The authors screen for compounds that show synergistic antifungal activity when combined with the widely-used fungistatic drug fluconazole. Chemogenomic profiling explains the mode of action of synergistic drugs and allows the prediction of additional drug synergies.
Keywords: antifungal, combination, pathogen, resistance, synergism
Abstract
Resistance to widely used fungistatic drugs, particularly to the ergosterol biosynthesis inhibitor fluconazole, threatens millions of immunocompromised patients susceptible to invasive fungal infections. The dense network structure of synthetic lethal genetic interactions in yeast suggests that combinatorial network inhibition may afford increased drug efficacy and specificity. We carried out systematic screens with a bioactive library enriched for off-patent drugs to identify compounds that potentiate fluconazole action in pathogenic Candida and Cryptococcus strains and the model yeast Saccharomyces. Many compounds exhibited species- or genus-specific synergism, and often improved fluconazole from fungistatic to fungicidal activity. Mode of action studies revealed two classes of synergistic compound, which either perturbed membrane permeability or inhibited sphingolipid biosynthesis. Synergistic drug interactions were rationalized by global genetic interaction networks and, notably, higher order drug combinations further potentiated the activity of fluconazole. Synergistic combinations were active against fluconazole-resistant clinical isolates and an in vivo model of Cryptococcus infection. The systematic repurposing of approved drugs against a spectrum of pathogens thus identifies network vulnerabilities that may be exploited to increase the activity and repertoire of antifungal agents.
Introduction
The recent increase in fungal infection rates presents a serious clinical challenge (Arendrup et al, 2009; Gullo, 2009; Shorr et al, 2009). Immune-suppressed individuals, including transplant, cancer chemotherapy and HIV-infected patients, often succumb to opportunistic fungal pathogens from the genera Candida, Cryptococcus, Aspergillus and others (Groll et al, 1996; Baddley et al, 2001; Clark and Hajjeh, 2002; Richardson and Warnock, 2003). Unlike bacterial infections that can be treated with multiple antibiotic classes, therapeutic options for fungal infections are limited. The polyene amphotericin B, discovered in 1955, remains a front line fungicidal drug; however, amphotericin B non-specifically disrupts cell membrane integrity, with concomitant severe patient toxicity. Synthetic azole antifungals such as fluconazole were introduced 40 years ago and inhibit lanosterol 14α-demethylase, the gene product of ERG11, an essential cytochrome P450 enzyme in the ergosterol biosynthetic pathway (Groll et al, 1998; Revankar et al, 2004). Fluconazole binds to the heme Fe(III) of Erg11, resulting in depletion of ergosterol, the accumulation of C-14 methyl sterols and cell membrane disruption. The crossreactivity of azoles toward human P450 enzymes also results in toxicity and, moreover, clinical resistance is prevalent (Cannon et al, 2009; Marie and White, 2009). Finally, the echinocandins, which include caspofungin, micafungin and anidulafungin, were introduced 10 years ago and inhibit the cell wall biosynthesis enzyme β-(1,3)-D-glucan synthase; however, these agents have a restricted antifungal spectrum (Sucher et al, 2009). The dearth of selective agents and emerging patterns of clinical resistance demand new antifungal strategies.
A primary challenge in antifungal drug discovery is the paucity of fungal-specific molecular targets that are essential for cell growth, due to the conserved biochemical and molecular biological networks of all eukaryotes. This problem is exacerbated by the observation that many essential yeast genes can provide sufficient function at a fraction of wild-type dosage (Yan et al, 2009). Although only ∼1100 of the ∼6000 genes in yeast are essential under nutrient-rich growth conditions (Winzeler et al, 1999), almost all genes become essential in specific genetic backgrounds in which another non-essential gene has been deleted or otherwise attenuated, an effect termed synthetic lethality (Tong et al, 2001). Genome-scale surveys suggest that over 200 000 binary synthetic lethal gene combinations dominate the yeast genetic landscape (Costanzo et al, 2010). The genetic buffering phenomenon is also manifest as a phalanx of differential chemical–genetic interactions in the presence of sublethal doses of bioactive compounds (Hillenmeyer et al, 2008). These observations illuminate the inherent redundancy of genetic networks, and frame the problem of interdicting network functions with single agent therapeutics (Hopkins, 2008).
This genetic network organization suggests that judicious combinations of small molecule inhibitors of both essential and non-essential targets may elicit additive or synergistic effects on cell growth (Sharom et al, 2004; Agoston et al, 2005; Fitzgerald et al, 2006; Lehar et al, 2007, 2008; Hopkins, 2008). Indeed, ad hoc combinations of anti-infective drugs are frequently used to treat fungal infections (Eliopoulos and Moellering, 1991; Johnson and Perfect, 2010). However, this chance approach fails to exploit richness of the chemical–genetic landscape (Sharom et al, 2004; Hopkins, 2008; Lehar et al, 2008). Instead, unbiased screens for synergistic enhancers of a specific bioactivity that are not themselves active, sometimes termed syncretic combinations, are needed to fully explore chemical space (Keith et al, 2005). Compounds that enhance the activity of known agents in model yeast and cancer cell line systems have been identified both by focused small molecule library screens (Borisy et al, 2003; Zhang et al, 2007; Zhai et al, 2010) and by computational methods (Lehar et al, 2007; Nelander et al, 2008; Jansen et al, 2009; Zinner et al, 2009). Furthermore, direct tests of synergistic compounds have successfully yielded combinations that are active against pathogenic fungi, including the combination of fluconazole with chemical inhibitors of Hsp90, calcineurin or ARF (Cowen et al, 2009; Singh et al, 2009; Epp et al, 2010) and the antibiotic polymyxin B (Zhai et al, 2010).
To extend the strategy of chemical synthetic lethality to clinically relevant fungal pathogens, we interrogated a focused bioactive library of known drugs for synergistic enhancers of the fungistatic drug fluconazole in systematic screens against Candida albicans, Cryptococcus neoformans and Cryptococcus gattii, as well as the genetically tractable budding yeast Saccharomyces cerevisiae. Compounds not previously recognized in the clinic as antifungal agents caused potent growth inhibition in conjunction with fluconazole, often in a genus- or species-specific manner. Selected combinations were characterized for mechanism of action and shown to be active against fluconazole-resistant isolates and efficacious in an in vivo infection model. The combinatorial redeployment of known drugs defines a powerful antifungal strategy and establishes a number of potential lead combinations for future clinical assessment.
Results
Systematic antifungal potentiation screens in model and pathogenic fungi
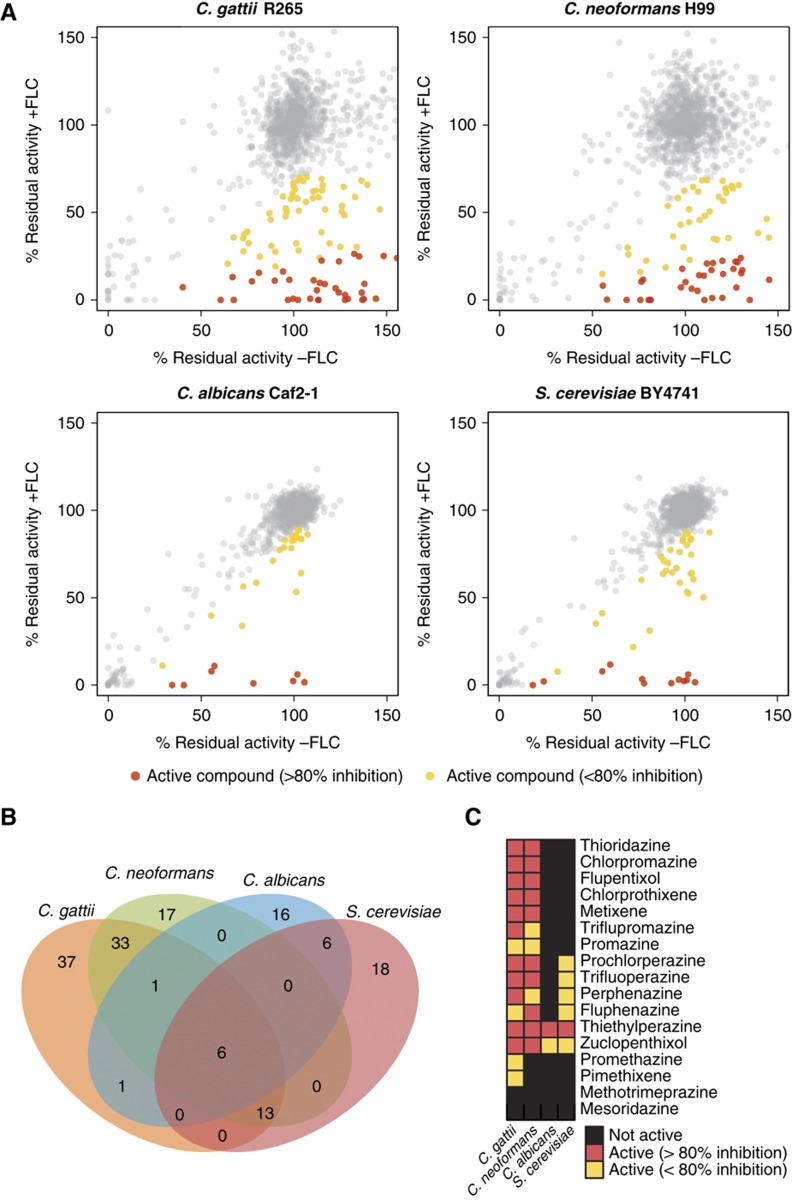
Cell-based high-throughput screens were performed on a panel of four fungal strains to identify small molecules that potentiate fluconazole across a range of genera and species. The human pathogens C. neoformans (H99), C. gattii (R265) and C. albicans (Caf2-1) as well as the model fungus S. cerevisiae (BY4741) were screened in duplicate against the Prestwick library, which consists of 1120 off-patent drugs and other bioactive agents (http://www.prestwickchemical.com). To identify compounds that potentiate the effect of fluconazole, yet have minimal antifungal activity on their own, each screen was performed in the presence and absence of 0.5 minimal inhibitory concentration (MIC) of fluconazole at a single compound concentration of 30 μM. Residual activity was calculated for each compound and all data were normalized for plate- and row/column-specific effects (Supplementary Figure S1; see Supplementary Table S1 for screen data). Hits were determined using median absolute deviation (MAD) statistics. By this criterion, 43 compounds were active against S. cerevisiae, 30 against C. albicans, 70 against C. neoformans and 91 against C. gattii (Figure 1A and B).
