This study determines the subunit stoichiometry of the tetradecameric caseinolytic protease (Clp) protease, the most abundant soluble protease in plant plastids. Using tagged Clp subunits and stable isotope-labeled peptides for mass spectrometry-based quantification, this analysis provides information that is critical for understanding the interaction of Clp protease with Clp chaperones and substrate delivery systems.
Abstract
The caseinolytic protease (Clp) protease system has been expanded in plant plastids compared with its prokaryotic progenitors. The plastid Clp core protease consists of five different proteolytic ClpP proteins and four different noncatalytic ClpR proteins, with each present in one or more copies and organized in two heptameric rings. We determined the exact subunit composition and stoichiometry for the intact core and each ring. The chloroplast ClpP/R protease was affinity purified from clpr4 and clpp3 Arabidopsis thaliana null mutants complemented with C-terminal StrepII-tagged versions of CLPR4 and CLPP3, respectively. The subunit stoichiometry was determined by mass spectrometry-based absolute quantification using stable isotope-labeled proteotypic peptides generated from a synthetic gene. One heptameric ring contained ClpP3,4,5,6 in a 1:2:3:1 ratio. The other ring contained ClpP1 and ClpR1,2,3,4 in a 3:1:1:1:1 ratio, resulting in only three catalytic sites. These ClpP1/R1-4 proteins are most closely related to the two subunits of the cyanobacterial P3/R complex and the identical P:R ratio suggests conserved adaptation. Furthermore, the plant-specific C-terminal extensions of the ClpP/R subunits were not proteolytically removed upon assembly, suggesting a regulatory role in Clp chaperone interaction. These results will now allow testing ClpP/R structure–function relationships using rationale design. The quantification workflow we have designed is applicable to other protein complexes.
INTRODUCTION
The Clp (caseinolytic protease) family has been found in almost all bacterial species and eukaryotic organelles (Yu and Houry, 2007). The Clp machinery has two oligomeric components: A barrel-shaped tetradecameric protease core with the catalytic sites sequestered inside the complex and hexameric ring-like ATP-dependent chaperones. The chaperones recognize specific substrates with or without the aid of adaptors, unfold these substrates, and translocate them into the proteolytic core for degradation (Baker and Sauer, 2006; Striebel et al., 2009). Compartmentalization of the proteolytic sites within the core complex, and coupling with chaperones and associated factors for substrate delivery, enable targeted protein degradation within the cell. The Clp proteolytic core in Escherichia coli consists of 14 identical Ser-type ClpP subunits, organized as a barrel-like structure with two stacked heptameric rings (Wang et al., 1997). Photosynthetic bacteria possess three ClpP proteins (ClpP1-P3) and one ClpR protein, which is structurally similar to ClpP but lacks the catalytic residues for peptide bond hydrolysis (Schelin et al., 2002). These cyanobacterial proteins assemble into an essential ClpR/P3 complex and a nonessential ClpP1/P2 complex (Stanne et al., 2007; Andersson et al., 2009).
The Clp system in plastids of plants and green algae has greatly expanded and diversified compared with its prokaryotic progenitors. The Clp family is the largest plastid-localized protease system in plants with nine different subunits (ClpP1, ClpP3-6, and ClpR1-4) comprising the core protease complex and six others (ClpC1-2, ClpD, ClpT1-2, and ClpS) involved in substrate recognition and delivery (reviewed in Kato and Sakamoto, 2010; Olinares et al., 2011). Except for the plastid-encoded ClpP1, all Clp proteins in plants are nucleus-encoded (Sokolenko et al., 2002; Peltier et al., 2004). Amino acid identities between the processed ClpP/R subunits range from 24 to 48% (Kim et al., 2009). Genetic and phenotypic analyses of various ClpP/R mutants in plants showed that the proteolytic and nonproteolytic subunits have differential functional contributions and all of them, except ClpR1, are essential for embryo or seedling development (Rudella et al., 2006; Sjögren et al., 2006; Zheng et al., 2006; Koussevitzky et al., 2007; Kim et al., 2009; Zybailov et al., 2009b) (reviewed in Kato and Sakamoto, 2010; Olinares et al., 2011).
The diversity of the subunits of the plastid ClpP/R core is puzzling, and given that most subunits are essential for Clp function and viability, it is critical to precisely determine the subunit stoichiometry for the intact core and the individual rings (for an extensive discussion, see Olinares et al., 2011). Once these stoichiometries are known, it will be more feasible to address ClpP/R structure–function relationships and to determine how the two rings interact with substrate delivery systems. We expect also that this will provide important information as to how the Clp system has adapted or coevolved with the plastid proteome. Based on native gel analysis, combined with Coomassie blue staining, and taking into account protein size and amino acid composition, we previously estimated a subunit stoichiometry of the ClpP/R core for P1, R2, R4, (R1/R3/P3), P4, P5, and P6 of 1:1:2:(3):3:3:1 (Peltier et al., 2004). Using native gels combined with immunoblotting, it was subsequently proposed that the core complex consists of a 230-kD ring containing ClpP1 and ClpR1-4, and a 180-kD ring containing ClpP3-6 (Sjögren et al., 2006). Combining this information with the stoichiometry estimated from Peltier et al. (2004), it was proposed that the ClpP1/ClpR1-4 heptamer would have a stoichiometry of P1, (R1+R3), R2, and R4 of 1:(3):1:2, and the ClpP3-6 heptamer would have P3, P4, P5, and P6 = 1:3:2:1 (Sjögren et al., 2006). The current study describes a systematic analysis to determine the precise stoichiometry in each purified ring and for the intact purified core complex, involving complementation of Clp mutants with StrepII-tagged subunits, affinity purification of individual rings, and quantification using isotope-labeled peptides. We show that both previous stoichiometry estimates are incorrect. In addition, we show that the plant ClpP3-5 and ClpR1-4 have extended C termini (up to 52 residues beyond the E. coli ClpP C terminus) and these extensions are predicted to form α-helical fragments that can fold over the adaxial surfaces, possibly affecting the docking of chaperones (Peltier et al., 2004). However, it is not known if these C termini are indeed stable when the ClpP/R proteins are assembled in the core complex.
Various mass spectrometry (MS)-based strategies have characterized the stoichiometry and interaction dynamics of protein complexes (Sharon and Robinson, 2007; Heck, 2008; Kaake et al., 2010). MS is not inherently quantitative, but this limitation can be overcome by including predetermined amounts of internal peptide standards that are stable isotope-labeled versions of representative peptides (from here on assigned proteotypic peptides) for each protein in the complex (reviewed in Brun et al., 2009). These proteotypic peptides can be generated by in vitro synthesis (Schmidt et al., 2010) or by in vivo synthesis of a concatamer of the proteotypic peptides (QconCAT) (Beynon et al., 2005). This latter approach involves the chemical synthesis of an artificial gene encoding the concatamer and its overexpression in a recombinant bacterial system grown in the presence of heavy isotopes (Beynon et al., 2005). Protease digestion of the stable isotope-labeled QconCAT protein releases equimolar amounts of peptide standards, allowing for multiplexed absolute protein quantification. This technique has been applied to analysis of complexes in yeast (Kito et al., 2007) and other species, but not yet to plants.
In this study, the chloroplast ClpP/R protease core complex and component rings were affinity purified from Arabidopsis thaliana transgenic lines harboring StrepII-tagged Clp subunits. By manipulating the buffer compositions, we were able to split each affinity-purified complex into its component heptameric rings. Subunit stoichiometries of the purified intact Clp complex, as well as the individual rings, were determined by MS-based absolute quantification using the QconCAT strategy. We also determined that the C-terminal extensions of the plastid ClpP/R subunits are conserved in plants and are not proteolytically removed during Clp complex assembly. Moreover, we inferred the evolutionary relationships of the Clp proteins from the plastid and their prokaryotic progenitors on the level of individual subunits and their oligomeric states. Finally, the workflow for detailed analysis of a heterooligomeric complex in plants developed and implemented for this study is generally applicable to other plant complexes. We believe that this will facilitate much needed protein–protein interaction studies in Arabidopsis and other plant model systems.
RESULTS
Affinity Purification and MS-Based Characterization of Chloroplast Clp Assemblies
Native gel separation of the chloroplast stroma proteome indicated that the Clp core complex migrates at 350 kD (Peltier et al., 2001, 2004) and its component rings at 180 to 230 kD (Sjögren et al., 2006). Native isoelectric focusing further showed that the plastid ClpPR subunits form a single core complex (Peltier et al., 2004). To facilitate purification of the ClpP/R core, as well as its individual rings, we generated two different transgenic plant lines expressing tagged ClpR4 or ClpP3 subunits by complementing homozygous CLPR4 and CLPP3 null mutants (clpr4-1 and clpp3-1) with, respectively, a 1× 35S-driven CLPR4 or CLPP3 cDNA fused to C-terminal StrepII tags (Figure 1). The eight-residue StrepII tag (Schmidt and Skerra, 2007) was attached to the C terminus rather than the N terminus to prevent interference with the N-terminal chloroplast-targeting peptide. A C-terminal tag also likely minimizes interference with the Clp core function because the N-terminal domains of ClpP directly modulate chaperone interactions for substrate delivery in the E. coli system (Gribun et al., 2005). The complemented mutant lines did grow on soil and, in contrast to the null mutants (Kim et al. 2009; Olinares et al. 2011), exhibited a wild-type phenotype (Figure 1A). Moreover, complementation was achieved at the protein level, with chloroplast proteins accumulating at wild-type levels (Figure 1B). We also employed other C-terminal tagging systems, including a polyhistidine tag (His6) and a tandem affinity tag (Rubio et al., 2005); whereas partial or complete phenotypic complementation was achieved, the isolated Clp complex was never sufficiently pure, as judged by MS/MS analysis. Therefore, these efforts will not be further discussed.
