This study shows the identification of a plant-specific actin binding protein, STOMATAL CLOSURE-RELATED ACTIN BINDING PROTEIN1, which associates with and stabilizes microfilaments, and regulates microfilament reorganization during stomatal closure in response to drought stress.
Abstract
Microfilament dynamics play a critical role in regulating stomatal movement; however, the molecular mechanism underlying this process is not well understood. We report here the identification and characterization of STOMATAL CLOSURE-RELATED ACTIN BINDING PROTEIN1 (SCAB1), an Arabidopsis thaliana actin binding protein. Plants lacking SCAB1 were hypersensitive to drought stress and exhibited reduced abscisic acid-, H2O2-, and CaCl2-regulated stomatal movement. In vitro and in vivo analyses revealed that SCAB1 binds, stabilizes, and bundles actin filaments. SCAB1 shares sequence similarity only with plant proteins and contains a previously undiscovered actin binding domain. During stomatal closure, actin filaments switched from a radial orientation in open stomata to a longitudinal orientation in closed stomata. This switch took longer in scab1 plants than in wild-type plants and was correlated with the delay in stomatal closure seen in scab1 mutants in response to drought stress. Our results suggest that SCAB1 is required for the precise regulation of actin filament reorganization during stomatal closure.
INTRODUCTION
Plants must cope with various environmental changes during their life cycle. For example, during periods of drought, plants close their stomata to prevent excessive water vapor loss from their leaves. Many cellular signaling molecules and ions, including abscisic acid, H2O2, Ca2+, and NO, regulate stomatal closure by controlling K+ and anion channel activities, leading to a reduction in guard cell turgor (Pei et al., 2000; Hosy et al., 2003; Desikan et al., 2004; Hirayama and Shinozaki, 2007). Guard cell movement involves rearrangement of the actin cytoskeletal network (Kim et al., 1995; Eun and Lee, 1997; Liu and Luan, 1998; Hwang and Lee, 2001; Lemichez et al., 2001; MacRobbie and Kurup, 2007; Choi et al., 2008; Gao et al., 2008; Higaki et al., 2010). During stomatal closure and opening, microfilaments (MFs) undergo dynamic changes that are associated with changes in the activity of osmosensitive and stretch-activated Ca2+-permeable channels (Zhang et al., 2007) or inward K+ channels (Hwang et al., 1997). Consistent with this, the inhibition of MF reorganization via pharmacological means interrupts stomatal closure and opening (Kim et al., 1995; MacRobbie and Kurup, 2007).
Actin is a highly conserved globular protein in eukaryotic cells. Less than 10% of the actin in plant cells is in the filamentous form, whereas the majority of the actin in yeast or animal cells is filamentous, suggesting that the MFs of plant cells are remarkably dynamic, likely enabling plants to efficiently cope with environmental changes (Karpova et al., 1995; Gibbon et al., 1999; Snowman et al., 2002; Wang et al., 2005). Besides their function in regulating stomatal movement, MF dynamics are involved in many other cellular processes, including cell division, expansion, motility, organelle trafficking, endocytosis, exocytosis, and signal transduction (Thomas et al., 2009). To perform these functions, MF dynamics must be precisely regulated, and this requires many specific actin-associated factors, including actin binding proteins.
Actin binding proteins are essential for normal functioning of the actin cytoskeleton during plant growth and development (Barrero et al., 2002; Jedd and Chua, 2002; Oikawa et al., 2003; Ketelaar et al., 2004b; Ingouff et al., 2005; Szymanski, 2005; Djakovic et al., 2006; Xiang et al., 2007; Holweg and Nick, 2008; Peremyslov et al., 2008; Prokhnevsky et al., 2008; Sparkes et al., 2008; Tian et al., 2009; Li et al., 2010; Peremyslov et al., 2010; Suetsugu et al., 2010; Treitschke et al., 2010; Ueda et al., 2010; Vidali et al., 2010). Although pharmacological data suggest that MF reorganization plays important roles in plant stress responses, the molecular mechanisms underlying these effects are not well understood. Two genetic studies have suggested that stomatal movement and MF disruption are correlated (Dong et al., 2001; Lemichez et al., 2001); however, the functional significance of the interaction between MFs and stomatal movement is unclear. In this study, we identified a new actin binding protein, STOMATAL CLOSURE-RELATED ACTIN BINDING PROTEIN1 (SCAB1), that associates with and stabilizes MFs, and that regulates MF reorganization during stomatal closure in response to drought stress.