Figure 1.
Unbiased screens for bioactive compounds that potentiate the antifungal activity of fluconazole. (A) Scatter plots for Prestwick library screens for four fungal species. Growth inhibition caused by compounds in the absence (x axis) and presence of fluconazole (y axis) is represented by residual activity after treatment. Yellow and red filled circles indicate compounds that were classified as active (2 median absolute deviations below the diagonal). Compounds that inhibited growth in the presence of fluconazole by at least 80% compared with the effect of that compound alone are highlighted in red; FLC, fluconazole. (B) Overlap of hits between different fungal species. (C) Activity of 17 phenothiazine/thioxathene compounds in different fungal species.
The set of 148 compounds that potentiated the antifungal action of fluconazole in one or more of the screens (Supplementary Figure S2) was structurally diverse and represented a broad range of different therapeutic activities, including antiparasitics, cardiovascular protectives, dermatologicals, genitourinary tract anti-infectives, hormone modulators and a variety of neuroleptic drugs. Notably, 15 of the 17 tricyclic phenothiazine/thioxanthene antipsychotics present in the Prestwick library exhibited strong interactions with fluconazole against C. gattii and C. neoformans (Figure 1C). Derivatives of tricyclic phenothiazines inhibit fatty acid synthesis and disrupt lipid trafficking (Li et al, 2008).
A striking number of hits were species or genus specific (Figure 1B). Six compounds were hits in all screens: (i) the antidepressant sertraline (Zoloft®); (ii) the monoamine oxygenase inhibitor pirlindole, also known to have antidepressant activity; (iii) the allylamine antifungal naftifine; (iv) the antibiotic prodrug pivampicillin; (v) the antinausea drug thiethylperazine (Torecan®); and (vi) the antipsychotic drug zuclopenthixol. The latter two compounds are members of the large family of phenothiazines that have antipsychotic and other central nervous system (CNS) activities.
Synergy assessment and fungicidal activity
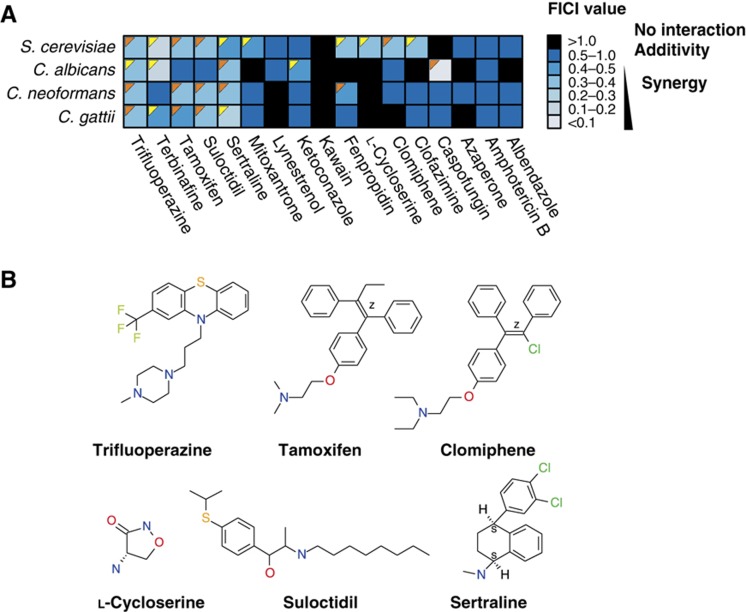
To determine whether hit compounds acted in a synergistic or additive manner with fluconazole, we selected 12 of the 148 hits (albendazole, azaperone, clofazimine, clomiphene, L-cycloserine, kawain, lynestrenol, mitoxantrone, sertraline, suloctidil, tamoxifen and trifluoperazine) for detailed studies in all four fungal species. We based this selection on an analysis of distinct chemical class, with one or two representative structures from families of similar agents that emerged in the screen, commercial availability of compound, therapeutic importance, and known mode of action (Supplementary Figure S3). These criteria yielded a tractable number of hit compounds for detailed downstream analysis. We also tested five known antifungal drugs, both as positive controls and to explore other potential interactions with fluconazole: amphotericin B, the ergosterol biosynthesis inhibitors ketoconazole, terbinafine (an allylamine analog of naftifine used in the clinic) and fenpropidin (an agricultural fungicide), and the echinocandin caspofungin. Dose-dependent MIC values for these 17 compounds were determined for each of the four species (Supplementary Table S2). The interaction of each compound with fluconazole was assessed by standard concentration matrix (checkerboard) analysis (Figure 2A; Supplementary Table S3). Data were quantified by calculation of the fractional inhibitory concentration index (FICI), the accepted method for drug interactions in infectious disease (Eliopoulos and Moellering, 1991; Odds, 2003). Only two compounds, sertraline and trifluoperazine, exhibited synergy with fluconazole against all four fungal species. A number of synergizers exerted effects exclusively on a particular species: S. cerevisiae was uniquely susceptible to four different compounds in the presence of fluconazole (clofazimine, clomiphene, L-cycloserine and mitoxantrone), while only C. albicans was susceptible to ketoconazole or caspofungin in combination with fluconazole. Neither Cryptococcus species exhibited any unique synergistic susceptibilities. Most hits from the screens were confirmed as synergistic with fluconazole, except for albendazole, azoperone and kawain in S. cerevisiae, and azaperone, L-cycloserine and lynestrenol in C. albicans (Supplementary Figure S4). Quantification of interactions at different drug concentrations revealed some additional synergies with fluconazole: trifluoperazine exhibited synergy against C. albicans, tamoxifen against C. gattii and C. neoformans, and suloctidil against C. neoformans and S. cerevisiae (Supplementary Figure S4). Based on the detailed analysis of these 12 compounds, the high-throughput screens proved a reliable means to identify synergistic drug interactions, with an estimated false positive rate of 0.20 and a false negative rate of 0.28. Importantly, and in contrast to the merely fungistatic effect of fluconazole alone, several combinations of fluconazole and different synergistic compounds were fungicidal, often in a species-dependent manner (Figure 2A). For example, trifluoperazine exhibited synergy with fluconazole and was fungicidal in all species with the exception of C. albicans.
Figure 2.
Synergistic drug interactions with fluconazole. (A) Heat map of drug interactions with fluconazole in each species. Dark blue indicates additive effects (FICI of 0.5–1); lighter shades of blue represent synergy (FICI <0.5). Orange triangles indicate fungicidal drug combinations; yellow triangles indicate fungistatic drug combinations. (B) Chemical structures of the six drugs chosen for detailed mode of action studies. Source data is available for this figure at www.nature.com/msb.
Chemical–genetic profiles of synergistic combinations
We explored the molecular basis for the synergy of trifluoperazine, tamoxifen, clomiphene, sertraline, suloctidil and L-cycloserine with fluconazole (Figure 2B), using established genome-wide methods in S. cerevisiae to identify gene deletion strains that are sensitive to drug treatment (Giaever et al, 1999; Parsons et al, 2006; Hillenmeyer et al, 2008). Genome-wide pools of deletion strains were grown in the presence of drugs, genomic DNA isolated from drug-treated and control cultures, and barcode sequence tags amplified and hybridized to barcode microarrays (Cook et al, 2008).