Figure 1.
Generation of Transgenic Plants Expressing a StrepII-Tagged Clp Subunit.
(A) Left panel: Homozygous clpr4-1, clpp3-1 could only develop on Suc-supplemented (2%) agar plants and show a pale green leaf phenotype with reduced growth. Plants were grown for 2 months under a 10/14-h light/dark cycle at 40 μmol photons m−2 s−1. Right panel: Comparative analysis of wild-type (wt), clpr4-1 complemented with CLPR4-StrepII and clpp3-1 complemented with CLPP3-StrepII grown on soil for ~5 weeks under a 10/14-h light/dark cycle at 100 μmol photons m−2 s−1. The complemented lines exhibit wild-type phenotype and are autotrophic.
(B) Immunoblot analysis of titrations (indicated as 1/10, 1/2, and 1) of total leaf protein extracts from wild-type, clpr4-1 complemented with CLPR4StrepII and homozygous clpr4-1 plants grown for 5 weeks on agar plates containing 0.5× Murashige and Skoog medium plus 2% Suc under a 10/14-h light/dark cycle at 40 μmol photons m−2 s−1. Membranes were probed with antibodies generated against different chloroplast proteins: The stromal RNA helicase 3; PsaF of photosystem I; OEC23; and OEC33 of the photosystem II. RNA helicase 3 was upregulated, whereas the thylakoid-bound PsaF, OEC23, and OEC33 were downregulated in clpr4-1. Accumulation of these proteins in the complemented plants was restored to wild-type levels, indicating successful complementation.
[See online article for color version of this figure.]
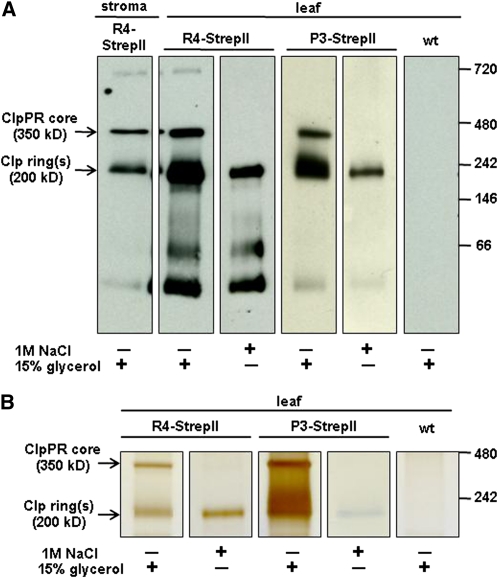
After optimization of purification conditions, immunoblot analyses with anti-StrepII serum showed that we obtained a tagged ~350-kD Clp core complex and a tagged ~200-kD Clp ring from the ClpR4 and ClpP3 StrepII-tagged lines (Figure 2A). High-resolution and high-accuracy MS analyses of the ~350- and ~200-kD protein gel bands (Figure 2B; see Supplemental Figure 1 and Supplemental Table 1 online) identified all nine ClpP and ClpR proteins in both bands. This suggested that the ~200-kD band contained a mixture of heptameric rings resulting from destabilization of the Clp core. To obtain highly purified individual heptameric rings, we incubated the column-bound tagged 350-kD Clp complexes in 1M NaCl without glycerol. After subsequent column washing, elution with buffer containing 2.5 mM desthiobiotin and 15% glycerol released a single StrepII-tagged Clp ring at ~200 kD for R4- and P3-tagged lines (Figure 2). MS/MS analyses showed that the purified 200-kD Clp ring from the StrepII-tagged ClpR4 plants contained essentially only ClpP1 and ClpR1-4, whereas the purified ring from StrepII-tagged ClpP3 plants contained only ClpP3, P4, P5, and P6 (see Supplemental Table 1 online). We did note that we observed just two or three MS/MS spectra for P4 and P5 (but not P3 and P6) in the salt-eluted R4-StrepII sample. These few spectral counts for P4 and P5 in that particular ring were from hydrophobic (“sticky”) peptides that could not be fully removed even after two blank liquid chromatography (LC)-MS runs; they should thus be considered as minor contaminating peptides.
Figure 2.
Native PAGE Analysis of Affinity-Purified StrepII-Tagged Clp Assemblies (350 and 200 kD) from Soluble Chloroplast (Stroma) or Leaf Extracts of Transgenic Plants Expressing R4-StrepII or P3-StrepII.
(A) and (B) On-column salt incubation in the absence of glycerol dissociated the individual rings from the core complex, facilitating the purification of only the StrepII-tagged Clp ring. (A) Immunoblot analyses with an anti-StrepII antibody. The 30-kD bands observed in both transgenic lines correspond to unprocessed forms of R4-StrepII and P3-StrepII, and the 66-kD band corresponds to dimers. A faint band at 700 kD indicates cross-interaction of the antibody to the Cpn60 complex. (B) Silver stain of native PAGE with similar samples as shown in (A). The complete gel is shown in Supplemental Figure 1A. wt, wild type.
[See online article for color version of this figure.]
Clearly, the on-column salt incubation separated the tagged ring-containing tetradecameric Clp core into two individual rings, similar as for the homotetradecamer in E. coli (Maurizi et al., 1998; Maglica et al., 2009), and subsequent washing removed the untagged ring. Importantly, this firmly established that each ClpP/R subunit is only present in one of the rings.
The identical ClpP rings in E. coli reassociate when glycerol is added after initial incubation in high salt conditions, even at high salt concentrations (>2 M KCl) (Maglica et al., 2009). By contrast, no 350-kD complex was observed upon addition of glycerol to the purified R4- or P3-tagged rings from the salt incubation experiments (Figure 2), suggesting that identical plastid Clp rings do not form stable double-ring complexes. We were unable to generate sufficient material that would allow mixing the ClpP1/R1-4 and ClpP3-6 rings to test if these different rings reassociate in the presence of glycerol; such positive results would strengthen the notion that identical plastid Clp rings do not form stable double-ring complexes and also demonstrate spontaneous assembly of nonidentical Clp rings.
The plant-specific ClpT1,2 proteins (with high similarity to the N terminus of the ClpC chaperones) (Peltier et al., 2004) were consistently identified with the 350-kD core, but the on-column salt incubations removed these subunits (see Supplemental Table 1 online), supporting the peripheral nature of their interaction with the ClpP/R subunits, as previously suggested based on homology modeling (Peltier et al., 2004).
Determination of Clp Protease Core Stoichiometry
To quantify the amount of each Clp subunit within the purified Clp core and individual rings, and to determine the stoichiometry among the subunits, we implemented the QconCAT approach. This involved spiking the samples with an equimolar set of stable isotope-labeled proteotypic tryptic peptides generated from a synthetic gene expressed in E. coli (Beynon et al., 2005).
First, we generated a list of experimentally observed proteotypic peptides that uniquely matched each of the 15 members of the plastid-localized Clp family based on extensive MS analysis of Arabidopsis samples available at our Plant Proteome Database (PPDB; http://ppdb.tc.cornell.edu/). Moreover, to ensure the most accurate quantification, it is important to avoid proteotypic peptides that contain residues that are easily chemically modified or peptides that have flanking sequences resulting in unreliable or variable tryptic cleavages (see Discussion and Beynon et al., 2005; Pratt et al., 2006; Kito et al., 2007). The Supplemental Text online shows the sequence coverage of each of the ClpP/R subunits that were obtained from extensive MS analysis of Arabidopsis chloroplasts, leaf extracts, and purified Clp complexes (see also PPDB). Tryptic peptide sequences containing residues prone to modifications during sample preparation (e.g., Cys, Met, and Trp) and chemically unstable sequences (e.g., N-terminal Asn, N-terminal Gln, and Asn-Gly) were marked. We also checked for potential missed cleavages and post-translational modifications from PPDB and for phosphorylation from the PhosPhAt database (Durek et al., 2010) to fully account for possible modifications on the endogenous peptides. From the PhosPhAt database, no Ser/Thr/Tyr phosphorylation was observed in any of the Clp protease subunits. Applying these restrictions, one or two peptides for each Clp subunit could be selected with peptide lengths ranging from 7 to 23 residues (Table 1; see Supplemental Table 2 online). Because no other suitable proteotypic peptides for quantification were available, we included a few peptides containing labile amino acids, namely P3-2 (with Met), P5-1 (with Asn-Gly), and P1-2 (with Trp) (Table 1).
Table 1.
