/590(10)Em) using a Victor3V 1420 Multilabel Counter (Perkin-Elmer) plate reader.
Breast cancer pleural effusion collection, growth in 3D cultures, and drug response assays
All research involving human samples have been approved by the authors' institutional review board. With informed consent, breast tumor samples were collected from patients at the University of Utah. After obtaining written informed consent, pleural effusion samples were collected from excess fluid obtained at the time of therapeutic thoracenteses or solid tumor samples were collected from mastectomies or lumpectomies. Malignant cells from the pleural effusion or solid tumors were either used to isolate RNA or grown in 3D cultures to form organoid structures. Prior to embedding in BD Matrigel Matrix, Growth Factor Reduced (BD Biosciences), solid tumors were incubated overnight in tissue mix media, MEBM containing 10% fetal bovine serum, collagenase type III (1500 U/ml) (Worthington), and hyaluronidase (1000 U/ml) (Sigma). The single cells from either solid tumor or pleural effusion were then re-suspended in complete media and plated in a 24-well ultra low attachment tissue culture plate for 24 h at 37°C in 5% CO2 to form organoid structures. Cells were diluted in media and added to matrigel for a total volume of 4:1 matrix to media. A total of 30 μl was then seeded into a 96-well half-area assay plate at 104 cells/well. After 24 h, VPA was added at a range of dosages determined by our drug response assay. After 96 h, viability was determined as above. Images were taken using an Olympus IX81-ZDC DSU with ORCAER camera and Slidebook 5.0 software
In vivo drug treatment experiments
Triple negative breast cancers were obtained from Stanford University Hospital with informed consent. Tumor tissue was frozen on dry ice for RNA isolation and microarray analysis. Tumors were minced and then implanted into the number 2 and/or number 4 mammary fat pads of 5–10 female NOD/SCID mice (NOD.CB17–Prkdcscid/J, Jackson Laboratory West). When tumor xenografts grew >5 mm in diameter, mice were stratified by tumor size and randomized into two or three treatment groups of 5–10 mice each: (1) a VPA group, receiving daily intraperitoneal administration of VPA (400 mg/kg) for up to 4 weeks; (2) a control group, receiving daily intraperitoneal administration of control vehicle (sterile saline) for up to 4 weeks; and, in another set of experiments, (3) a doxorubicin group, receiving intraperitoneal administration of doxorubicin (2 mg/kg) at time zero and at 3 weeks. Tumors were measured twice a week with a caliper in two dimensions. Tumor size (M) was calculated as M=a2b/2, where a is the maximum width and b is the maximum length. Means of tumor volume in the same treatment group were calculated, and growth curves were established as a function of time. All animal care was in accordance with Stanford University and IACUC guidelines.
Microarray data sets
All novel microarray data sets are posted on GEO under accession GSE18331. CEL files for the cell lines used in the 2D culture experiments have previously been made available as GSE3156. All input files, program files, and output files are posted at http://io.genetics.utah.edu/files/bildres. Affymetrix U133A 2.0 microarrays were used according to the manufacturer's protocols.
Statistics
Means were compared with t-tests or with ANOVA with post-test t-tests using Bonferroni multiple comparison testing, except where indicated. Correlations were calculated using Spearman's r. Drug-response graphs are plotted as means with SEM, fitted to an exponential curve, and compared with MANOVA. We considered P<0.05 statistically significant. Calculations were done using MATLAB, R, and Graphpad Prism version 4.02.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Health (R01GM085601, AHB and SSJ); the Pharmaceutical Research and Manufacturers of America (AHB); a Multidisciplinary Cancer Research Training Program award (T32 CA93247, RS and AC), and an award from the MIDT cancer center support grant. The Breast Interdisciplinary Group of the University of Utah is acknowledged for their assistance in breast tumor collection.