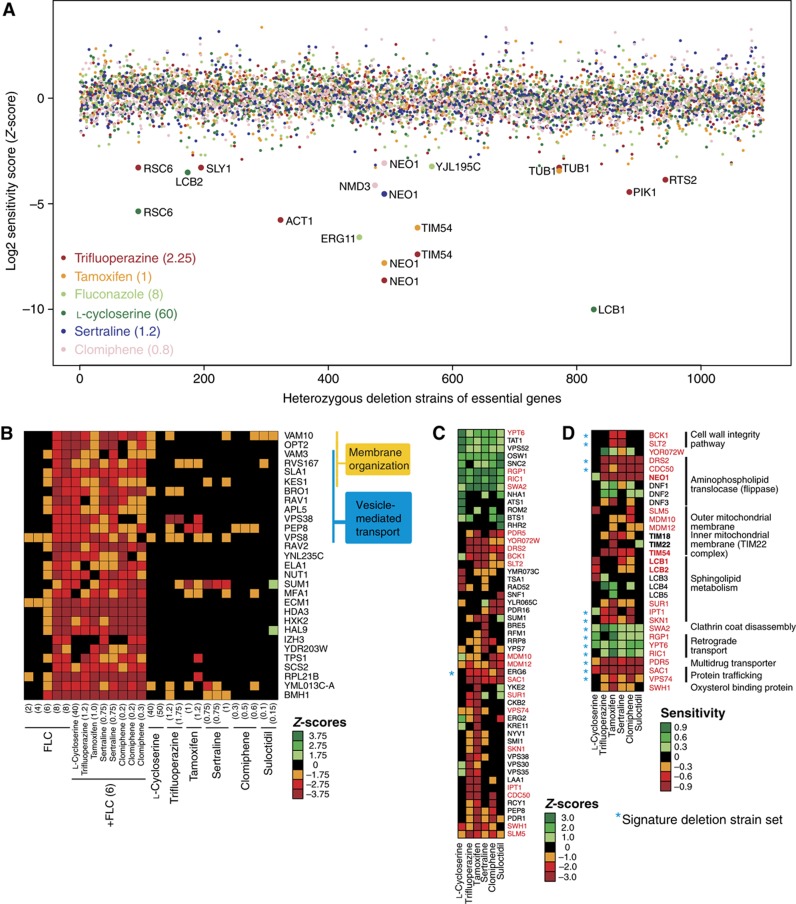
First, we profiled compound action in haplo-insufficiency screens, in which the ∼1000 deletion strains heterozygous for essential genes were tested for drug sensitivity to identify candidate drug targets (Giaever et al, 1999). As expected, deletion of one copy of ERG11 conferred sensitivity to fluconazole (Figure 3A; Supplementary Figure S5). Deletion of one copy of either LCB1 or LCB2, which encode subunits of the enzyme serine palmitoyltransferase that catalyzes the committed step of sphingolipid biosynthesis, caused sensitivity to L-cycloserine (Figure 3A; Supplementary Figure S5). In yeast membrane extracts, high concentrations of L-cycloserine (1 mM) partially inhibit serine palmitoyltransferase (Pinto et al, 1992). All five remaining compounds—trifluoperazine, tamoxifen, clomiphene, sertraline and suloctidil (referred to as the membrane active group)—conferred sensitivity to loss of one copy of the NEO1 gene (Figure 3A; Supplementary Figure S5), which encodes an essential aminophospholipid translocase required for membrane trafficking and vacuolar biogenesis. Deletion of the ortholog of NEO1 in C. neoformans (APT1) has recently been shown to result in hypersensitivity to amphotericin B and fluconazole, as well as attenuated virulence (Hu and Kronstad, 2010). In addition, deletion of one copy of TIM54, a translocase of the inner mitochondrial membrane, confers sensitivity to tamoxifen and trifluoperazine (Figure 3A), consistent with the potential membrane targets of these drugs.
Figure 3.
Chemical–genetic interactions of six syncretic synergizers. (A) Sensitivity of heterozygous essential deletion strains to five different syncretic drugs and fluconazole, as assessed by barcode microarray hybridization. (B) Core set of haploid deletion strains that are sensitive to fluconazole, as assessed by barcode microarray hybridization. Several concentrations of fluconazole were tested to correlate the signature with MIC. The effect of the six syncretic drugs on the core fluconazole profile was examined in the presence or absence of a threshold concentration of fluconazole (6 μg/ml). Genes implicated in membrane organization and vesicle-mediated transport are indicated. (C) Main cluster of haploid deletion strain sensitivities to the six syncretic drugs in the absence of fluconazole, as assessed by barcode microarray hybridization. Strains that have a Z-score more significant than ±3 for at least one of the drugs in duplicate profiles are shown. Gene names in red indicate deletion strains that were chosen for verification by quantitative growth curve assays. (D) Log-ratio scores calculated from individual growth curve assays to confirm chemical–genetic interactions of the six syncretic drugs. Gene names in bold indicate heterozygous deletion strains for essential genes. Values in parentheses indicate drug concentration in μg/ml. Negative Z-scores and log-ratios indicate sensitivity of a strain to a given drug, whereas positive scores represent resistance. Asterisks indicate 14 deletion strains that comprise the core signature set for membrane active compounds. Source data is available for this figure at www.nature.com/msb.
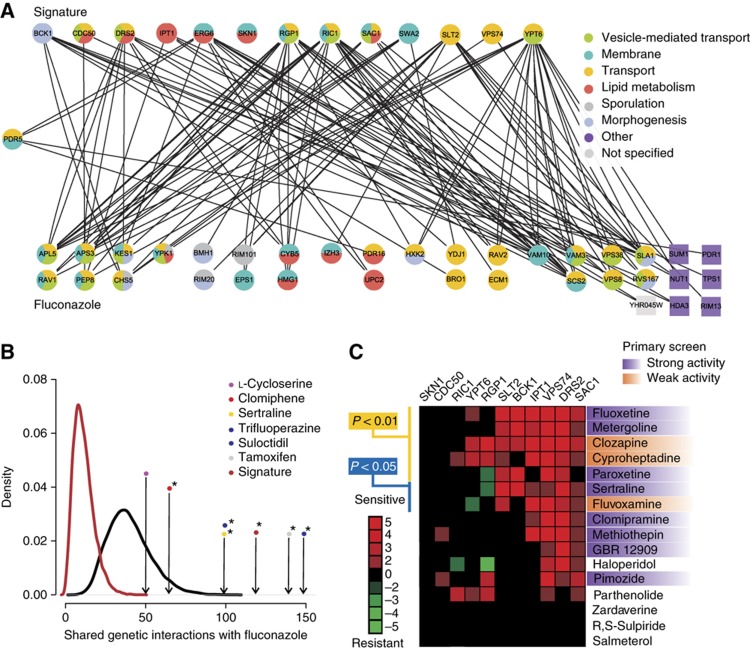
We then generated haploid chemical–genetic profiles for the syncretic compounds individually and in combination with fluconazole (Supplementary Figure S6). This profiling method reveals genes that buffer against drug toxicity and can identify compounds with similar bioactivities based on shared chemical–genetic interaction profiles (Hillenmeyer et al, 2008). Strains deleted for genes that function in vesicle-mediated transport and membrane organization were sensitive to fluconazole alone (Figure 3B). For drug combinations, we chose a concentration of fluconazole (6 μg/ml) that caused ∼20% growth inhibition compared with control and thereby minimized the selection against fluconazole-sensitive strains. Importantly, the syncretic drugs alone did not impair the growth of fluconazole-sensitive deletion strains (Figure 3B; Supplementary Figure S5), but significantly sensitized cells to low doses of fluconazole (Supplementary Table S4). To explore the potential mechanism of this sensitization further, we examined the chemical–genetic profiles of single compounds (Figure 3C; Supplementary Figure S7; Supplementary Table S5). The membrane active group of trifluoperazine, tamoxifen, clomiphene, sertraline and suloctidil caused growth inhibition of a core set of deletion strains that included genes that encode the post-Golgi-associated aminophospholipid translocase (flippase) Drs2 and its activating subunit Cdc50, the ergosterol biosynthesis enzyme Erg6, the protein trafficking factors Sac1 and Vps74, the mitochondrial outer membrane import factors Mdm10 and Mdm12, and the cell wall integrity MAPK kinase Slt2 and its upstream activating kinase Bck1. A number of genes implicated in downstream steps of sphingolipid metabolism, including IPT1, SUR1, SKN1, YPK1 and SWH1, were also required for cell survival in the presence of the membrane active compounds. Notably, strains disrupted for non-essential genes implicated in uncoating of clathrin vesicles (SWA2) and retrograde transport to the cis-Golgi network (YPT6, RGP1, RIC1 and VPS52) were resistant to all six fluconazole synergizers, suggesting that altered vesicle trafficking may compensate for membrane perturbation and/or Erg11 inhibition.
We confirmed the chemical–genetic interactions between these haploid deletion strains and each drug using quantitative growth curve assays (Figure 3D). We also assessed strains that were heterozygous for ERG11, NEO1, LCB1 and LCB2 as well as TIM18 and TIM22, which function in a complex with TIM54. In addition, we included haploid deletion strains for DNF1/2/3, the other three flippases in S. cerevisiae, and LCB3/4/5, which function downstream of LCB1/2. The quantitative growth curves corroborated the barcode microarray results, with the exception of the dubious ORF YOR072W. The range of drug concentrations tested in the growth curve assays revealed additional chemical–genetic interactions, such as TIM18, which were not recovered at the single drug concentrations used in the barcode profiles.