RESULTS
Genetic Screening for Arabidopsis thaliana Mutants Defective in Stomatal Movement
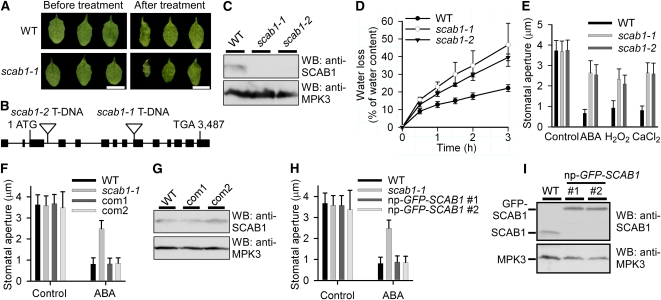
To identify components regulating stomatal movement and drought tolerance in Arabidopsis, we screened a T-DNA (Agrobacterium tumefaciens-transferred DNA) insertion pool by monitoring the rate of water loss in detached leaves. Rosette leaves at a similar developmental stage were detached and the degree of wilting was used as the screening standard. Several stomatal movement-defective mutants were recovered. One of them, scab1-1, was hypersensitive to drought stress (Figure 1A). Insertion of the T-DNA fragment was detected in exon 7 of At2g26770 by plasmid rescue (Figure 1B). To analyze the expression of SCAB1 in vivo, we generated anti-SCAB1 polyclonal antibodies. The antibodies recognized a specific band of ~55 kD in wild type but not in the scab1 mutants (Figure 1C). Water loss from detached leaves occurred much more rapidly in scab1-1 than in wild type (Figure 1D). Consistent with this observation, stomatal closure in the scab1-1 mutant was less sensitive to abscisic acid, H2O2, and CaCl2 compared with wild type (Figure 1E). The phenotype of scab1-2 (SAIL_193_E09), an allelic mutant obtained from the Arabidopsis Biological Resources Center (ABRC), was similar to that of scab1-1 (Figures 1B to 1E). Expression of a 7.5-kb SCAB1 genomic DNA fragment, including the 1.9-kb promoter and 2.1-kb 3′-untranslated region, complemented the stomatal phenotype of scab1-1 (Figure 1F). The expression level of SCAB1 in the complemented lines was similar to that in wild type (Figure 1G). The stomatal closure phenotype of the scab1-1 mutant was also rescued by the transgene of green fluorescent protein (GFP)-SCAB1 driven by the SCAB1 promoter (np-GFP-SCAB1) (Figure 1H), indicating that the chimeric protein was functional. The amount of GFP-SCAB1 was similar to that of SCAB1 in wild type (Figure 1I). Except for a defect in stomatal movement, no other significant difference in growth or development was observed between wild type and scab1.
Figure 1.
The scab1 Mutant Shows Impaired Stomatal Closure.
(A) Leaves detached from wild-type (WT; top panel) and scab1-1 (bottom panel) plants for 0 (left) and 3 h (right). Scale bars, 1 cm.
(B) Structure of SCAB1. The filled black boxes indicate exons, and the lines between the boxes indicate introns. The two T-DNA insertions are also indicated.
(C) Immunoblot analysis of 100 μg of total soluble protein extracted from 1-d-old wild-type and scab1 mutant seedlings with anti-SCAB1 or -MPK3 antibodies. The SCAB1 polyclonal antibodies specifically recognized a band corresponding to SCAB1, which was present in the wild type and absent from scab1. MPK3 was used as a loading control.
(D) Cumulative leaf transpirational water loss in wild-type (black circles), scab1-1 (open circles), and scab1-2 (black triangles) rosettes at the indicated times after detachment (means ± sd, n = 3).
(E) Stomatal closure in the scab1-1 and -2 mutants showed reduced sensitivity to abscisic acid, H2O2, and CaCl2. The data shown represent the stomatal apertures in response to 20 μM abscisic acid, 0.5 mM H2O2, and 2 mM CaCl2. The data represent the means ± sd of three independent experiments; 50 stomata were analyzed per line.
(F) The stomatal closure phenotype of scab1-1 was complemented by a SCAB1 genomic DNA fragment. The data shown represent the stomatal apertures in response to 20 μM abscisic acid. The data represent the means ± sd of three independent experiments; 50 stomata were analyzed per line. Com1 and com2: lines 1 and 2 of scab1-1 expressing SCAB1 under the control of the SCAB1 native promoter, respectively.
(G) Immunoblot analysis of SCAB1 expression in 100 μg of total soluble protein from 1-d-old wild-type, com1, and com2 seedlings using anti-SCAB1 or -MPK3 (loading control) antibodies.
(H) scab1-1 was complemented with a ProSCAB1:GFP-SCAB1 clone. The data shown represent the stomatal apertures in response to 20 μM abscisic acid. The data represent the mean ± sd of three independent experiments; 50 stomata were analyzed per line.
(I) Immunoblot analysis of 100 μg of total soluble protein extracted from 1-d-old wild-type and np-GFP-SCAB1 seedlings with anti-SCAB1 or -MPK3 (loading control) antibodies.
[See online article for color version of this figure.]
SCAB1 Associates with MFs in Vivo
SCAB1 encodes a protein of 496 amino acids in length with a calculated molecular mass of 55 kD. To determine the expression pattern of SCAB1 in Arabidopsis, the promoter region of SCAB1 (1.09 kb) was fused to a β-glucuronidase (GUS) reporter gene. The resulting construct was transferred to wild-type Arabidopsis (Columbia-0 [Col-0] background), and 12 independent T2 lines were analyzed by GUS staining. GUS signal was detected in the roots, stems, leaves, flowers, and guard cells (see Supplemental Figures 1A to 1G online). To confirm these results, total RNA was extracted from various tissues of 1-month-old plants and subjected to real-time PCR analysis. SCAB1 was widely expressed in a variety of tissues throughout plant development (see Supplemental Figure 1H online). These results are consistent with microarray data from the Nottingham Arabidopsis Stock Centre (Honys and Twell, 2004) and AtGeneExpress (Schmid et al., 2005) (see Supplemental Figures 1I and 1J online).