Peptides Comprising Clp Protease Core Subunits Selected for Absolute Quantificationa
| Accession | Peptide | Sequence | m/zb | Qc |
| ATCG00670.1 | P1-1 | IAFPHAR | 271.1573 (+3) | N |
| P1-2 | SPGEGDTSWVDIYNR | 848.3841 (+2) | Y | |
| AT1G49970.1 | R1-1 | YLQAQAAIDYGIADK | 820.4199 (+2) | N |
| R1-2 | TAPPDLPSLLLDAR | 739.9143 (+2) | Y | |
| AT1G12410.1 | R2-1 | IALQSPAGAAR | 527.8038 (+2) | Y |
| R2-2 | FNAEEAIEYGLIDK | 806.3987 (+2) | Y | |
| AT1G09130.1 | R3-1 | EPIYIYINSTGTTR | 814.4199 (+2) | N |
| R3-2 | DILVELLSK | 515.3132 (+2) | Y | |
| AT4G17040.1 | R4-1 | GSAHEQPPPDLASYLFK | 619.6441 (+3) | Y |
| R4-2 | YFSPTEAVEYGIIDK | 866.4274 (+2) | Y | |
| AT1G66670.1 | P3-1 | DNTNLPSER | 523.2491 (+2) | N |
| P3-2 | LPSFEELDTTNMLLR | 889.9533 (+2) | Y | |
| P3-2 (ox) | LPSFEELDTTNM(ox)LLR | 897.9508 (+2) | Y | |
| AT5G45390.1 | P4-1 | SFEQVLK | 425.7371 (+2) | Y |
| P4-2 | ADVSTIALGIAASTASIILGAGTK | 1101.5790 (+2) | N | |
| AT1G02560.1 | P5-1 | ANLNGYLAYHTGQSLEK | 626.9830 (+3) | Y |
| P5-1 (d) | ANLDGYLAYHTGQSLEK | 627.3110 (+3) | Y | |
| P5-2 | FQSIISQLFQYR | 765.4092 (+2) | Y | |
| AT1G11750.1 | P6-1 | IIFIGQPINAQVAQR | 834.4832 (+2) | Y |
| P6-2 | VISQLVTLASIDDK | 751.4272 (+2) | Y |
For details on the other peptides that were concatenated within the Clp-QconCAT construct (includes peptides for Clp chaperones and adaptors), see Supplemental Table 2 online.
The m/z for the most abundant peptide species (unlabeled) with the charge state indicated in parentheses.
Can be consistently detected and quantified. Y, yes; N, no.
A gene (designated here as Clp-QconCAT gene) encoding for 29 Clp-derived peptides (this includes peptides for ClpP/R subunits and Clp chaperones), plus a number of additional peptides as controls, and a C-terminal His6 tag for affinity purification (see Supplemental Table 2 online) was then synthesized. The resulting 1506-bp gene was expressed from a plasmid in E. coli cells grown in media containing [13C6]-Lys and [13C6]-Arg (for stable isotope-labeled Clp-QconCAT protein) or unlabeled Lys and Arg (for unlabeled Clp-QconCAT protein; in addition to all other unlabeled amino acids). The 55-kD Clp-QconCat protein was then isolated by Ni2+-NTA affinity purification and the identity was confirmed by MS/MS analysis.
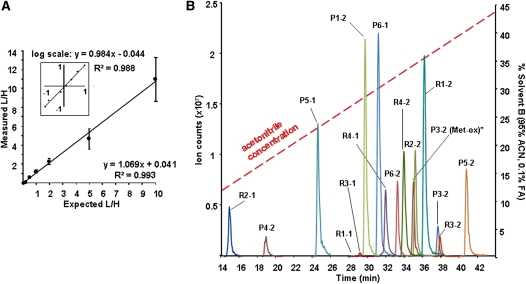
Quantitative LC-MS analysis was assessed and optimized using heavy (H, labeled) and light (L, unlabeled) affinity-purified E. coli Clp-QconCAT protein. L/H ratios for each peptide were calculated from peak areas of the corresponding extracted ion chromatograms (XIC). Peptide retention times deviated only within 0.2 min (2% coefficient of variation [CV]) across multiple LC-MS runs. Using 100 fmol heavy Clp-QconCAT protein and variable amounts of its light version, a linear MS instrument response was observed within two orders of magnitude (0.1–10 L/H ratios) with high precision for each of the identified Clp-QconCAT peptides (see Supplemental Table 3 online) and at the protein level (Figure 3A). Overall, 32 of the 37 QconCAT peptides could be detected and quantified, with 13 peptides representing the ClpP/R core subunits.
Figure 3.
Optimization and Accuracy of the Clp-QconCAT Quantification Method.
(A) The average linear MS instrument response for the Clp-QconCAT protein in varying concentrations with each sample being injected and analyzed by MS in triplicate. Samples consisted of 10 fmol to 1 pmol of light (L) Clp-QconCAT mixed with constant 100 fmol of heavy (H) Clp-QconCAT. In total, 32 peptides were quantified and the error bars indicate the sd. The inset shows the same plot on a logarithmic scale. For a detailed linear response for each peptide, see Supplemental Table 3 online.
(B) Representative XIC of the selected proteolytic peptides for Clp core subunits derived from 100 fmol Clp-QconCAT protein. Four of the nine Clp subunits could be quantified with two peptides and the rest with one peptide. An XIC plot of all quantifiable Clp-QconCAT peptides is shown in Supplemental Figure 2. All quantifiable Clp-QconCAT peptides eluted within the first 45 min of the gradient. ACN, acetonitrile.
[See online article for color version of this figure.]
Peak areas for peptides with modified and unmodified forms were summated. For example, peptide P3-2, used for the quantification of the ClpP3 subunit, contains a Met. Its mono-oxidized form (P3-2(ox)) was consistently two to three times more abundant than the unoxidized version and eluted ~2.6 min earlier (Figure 3B) as has been previously observed for Met-bearing peptides (Zybailov et al., 2009a).
We then used the 13C6-labeled QconCAT protein to quantify the subunits in the purified 350-kD core, as well as the mixed and purified individual 200-kD Clp rings. These gel-separated complexes were each in-gel digested, spiked with in-gel digested 13C6-labeled ClpQconCAT protein, and analyzed by MS in triplicate. The absolute amount of each selected proteotypic peptide was determined from the measured sample-to-standard (L/H) peak area ratios (see Supplemental Table 4 online). The average technical variation was 1.6% across all runs, which indicates very high reproducibility and accuracy. Four (ClpR2, R4, P5, and P6) of the nine Clp subunits could be quantified with two peptides. We then derived the relative stoichiometry of the Clp subunits for each ring (Table 2). ClpR4 and ClpP6 were chosen as the reference for the ClpP1/R and ClpP3-6 rings, respectively, because these proteins were reliably quantified with two peptides at high precision. The resulting stoichiometry for the ClpP1/R ring was ClpP1:R1:R2:R3:R4 = 3:1:1:1:1 and ClpP3:P4:P5:P6 = 1:2:3:1 for the other ring. Based on several biological replicates, we observed an average CV for the calculated stoichiometry of 15% (Table 2). Notably, the ratio of the total moles of each ring type within the core complex was 1.16 ± 0.2 (see Supplemental Table 4 online), confirming that the component rings indeed assembled in a 1:1 configuration. Moreover, the resulting sum of the calculated relative stoichiometries per ring validated the presence of seven subunits within each assembly and 14 subunits per complex.
Table 2.
Stoichiometry of the Clp Subunits within Each Clp Ringa
| Summary |
|||||||
| Clp | Peptide | Clp Coreb | Clp Ring (No Salt Added)c | Clp Ring (Salt Incubation)d | Average | % CV | Overall |
| ClpP1/R1-4 ring | |||||||
| P1 | P1-2 | 2.93 ± 0.38 | 3.32 ± 0.15 | 2.95 ± 0.38 | 3.12 ± 0.33 | 11 | 3 |
| R1 | R1-2 | 1.19 ± 0.23 | 1.27 ± 0.18 | 1.21 ± 0.12 | 1.23 ± 0.17 | 14 | 1 |
| R2 | R2-1 | 1.33 ± 0.24 | 1.55 ± 0.26 | 1.30 ± 0.11 | 1.42 ± 0.24 | 17 | 1 |
| R2-2 | 0.75 ± 0.07 | 0.84 ± 0.11 | 0.81 ± 0.12 | 0.80 ± 0.10 | 13 | ||
| R3 | R3-2 | 0.95 ± 0.12 | 1.04 ± 0.12 | 0.99 ± 0.36 | 1.00 ± 0.18 | 18 | 1 |
| R4 | R4-1 | 0.86 ± 0.16 | 0.94 ± 0.07 | 0.90 ± 0.02 | 0.9 ± 0.10 | 11 | 1 |
| R4-2 | 0.94 ± 0.10 | 1.06 ± 0.07 | 1.10 ± 0.02 | 1.03 ± 0.09 | 9 | ||
| n = 4 | n = 6 | n = 3 | n = 13 | ||||
| ClpP3-6 ring | |||||||
| P3 | P3-2 | 1.18 ± 0.16 | 0.98 ± 0.18 | 0.99 ± 0.12 | 1.04 ± 0.17 | 17 | 1 |
| P4 | P4-1 | 2.17 ± 0.04 | 2.53 ± 0.23 | 2.30 ± 0.28 | 2.37 ± 0.26 | 11 | 2 |
| P5 | P5-1 | 2.53 ± 0.59 | 2.52 ± 0.48 | 2.14 ± 0.45 | 2.40 ± 0.50 | 21 | 3 |
| P5-2 | 3.13 ± 0.80 | 3.11 ± 0.74 | 3.21 ± 0.67 | 3.15 ± 0.68 | 21 | ||
| P6 | P6-1 | 1.37 ± 0.26 | 1.13 ± 0.05 | 1.12 ± 0.02 | 1.20 ± 0.18 | 15 | 1 |
| P6-2 | 1.10 ± 0.29 | 0.87 ± 0.05 | 0.88 ± 0.02 | 0.95 ± 0.18 | 19 | ||
| n = 4 | n = 5 | n = 4 | n = 13 | ||||
Obtained from normalization against the absolute amounts of ClpR4 or ClpP6 within each Clp assembly (shown in Supplemental Table 4 online). SD is indicated for n samples spiked with 100 fmol of peptide standard with each sample injected and analyzed by MS in triplicate.