The results of these genome-wide chemical–genetic screens point to two related modes of action for the syncretic combinations tested. Trifluoperazine, tamoxifen, clomiphene, sertraline and suloctidil appear to cause general perturbation of membrane, vesicle trafficking and lipid biosynthesis functions, whereas L-cycloserine preferentially interferes with an early step in sphingolipid biosynthesis, consistent with its proposed mechanism of action (Pinto et al, 1992). To test the latter hypothesis, we examined the effects of myriocin, another known inhibitor of the first step of the sphingolipid biosynthesis pathway (Miyake et al, 1995), and found that it also potentiated the inhibition of cell growth by fluconazole (FICI=0.625).
Cell biological effects of synergistic combinations
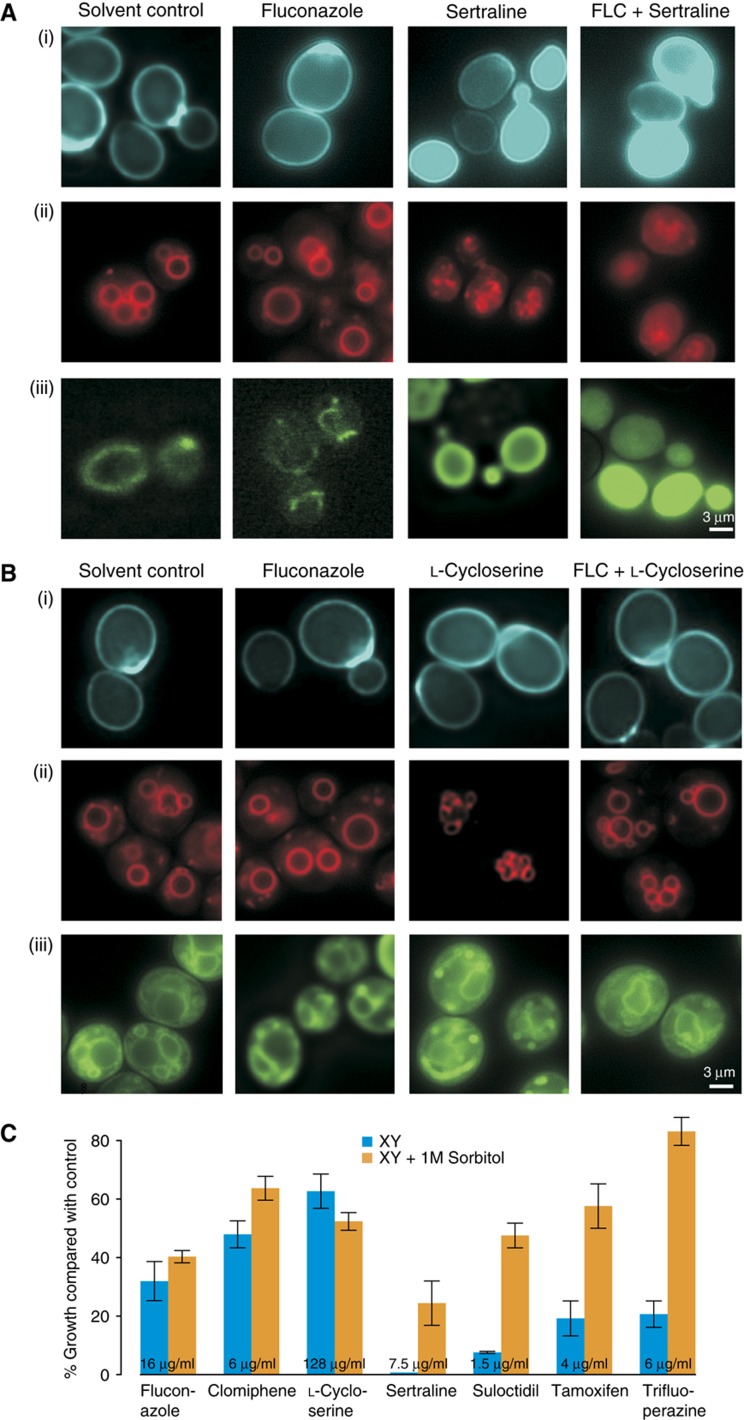
We assessed the effects of trifluoperazine, tamoxifen, sertraline and L-cycloserine alone and in combination with fluconazole on S. cerevisiae cell physiology. The diagnostic fluorescent dyes Calcofluor White, FM4-64 and Mitotracker Green were used to visualize cell wall and bud scars, vacuolar membranes and mitochondria, respectively. For each reporter dye, fluconazole produced staining patterns that were similar to solvent controls. In contrast, treatment with trifluoperazine, tamoxifen, clomiphene and sertraline caused a drastic loss of localization and strong intracellular accumulation of each dye (Figure 4A; Supplementary Figure S8). In particular, the disruption of vacuolar structure revealed by FM4-64 staining suggested severe loss of cell membrane integrity. Consistent with its different genetic target profile, treatment with L-cycloserine had no observable effects on the localization of any of the dyes (Figure 4B).
Figure 4.
Effects of syncretic drugs on membrane integrity. A wild-type S. cerevisiae strain was grown in the presence of the indicated drugs and stained with (i) Calcofluor White M2R, (ii) FM4-64 and (iii) Mitotracker Green FM, and imaged by fluorescence microscopy. (A) Sertraline (128 μg/ml) in the presence and absence of fluconazole (64 μg/ml). (B) L-Cycloserine (128 μg/ml) in the presence and absence of fluconazole (128 μg/ml). (C) Growth of wild-type S. cerevisiae compared with control wells in the presence of the indicated drugs with and without 1 M sorbitol. The mean of four independent measurements is shown; error bars represent standard error. Source data is available for this figure at www.nature.com/msb.
Lethal perturbation of the membrane and/or cell wall can often be rescued by osmotic stabilization. Sorbitol (1 M) effectively suppressed the syncretic growth inhibitory effects of trifluoperazine, tamoxifen, clomiphene, sertraline and suloctidil, again supporting a common membrane perturbation mechanism for these compounds, but had no such protective effect on cells treated with L-cycloserine and fluconazole (Figure 4C). These cell biological results affirm the different mechanisms of action of the two compound classes.
Integration of chemical–gene interactions with genetic interaction networks
A primary challenge in the discovery of synergistic drug combinations is the vast number of possible combinations of drug pairs (Sharom et al, 2004; Hopkins, 2008; Lehar et al, 2008). Integration of drug-induced gene expression profiles (Lum et al, 2004) and chemical–genetic profiles (Hillenmeyer et al, 2008) with comprehensive genetic interaction networks (Costanzo et al, 2010) can allow computational prediction of synergistic drug pairs (Lehar et al, 2007; Nelander et al, 2008; Jansen et al, 2009). To assess whether the individual profiles of fluconazole and each of the syncretic drugs could rationalize drug interactions, we integrated the chemical–genetic profiles generated above for each syncretic compound with a global genetic interaction network composed of both high-throughput (HTP) and low-throughput (LTP) data compiled from the primary literature (Breitkreutz et al, 2008; Costanzo et al, 2010). Deletion strains that were sensitive to treatment with single drugs were used to assess the number of genetic interactions linked to the chemical–genetic space (CGS) of fluconazole and each of the synergizers. A core set of haploid deletion strains affected by the membrane active group of compounds, referred to as the signature strain set (Figure 3D), exhibited many genetic interactions with the top 50 fluconazole-sensitive strains (Figure 5A). The top 50 most sensitive deletion strains for each individual drug (Z-scores above ∼2.0) also showed many genetic interactions with the fluconazole profile. We tested the significance of the genetic connections between the profiles using simulations of genetic interactions shared between randomly chosen gene sets of a specific size, based on the known chemical sensitivities of 1143 non-essential deletion strains that respond to various drug treatments (Hillenmeyer et al, 2008). The signature deletion set shared by the membrane active group was significantly enriched for genetic interactions with fluconazole-sensitive deletion strains (P-value <10−7). The individual profiles for tamoxifen, trifluoperazine, clomiphene, sertraline and suloctidil also showed a significant enrichment of genetic interactions with the fluconazole-sensitive strain profile (all P-values <0.05), whereas L-cycloserine did not (Figure 5B; Supplementary Table S6). As a more conservative measure of pathway separation (Kelley and Ideker, 2005), we applied a parallel pathway permutation (PPP) test, in which the chemical–genetic interactors of fluconazole and each of the synergistic drugs were pooled and randomly assigned to two groups (Supplementary Figure S9). By this stringent method, the signature deletion set and the top 50 most sensitive deletion strains from the trifluoperazine profile also exhibited significant enrichment (both P-values <0.05; Supplementary Figure S9; Supplementary Table S6). The genetic interactions that link the chemogenomic profiles of synergistic compound pairs thus provide a rational basis for synergism.
Figure 5.