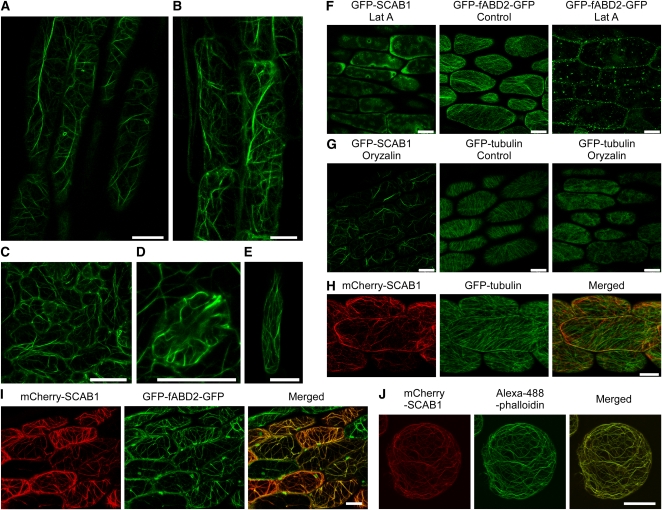
To determine the subcellular localization of SCAB1, GFP was fused to the N terminus of SCAB1 under the control of the SCAB1 promoter, and this construct was transformed into scab1-1 mutant plants (Figures 1H and 1I). GFP-labeled SCAB1 was detected in fibrous structural networks in the cytoplasm. Some of the GFP-SCAB1 filaments formed bundles at the cell cortex, while others formed a fine network. These filamentous structures were observed in hypocotyl epidermal cells (Figure 2A), root epidermal cells (Figure 2B), leaf epidermal cells (Figure 2C), guard cells (Figure 2D), and root hairs (Figure 2E).
Figure 2.
SCAB1 Forms Fibrous Networks and Decorates MFs in Vivo.
(A) to (E) Confocal images of epidermal cells taken from the hypocotyls (A), roots (B), leaves (C), guard cells (D), and root hairs (E) of GFP-SCAB1 driven by the SCAB1 native promoter in a scab1-1 background.
(F) ProSCAB1:GFP-SCAB1 and GFP-fABD2-GFP transgenic seedlings were treated with 1 μM Lat A for 1 h.
(G) ProSCAB1:GFP-SCAB1 and GFP-tubulin transgenic seedlings were treated with 10 μM oryzalin for 1 h.
(H) mCherry-SCAB1 (left) and GFP-labeled MTs (middle) in hypocotyl epidermal cells. The merged image is shown on the right.
(I) mCherry-SCAB1 (left) and GFP-fABD2-GFP-labeled MFs (middle) in hypocotyl epidermal cells. The merged image is shown on the right.
(J) mCherry-SCAB1 (left) and Alexa-488-phalloidin-labeled MFs (middle) in Arabidopsis suspension cells. The merged image is shown on the right. Scale bars, 20 μm.
The SCAB1-labeled filamentous structure resembled that of the plant cytoskeletal system. To determine the spatial relationship of the filamentous structure with microtubules (MTs) and MFs, 4-d-old ProSCAB1:GFP-SCAB1 transgenic seedlings were treated with latrunculin A (Lat A) or oryzalin. Lat A is an inhibitor of actin polymerization that disrupts actin filaments by binding actin monomers. After 1 h of treatment with 1 μM Lat A, the GFP-SCAB1 network and GFP-fABD2-GFP–labeled MFs (Wang et al., 2008b) were disrupted in most of the cells (Figure 2F). By contrast, after 1 h of treatment with 10 μM oryzalin, which disrupts MTs by binding α-tubulin, the filamentous structure remained intact in most of the cells, whereas the GFP-labeled MTs (Ueda et al., 1999) were disrupted (Figure 2G). These results suggest that GFP-SCAB1 decorates MFs rather than MTs. To confirm this, mCherry-SCAB1 was introduced into transgenic plants harboring Pro35S:GFP-Tubulin or Pro35S:GFP-fABD2-GFP. The hypocotyl epidermal cells of the double-labeled plants were then analyzed by confocal microscopy. The SCAB1 filaments did not show obvious colocalization with MT (Figure 2H). For example, we did not detect a 1:1 coalignment between the SCAB1 filaments and more numerous cortical MTs, although, as seen in Figure 2H, they tended to share the same general orientation. By contrast, obvious and extensive overlapping was observed between the mCherry-SCAB1 and GFP-fABD2-GFP signals (Figure 2I), particularly in the thick, subcortical actin strands, which do not generally contain MTs. In F-actin staining assays, mCherry-SCAB1 colocalized with Alexa-488-phalloidin–stained MFs in Arabidopsis suspension cells (Figure 2J). Taken together, our results demonstrate that SCAB1 is an MF-associated protein.
SCAB1 Binds F-Actin in Vitro
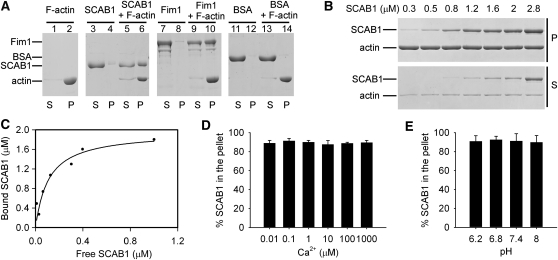
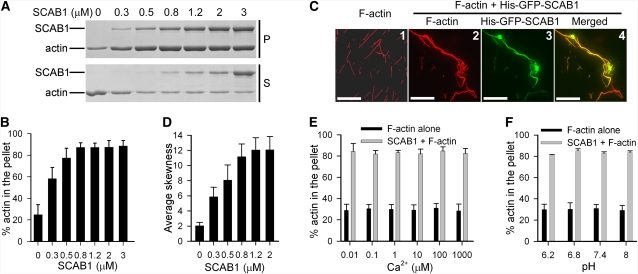
Given that GFP-SCAB1 associates with the MF network in Arabidopsis cells, we used an actin cosedimentation assay to investigate whether SCAB1 binds F-actin directly in vitro. SCAB1 or Fim1, a known F-actin binding protein (Kovar et al., 2000), was incubated with preformed F-actin and then pelleted by centrifugation at 150,000g. In the F-actin–free controls, only a small amount of SCAB1 or FIM1 was detected in the pellets (Figure 3A, lanes 4 and 8); however, SCAB1 and FIM1 was obviously coprecipitated in the presence of F-actin (Figure 3A, lanes 6 and 10). The control (BSA) remained in the supernatant in the absence (Figure 3A, lane 11) and presence of F-actin (Figure 3A, lane 13). Thus, SCAB1 binds F-actin directly.