Affinity-purified 350-kDa Clp core complex from R4-StrepII and ClpP3-StrepII transgenic lines.
The 200-kDa Clp rings from the affinity purifications without salt incubation.
The ClpP1/R1-4 ring was purified from the ClpR4-StrepII line and the ClpP3-6 ring was obtained from the ClpP3-StrepII line.
Mapping the C-Terminal Extensions of Clp Subunits
The extended C termini of ClpP3-6 and ClpR1-4 (compared with E. coli ClpP) may form two to four α-helical fragments that can fold over the adaxial side of the core, thereby potentially interfering with docking of the ClpC/D chaperones and/or ClpT1,2 (Peltier et al., 2004). This could be particularly significant for the longer extensions of ClpP3 (52 amino acids) and P4 (41 amino acids). Sequence alignments to other recently fully sequenced plant species showed that these extensions are conserved in higher plants (Figure 4). The C-terminal Strep-II tags attached to the ClpP3 and ClpR4 subunits in the complemented lines used in this study were detected in the immunoblot analyses of the Clp core complex and the individual rings (Figure 2), indicating that their C termini were not proteolytically cleaved during Clp complex assembly. We also mapped all MS/MS-identified peptides in the Clp rings and core to their protein sequences and showed that the extended C termini are indeed part of the assembled ClpP/R complexes (Figure 4). Multiplying the subunit stoichiometries with the length of each C-terminal extension shows a twofold longer total sequence extension (174 versus 89 amino acids) in the ClpP3-6 ring than in the ClpP1/R ring; this suggests potentially stronger effects on adaxial docking for the ClpP3-6 ring.
Figure 4.
Sequence Analysis of the C-Terminal Extensions of Several Clp Subunits from Plants, Including the Bryophyte Physcomitrella patens (Pp), the Lycopod S. moellendorffii (Sm), the Dicots Arabidopsis (At) and Populus trichocarpa (Pt), and Monocots Maize (Zm) and Rice (Os).
Conserved residues are in purple. These C-terminal extensions are not excised in assembled Clp core complexes and Clp rings as observed from MS-based characterization (sequence coverage outlined in orange) of affinity-purified Clp core assemblies from Arabidopsis (see PPDB at http://ppdb.tc.cornell.edu/).
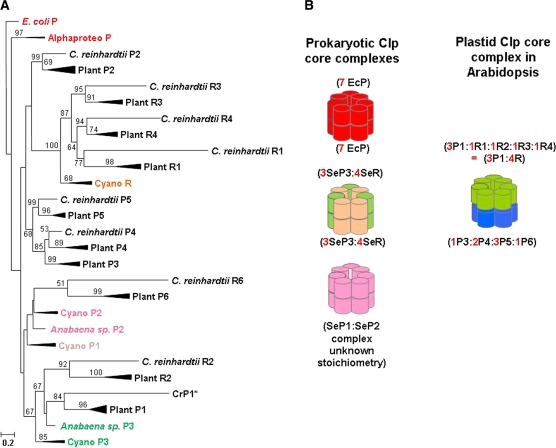
Evolutionary Relationships among Clp Subunits
Previous phylogenetic analyses of the Clp family (Peltier et al., 2001; Majeran et al., 2005) highlighted the evolutionary linkages among the Clp subunits in cyanobacterial and algal species with Arabidopsis and tobacco (Nicotiana tabacum). To better understand the evolution and adaptation of the Clp core complex in plants, we took advantage of the recently sequenced plant genomes from the monocotelydons maize (Zea mays) and rice (Oryza sativa) and dicotyledons Arabidopsis and poplar (both Rosales), in addition to the moss Physcomitrella patens (Bryophyta) and the spikemoss Selaginella moellendorffii (Lycopodiophyta). The resulting phylogenetic analysis of the ClpP/R proteins (Figure 5A) included 118 proteins from proteobacteria, photosynthetic bacteria, and plastids in green algae, bryophytes, lycopod, ferns, pine, monocots, and dicots (see Supplemental Table 5 online for all accession numbers). Several of the subtrees in Figure 5A were compressed (indicated as thick branch lines) for simplicity (see Supplemental Figure 3 online for the full version).The prokaryotic Clp proteins include the E. coli ClpP, and the ClpP from two alphaproteobacterial species that are closely related to the ancestors of mitochondria (indicated as the Alphaproteo subtree). The cyanobacterial Clps (indicated as the Cyano subtree) included five species. It should be noted that ClpP2 and ClpP3 for the cyanobacterial species Anabaena sp. (strain PCC 7120) did not fit in the cyanobacterial subtree and are indicated separately. The plastid-localized Clp proteins include those from the green alga Chlamydomonas reinhardtii and representative plant species (indicated as the Plant subtree), including a bryophyte, a lycopod, two monocotyledons, and two dicotyledons. The plastid ClpP1 homologs for the ferns Psilotum nodum and Adiantum capillus-veneris and the gymnosperm Pinus thunbergii (a pine) were also included in the Plant ClpP1 subtree.
Figure 5.
Evolutionary Relationships among Clp Assemblies.
(A) Compressed version of a phylogenetic analysis of 118 ClpP/R proteins from proteobacteria, photosynthetic bacteria, and plastids in green algae, bryophytes, lycopod, ferns, pine, monocots, and dicots, employing the ML approach (Felsenstein, 1981) with the GTR model for nucleotide substitution (Tavar, 1986) and a gamma model of substitution rate heterogeneity (Yang, 1996). Clp cDNA sequences (118) were aligned and analyzed (see Supplemental Data Set 1 online for the sequences and alignment, and Supplemental Table 5 online for all accession numbers). Several of the subtrees were compressed (indicated as thick branch lines) for simplicity and Supplemental Figure 3 online shows the full version). Support values are shown from 1000 nonparametric bootstrap replicates. Significant bootstrap support (>50%) is shown next to the respective nodes. The E. coli ClpP was designated as the outgroup. The truncated sequence (without the large insertion [IS1] sequence) of C. reinhardtii ClpP1 (CrP1*) was used.
(B) Oligomeric states of Clp protease complexes corresponding to homo- or heterooligomeric assembly states: E. coli and proteobacterial ClpP (red) and cyanobacterial ClpP1,2 (fuchsia), ClpP3 (green) and ClpR (orange), as well as the Arabidopsis plastid-localized Clp complex with the ring containing ClpP1-ClpR1-2-3-4 (green) or ClpP3-4-5-6 (blue).
Homology searches against these plant genomes and the green algae C. reinhardtii revealed that all plastid ClpP/R core subunits in Arabidopsis have orthologs in other plants and in green alga, except for CLPP3, which is not found in C. reinhardtii (Figure 5A; see Supplemental Figures 3 and 4 online). This indicates that the expansion of the Clp family occurred early in the green lineage and was maintained throughout the evolution of land plants. Moreover, the duplication event that yielded CLPP3 and CLPP4 in land plants occurred early in the terrestrial adaptation as both genes are already found in moss (see Supplemental Figure 3 online). Unlike the essential ClpP3/R core in Synechococcus, the two heptameric rings of the essential plastid ClpP/R core complex in Arabidopsis are composed of completely different subunits, and consequently, the adaxial sides (providing docking for Clp chaperones and ClpT1,2) are different. Careful phylogenetic analysis shows that the subunits of the ClpP1,R1-4 ring originate from the essential ClpP3-ClpR genes (Figure 5; see Supplemental Figure 3 online) (Majeran et al., 2005). In particular, the plant ClpP1 and ClpR2 genes originate from an ancestral cyanobacterial ClpP3, whereas ClpR1, ClpR3, and ClpR4 arose from duplications of the ancestral cyanobacterial ClpR (Figure 5; see Supplemental Figure 3 online). ClpP1 is the only plastid-encoded Clp gene in all plant and algal species, whereas ClpR2 is nuclear encoded. This suggests that divergence of ClpR2 involved lateral gene transfer of the ancestral cyanobacterial ClpP3 into the nuclear genome after which it underwent further modifications (e.g,. loss of catalytic residues). The origins of the subunits of the plastid ClpP3-6 ring is unclear; phylogenetic analysis suggests that ClpP6 is related to cyanobacterial ClpP2 (Figure 5; see Supplemental Figure 3 online).
DISCUSSION
The chloroplast-localized Clp protease core complex and its constituent rings were affinity purified and further characterized by MS-based absolute quantification, stoichiometry determination, and C-terminal mapping of the corresponding Clp subunits. Our results firmly established that the plastid-localized ClpP and ClpR proteins assemble into one asymmetric core complex comprised of a ClpP1/R1/R2/R3/R4 and a ClpP3/P4/P5/P6 ring, consistent with the essential nature of most ClpP/R subunits and earlier observations (Sjögren et al., 2006; Kim et al., 2009). Furthermore, results from salt-incubation treatment in the affinity purification of Clp complexes revealed that inter-ring associations within the plastid Clp core are primarily ionic and can be perturbed by high salt concentrations, similar to the situation in the bacterial Clp complex (Maurizi et al., 1998; Maglica et al., 2009). However, unlike in bacteria, the two heptameric rings within the plastid core complex are not identical (Figure 5B). The intact Clp core contained ClpT1 and ClpT2, similar to observations in our previous studies for wild-type plants, as well as in ClpR1 and ClpR2 mutants (Peltier et al., 2004; Rudella et al., 2006; Kim et al., 2009). However, ClpT1,T2 were consistently removed from the rings by on-column salt incubations, supporting the peripheral nature of their interaction with the ClpP/R subunits, as previously suggested based on homology modeling (Peltier et al., 2004).