Rationalization of synergistic interactions by integration of chemical–genetic and genetic interaction networks. (A) Bipartite graph of genetic interactions between top 50 chemical–genetic interactors of fluconazole and the signature deletion strains sensitive to the five membrane active compounds. As PDR5 was a member of both sets, it is positioned midway between the two sets. Enriched Gene Ontology (GO) SLIM biological processes are indicated (adjusted P-value <0.05). GO enrichment was calculated and visualized using GOlorize (Garcia et al, 2007). (B) Chemical–genetic space (CGS) simulation with the 50 most sensitive deletion strains for each of the synergistic drugs as well as the signature deletion strain set. Arrows indicate the number of actual genetic interactions for the different drugs; black curve represents the background distribution of genetic interactions between two random sample sets of 50 non-essential deletion strains chosen from 1143 strains that respond to a variety of different chemicals and drugs (Hillenmeyer et al, 2008); dark red curve depicts the same background distribution except that the second sample set size was chosen to match the size of signature deletion set. Asterisks indicate a P-value <0.05. (C) Drug sensitivity of 11 of the 14 signature deletion strains identified in this study for 16 previously profiled psychiatric drugs present in the Prestwick library (Ericson et al, 2008). Strong activity refers to compounds that were hits in the primary screens, that is, at least 2 MAD away from the diagonal, whereas weak activity refers to compounds that showed at least 20% growth inhibition and were >1 MAD away from the diagonal. Source data is available for this figure at www.nature.com/msb.
To assess the predictive power of the signature deletion set derived from the membrane active compounds, we retrospectively analyzed chemical–genetic profiles for 81 psychoactive drugs known to impair yeast growth (Ericson et al, 2008). Of this set, 16 compounds were represented in the Prestwick library, 7 of which were predicted to synergize with fluconazole based on their effect on deletion strains in the signature set. In our primary screens against the four fungal species, four of these seven compounds were indeed hits in our screen, while the other three compounds showed weak activity (Figure 5C; Supplementary Table S1). These results demonstrate that chemical–genetic interaction profiles can predict synergistic drug combinations.
Species-specific effects of ergosterol pathway inhibition
As the psychiatric drugs trifluoperazine and sertraline exhibited synergy with fluconazole against each fungal species, we tested whether other ergosterol biosynthesis inhibitors might exhibit synergy with these compounds. We assessed interactions with ketoconazole, an imidazole inhibitor of ERG11, and terbinafine, an inhibitor of the Erg1 squalene epoxidase in Cryptococcus, Candida and Saccharomyces. Ketoconazole was synergistic with both psychiatric drugs in all fungal species, whereas terbinafine showed synergies with sertraline in Candida and Saccharomyces but not in Cryptococcus, and synergized with trifluoperazine only in Saccharomyces (Table IA). These findings suggest that while mechanisms of synergy are conserved between different inhibitors of the same enzymes/pathways, species-specific differences readily emerge with different compounds, likely due to subtle differences in genetic network structure (Kuo et al, 2010).
Table 1. Combinations of syncretic drugs exhibit species-specific synergism and higher order interactions with fluconazole.
| Terbinafine | Terbinafine | Ketoconazole | Ketoconazole | |
|---|---|---|---|---|
| Trifluoperazine | Sertraline | Trifluoperazine | Sertraline | |
| (A) FICI values for drug combinations in different fungal species | ||||
| C. neoformans (H99) | 2 | 0.75 | 0.25 | 0.38 |
| C. albicans (Caf2-1) | 2 | 0.5 | 0.38 | 0.16 |
| S. cerevisiae (BY4741) | 0.38 | 0.52 | 0.38 | 0.31 |
| Sert (32) | Sert (64) | Tri | Tri+FLC (4) | Tam | Tam+FLC (4) | Suloc | Suloc+FLC (4) | |
|---|---|---|---|---|---|---|---|---|
| (A) FICIs from combination matrix analysis of sertraline, trifluoperazine, with the ergosterol inhibitors terbinafine and ketoconazole in different species. | ||||||||
| (B) FICIs from combination matrix analysis of syncretic drugs and higher order combinations. Trifluoperazine (Tri), tamoxifen (Tam), suloctidil (Suloc) and L-cycloserine were combined as indicated, in the presence or absence of 1/8 MIC fluconazole (4 μg/ml) and assayed for growth inhibition of a S. cerevisiae strain (BY4741). FICI values <0.5 indicate synergy, values between 0.5 and 1 indicate additivity and values >1 indicate no interaction. Drug concentrations are given in μg/ml. | ||||||||
| (B) FICI values for double and triple drug combinations in S. cerevisiae | ||||||||
| Trifluoperazine | 0.38 | 0.50 | ||||||
| Tamoxifen | 0.50 | 0.63 | 0.50 | 0.75 | ||||
| Suloctidil | 1.25 | 1.50 | 2.00 | 0.56 | 1.00 | 0.75 | ||
| L-Cycloserine | 1.25 | 0.63 | 1.00 | 2.00 | 1.00 | 0.52 | 2.00 | 0.31 |
Higher order combinations of synergizers
Compounds that act in an identical manner are in principle expected not to exhibit synergy but instead should show only additive dosage effects. We examined pairwise combinations within four members of the membrane active group (sertraline, trifluoperazine, suloctidil and tamoxifen), as well as with the sphingolipid-selective synergizer L-cycloserine (Table IB). Despite their partially overlapping genetic profiles, synergistic interactions in S. cerevisiae were observed between tamoxifen and trifluoperazine (FICI=0.5), sertraline and trifluoperazine (FICI=0.4) and sertraline and tamoxifen (FICI=0.5). These synergies suggested that in addition to the core effects on membrane permeability, each compound likely elicits one or more specific effects that contribute to overall mechanism of action, and that combining these effects results in further synergism. This observation predicted that higher order combinations between synergizers might lead to even stronger growth inhibition. When compound pairs were tested in the presence of fluconazole in three-way combinations, fungal growth was often potently inhibited (Table IB). Titration of fluconazole concentrations revealed exquisite sensitivity to the suloctidil/trifluoperazine, L-cycloserine/suloctidil and L-cycloserine/tamoxifen combinations in the presence of just 1/8 MIC fluconazole (Table IB). The L-cycloserine/suloctidil pair exhibited the most potent synergy with fluconazole with an FICI of 0.31. These results demonstrate that it is possible to incrementally build higher order synergistic combinations based on the subtly different properties of individual synergizers, even within in the same class.
In vivo synergy in an insect model of infection
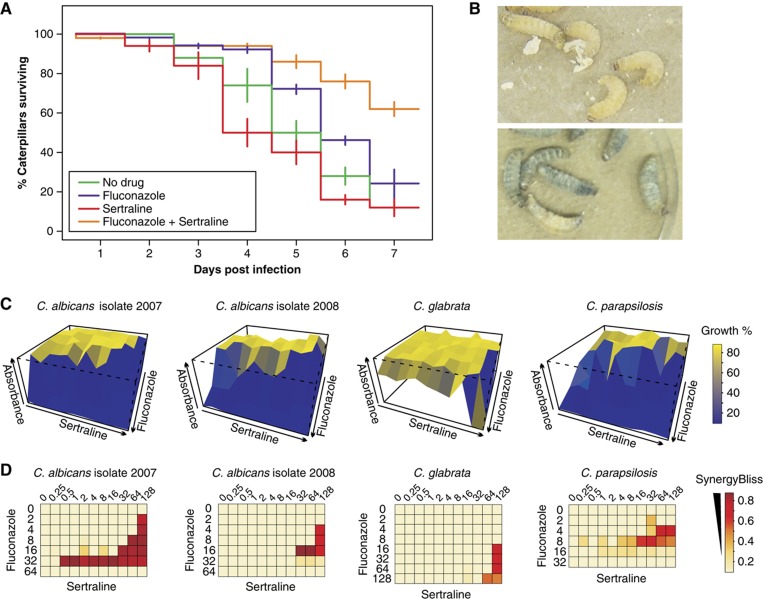
The caterpillar of the greater wax moth Galleria mellonella is a validated in vivo infection model for study of Cryptococcal virulence, host immune responses to infection and the effects of antifungal compounds (Mylonakis et al, 2005; Scully and Bidochka, 2006; Cowen et al, 2009). We assessed the effect of a synergistic drug combination on the survival rate of G. mellonella infected with C. neoformans because of the prevalence of this virulent pathogen in immune-suppressed individuals. G. mellonella was inoculated with C. neoformans H99 and subsequently treated with fluconazole and sertraline, individually and in combination. The synergistic action of fluconazole and sertraline was evident in G. mellonella survival rates when compared with either drug alone (Figure 6A and B). Survival increased by 40% when treated with fluconazole and sertraline in combination, with an overall 60% survival rate after seven days of infection (P<0.02 for fluconazole versus fluconazole/sertraline). These results demonstrate that potentiators of fluconazole activity identified in vitro exhibit comparable activity in an animal model of infection.
Figure 6.
Synergistic activity of sertraline and fluconazole in an in vivo infection model and against clinical isolates. (A) G. mellonella caterpillars were injected with 8 × 103 cfu C. neoformans H99 on day 0 and drugs alone or in combination (1 μg fluconazole; 26 μg sertraline) on the first day and incubated for 1 week at 37°C. Values are mean of three independent experiments; error bars indicate standard deviation of the mean. (B) Uninfected G. mellonella caterpillars (top); melanization of infected G. mellonella caterpillars without drug treatment (bottom). (C) Combination matrix assays against drug-resistant Candida strains. Residual growth was plotted as a function of combinations of two-fold dilutions of each drug. (D) Bliss synergy analysis for combination assays shown in panel (C). The apparent absence of synergy at the highest fluconazole concentrations for C. albicans and C. parapsilosis is due to growth inhibition caused by fluconazole alone. Drug concentrations are in μg/ml. Source data is available for this figure at www.nature.com/msb.