Figure 3.
SCAB1 Binds F-Actin in Vitro.
(A) A high-speed cosedimentation assay was used to assess SCAB1 binding to F-actin. Cosedimentation experiments were performed with 3 μM SCAB1, BSA, or Fim1 in the presence or absence of 2 μM F-actin. After centrifugation at 150,000g, the proteins in the supernatant (S) and pellet (P) were resolved by SDS-PAGE and visualized by Coomassie Blue staining. BSA and Fim1 were used as negative and positive controls, respectively.
(B) Increasing concentrations of SCAB1 (0.3–2.8 μM) were cosedimented with 2 μM F-actin.
(C) The experiments described in (B) were repeated six times. After gel quantification, the concentration of bound SCAB1 was plotted against the concentration of free SCAB1 and fitted with a hyperbolic function. In this representative experiment, the Kd was 0.12 μM with a stoichiometry at saturation of 0.98 molecules of SCAB1 bound per actin subunit.
(D and E) High-speed cosedimentation assays were used to examine the effects of Ca2+ (D) and pH (E) on SCAB1 binding to F-actin. The experiments were performed with 0.5 μM SCAB1 and 2 μM F-actin. The percentage of SCAB1 in the pellet was quantified by densitometry and plotted as a function of the [Ca2+]free (D) and pH (E) (means ± sd, n = 3).
To determine the binding affinity of SCAB1 for F-actin, increasing concentrations of SCAB1 were incubated with preformed F-actin as described previously (Huang et al., 2005). The supernatants and pellets were separated by centrifugation at 150,000g and analyzed by SDS-PAGE (Figure 3B). The amount of SCAB1 in the supernatants and pellets was quantified by densitometry. The concentration of F-actin–bound SCAB1 was plotted against the concentration of free SCAB1 and fitted to a hyperbolic function (Figure 3C). In this representative experiment, the calculated dissociation constant (Kd) was 0.12 μM with a stoichiometry at saturation of 0.98 mole of SCAB1 bound per mole of actin. From six independent experiments, a mean Kd value (± sd) of 0.18 ± 0.06 μM (n = 6) for SCAB1 binding to F-actin was calculated. At saturation, the stoichiometry of the interaction was 1:1. Investigating the SCAB1 and actin protein levels in planta, we found that the ratio of actin to SCAB1 was ~700:1 by immunoblot analyses (see Supplemental Figure 2 online). In wild-type seedlings, the actin concentration was 10.0 ± 1.1 ng and SCAB1 was 12.7 ± 1.8 pg per 1 mg of total protein.
The actin binding activity of actin binding proteins is tightly regulated by many cellular parameters, including Ca2+ and pH (Khurana et al., 2010; Papuga et al., 2010; Zhang et al., 2010). Therefore, we performed high-speed cosedimentation assays at different concentrations of free Ca2+ ([Ca2+]free) or pH values to determine whether the activity of SCAB1 is regulated by Ca2+ or pH. Our results suggest that the binding of SCAB1 to F-actin is insensitive to Ca2+ and pH (Figures 3D and 3E).
SCAB1 Is a Plant-Specific Actin Binding Protein
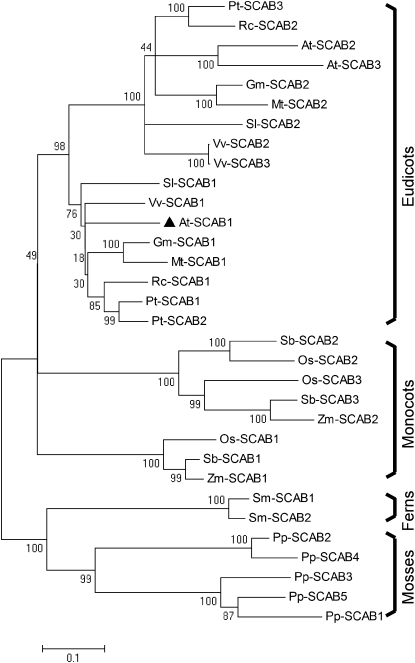
SCAB1 does not exhibit obvious similarities to other known actin binding proteins. Proteins with strong sequence similarity to SCAB1 have been identified in several plant species (E value < 1e-90), including Arabidopsis, soybean (Glycine max), alfalfa (Medicago truncatula), grape (Vitis vinifera), poplar (Populus trichocarpa), castor (Ricinus communis), tomato (Solanum lycopersicum), rice (Oryza sativa), maize (Zea mays), sorghum (Sorghum bicolor), Selaginella moellendorffii, and Physcomitrella patens (see Supplemental Table 1 online). These sequences were aligned using ClustalX 2.0.5 and the result (available as Supplemental Data Set 1 online) was analyzed using MEGA 4 (Figure 4). SCAB1 homologs were identified in eudicots, monocots, ferns, and mosses, but not in green algae or other nonplant species (E value > 1e-2). SCAB1 showed greater similarity to proteins from eudicots, and lower similarity to proteins from ferns and mosses. These data indicate that SCAB1 is a previously undiscovered F-actin binding protein in plants.
Figure 4.
Phylogenetic Tree Showing SCAB1 and Its Homologs.