Author contributions: AHB, AC, AS, and SSJ designed the study. AHB, AC, and SSJ wrote the majority of the manuscript. AC and AG performed the microarray analyses. RW and RS performed two- and three-dimensional dose-response assays. BEW captured the patient tumor images and assisted in growth of tumors in matrigel. HZ performed the mouse xenograft experiments. JTC and EJ developed statistical and computational resources.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adjei AA, Christian M, Ivy P (2009) Novel designs and end points for phase II clinical trials. Clin Cancer Res 15: 1866–1872 [DOI] [PubMed] [Google Scholar]
- Alexe G, Dalgin GS, Scanfeld D, Tamayo P, Mesirov JP, DeLisi C, Harris L, Barnard N, Martel M, Levine AJ, Ganesan S, Bhanot G (2007) High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res 67: 10669–10676 [DOI] [PubMed] [Google Scholar]
- Barron JJ, Cziraky MJ, Weisman T, Hicks DG (2009) HER2 testing and subsequent trastuzumab treatment for breast cancer in a managed care environment. Oncologist 14: 760–768 [DOI] [PubMed] [Google Scholar]
- Bast RC Jr, Hortobagyi GN (2004) Individualized care for patients with cancer—a work in progress. N Engl J Med 351: 2865–2867 [DOI] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA Jr, Marks JR, Dressman HK, West M, Nevins JR (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439: 353–357 [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Labarge MA (2005) Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell 7: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, Mohsin S, Osborne CK, Chamness GC, Allred DC, O'Connell P (2003) Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 362: 362–369 [DOI] [PubMed] [Google Scholar]
- Coulson SG, Kumar VS, Manifold IM, Hatton MQ, Ramakrishnan S, Dunn KS, Purohit OP, Bridgewater C, Coleman RE (2010) Review of testing and use of adjuvant trastuzumab across a cancer network—are we treating the right patients? Clin Oncol (R Coll Radiol) 22: 289–293 [DOI] [PubMed] [Google Scholar]
- Dam V, Morgan BT, Mazanek P, Hogarty MD (2006) Mutations in PIK3CA are infrequent in neuroblastoma. BMC Cancer 6: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancey JE, Dobbin KK, Groshen S, Jessup JM, Hruszkewycz AH, Koehler M, Parchment R, Ratain MJ, Shankar LK, Stadler WM, True LD, Gravell A, Grever MR (2010) Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clin Cancer Res 16: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Desmedt C, Ruiz-Garcia E, Andre F (2008) Gene expression predictors in breast cancer: current status, limitations and perspectives. Eur J Cancer 44: 2714–2720 [DOI] [PubMed] [Google Scholar]
- Di Cosimo S, Baselga J (2010) Management of breast cancer with targeted agents: importance of heterogeneity [corrected]. Nat Rev Clin Oncol 7: 139–147 [DOI] [PubMed] [Google Scholar]
- Downward J (2006) Cancer biology: signatures guide drug choice. Nature 439: 274–275 [DOI] [PubMed] [Google Scholar]
- Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, Maglathlin RL, Lewis TA, Liau LM, Nghiemphu P, Mellinghoff IK, Louis DN, Loda M, Carr SA, Kung AL, Golub TR (2009) Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol 27: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta PR, Maity A (2007) Cellular responses to EGFR inhibitors and their relevance to cancer therapy. Cancer Lett 254: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva FJ, Guo H, Zhang S, Santa-Maria C, Stone S, Lanchbury JS, Sahin AA, Hortobagyi GN, Yu D (2010) PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol 177: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R, Nau H, Gandolfi AJ, Putnam CW, Brendel K (1991) Valproic acid hepatotoxicity in human liver slices. Drug Chem Toxicol 14: 375–394 [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB (2010) Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foekens JA, Wang Y, Martens JW, Berns EM, Klijn JG (2008) The use of genomic tools for the molecular understanding of breast cancer and to guide personalized medicine. Drug Discov Today 13: 481–487 [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. N Engl J Med 363: 1938–1948 [DOI] [PubMed] [Google Scholar]
- Freedman AN, Sansbury LB, Figg WD, Potosky AL, Weiss Smith SR, Khoury MJ, Nelson SA, Weinshilboum RM, Ratain MJ, McLeod HL, Epstein RS, Ginsburg GS, Schilsky RL, Liu G, Flockhart DA, Ulrich CM, Davis RL, Lesko LJ, Zineh I, Randhawa G et al. (2010) Cancer pharmacogenomics and pharmacoepidemiology: setting a research agenda to accelerate translation. J Natl Cancer Inst 102: 1698–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau CS, Chang CJ, Tsai FJ, Chao PF, Gau SS (2010) Association between mood stabilizers and hypothyroidism in patients with bipolar disorders: a nested, matched case-control study. Bipolar Disord 12: 253–263 [DOI] [PubMed] [Google Scholar]