Synergistic activity against fluconazole-resistant Candida isolates
To address whether syncretic compounds can act on clinically resistant strains, we investigated whether the combination of fluconazole and sertraline is effective against fluconazole-resistant clinical isolates of C. albicans (F-07-2007, F-01-2008), C. glabrata and a resistant control strain C. parapsilosis (ATCC 22019). Sertraline increased the susceptibility of resistant strains to fluconazole by up to 32-fold (Figure 6C and D; Supplementary Table S7). In the presence of sertraline, fluconazole MIC values ranged from 2 to 8 μg/ml, comparable to wild-type (Caf2-1, MIC=8 μg/ml) and drug pump-deficient (MIC=2 μg/ml) strains of C. albicans. The sertraline/fluconazole combination was synergistic in both C. albicans clinical isolates as well as the C. parapsilosis reference strain, but not in the C. glabrata strain. As noted above, this differential sensitivity may indicate strain-specific drug effects, or different mechanisms of drug resistance (Kuo et al, 2010). We conclude that syncretic activities in reference laboratory strains can be transposed to drug-resistant pathogens derived from clinical environments.
Discussion
Systematic screens for syncretic combinations reveal new antifungal chemical space
The combination of known antifungal agents is an established therapeutic tactic in infectious disease control (Johnson and Perfect, 2010). Here, we show that antifungal chemical space can be systematically expanded through the combination of a known antifungal drug with other bioactive compounds that do not have antifungal activity per se, including off-patent drugs previously approved for other indications. Novel syncretic drug combinations were readily identified in systematic screens against different fungal pathogens in the presence of subtherapeutic concentrations of fluconazole. These chemically diverse drug hits derive from a broad spectrum of human therapeutic areas that otherwise would not have been explored by infectious disease clinicians. Although hits from screens for fluconazole potentiation in the S. cerevisiae model system can be transposed to pathogenic fungi (Borisy et al, 2003; Zhang et al, 2007; Jansen et al, 2009; Epp et al, 2010), our primary screen data reveals considerable species specificity that by definition cannot be predicted from model organism drug-gene interactions. Indeed, in our primary screen, while 58% (25/43) of hits against S. cerevisiae exhibited synergistic activity against one or more fungal pathogens, only 19% (25/130) of the total hits against pathogenic species were detected in S. cerevisiae. This observation underscores the need to undertake primary screens in the pathogen of interest.
Many syncretic combinations exhibited fungicidal activity, a highly desirable feature for neutropenic or otherwise immune-compromised patients. The fungicidal combination of fluconazole and the antidepressant sertraline (Zoloft®) was effective against all species tested, including drug-resistant clinical isolates of Candida, and in an in vivo insect model of C. neoformans infection. Therapeutic intervention for fungal infections of the CNS is a particular clinical challenge because of the stringent requirement to breach the blood brain barrier. The fact that sertraline targets serotonin receptors in the CNS suggests that the sertraline-fluconazole combination may be effective in the treatment of fungal meningitis.
Molecular mechanisms and prediction of drug synergism
Genome-wide chemical–genetic profiles of a selected set of six fungicidal synergizers revealed two different patterns of synergy. Five compounds—trifluoperazine, tamoxifen, clomiphene, sertraline and suloctidil—elicited genetic sensitivities and cell biological phenotypes associated with a loss of membrane integrity. The membrane perturbation caused by these compounds may increase susceptibility to accumulation of ergosterol pathway intermediates, impair fluconazole export by drug efflux pumps and/or impair import of exogenous ergosterol (Kuo et al, 2010). It is also possible that the synergizers affect active import of azoles through altered localization of drug transporters or general membrane perturbation (Mansfield et al, 2010). Notably, all five membrane active compounds are cationic amphiphilic drugs (CADs) that intercalate preferentially into one side of the lipid bilayer, thereby causing membrane expansion and cell wall stress (Sheetz and Singer, 1974), consistent with the observed chemical–genetic interactions with NEO1, DRS2, SLT2 and BCK1. Moreover, genetic resistance to CADs is conferred by perturbation of vesicular membrane biogenesis and/or trafficking (Rainey et al, 2010). The synthetic lethal genetic interactions that occur between strains in the fluconazole and membrane active chemical–genetic profiles retrospectively predicted the synergistic effects of other hits in our primary screens. Moreover, when combined with another source of chemical–genetic interaction data (Ericson et al, 2008), the membrane active signature strain set correctly identified further synergistic hits in our primary screen data. In addition, the genetic interaction profile of L-cycloserine correctly predicted a novel synergistic interaction between the sphingolipid biosynthesis inhibitor myriocin and fluconazole. The potentiation of fluconazole activity by CADs and/or inhibition of sphingolipid biosynthesis may allow new general approaches to antifungal therapy in the clinic. As genetic and chemical–genetic space is elaborated, mechanism-based predictive approaches should become a powerful means of identifying new synergistic combinations.
Species-specific syncretic effects
We observed many genus- and species-specific syncretic interactions, which reflects differences in the genetic networks that dictate cellular responses to each compound (Perlstein et al, 2007). Since divergence from a common ancestor over 100 million years ago, different pathogenic species have adapted to particular host environments. For example, genetic plasticity of the fungal mating-type locus affects survival in mammalian hosts (Nielsen and Heitman, 2007). Developmental system drift (True and Haag, 2001) can also affect drug susceptibility, as shown by the differential effects of nikkomycin Z on chitin synthase paralogs in Saccharomyces and Candida (Gaughran et al, 1994; Sudoh et al, 2000). Marked differences in the transcriptional response of Saccharomyces, Candida and Kluyveromyces to fluconazole treatment underscore the quite distinct mechanisms whereby different species can respond to the same drug (Kuo et al, 2010). More generally, species differences in the response to chemical perturbation may reflect the evolutionary plasticity of genetic interaction networks (Kapitzky et al, 2010). Species-selective antifungal combinations may afford a means to both increase efficacy and decrease host toxicity. Systematic analysis of drug–drug interactions may also provide a means to classify and predict drug mechanism of action (Yeh et al, 2006; Hopkins, 2008).
Higher order drug–drug interactions
The densely connected structure of genetic networks predicts that it should be possible to devise higher order drug combinations with greater selectivity and potency (Sharom et al, 2004; Agoston et al, 2005; Lehar et al, 2007). That is, compounds that target multiple genetically redundant parallel pathways may exhibit n-way synergies. In an initial elaboration of this concept, we found that the combination of a non-synergistic pair (suloctidil and L-cycloserine, drawn from the membrane active and sphingolipid target classes, respectively) with a low dose of fluconazole resulted in a highly potent three-way synergism. Somewhat unexpectedly given their shared core genetic profiles, pairwise tests of four compounds in the membrane active class also revealed synergistic interactions in the absence of fluconazole. This type of drug–drug interaction, which has been observed previously with bacteria and yeast (Yeh et al, 2006; Jansen et al, 2009), suggests that, aside from the common core profile, each drug must have additional specific targets that contribute to overall synergism. The complex genetic profiles of each drug reflect effects on primary and secondary targets in the cell, drug metabolism and detoxification, and genetic feedback between different network elements (Sharom et al, 2004; Kitano, 2007; Lehar et al, 2008). Other documented interactions between fluconazole, reactive oxygen species, Hsp90, calcium metabolism and vesicle trafficking may contribute to these complex interactions (Cowen et al, 2009; Xu et al, 2009; Epp et al, 2010; Gamarra et al, 2010). We note that although shared drug profiles have been suggested to be predictive of synergistic interactions (Jansen et al, 2009), in many instances this is not the case (Yeh et al, 2006). Even drugs with well-documented mechanisms of action can have substantially different genetic interaction profiles compared with their presumptive targets. For example, although the genetic interaction profiles of fluconazole and its known target ERG11 exhibit significant overlap, more than half of the interactions are not shared (Parsons et al, 2004). Recently, it has been shown that drug combinations can exhibit remarkably selective but unpredictable effects on the abundance of many different proteins (Geva-Zatorsky et al, 2010).
Therapeutic implications
The benefits of combinatorial anti-infective therapies include a decrease in the rate of selection of resistant strains, a lower required dosage of individual drugs, a decrease in host toxicity and enhanced antimicrobial activity (Sharom et al, 2004; Hopkins, 2008; Lehar et al, 2008). As shown in this study and elsewhere (Zhang et al, 2007; Jansen et al, 2009; Epp et al, 2010), syncretic combinations of drugs with improved antifungal properties can be readily identified in both model fungal species and highly pathogenic clinical isolates. Importantly, while it is a potential concern that undesirable side effects may arise from drug combinations, as occurs for example with known contraindicated drugs, it has recently been shown that synergistic combinations usually yield enhanced selectivity without adverse side effects (Lehar et al, 2009). As noted above, these benefits may include improved activity in therapeutically recalcitrant tissues, such as the CNS. These combinatorial principles apply equally to viral and bacterial pathogens, cancer and other genetic diseases (Borisy et al, 2003; Fitzgerald et al, 2006; Hopkins, 2008; Lehar et al, 2009).