MS-based absolute protein quantification strategies such as the QconCAT approach require peptide standards that exhibit excellent chromatographic behavior, high MS instrument response, and, ideally, absence of labile amino acids (e.g., Cys, Met, Trp, and N-terminal Asn or Gln) (Kuster et al., 2005). As such, peptide selection requires rigorous and exhaustive analysis of all available MS-derived information to fully characterize the behavior of these peptides within the sample preparation and LC-MS workflow. For equalizing cleavage efficiency and release of peptides generated from QconCAT and endogenous proteins during proteolytic digestion, inclusion of flanking C- and N-terminal sequences from the selected endogenous peptides has been recommended (Kito et. al., 2007). This is especially important for peptides with ragged ends, that is, in which the N- or C-terminal flanking sequence had multiple Lys or Arg, resulting in variable missed cleavages (if trypsin digestion is employed). Modifications on the endogenous peptides (phosphorylation, deamidation, and oxidation) also have to be accounted for, as these have to be recapitulated in the standard peptides for accurate quantification.
The use of multiple peptide standards per protein is required to increase the confidence in target protein quantification and to account for potential modifications and variations among individual peptide standards. We chose two peptides per subunit for concatenation into the Clp-QconCAT standard. In cases where only a single peptide can be found, we relaxed some of the restrictions to select a second proteotypic peptide. In as much as we would want to include more than two peptides for reliable and accurate quantification, we were practically limited by the available detectable and quantifiable tryptic peptides inherent in the Clp protein sequences (see Supplemental Text online). In addition, a few of the peptides chosen for quantification were either not detected (P1-1 and P3-1) due to short peptide length and instability or were detected but could not be reliably used for quantification due to low MS intensity response (R1-1 and R3-1) or aberrant chromatographic behavior (P4-2 is long and too hydrophobic). As such, we were left with only one peptide for quantification of several proteins. Nevertheless, we were able to overcome technical challenges related to accounting for chemical modifications of some of the peptides (oxidation and deamidation) because of the high mass resolution and accuracy of the Orbitrap and by summation of peak areas for unmodified and modified peptide forms.
We were able to quantify the nine ClpP/R subunits using 13 selected proteotypic peptides. From the absolute amounts of the Clp subunits within each Clp assembly that was analyzed, we determined a 1:2:3:1 subunit stoichiometry for the ClpP3,4,5,6 ring (the P ring) and 3:1:1:1:1 for the ClpP1,R1,2,3,4 in the other ring (the P1-R ring). However, in the case of the P ring, the ratio between P4 and P5 was not precisely 2:3. ClpP4 was quantified with one peptide in the 350-kD core and in the different ring preparations, whereas ClpP5 was quantified with two peptides also in three different types of preparations. The ClpP4 copy number was, on average, 2.37, whereas the ClpP5 copy number was, on average, 2.78. Importantly, when measured in the intact purified 350-kD core, the ratio between P4 and P5 was 2.17:2.83. Assuming a fixed stoichiometry within the core, i.e., a homogeneous population of Clp core complexes, our data best support a 1:2:3:1 subunit stoichiometry for the ClpP3,4,5,6 ring (the P ring). Alternatively, if there was no fixed stoichiometry within the 350-kD Clp core complex, then one could possibly suggest a mixed population of Clp complexes with 2P4 and 3P5 or 3P4 and 2P5. However, all published experimental evidence in the literature supported a fixed stoichiometry—this includes native gel electrophoresis and native isoelectric focusing (Peltier et al., 2004; Sjögren et al., 2006; for more discussion, see Olinares et al., 2011). Moreover, null mutants for CLPP4 (J. Kim and K.J. van Wijk, unpublished data) and CLPP5 (Kim et al., 2009) are embryo lethal, unlike ClpP3 (J. Kim and K.J. van Wijk, unpublished data), ClpR2, and ClpR4 (Kim et al., 2009), which are present in one copy per complex. This suggests that the loss of ClpP proteins normally present in multiple copies per complex is detrimental to plant viability.
Using our calculated stoichiometry and the predicted mass, the ClpP1/R1-4 ring and P3-6 ring are ~189 and 177 kD. Clearly, this cannot be fully resolved by native gel electrophoresis, consistent with our observations (we used a Bis-Tris native gel system and light blue-native gel separation). In the study by Sjögren et al. (2006), the Clp core complex mostly destabilized into partially separated gel bands when the stromal proteome was analyzed using a Tris-borate native gel system. Immunoblot analysis suggested that these partially resolved bands each contained one of the Clp heptameric rings. Moreover, additional band(s) representing the ClpP3-6 ring subcomplex with ClpT1-T2 (each ~20 kD) attached was also observed (Sjögren et al., 2006; Sjögren and Clarke, 2011). We consistently identified the ClpT1-T2 proteins in the 350-kD core complex but the on-column salt incubations removed these subunits (see Supplemental Table 1 online). Overall, the difference in native gel electrophoresis systems employed and the sample context (purified core or rings versus stromal proteome) might have yielded the slightly different migration profiles and resolution observed.
Given its ring composition and stoichiometry, the asymmetric plastid Clp core complex can be expected to exhibit different proteolytic capacities within each ring because the P ring has seven catalytic subunits, whereas the P1-R ring has only three. The lower concentration of catalytic sites in the P1-R ring is likely to show a lower rate of proteolysis, which may or may not be the rate-limiting step in the sequence of substrate selection, delivery, and unfolding. In cyanobacteria, the inclusion of the catalytically inactive ClpR in the ClpP3/R complex appeared not to be rate limiting because restoration of the catalytic sites and replacement of the whole internal domain of ClpR with that of ClpP3 did not enhance the proteolytic activity (Andersson et al., 2009). Asymmetric Clp protease core particles derived from combinations of proteolytically inactive (mutation in catalytic site of ClpP) and chaperone binding-incompetent (N-terminal mutation) Clp rings from E. coli have been assembled in vitro (Maglica et al., 2009). Surprisingly, complexes harboring one proteolytically inactive Clp ring stimulated the ATPase activity of ClpA to a similar degree, irrespective as to whether ClpA was bound proximal or distal to the inactive ring. This indicated allosteric communication between the two different rings and concerted conformational changes throughout the whole Clp core barrel (Maglica et al., 2009).
It is striking that the chloroplast P1-R ring has an overall 3:4 ClpP:R ratio similar to that in the cyanobacterial ClpP3/R core complex. Intriguingly, the 3:4 active:inactive composition is also observed in heptameric component rings of the eukaryotic proteasome (Groll et al., 1997), suggesting a functional restraint on the composition of these various proteolytic assemblies. Intact MS analysis of the dissociated subcomplexes of the cyanobacterial ClpP3/R core revealed mainly heterodimers and other heteromeric combinations, indicating that the ClpP3 subunit is localized between two ClpR subunits within the ring (Andersson et al., 2009). This organization might also apply to the chloroplast-localized ClpP1-R ring, with the ClpP1 arranged between two ClpR subunits. This would result in a P1-R-P1-R-P1-R-R order; thus, two of the ClpR subunits are directly next to each other in the ring but we do not know which ones. Interestingly, complete loss of CLPR2 and CLPR4 results in seedling lethality, whereas ClpR1 was redundant to ClpR3 (Kim et al., 2009).
The essential ClpP3/R complex in cyanobacteria only interacts with ClpC, a homolog of E. coli ClpA, whereas the nonessential ClpP1/P2 complex only interacts with ClpX (Stanne et al., 2007). Higher plant plastids do not contain any of the three predicted ClpX proteins (instead, these ClpX proteins are confined to the plant mitochondria) (Halperin et al., 2001; Peltier et al., 2004), but they do contain three ClpA homologs, namely ClpC1, C2, and D. Therefore, we speculate that the chloroplast ClpP1/R ring provides the docking surface for ClpC/D interaction. By contrast, the ClpP3-6 ring with unclear phylogenetic origin is most likely the preferred interaction partner for ClpT1,2 because these peripheral subunits are only observed in chloroplasts. It has been suggested that ClpT1 initially interacts with the ClpP3-P6 ring, followed by ClpT2, and the resulting complex subsequently associates with the ClpP1/R ring to form the core complex (Sjögren and Clarke, 2011). We speculate that the ClpP3-6 ring coevolved with ClpPT1,2 and together they represent a specific adaptation to the plastid proteome (for a discussion on the possible roles of ClpT1,2 in substrate selection, see Peltier et al., 2004). The extended C-terminal sequences of ClpP3 and ClpP4 in particular in the ClpP3-P6 ring may play a significant role in regulating the interaction with ClpT1,2 and ClpC1,2D, and thereby in contributing to substrate selection and delivery. In this case, ClpT1,T2 might further play a direct or indirect role in substrate delivery (for discussion, see Olinares et al., 2011), in addition to its involvement in core assembly as suggested recently (Sjögren and Clarke, 2011).