The sequences were aligned using ClustalX 2.0.5, and the result was analyzed using MEGA 4. At, Arabidopsis; Gm, G. max; Mt, M. truncatula; Os, O. sativa; Pp, P. patens; Pt, P. trichocarpa; Rc, R. communis; Sb, S. bicolor; Sl, S. lycopersicum; Sm, S. moellendorffii; Vv, V. vinifera; and Zm, Z. mays. SCAB1 (black triangles) homologs were identified in eudicots, monocots, ferns, and mosses, but not in green algae or other nonplant species. The numbers on the nodes indicate the confidence values using 1000 replications. Accession numbers are shown in Supplemental Table 1 online and the alignment is available as Supplemental Data Set 1 online.
SCAB1 Bundles Actin Filaments
The 1:1 binding stoichiometry of SCAB1 to actin suggested that SCAB1 is an actin filament side binding protein. Given that some side binding proteins also bundle actin filaments, we examined the effect of SCAB1 on F-actin bundling by low-speed centrifugation (13,000g) analysis. Preassembled F-actin (2 μM) was incubated with increasing concentrations of SCAB1 (0–3 μM) and centrifuged at 13,000g for 30 min. The supernatants and pellets were analyzed by SDS-PAGE (Figure 5A) and the amount of SCAB1 in the pellets was quantified by densitometry (Figure 5B). As shown in Figure 5B, most of the F-actin appeared in the supernatant in the absence of SCAB1 following low-speed centrifugation. However, the amount of F-actin in the pellet fraction increased in proportion to the SCAB1 concentration. These results demonstrate that SCAB1 bundles actin filaments in vitro.
Figure 5.
SCAB1 Bundles F-Actin in Vitro.
(A) A low-speed cosedimentation assay was used to assess the bundling activity of SCAB1. Increasing concentrations of SCAB1 (0.3–3 μM) were incubated with 2 μM F-actin and the reactions were centrifuged at 13,000g. Equivalent amounts supernatant (S) and pellet (P) were separated by SDS-PAGE.
(B) The experiments described in (A) were repeated five times. The amount of actin present in the pellet fraction was determined. The data given are the means ± sd.
(C) Bundling of actin filaments by SCAB1 as visualized by fluorescence microscopy. Images from left to right: 1, micrograph of actin filaments stained with rhodamine-phalloidin in the absence of SCAB1; 2, micrograph of actin filaments in the presence of His-GFP-SCAB1 stained with rhodamine-phalloidin; 3, the corresponding GFP channel of (2); and 4, merged images of (2) and (3). Scale bars, 20 μm.
(D) Bundling of actin filaments in vitro by SCAB1 as shown by skewness analysis. The data represent the mean ± sd from three independent experiments; 20 micrographs were measured per concentration.
(E and F) Low-speed cosedimentation assays were used to assess the effects of Ca2+ (E) and pH (F) on the bundling activity of SCAB1. The experiments were performed with 2 μM F-actin in the presence or absence of 0.5 μM SCAB1. The percentage of actin in the pellet was plotted as a function of the [Ca2+]free (E) and pH (F) (means ± sd, n = 3).
To confirm the effect of SCAB1 on F-actin bundling, actin filaments were incubated with or without 1 μM His-GFP-SCAB1, stained with rhodamine-phalloidin, and visualized by fluorescence microscopy. In the absence of GFP-SCAB1, actin filaments were observed as thin fibers. In the presence of GFP-SCAB1, the GFP signal overlapped obviously with F-actin and the F-actin formed thicker strands (Figure 5C), indicating that SCAB1 associates directly with actin filaments and organizes them into bundles. To evaluate the actin-bundling activity of SCAB1, we performed a quantitative skewness analysis using an algorithm for examining the degree of actin bundling based on the asymmetry of fluorescence intensity distribution (Higaki et al., 2010), using phalloidin-decorated F-actin in the presence or absence of SCAB1. As shown in Figure 5D, the average skewness values increased in proportion to the SCAB1 concentration. These results are consistent with our low-speed centrifugation results, indicating that SCAB1 is an actin filament-bundling protein. We also performed low-speed cosedimentation assays using various [Ca2+]free or pH values; our results suggest that the bundling activity of SCAB1 toward F-actin is insensitive to Ca2+ and pH (Figures 5E and 5F).
SCAB1 Stabilizes Actin Filaments in Vitro
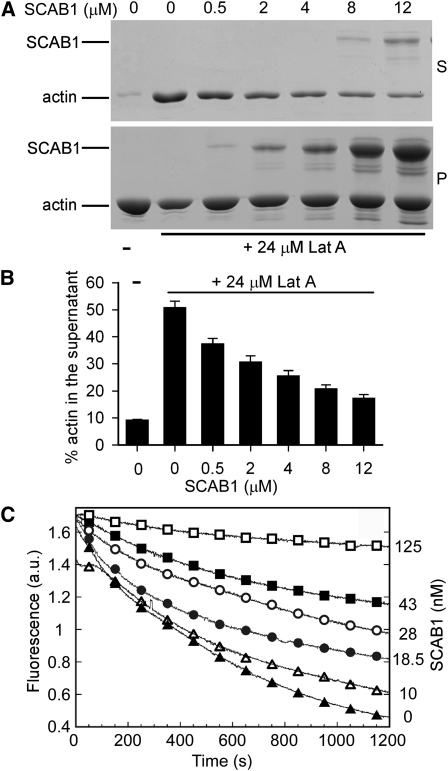
We next used Lat A to test the effect of SCAB1 on F-actin stability. Preformed F-actin was incubated with varying concentrations of SCAB1 and then treated with Lat A. The supernatants and pellets were separated by centrifugation at 150,000g, analyzed by SDS-PAGE (Figure 6A), and the amount of actin in the supernatants was quantified (Figure 6B). Less than 10% of the actin was detected in the supernatant without Lat A treatment; by contrast, the addition of Lat A increased the amount of actin in the supernatant to more than 50%. When SCAB1 was incubated with actin prior to the addition of Lat A, the proportion of actin in the supernatant decreased corresponding to the SCAB1 concentration (Figures 6A and 6B), demonstrating that SCAB1 stabilizes F-actin in vitro.