Materials and methods
Chemicals, high-throughput screens and MIC determination
Fluconazole was purchased from Sandoz (Quebec, Canada). All other compounds were obtained from Sigma (St Louis) or Prestwick Chemicals (Illkirch, France). The Prestwick Chemical library was screened in duplicate in the presence and absence of 1/2 MIC fluconazole at a final concentration of 30 μM in 384-well flat bottom microtitre plates. OD600 was determined after 48 h at 30 or 37°C. MIC determinations were based on Clinical and Laboratory Standards Institute (CLSI) protocols (Eliopoulos and Moellering, 1991; Odds, 2003), with the exception that yeast SC medium was used instead of mammalian cell RPMI 1640 medium. Overnight cultures in synthetic complete media (SC: 0.67% Difco™ yeast nitrogen base w/o amino acids, 0.08% amino acid add back and 2% glucose) were diluted in 0.85% NaCl to an OD530 of 0.11, followed by a 1:100 dilution in 0.85% NaCl, and a final 1:20 dilution in SC media. Two-fold dilution series (0–128 μg/ml) of fluconazole and other antifungal drugs were added to 200 μl of diluted culture in 96-well plates and OD600 determined after 48 h at 30 or 37°C. For fluconazole, MIC was set at the lowest concentration that caused 80% reduction in growth, corresponding to two on the azole MIC numerical scale. For other drugs, MIC was set as the lowest concentration that yielded no growth.
Time kill MIC assays were performed at six different concentrations of compound and fluconazole (fluconazole at MIC, fluconazole at 1/4 MIC, compound at MIC, compound at 1/4 MIC, both fluconazole and compound at MIC, and both fluconazole and compound at 1/4 MIC). At 0, 24 and 48 h dilutions from each well were spotted on an SC agar plate, incubated for 48 h and colony counts determined. A fungicidal effect was defined as >3log10 (99.9% killing) reduction in CFU/ml at synergistic concentrations after 24 h incubation.
Synergy matrix assays
Fluconazole and syncretic compounds were two-fold serially diluted across the rows and columns of a 96-well plate (0–128 μg/ml; for daunorubicin HCl, terbinafine, trifluoroperazine dihydrochloride and ellipticine dilutions were from 0–64 μg/ml), incubated with fungal cultures and OD600 determined after 48 h. The FIC index of each drug combination was determined by adding the individual FIC values, as calculated by standard CLSI protocols (Eliopoulos and Moellering, 1991; Odds, 2003). To probe chemical interactions between sertraline, trifluoperazine, L-cycloserine, suloctidil and tamoxifen, checkerboard assays were carried out between these five compounds in the absence and presence of 1/2 and 1/8 MIC fluconazole (16 and 4 μg/ml, respectively).
Chemical–genetic profiles and secondary assays
S. cerevisiae deletion collections (MATa haploid and heterozygous essential deletion strains) were obtained from Research Genetics (Germany). Compounds were screened at a concentration that caused ∼30% growth inhibition at a final DMSO concentration of 0.2% (Giaever et al, 1999). Deletion pools were grown for 10 generations, gDNA extracted and barcode tags amplified with fluorescently labeled UP and DN primers, followed by hybridization of PCR products to spotted barcode microarrays (Cook et al, 2008). Arrays were scanned on a GenePix 4200AL and analyzed with GenePix Pro 6.0 software. Data sets are available at ArrayExpress (E-MTAB-394). Chemical–genetic interactions were confirmed in quantitative growth assays at 30°C with continuous shaking at 564 r.p.m. on a Sunrise shaker/reader (Tecan); OD600 readings were taken every 15 min and values at the end of logarithmic phase used to calculate the log ratio between deletion and wild-type strains. For sorbitol rescue, wild-type strains were grown in the presence of indicated compounds and 1 M sorbitol. For microscopy, cells were embedded in 1% low melt agarose and stained with Calcofluor White M2R (Sigma), Mitotracker Green FM (Molecular Probes) or FM4-64 (Molecular Probes) and imaged at × 100 on a Leica DMI 6000 B microscope with a Hamamatsu Orca ER-AG camera and Volocity 4 software. Images were deconvolved using AutoDeblur Gold CWF using 2-D blind deconvolution and 10 iterations per image.
Computational analysis of gene-drug network interactions
The 50 most sensitive deletion strains from duplicate chemical–genetic profiles for clomiphene, L-cycloserine, sertraline, suloctidil, tamoxifen and trifluoperazine were tested against the top 50 fluconazole-sensitive deletion strains (from replicate arrays at 8 μM). Shared genetic interactions between the sets of deletion strains were determined based on genetic interaction data obtained from BioGRID (Breitkreutz et al, 2008; BIOGRID release 2.62, http://www.thebiogrid.org). Visualization of bipartite graphs and simulations was performed with an online tool available at http://tyerslab.bio.ed.ac.uk/tools/genelookup_bipartite.php. Simulations based on CGS were derived from 1143 non-essential deletion strains that respond to various drug treatments (Hillenmeyer et al, 2008). For each drug pair, control gene sets of the same size were picked at random and the number of genetic interactions counted to generate a background distribution of the number of interactions that would occur by chance, based on the compiled genetic interaction data. This distribution was used to calculate the P-value for each drug pair. For PPP, the chemical–genetic interactors of fluconazole and each of the synergistic drugs were pooled, randomly assigned to two groups and genetic interactions counted to obtain a background distribution for each drug pair. The definition of the signature deletion strain set was based on confirmatory quantitative growth curve assays (Figure 3D). For both the CGS and PPP methods, 10 000 simulations were conducted for each drug pair. To predict potential synergistic candidates based on overlap with published chemical–genetic profiles (Ericson et al, 2008), we used a binary data matrix based on a Z-score cutoff of ±3. The significance of enrichment was calculated based on the number of genes that overlapped with the signature strain set; a subset of 4 out of 11 genes was significant with a P-value <0.05.
Insect larvae assays
Ten weight matched (250–400 mg/worm) G. mellonella caterpillars per dish were inoculated with C. neoformans H99 and subsequently injected with different combinations of compound, fluconazole and/or control solutions (Mylonakis et al, 2005). Over a 7-day period, caterpillars were examined visually for discoloration due to melanization and for failure to respond to touch as an inviability end point.
Supplementary Material
Supplementary Methods, Figures S1-S9, Tables S2-S7
Supplementary Table S1. Normalized primary screen data.
Z-scores from all haploid deletion strain sensitivity profiles
Z-scores from all haploid deletion strain sensitivity profiles
Acknowledgments
We thank J Blanchard and C Murphy for helpful discussions on small molecule screens, L Scully for advice on the G. mellonella infection model, JW Kronstad for the C. neoformans H99 and C. gattii R265 strains, M Whiteway for the C. albicans Caf-2 strain and D Yamamura for the fluconazole-resistant C. glabrata, C. parapsilosis and C. albicans clinical isolates. This research was supported by the Canada Research Chair program (EDB, GDW), a Royal Society Wolfson Research Merit Award (MT), a Scottish Universities Life Sciences Alliance Research Chair (MT), and by grants from the Canadian Institutes for Health Research (FRN79488 to GDW and MT) and the European Research Council (2007-223411 to MT).