METHODS
Plant Growth Conditions
Arabidopsis thaliana (Columbia-0) plants were grown on agar plates with 0.5× Murashige and Skoog medium with 2% Suc under a 10/14-h light/dark cycle at 40 μmol photons m−2 s−1. Plants were grown on soil under a 10/14-h light/dark cycle at 100 to 120 μmol photons m−2 s−1.
Complementation of clpr4-1 with CLPR4-StrepII and clpp3-1 with CLPP3-StrepII
CLPR4 and CLPP3 cDNAs were PCR amplified and the C-terminal StrepII sequence was attached using Platinum Pfx polymerase (Invitrogen) or GOTaq Polymerase (Promega). The PCR products CLPR4-StrepII and CLPP3-StrepII were then subcloned into pENTR-D-TOPO (Invitrogen) or pCR8-TOPO (Invitrogen), respectively. The resulting pENTR-D-TOPO construct was then digested with MluI (New England Biolabs) and the CLPR4-StrepII-containing fragment was gel purified and subcloned into the Gateway destination vector pEARLEYGATE100 (Earley et al., 2006) using LR Clonase™ Enzyme Mix (Invitrogen). The same subcloning procedure was employed with the CLPP3-StrepII sequence in pCR8-TOPO without any restriction digestion step. Competent cells of Agrobacterium tumefaciens strain GV3101 were transformed with pEarleyGate100-CLPR4-StrepII or pEarleyGate 100-CLPP3-StrepII. Heterozygous clpr4-1 (JP7_7HO7l; see Kim et al. 2009) or clpp3-1 (SALK_065330) plants were used for Agrobacterium-mediated plant transformation by the floral dip method (Zhang et al., 2006). Transformants were screened using 10 μg/mL DL-phosphinothricin (BioWorld). Complemented plants that are homozygous to the T-DNA insertion were selected and verified by PCR genotyping. The primers used in this study are listed in Supplemental Table 6 online.
Affinity Purification of StrepII-Tagged Clp Core and Clp Ring
Total leaf material from transgenic plants was ground in liquid nitrogen and solubilized in 50 mM HEPES-KOH, pH 8.0, 15% glycerol, and 10 mM MgCl2 (extraction buffer, EB) with protease inhibitor cocktail. The suspension was then filtered in Miracloth and spun at 100,000g. The supernatant was concentrated, loaded on a Strep-Tactin column (IBA), and washed with EB for isolation of the Clp core or washed with EB without glycerol for purification of Clp rings. For isolating Clp rings, the column-bound Clp core was incubated in EB with 1 M NaCl for 1 h and was then subsequently eluted with EB lacking glycerol. Elution was performed using EB with 2.5 mM desthiobiotin. Light-blue native PAGE was performed with the NativePAGE Novex gel system (Invitrogen) using precast 4 to 16% acrylamide Bis-Tris gels (Invitrogen). Protein bands were stained with Coomassie dye or silver. For immunoblots, proteins were blotted onto polyvinylidene difluoride membranes and probed with anti-StrepII antibody (Genscript) using chemiluminescence for detection, following standard procedures.
Clp-QconCAT Design, Expression, and Purification
The selected Clp-QconCAT peptides (see Supplemental Table 1 online) were concatenated and flanked by a leader N-terminal sequence (MAGKVIR-) and a C-terminal sequence (AGKVICSAEGSK-) as in Beynon et al. (2005). Subsequent gene synthesis, cloning, transformation, protein expression, and protein purification was performed as previously described (Beynon et al., 2005; Pratt et al., 2006). Briefly, the corresponding Clp-QconCAT DNA sequence was designed, synthesized (Entelechon), and subcloned into pCR4-TOPO vector (Invitrogen). The resulting construct was confirmed by sequencing and was excised from the plasmid by restriction digestion and ligated into the NdeI and Hind III sites of the pET21a expression vector to yield the pET21a-Clp-QConCAT plasmid. Fusion to the vector also attaches a C-terminal His6 tag for subsequent purification. Overall, the full Clp-QconCAT protein sequence is: MAGKVIRVLVVANPANTNALILKLLSSGELYDIVGIPTSKVLDVVYNASNNELVRFNAEEAIEYGLIDKVAEIPDIDLSQVGVTKSPGEGDTSWVDIYNRDNTNLPSERSFEQVLKFQSIISQLFQYRVISQLVTLASIDDKYLQAQAAIDYGIADKEPIYIYINSTGTTRDETLSLLGKYFSPTEAVEYGIIDKILATLGFTDEKGGGVLDKPIIEKVLENLGADPSNIRVVGQDEAVAAISRIAFPHARLPSFEELDTTNMLLRADVSTIALGIAASTASIILGAGTKANLNGYLAYHTGQSLEKIIFIGQPINAQVAQRTAPPDLPSLLLDARIALQSPAGAARDILVELLSKGSAHEQPPPDLASYLFKAIAWAIDEKALDSALDQNLKVILHNDNFNKVPEPTVDETIQILKGSGFVAVEIPFTPRVFEAAVEYSRLSVPVSDVKSLGIPLVGLDTHPRSAIVQVDAAPFKIYGNTLSISEIKAGKVICSAEGSKLAAALEHHHHHH.
The pET21aClp-QConCAT plasmid was then transformed in BL21DE cells. For protein expression, the cells were initially grown overnight in minimal media consisting of M9 salts (NaCl and NH4Cl in phosphate buffer), Glc, thiamine, CaCl2, and MgSO4 and was then diluted in minimal media with amino acid mix with or without L-[13C6]Lys and L-[13C6] Arg (97–99% enrichment; Cambridge Isotope Laboratories) to a starting O.D. of 0.06 to 0.1. Expression of labeled or unlabeled Clp-QconCAT was induced with isopropyl β-d-thiogalactoside addition at an OD of 0.6 to 0.8 for 5 h. The cell culture was then centrifuged at 1450g for 10 min at 4°C and the cell pellets were stored at −80°C until needed.
Cells were then lysed using the Bugbuster Protein Extraction Reagent (Novagen) according to the manufacturer’s instructions and from Pratt et al. (2006). Briefly, the harvested cell pellet was resuspended in Bugbuster reagent and incubated at room temperature for 20 min. The suspension was then centrifuged to yield inclusion bodies containing the overexpressed Clp-QconCAT protein. The inclusion bodies were then resuspended in Bugbuster reagent and were treated with lysozyme (0.2 mg/mL final concentration) for 5 min at room temperature. A 1:10 diluted Bugbuster reagent was added and the mixture was centrifuged at 5000g for 5 min at 4°C. The resulting pellet was washed twice with 1:10 diluted Bugbuster reagent and resuspended in 20 mM phosphate buffer, pH 7.4, 0.5M NaCl, and 6 M guanidinium chloride (Buffer W) with 20 mM imidazole for nickel affinity purification. The sample was loaded in a HisTrap column (Amersham Biosciences) by a peristaltic pump, washed with buffer W with 20 mM imidazole, and bound protein was eluted with Buffer W and 250 mM imidazole.
Prior to SDS-PAGE separation, guanidine was removed from the purified Clp-QconCAT as in Mirzaei et al. (2008). Equal volumes of 20% trichloroacetic acid and purified Clp-QconCAT were mixed and incubated on ice for 30 min. The acidified solution was then centrifuged at 6000g at 4°C for 15 min, and the supernatant was carefully removed. The pellet was washed twice with 1 mL of 80% cold acetone, vortexed, centrifuged at 18,000g at 4°C for 5 min, and acetone was decanted. The pellet was then dried in a speed vacuum concentrator to fully remove acetone and was resuspended with a brief sonication on ice in 50 mM Tris, pH 6.8, and 1% SDS. Protein concentration was determined by the BCA Protein Assay Kit (ThermoScientific). Samples were then separated on a precast 10.5 to 14% gradient acrylamide Laemmli gel (Bio-Rad) and protein bands were stained with Coomassie dye.
LC-MS Analysis and Data Processing
For sample preparation for MS analysis, visible protein bands from native PAGE separation of Clp assemblies and from SDS-PAGE of labeled Clp-QconCAT protein were excised and in-gel digested separately with trypsin as previously described (Shevchenko et al., 1996). Trypsin digestion was performed overnight (typically for 19 h) at 37°C. Peptide extracts were dried using the speed vacuum concentrator and stored at −80°C until needed. The extracted peptides were resuspended in 2% formic acid (FA) and were mixed accordingly (e.g., peptides from in-gel–digested Clp assemblies spiked with peptides from in-gel–digested stable isotope-labeled Clp-QconCAT). An aliquot of the peptide mixture (typically 5 μL) was automatically loaded on a guard column (LC Packings MGU-30-C18PM) via an autosampler followed by separation on a PepMap C18 reverse-phase nanocolumn (LC Packings nan75-15-03-C18PM) using 85-min gradients with 95% water, 5% acetonitrile, and 0.1% FA (solvent A), and 95% acetonitrile, 5% water, and 0.1% FA (solvent B) at a flow rate of 200 nL/min. The gradient proceeds as follows for solvent B: 0 min – 10%, 6 min – 10%, 40 min – 45%, 48 min – 95%, 50 min – 95%, 51 min – 10%, 57 min – 10%, 65 min – 95%, 67 min – 95%, 68 to 85 min – 10%. Two blanks were run after every sample. Each sample was analyzed in triplicate. The MS acquisition cycle consisted of a survey MS scan in the Orbitrap with a set mass range from 250 to 1800 m/z at resolution 60,000 followed by five data-dependent MS/MS scans acquired in the LTQ. Dynamic exclusion was used with the following parameters: exclusion size, 500; repeat count, 2; repeat duration, 30 s; exclusion time, 90 s; exclusion window, ±0.6 m/z. Target values were set at 5 × 105 and 104 for the survey and tandem MS scans, respectively. Regular scans were used for the precursor and tandem MS with no averaging.