Figure 6.
SCAB1 Stabilizes Actin Filaments in Vitro.
(A and B) SCAB1 inhibited the Lat A-induced depolymerization of F-actin. (A) Preassembled F-actin (7 μM) was incubated with various amounts of SCAB1 (0–12 μM) and subsequently treated with 24 μM Lat A. The samples were centrifuged at 150,000g for 40 min and the resulting pellets (P) and supernatants (S) were analyzed by SDS-PAGE. (B) The experiments described in (A) were repeated three times. The amount of actin present in the supernatant fraction was determined. The data given are the means ± sd.
(C) SCAB1 prevents dilution-mediated actin depolymerization. F-actin depolymerization was monitored by tracking the decrease in pyrene-actin fluorescence. Actin filaments (5 μM) were diluted 25-fold in Buffer G in the absence (black triangle) or presence of 10 (open triangle), 18.5 (black circle), 28 (open circle), 43 (black square), and 125 nM SCAB1 (open square). a.u., arbitrary units.
The stabilizing activity of SCAB1 was also measured with a dilution-mediated depolymerization assay. Pyrene-labeled F-actin was diluted with Buffer G and the F-actin level was monitored by measuring the decrease in fluorescence. In the absence of SCAB1, the fluorescence decreased dramatically after dilution. When increasing concentrations of SCAB1 were preincubated with F-actin, the decrease in fluorescence was obviously reduced (Figure 6C). These results demonstrate that SCAB1 plays a role in stabilizing actin filaments.
Many actin-bundling and -stabilizing proteins have nucleation and capping activities (Huang et al., 2003; Li et al., 2010; Yang et al., 2011; Zhang et al., 2011). Therefore, we investigated whether SCAB1 has these activities. As shown in Supplemental Figure 3 online, increasing concentrations of SCAB1 did not change the initial actin polymerization rate or the initial rate of actin elongation; however, recombinant Os-FH5 FH2 (Yang et al., 2011; Zhang et al., 2011), which was included as a positive control, decreased the initial lag phase corresponding to nucleation (see Supplemental Figure 3A online) and the initial elongation rate corresponding to capping (see Supplemental Figure 3B online). These results suggest that SCAB1 does not possess nucleation or capping activity.
SCAB1 Stabilizes Actin Filaments in Vivo
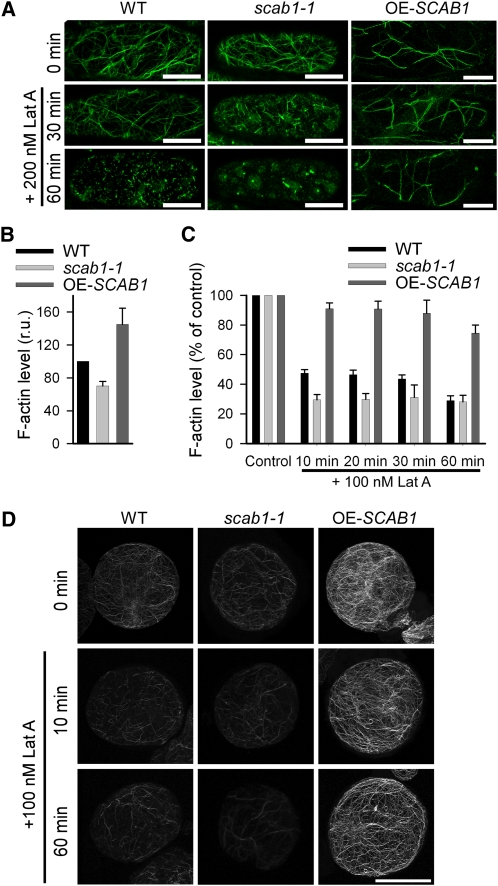
To determine whether SCAB1 stabilizes actin filaments in Arabidopsis cells, we transformed Pro35S:fABD2-GFP (for expression of the second actin binding domain of At-Fim1 fused to GFP) (Wang et al., 2004) into wild-type plants, scab1-1, and a SCAB1 overexpression line (OE-SCAB1) that expressed Pro35S:SCAB1 in a Col-0 background. The transgenic lines were treated with 200 nM Lat A. As shown in Figure 7A, actin filament depolymerization in hypocotyl epidermal cells was assessed by monitoring the fABD2-GFP signal. After 30 min of Lat A treatment, actin filament shortening was detected in 81% of the cells in the scab1-1 mutant. By contrast, the actin network was slightly affected in 68% of the cells from wild-type plants and 95% of the cells from OE-SCAB1 transgenic plants. After 60 min of treatment, the actin cytoskeleton in 93% of the scab1-1 cells and 81% of the wild-type cells was depolymerized; however, thick actin bundles were still present in ~80% of the cells in the OE-SCAB1 transgenic plants. These observations clearly demonstrate the bundling and stabilizing effect of SCAB1 on actin filaments in Arabidopsis.
Figure 7.
SCAB1 Stabilizes Actin Filaments in Vivo.
(A) Actin filament organization in hypocotyl epidermal cells from wild-type (WT; left column), scab1-1 mutant (middle column), and OE-SCAB1 (right column) plants expressing 35S:fABD2-GFP before and after 30 or 60 min of Lat A treatment.