- Griffith LG, Swartz MA (2006) Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol 7: 211–224 [DOI] [PubMed] [Google Scholar]
- Gusterson B (2009) Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer 9: 128–134 [DOI] [PubMed] [Google Scholar]
- Haibe-Kains B, Desmedt C, Piette F, Buyse M, Cardoso F, Van't Veer L, Piccart M, Bontempi G, Sotiriou C (2008) Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics 9: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D et al. (2006) The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Reeves K, Han X, Fairchild C, Platero S, Wong TW, Lee F, Shaw P, Clark E (2007) Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res 67: 2226–2238 [DOI] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, Tsai J, Chapman PB, Bollag G, Solit DB, Rosen N (2010) The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci USA 107: 14903–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ (2007) The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol 1: 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetz S, Desai J, Chan E, Hecht JR, O'Dwyer PJ, Lee RJ, Nolop KB, Saltz L (2010) PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J Clin Oncol 28 (Suppl): 3534 [Google Scholar]
- Lamb J (2007) The connectivity Map: a new tool for biomedical research. Nat Rev Cancer 7: 54–60 [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR (2006) The connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929–1935 [DOI] [PubMed] [Google Scholar]
- Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, Lebigot I, Djelti F, Tourdes A, Gestraud P, Hupe P, Barillot E, Cruzalegui F, Tucker GC, Stern MH, Thiery JP, Hickman JA, Dubois T (2008) Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res 10: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J (2007) Sorting out breast-cancer gene signatures. N Engl J Med 356: 294–297 [DOI] [PubMed] [Google Scholar]
- Michaelis M, Doerr HW, Cinatl J Jr (2007) Valproic acid as anti-cancer drug. Curr Pharm Des 13: 3378–3393 [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM et al. (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- PhRMA (2009) Medicines in development for cancer. In http://www.phrma.org/files/attachments/meds_in_dev/09-046PhRMACancer09_0331.pdf
- Ravi RK, Weber E, McMahon M, Williams JR, Baylin S, Mal A, Harter ML, Dillehay LE, Claudio PP, Giordano A, Nelkin BD, Mabry M (1998) Activated Raf-1 causes growth arrest in human small cell lung cancer cells. J Clin Invest 101: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, Barrette TR, Ghosh D, Varambally S, Chinnaiyan AM (2007) Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia 9: 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D, Engels K, Karn T, Kaufmann M (2009) T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res 11: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360: 790–800 [DOI] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung MC, van de Vijver MJ, Valero V, Gray JW, Bernards R et al. (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68: 6084–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker AV, Boussioutas A, Bowtell DD (2006) The challenges of gene expression microarrays for the study of human cancer. Cancer Cell 9: 333–339 [DOI] [PubMed] [Google Scholar]
- Traynor AM, Dubey S, Eickhoff JC, Kolesar JM, Schell K, Huie MS, Groteluschen DL, Marcotte SM, Hallahan CM, Weeks HR, Wilding G, Espinoza-Delgado I, Schiller JH (2009) Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol 4: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteenkiste J, Van Cutsem E, Dumez H, Chen C, Ricker JL, Randolph SS, Schoffski P (2008) Early phase II trial of oral vorinostat in relapsed or refractory breast, colorectal, or non-small cell lung cancer. Invest New Drugs 26: 483–488 [DOI] [PubMed] [Google Scholar]
- Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schutz F, Goldstein DR, Piccart M, Delorenzi M (2008) Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 10: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JR, Nelson-Degrave VL, Jansen E, McAllister JM, Mosselman S, Strauss JF III (2005) Valproate-induced alterations in human theca cell gene expression: clues to the association between valproate use and metabolic side effects. Physiol Genomics 20: 233–243 [DOI] [PubMed] [Google Scholar]
- Xing J, Liu R, Xing M, Trink B (2011) The BRAFT1799A mutation confers sensitivity of thyroid cancer cells to the BRAFV600E inhibitor PLX4032 (RG7204). Biochem Biophys Res Commun 404: 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Ganesan K, Tan LK, Laban M, Wu J, Zhao XD, Li H, Leung CH, Zhu Y, Wei CL, Hooi SC, Miller L, Tan P (2008) A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet 4: e1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.