The XIC were calculated using the XISee software (Fusaro et al., 2009) with a retention time tolerance of ±2.5 min and m/z tolerance of ±10 ppm. For the deamidated P5-1 species, the m/z tolerance was set at ±7.5 ppm to distinguish the mass shift due to deamidation (0.98402 D) and the 13C peak of the unmodified P5-1 (mass difference of 1 D). The retention times were initially determined manually from examination of several raw files using the Thermo QualBrowser software, version 2.0.7 (ThermoFisher Scientific). Peak area quantification was done on SILAC mode with [13C6]Lys and [13C6]Arg. Peak smoothing was enabled and default parameters for peak detection were used. We also checked for possible modified forms and charge states of the peptide standards to ensure accurate quantification. For His-containing peptides R4-1 and P5-1 that exhibit multiple charge states, the dominant species were triply charged and quantification was based on only these species. Peak area analysis of the Trp-bearing peptide (P1-2) and its various oxidized forms (e.g., kynurenine, hydroxytryptophan, and dihydroxytryptophan; see also Perdivara et al., 2010) revealed that ~98% of P1-2 remained unoxidized. As such, only the peak areas from the +3 species of R4-1 and P5-1 and from the unmodified P1-2 were used for further quantification of the respective peptides. Deamidation of Asn, most common in -Asn-Gly sequences, yields Asp and βAsp and is usually observed in proteomic workflows involving overnight tryptic digestion (Krokhin et al., 2006). Two peaks were observed for the deamidated species (Asp and βAsp are isomeric) of P5-1 and one species eluted closely with the unmodified P5-1 (not shown). Previous studies revealed that reversed-phase LC using FA as an ion-pairing modifier separated native and deamidated peptides in the order of -Asn−/−βAsp−/−Asp- (Krokhin et al., 2006). In addition, the only reference peptide for quantification of the ClpP3 subunit (P3-2) contains a Met, which can readily undergo various states of oxidation during sample preparation. To allow inclusion of P5-1 and P3-2 peptides for quantification, peak area calculations included both from the native and modified forms. To generate the XIC for all Clp-QconCAT peptides in one plot, the peak profiles were obtained from an MS raw file using MASIC (Monroe et al., 2008; http://www.pnl.gov/), exported in Microsoft Excel and then reconstructed. Stable isotope-label incorporation was 97.5%.
For peptide identifications, peak lists (in .mgf format) were generated using DTA supercharge, version 1.19, software (http://msquant.sourceforge.net/) and searched with Mascot v2.2 (Matrix Science). For off-line calibration, first, a preliminary search against The Arabidopsis Information Resource ATH database, version 8, was performed, including sequences for known contaminants (e.g. keratin and trypsin; a total of 33,013 entries) with the precursor tolerance window set at ±30 ppm. Peptides with the ion scores above 33 were chosen as benchmarks to determine the offset for each LC-MS/MS run (Zybailov et al., 2009a). This offset was then applied to adjust precursor masses in the peak lists of the respective .mgf file for recalibration. The recalibrated peak lists were then searched against the The Arabidopsis Information Resource ATH database, including sequences for known contaminants (e.g. keratin and trypsin; a total of 33,013 entries) with or without a concatenated decoy database (made up of all the sequences in reverse orientation). Three searches were performed: a tryptic search with precursor ion tolerance window set at ±6 ppm, semi-tryptic search with N-terminal acetylation with precursor ion tolerance window set at ±3 ppm, and error-tolerant search at ±3 ppm (see Zybailov et al., 2009a). Fixed Cys carbamido-methylation modification was set, as well as Met oxidation and N-terminal Gln-to-pyroglutamate conversion for variable modifications. For samples with spiked stable isotope-labeled QconCAT, [13C6]Lys- and [13C6]Arg-labeling were included in the first two searches as variable modifications. To reduce the false protein identification rate for proteins identified by one peptide, the Mascot search results were further filtered as follows: the ion score threshold was increased to 35, and mass accuracy on the precursor ion was required to be within ±3 ppm. Furthermore, all matched precursor ions outside the 700to 3500 D range were disregarded. Overall, this yielded a peptide false discovery rate of 1%, with peptide false discovery rate calculated as: 2*(decoy_hits)/total_hits derived from searches against the database with decoy. All filtered results were uploaded into the PPDB (http://ppdb.tc.cornell.edu/) (Sun et al., 2009).
Total Leaf Extraction for Protein Immunoblot Analysis
Leaf material from wild-type, clpr4-1, and clpr4-1 complemented with CLPR4-StrepII was ground in liquid nitrogen, resuspended in 50 mM Tris, pH 6.8, 2% SDS, and protease inhibitor cocktail and was filtered through a 0.8-frit column (ThermoFisher Scientific) by a quick centrifugation step (1 min at 9000g rpm at 4°C). The filtrate was collected and protein concentration was determined using the BCA Protein Assay Kit (ThermoScientific). Proteins were separated on a precast 10.5 to 14% gradient acrylamide Laemmli gel (Bio-Rad), blotted unto polyvinylidene difluoride membranes, and probed with specific antibodies using chemiluminescence for detection, following standard procedures. The antibodies used were anti-RH3 and anti-OEC23 (from Dr. Alice Barkan) and anti-PsaF (Dr. Hendrik Scheller).
Phylogenetic Tree Construction
A phylogenetic analysis of 118 ClpP/R proteins from proteobacteria, photosynthetic bacteria, and plastids in green algae, bryophytes, lycopod, ferns, pine, monocots, and dicots was performed employing the Maximum Likelihood (ML) approach (Felsenstein, 1981) with the General Time Reversible (GTR) model for nucleotide substitution (Tavar, 1986) and a gamma model of substitution rate heterogeneity (Yang, 1996). Clp cDNA sequences (118) were aligned and analyzed (see Supplemental Data Set 1 and Supplemental Figure 4 online for the sequences and alignment and Supplemental Table 5 online for all accession numbers). The Escherichia coli ClpP was designated as the outgroup. The truncated sequence (without the large insertion [IS1] sequence) of Chlamydomonas reinhardtii ClpP1 (CrP1*) was used. The prokaryotic Clp proteins include the E. coli ClpP; the ClpP from two alphaproteobacterial species that are closely related to the ancestors of mitochondria (indicated as the Alphaproteo subtree): Rickettsia prowazekii and Wolbachia wMel; the cyanobacterial Clps (indicated as the Cyano subtree): Synechocystis sp. (strain PCC 6803), Synechococcus elongatus (strain PCC 7942), Microcystis aeruginosa, and Prochlorococcus marinus MED4. Note that ClpP2 and ClpP3 for the cyanobacterial species Anabaena sp. (strain PCC 7120) did not fit in the cyanobacterial subtree and are indicated separately. The plastid-localized Clp proteins include those from the green alga C. reinhardtii and representative plant species (indicated as the Plant subtree): the bryophyte Physcomitrella patens, the lycopod Selaginalla moellendorffii, the monocotyledons maize (Zea mays) and rice (Oryza sativa), as well as dicotyledons Arabidopsis and Populus trichocarpa. The plastid ClpP1 homologs for the ferns Psilotum nodum and Adiantum capillus-veneris and the gymnosperm Pinus thunbergii (a pine) were also included in the Plant ClpP1 subtree. The cDNA and protein sequences for the prokaryotic Clp proteins and for C. reinhardtii, P. trichocarpa, and P. patens Clps were obtained from KEGG (http://www.genome.jp/kegg/), Uniprot database (http://www.uniprot.org/), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), and European Molecular Biology Laboratory (http://www.ebi.ac.uk). The genome sequencing databases for Arabidopsis (http://www.Arabidopsis.org/), rice (http://rice.plantbiology.msu.edu/), maize (http://www.maizesequence.org/index.html), and Selaginella moellendorffii (http://genome.jgi-psf.org) were used to obtain the protein and cDNA sequences for the Clp protein family in these plants. Protein sequences were initially aligned using TCOFFEE (http://toolkit.tuebingen.mpg.de/t_coffee; Notredame et al., 2000) employing MLalign_id_pair and mslow_pair alignments. The resulting protein alignment (see Supplemental Data Set 1 and Supplemental Figure 4 online) was used as a template to align the corresponding cDNA sequences. The phylogenetic tree was generated using the Randomized Axelerated Maximum Likelihood (RAxML) software, version 7.0.3 (Stamatakis, 2006). The ML approach (Felsenstein, 1981) was employed with the GTR model for nucleotide substitution (Tavar, 1986) and a gamma model for substitution rate heterogeneity (Yang, 1996). The likelihood trees were generated with 100 replicates using the GTRGAMMA model using the script: RAxML.exe -n Clp_Nov2010ML100 -s Clp_Nov2010v6.txt -m GTRGAMMA -f d -p 20,903 -# 100. Multiple nonparametric bootstrapping was performed with 1000 replicates as such: raxml -n Clp_Nov2010v6btstrap1000 -s Clp_Nov2010v6.txt -m GTRGAMMA -f d -b 53,975 -# 1000. The resulting trees were visualized and edited using MEGA, versions 4 or 5 (Tamura et al., 2007). Sequence alignments of the C-terminal extensions were visualized using Jalview, version 2.6.1 (Waterhouse et al., 2009).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers: AT1G02560.1 (ClpP5), AT1G11750.1 (ClpP6), AT1G66670.1 (ClpP3), AT5G45390.1 (ClpP4), ATCG00670.1 (ClpP1), AT1G09130.1 (ClpR3), AT1G12410.1 (ClpR2) AT1G49970.1 (ClpR1), and AT4G17040.1 (ClpR4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Affinity Purification and MS-Based Protein Component Identification of StrepII-Tagged Clp Assemblies.