(B) The relative level of F-actin in suspension cells of the wild type, scab1-1, and OE-SCAB1 was quantified before Lat A treatment. The F-actin level in the wild type was arbitrarily set to 100%. The data given are the means ± sd (n = 3). r.u., relative unit.
(C) Filamentous actin in suspension cells of the wild type, scab1-1, and OE-SCAB1 was quantified at different time points after the addition of Lat A. Each bar represents the mean value (± sd) of three independent experiments. The level of F-actin without Lat A treatment was set at 100% for each cell type.
(D) Alexa-488-phalloidin-stained MFs in suspension cells of the wild type (left column), scab1-1 (middle column), and OE-SCAB1 (right column) before and after 10 or 60 min of Lat A treatment. Scale bars, 20 μm.
[See online article for color version of this figure.]
To examine further the stabilizing effect of SCAB1 on actin filaments in vivo, we quantified the amount of filamentous actin after Lat A treatment. To evaluate the F-actin levels, suspension cell lines were generated from wild-type, OE-SCAB1, and scab1-1 seedlings. The F-actin level in the wild-type cells was higher than that in scab1-1 but lower than that in OE-SCAB1, indicating that the abundance of SCAB1 is correlated with the F-actin level in suspension cells (Figure 7B). After treatment with 100 nM Lat A, the level of F-actin decreased more quickly in scab1 and much more slowly in OE-SCAB1 compared with wild type (Figures 7C and D). These results confirm that SCAB1 stabilizes actin filaments in vivo.
Our data suggested that SCAB1 stabilizes actin filaments in hypocotyl epidermal cells and suspension cells (Figure 7). To investigate this effect in guard cells, we treated transgenic seedlings harboring 35S:GFP-fABD2-GFP with Lat A. GFP fluorescence-labeled dot-like structures were detected, which were assumed to be depolymerized F-actin (see Supplemental Figure 4A online). The percentage of guard cells with depolymerized MFs was analyzed after Lat A treatment. As shown in Supplemental Figure 4B online, F-actin depolymerization in scab1-1 occurred more rapidly compared with wild type, suggesting that SCAB1 also plays a role in stabilizing actin filaments in guard cells. To investigate the effect of SCAB1 overexpression on F-actin stability in guard cells, we generated GFP-SCAB1 overexpression (OE-GFP-SCAB1) lines by transforming Pro35S:GFP-SCAB1 into Col-0 plants. The protein abundance of SCAB1 compared with wild type was increased up to 140 times in the OE-GFP-SCAB1 #3 and 180 times in the OE-GFP-SCAB1 #11 based on immunoblot analyses (see Supplemental Figure 5 online). GFP-SCAB1-labeled MFs depolymerized much more slowly in OE-GFP-SCAB1 transgenic plants compared with np-GFP-SCAB1 (see Supplemental Figure 4C online), suggesting that SCAB1 stabilizes actin filaments in guard cells.
The Mutation of SCAB1 Delays Stomatal Closure-Associated Actin Reorganization
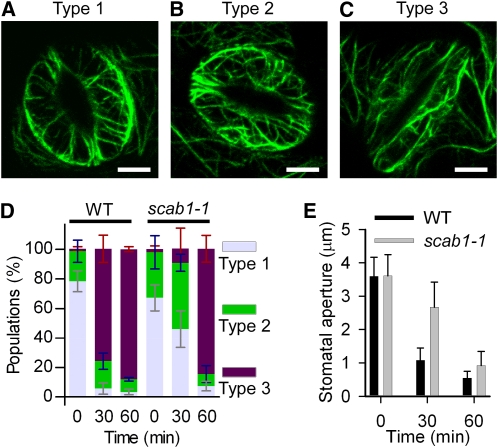
Stomatal closure in scab1 displayed reduced sensitivity to abscisic acid. To examine whether the mutation of SCAB1 alters stomatal movement-associated actin reorganization, stomata from wild-type and scab1-1 mutant plants expressing GFP-fABD2-GFP were opened in 50 mM KCl (aperture > 4) and then treated with 20 μM abscisic acid. As shown in Figure 8 and Supplemental Figure 6 online, abscisic acid treatment caused the stomatal closure-associated reorganization of actin filaments in wild type and scab1. When stomata were opened wide (aperture > 3), the actin filaments were well-organized and radiated out from the stomatal pore (Figure 8A; see Supplemental Figure 6A online, type 1). During stomatal closure (aperture > 2 and < 3), the actin filaments became randomly organized (Figure 8B; see Supplemental Figure 6B online, type 2). In closed stomata (aperture < 1), the actin filaments were rearranged and bundled preferentially as long cables in the longitudinal direction (Figure 8C; see Supplemental Figure 6C online, type 3). Our results are consistent with recently published results on the classification of MF configurations in guard cells of GFP-fABD2 transgenic lines during stomatal movement (Higaki et al., 2010).
Figure 8.
The scab1 Mutation Delays Actin Reorganization during Stomatal Closure.
(A) to (C) Confocal images of guard cells in rosette leaves from Pro35S:GFP-fABD2-GFP transgenic plants in a wild-type background. Actin organization was classified into three groups. Representative images of type 1, radial array (A); type 2, random meshwork (B); and type 3, longitudinal array (C) are shown. Scale bars, 5 μm.
(D) Histograms showing the actin organization in guard cells from wild-type (WT) and scab1-1 plants at the indicated times after abscisic acid treatment. The guard cells were classified into three groups: type 1, type 2, and type 3. The data represent the mean ± sd of six independent experiments; 100 guard cells per line were measured at the indicated times.