Author contributions: EG, MS, KMB, JW, GDP, MT and GDW designed experiments; EG, KMB, LE and LR performed chemical screens; MS performed genome-wide genetic profiles and growth curves; GDP, JC and LR performed microscopy; EG, MS and JW performed data analysis; EG, MS, JW, MT and GDW wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agoston V, Csermely P, Pongor S (2005) Multiple weak hits confuse complex systems: a transcriptional regulatory network as an example. Phys Rev E Stat Nonlin Soft Matter Phys 71: 051909. [DOI] [PubMed] [Google Scholar]
- Arendrup MC, Fisher BT, Zaoutis TE (2009) Invasive fungal infections in the paediatric and neonatal population: diagnostics and management issues. Clin Microbiol Infect 15: 613–624 [DOI] [PubMed] [Google Scholar]
- Baddley JW, Stroud TP, Salzman D, Pappas PG (2001) Invasive mold infections in allogeneic bone marrow transplant recipients. Clin Infect Dis 32: 1319–1324 [DOI] [PubMed] [Google Scholar]
- Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, Keith CT (2003) Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci USA 100: 7977–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M (2008) The BioGRID Interaction Database: 2008 update. Nucleic Acids Res 36: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC (2009) Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22: 291–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Hajjeh RA (2002) Recent trends in the epidemiology of invasive mycoses. Curr Opin Infect Dis 15: 569–574 [DOI] [PubMed] [Google Scholar]
- Cook MA, Chan CK, Jorgensen P, Ketela T, So D, Tyers M, Ho CY (2008) Systematic validation and atomic force microscopy of non-covalent short oligonucleotide barcode microarrays. PLoS One 3: e1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M et al. (2010) The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Singh SD, Kohler JR, Collins C, Zaas AK, Schell WA, Aziz H, Mylonakis E, Perfect JR, Whitesell L, Lindquist S (2009) Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci USA 106: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos GM, Moellering RC (1991) Antimicrobial Combinations. Baltimore: Williams and Wilkins [Google Scholar]
- Epp E, Vanier G, Harcus D, Lee AY, Jansen G, Hallett M, Sheppard DC, Thomas DY, Munro CA, Mullick A, Whiteway M (2010) Reverse genetics in Candida albicans predicts ARF cycling is essential for drug resistance and virulence. PLoS Pathog 6: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson E, Gebbia M, Heisler LE, Wildenhain J, Tyers M, Giaever G, Nislow C (2008) Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet 4: e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK (2006) Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2: 458–466 [DOI] [PubMed] [Google Scholar]
- Gamarra S, Rocha EM, Zhang YQ, Park S, Rao R, Perlin DS (2010) Mechanism of the synergistic effect of amiodarone and fluconazole in Candida albicans. Antimicrob Agents Chemother 54: 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia O, Saveanu C, Cline M, Fromont-Racine M, Jacquier A, Schwikowski B, Aittokallio T (2007) GOlorize: a Cytoscape plug-in for network visualization with Gene Ontology-based layout and coloring. Bioinformatics 23: 394–396 [DOI] [PubMed] [Google Scholar]
- Gaughran JP, Lai MH, Kirsch DR, Silverman SJ (1994) Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J Bacteriol 176: 5857–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Dekel E, Cohen AA, Danon T, Cohen L, Alon U (2010) Protein dynamics in drug combinations: a linear superposition of individual-drug responses. Cell 140: 643–651 [DOI] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW (1999) Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21: 278–283 [DOI] [PubMed] [Google Scholar]
- Groll AH, De Lucca AJ, Walsh TJ (1998) Emerging targets for the development of novel antifungal therapeutics. Trends Microbiol 6: 117–124 [DOI] [PubMed] [Google Scholar]
- Groll AH, Shah PM, Mentzel C, Schneider M, Just-Nuebling G, Huebner K (1996) Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J Infect 33: 23–32 [DOI] [PubMed] [Google Scholar]
- Gullo A (2009) Invasive fungal infections: the challenge continues. Drugs 69(Suppl 1): 65–73 [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G (2008) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 4: 682–690 [DOI] [PubMed] [Google Scholar]
- Hu G, Kronstad JW (2010) A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryot Cell 9: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Lee AY, Epp E, Fredette A, Surprenant J, Harcus D, Scott M, Tan E, Nishimura T, Whiteway M, Hallett M, Thomas DY (2009) Chemogenomic profiling predicts antifungal synergies. Mol Syst Biol 5: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Perfect JR (2010) Use of antifungal combination therapy: agents, order, and timing. Curr Fungal Infect Rep 4: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitzky L, Beltrao P, Berens TJ, Gassner N, Zhou C, Wuster A, Wu J, Babu MM, Elledge SJ, Toczyski D, Lokey RS, Krogan NJ (2010) Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol 6: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CT, Borisy AA, Stockwell BR (2005) Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 4: 71–78 [DOI] [PubMed] [Google Scholar]
- Kelley R, Ideker T (2005) Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol 23: 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2007) A robustness-based approach to systems-oriented drug design. Nat Rev Drug Discov 6: 202–210 [DOI] [PubMed] [Google Scholar]
- Kuo D, Tan K, Zinman G, Ravasi T, Bar-Joseph Z, Ideker T (2010) Evolutionary divergence in the fungal response to fluconazole revealed by soft clustering. Genome Biol 11: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ER, Rickles RJ, Short GF III, Staunton JE, Jin X, Lee MS, Zimmermann GR, Borisy AA (2009) Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol 27: 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar J, Stockwell BR, Giaever G, Nislow C (2008) Combination chemical genetics. Nat Chem Biol 4: 674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar J, Zimmermann GR, Krueger AS, Molnar RA, Ledell JT, Heilbut AM, Short GF III, Giusti LC, Nolan GP, Magid OA, Lee MS, Borisy AA, Stockwell BR, Keith CT (2007) Chemical combination effects predict connectivity in biological systems. Mol Syst Biol 3: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Black PN, Chokshi A, Sandoval-Alvarez A, Vatsyayan R, Sealls W, DiRusso CC (2008) High-throughput screening for fatty acid uptake inhibitors in humanized yeast identifies atypical antipsychotic drugs that cause dyslipidemias. J Lipid Res 49: 230–244 [DOI] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, Roberts CJ, Shoemaker DD (2004) Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116: 121–137 [DOI] [PubMed] [Google Scholar]
- Mansfield BE, Oltean HN, Oliver BG, Hoot SJ, Leyde SE, Hedstrom L, White TC (2010) Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog 6: e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie C, White TC (2009) Genetic basis of antifungal drug resistance. Curr Fungal Infect Rep 3: 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T (1995) Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun 211: 396–403 [DOI] [PubMed] [Google Scholar]
- Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A (2005) Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73: 3842–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelander S, Wang W, Nilsson B, She QB, Pratilas C, Rosen N, Gennemark P, Sander C (2008) Models from experiments: combinatorial drug perturbations of cancer cells. Mol Syst Biol 4: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Heitman J (2007) Sex and virulence of human pathogenic fungi. Adv Genet 57: 143–173 [DOI] [PubMed] [Google Scholar]
- Odds FC (2003) Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52: 1. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C (2004) Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol 22: 62–69 [DOI] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B et al. (2006) Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126: 611–625 [DOI] [PubMed] [Google Scholar]
- Perlstein EO, Ruderfer DM, Roberts DC, Schreiber SL, Kruglyak L (2007) Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat Genet 39: 496–502 [DOI] [PubMed] [Google Scholar]
- Pinto WJ, Wells GW, Lester RL (1992) Characterization of enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity. J Bacteriol 174: 2575–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey MM, Korostyshevsky D, Lee S, Perlstein EO (2010) The antidepressant sertraline targets intracellular vesiculogenic membranes in yeast. Genetics 185: 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revankar SG, Fu J, Rinaldi MG, Kelly SL, Kelly DE, Lamb DC, Keller SM, Wickes BL (2004) Cloning and characterization of the lanosterol 14alpha-demethylase (ERG11) gene in Cryptococcus neoformans. Biochem Biophys Res Commun 324: 719–728 [DOI] [PubMed] [Google Scholar]
- Richardson M, Warnock D (2003) Fungal Infection: Diagnosis and Management. Oxford: Blackwell Publishing [Google Scholar]
- Scully LR, Bidochka MJ (2006) The host acts as a genetic bottleneck during serial infections: an insect-fungal model system. Curr Genet 50: 335–345 [DOI] [PubMed] [Google Scholar]
- Sharom JR, Bellows DS, Tyers M (2004) From large networks to small molecules. Curr Opin Chem Biol 8: 81–90 [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Singer SJ (1974) Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci USA 71: 4457–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorr AF, Tabak YP, Johannes RS, Sun X, Spalding J, Kollef MH (2009) Candidemia on presentation to the hospital: development and validation of a risk score. Crit Care 13: R156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE (2009) Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog 5: e1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher AJ, Chahine EB, Balcer HE (2009) Echinocandins: the newest class of antifungals. Ann Pharmacother 43: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Sudoh M, Yamazaki T, Masubuchi K, Taniguchi M, Shimma N, Arisawa M, Yamada-Okabe H (2000) Identification of a novel inhibitor specific to the fungal chitin synthase. Inhibition of chitin synthase 1 arrests the cell growth, but inhibition of chitin synthase 1 and 2 is lethal in the pathogenic fungus Candida albicans. J Biol Chem 275: 32901–32905 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES (2001) Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 3: 109–119 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang Y, Yan L, Liang RM, Dai BD, Tang RJ, Gao PH, Jiang YY (2009) Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J Proteome Res 8: 5296–5304 [DOI] [PubMed] [Google Scholar]
- Yan Z, Berbenetz NM, Giaever G, Nislow C (2009) Precise gene-dose alleles for chemical genetics. Genetics 182: 623–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P, Tschumi AI, Kishony R (2006) Functional classification of drugs by properties of their pairwise interactions. Nat Genet 38: 489–494 [DOI] [PubMed] [Google Scholar]
- Zhai B, Zhou H, Yang L, Zhang J, Jung K, Giam CZ, Xiang X, Lin X (2010) Polymyxin B, in combination with fluconazole, exerts a potent fungicidal effect. J Antimicrob Chemother 65: 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yan K, Zhang Y, Huang R, Bian J, Zheng C, Sun H, Chen Z, Sun N, An R, Min F, Zhao W, Zhuo Y, You J, Song Y, Yu Z, Liu Z, Yang K, Gao H, Dai H et al. (2007) High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc Natl Acad Sci USA 104: 4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinner RG, Barrett BL, Popova E, Damien P, Volgin AY, Gelovani JG, Lotan R, Tran HT, Pisano C, Mills GB, Mao L, Hong WK, Lippman SM, Miller JH (2009) Algorithmic guided screening of drug combinations of arbitrary size for activity against cancer cells. Mol Cancer Ther 8: 521–532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Figures S1-S9, Tables S2-S7
Supplementary Table S1. Normalized primary screen data.
Z-scores from all haploid deletion strain sensitivity profiles
Z-scores from all haploid deletion strain sensitivity profiles