Supplemental Figure 2. Representative XIC of the Peptides Derived from 100 fmol Clp-QconCAT Protein.
Supplemental Figure 3. Phylogenetic Analysis of the ClpP/R Proteins from Proteobacteria, Photosynthetic Bacteria, Green Algae, and Plants.
Supplemental Figure 4. Protein and cDNA Sequence Alignments of Prokaryotic and Eukaryotic Clp Sequences.
Supplemental Table 1. MS/MS Analyses of the Affinity-Purified Clp Assemblies.
Supplemental Table 2. The Peptides Comprising Clp-QconCAT and Their Corresponding Properties.
Supplemental Table 3. Linear Instrument Response for Individual Clp-QconCAT Peptides.
Supplemental Table 4. Quantification of Clp Subunits and Determination of Subunit Stoichiometry in the Clp Core Complex and Constituent Clp Rings.
Supplemental Table 5. Accession Numbers for Protein and cDNA Sequences of the Clp Proteins from Prokaryotes, Green Algae, and Plants.
Supplemental Table 6. Primers Used in This Study.
Supplemental Text. Selection of the Clp-QconCAT Peptides.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 5A.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-1021963 and MCB-0718897) to K.J.v.W. We thank the members of the van Wijk lab for critical suggestions and helpful discussions.
AUTHOR CONTRIBUTIONS
P.D.B.O. performed most experiments and data analysis. J.K. provided the clpp3-1 null mutant line and generated the CLPP3-StrepII tagged transgenic line. J.I.D. provided expertise for the phylogenetic analysis. K.J.v.W., together with P.D.B.O., designed the experiments and wrote the manuscript. K.J.v.W. obtained the funding for this project and provided general oversight.
References
- Andersson F.I., et al. (2009). Structure and function of a novel type of ATP-dependent Clp protease. J. Biol. Chem. 284: 13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.A., Sauer R.T. (2006). ATP-dependent proteases of bacteria: Recognition logic and operating principles. Trends Biochem. Sci. 31: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon R.J., Doherty M.K., Pratt J.M., Gaskell S.J. (2005). Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2: 587–589 [DOI] [PubMed] [Google Scholar]
- Brun V., Masselon C., Garin J., Dupuis A. (2009). Isotope dilution strategies for absolute quantitative proteomics. J. Proteomics 72: 740–749 [DOI] [PubMed] [Google Scholar]
- Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X. (2010). PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38(Database issue): D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1981). Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17: 368–376 [DOI] [PubMed] [Google Scholar]
- Fusaro V.A., Mani D.R., Mesirov J.P., Carr S.A. (2009). Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 27: 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribun A., Kimber M.S., Ching R., Sprangers R., Fiebig K.M., Houry W.A. (2005). The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 280: 16185–16196 [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H.D., Huber R. (1997). Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Halperin T., Zheng B., Itzhaki H., Clarke A.K., Adam Z. (2001). Plant mitochondria contain proteolytic and regulatory subunits of the ATP-dependent Clp protease. Plant Mol. Biol. 45: 461–468 [DOI] [PubMed] [Google Scholar]
- Heck A.J. (2008). Native mass spectrometry: A bridge between interactomics and structural biology. Nat. Methods 5: 927–933 [DOI] [PubMed] [Google Scholar]
- Kaake R.M., Wang X., Huang L. (2010). Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol. Cell. Proteomics 9: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2010). New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D., van Wijk K.J. (2009). Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito K., Ota K., Fujita T., Ito T. (2007). A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J. Proteome Res. 6: 792–800 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Stanne T.M., Peto C.A., Giap T., Sjögren L.L., Zhao Y., Clarke A.K., Chory J. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63: 85–96 [DOI] [PubMed] [Google Scholar]
- Krokhin O.V., Antonovici M., Ens W., Wilkins J.A., Standing K.G. (2006). Deamidation of -Asn-Gly- sequences during sample preparation for proteomics: Consequences for MALDI and HPLC-MALDI analysis. Anal. Chem. 78: 6645–6650 [DOI] [PubMed] [Google Scholar]
- Kuster B., Schirle M., Mallick P., Aebersold R. (2005). Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 6: 577–583 [DOI] [PubMed] [Google Scholar]
- Maglica Z., Kolygo K., Weber-Ban E. (2009). Optimal efficiency of ClpAP and ClpXP chaperone-proteases is achieved by architectural symmetry. Structure 17: 508–516 [DOI] [PubMed] [Google Scholar]
- Majeran W., Friso G., van Wijk K.J., Vallon O. (2005). The chloroplast ClpP complex in Chlamydomonas reinhardtii contains an unusual high molecular mass subunit with a large apical domain. FEBS J. 272: 5558–5571 [DOI] [PubMed] [Google Scholar]
- Maurizi M.R., Singh S.K., Thompson M.W., Kessel M., Ginsburg A. (1998). Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and clpP. Biochemistry 37: 7778–7786 [DOI] [PubMed] [Google Scholar]
- Mirzaei H., McBee J.K., Watts J., Aebersold R. (2008). Comparative evaluation of current peptide production platforms used in absolute quantification in proteomics. Mol. Cell. Proteomics 7: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe M.E., Shaw J.L., Daly D.S., Adkins J.N., Smith R.D. (2008). MASIC: A software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 32: 215–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Olinares P.D., Kim J., van Wijk K.J. (2011). The Clp protease system: A central component of the chloroplast protease network. Biochim. Biophys. Acta 1807: 999–1011 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ripoll D.R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants: Their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg J., Liberles D.A., Roepstorff P., van Wijk K.J. (2001). Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276: 16318–16327 [DOI] [PubMed] [Google Scholar]
- Perdivara I., Deterding L.J., Przybylski M., Tomer K.B. (2010). Mass spectrometric identification of oxidative modifications of tryptophan residues in proteins: Chemical artifact or post-translational modification? J. Am. Soc. Mass Spectrom. 21: 1114–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J.M., Simpson D.M., Doherty M.K., Rivers J., Gaskell S.J., Beynon R.J. (2006). Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 1: 1029–1043 [DOI] [PubMed] [Google Scholar]
- Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh-Kumar S.P., Deng X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Rudella A., Friso G., Alonso J.M., Ecker J.R., van Wijk K.J. (2006). Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelin J., Lindmark F., Clarke A.K. (2002). The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus. Microbiology 148: 2255–2265 [DOI] [PubMed] [Google Scholar]
- Schmidt C., Lenz C., Grote M., Lührmann R., Urlaub H. (2010). Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal. Chem. 82: 2784–2796 [DOI] [PubMed] [Google Scholar]
- Schmidt T.G., Skerra A. (2007). The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2: 1528–1535 [DOI] [PubMed] [Google Scholar]
- Sharon M., Robinson C.V. (2007). The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 76: 167–193 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Sjögren L.L., Clarke A.K. (2011). Assembly of the chloroplast ATP-dependent Clp protease in Arabidopsis is regulated by the ClpT accessory proteins. Plant Cell 23: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L., Stanne T.M., Zheng B., Sutinen S., Clarke A.K. (2006). Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko A., Pojidaeva E., Zinchenko V., Panichkin V., Glaser V.M., Herrmann R.G., Shestakov S.V. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41: 291–310 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stanne T.M., Pojidaeva E., Andersson F.I., Clarke A.K. (2007). Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J. Biol. Chem. 282: 14394–14402 [DOI] [PubMed] [Google Scholar]
- Striebel F., Kress W., Weber-Ban E. (2009). Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 19: 209–217 [DOI] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37(Database issue): D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tavar S. (1986). Some probabilistic and statistical problems on the analysis of DNA sequences. Some Mathematical Questions in Biology: DNA Sequence Analysis, Miura R.M., (Providence, RI: American Mathematical Society; ), pp. 57–86 [Google Scholar]
- Wang J., Hartling J.A., Flanagan J.M. (1997). The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91: 447–456 [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. (2009). Jalview Version 2: A multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1996). Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. (Amst.) 11: 367–372 [DOI] [PubMed] [Google Scholar]
- Yu A.Y., Houry W.A. (2007). ClpP: A distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581: 3749–3757 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zheng B., MacDonald T.M., Sutinen S., Hurry V., Clarke A.K. (2006). A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224: 1103–1115 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Friso G., Kim J., Rudella A., Rodríguez V.R., Asakura Y., Sun Q., van Wijk K.J. (2009b). Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics 8: 1789–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Sun Q., van Wijk K.J. (2009a). Workflow for large scale detection and validation of peptide modifications by RPLC-LTQ-Orbitrap: Application to the Arabidopsis thaliana leaf proteome and an online modified peptide library. Anal. Chem. 81: 8015–8024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.