(E) Stomatal aperture in wild-type and scab1-1 mutant leaves at the indicated times after abscisic acid treatment. The data represent the mean ± sd of three independent experiments. At least 50 stomata were analyzed per line.
In wild-type and scab1 plants, actin filaments underwent a similar reorganization process during stomatal closure (see Supplemental Figure 6 online). However, the switch from type 1 to type 3 took more time in the scab1 mutants than in wild type (Figure 8D). After 30 min of treatment with 20 μM abscisic acid, 76% of the wild-type guard cells showed a change in actin organization to type 3. By contrast, the MFs in most of the scab1-1 mutant guard cells stayed as type 1 or 2; only 10% of the guard cells exhibited a type 3 actin organization. However, when the treatment period was extended to 60 min, the percentage of guard cells with a type 3 actin configuration in the scab1-1 mutant and wild type increased up to 85%. Consistent with the slower reorganization of actin in scab1-1, stomatal closure in the scab1-1 mutant was also slower than that in wild type (Figure 8E).
Previous studies suggested that changes in MF bundling are important for stomatal opening (Higaki et al., 2010). To test whether MF bundling is also essential for stomatal closure, we analyzed the skewness values of fABD2-labeled MFs in the guard cells of abscisic acid-treated wild-type and scab1-1 mutant plants. Analyses of more than 100 stomatal micrographs at each time point during abscisic acid treatment did not reveal an obvious correlation between stomatal closure and the skewness values (see Supplemental Figure 7 online), suggesting that the MF bundles indicated by the skewness values might not be directly associated with a particular stage of stomatal closure.
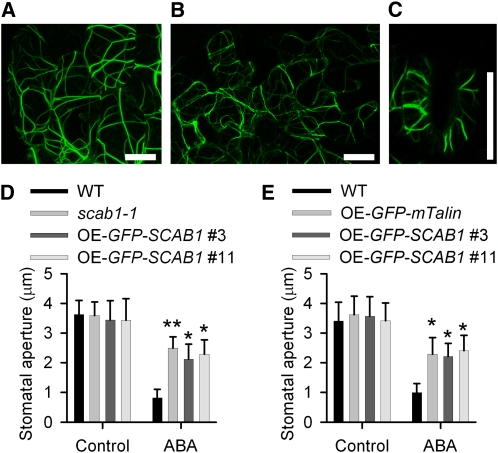
SCAB1 Overexpression Causes Actin Filament Bundling in Vivo and Delays Stomatal Closure
SCAB1 stabilizes MFs in guard cells, and plants lacking SCAB1 exhibit retardation in abscisic acid-mediated stomatal closure. To investigate the effect of SCAB1 overexpression on stomatal movement, we analyzed GFP-SCAB1 overexpression (OE-GFP-SCAB1) lines (see Supplemental Figure 5 online). Consistent with the results shown in Figure 7A, the number of actin cables labeled by GFP-SCAB1 in the OE-GFP-SCAB1 transgenic plants was decreased in hypocotyl epidermal cells (Figure 9A), leaf cells (Figure 9B), and guard cells (Figure 9C) compared with that in the plants expressing ProSCAB1:GFP-SCAB1 (Figure 2), and only thick actin bundles were observed (Figures 9A to 9C). To confirm the bundling activity of SCAB1, we generated OE-GFP-fABD2-GFP and OE-mCherry-SCAB1 double-labeled plants. Compared with the actin filaments in wild type (see Supplemental Figure 8A online), the GFP-fABD2-GFP-labeled MFs in the mCherry-SCAB1 transgenic plants were more frequently observed as bundled forms (see Supplemental Figures 8B to 8D online). These results were further supported by actin staining assays using rhodamine-phalloidin (see Supplemental Figures 8E to 8H online). However, SCAB1 did not alter the MT array in hypocotyl epidermal cells (Figures 2G and 2H).
Figure 9.
GFP-SCAB1 Overexpression Impairs Stomatal Closure.
(A) to (C) Confocal images of epidermal cells taken from the hypocotyls (A), leaves (B), and guard cells (C) of Pro35S:GFP-SCAB1 transgenic plants. Scale bars, 10 μm.
(D) Stomatal closure in the OE-GFP-SCAB1 lines showed reduced sensitivity to abscisic acid. The data shown represent the stomatal apertures in response to 30 min of treatment with 20 μM abscisic acid. The data represent the mean ± sd of three independent experiments; 50 stomata were analyzed per line. Asterisks represent significant differences between the wild-type (WT) and mutant or overexpression lines (*P < 0.05, **P < 0.01, one-way analysis of variance; post hoc: Tukey’s honestly significant difference).
(E) Stomatal closure in the OE-GFP-mTalin line showed reduced sensitivity to abscisic acid. The data shown represent the stomatal apertures in response to 30 min of treatment with 20 μM abscisic acid. The data given are the means ± sd (n = 3); 50 stomata were analyzed per line. Asterisks represent significant differences between the wild-type and transgenic lines (*P < 0.05, **P < 0.01, one-way analysis of variance; post hoc: Tukey’s honestly significant difference).
[See online article for color version of this figure.]
To determine whether the thick actin bundles in the OE-GFP-SCAB1 transgenic lines altered stomatal closure, stomata of wild-type, scab1-1, and OE-GFP-SCAB1 plants were opened in 50 mM KCl and then treated with 20 μM abscisic acid for 30 min. Stomatal closure in the two transgenic lines (2.1 ± 0.5 μm in #3 and 2.3 ± 0.5 μm in #11) was less sensitive to abscisic acid treatment compared with that in wild type (0.8 ± 0.3 μm) and was similar to that in scab1-1 (2.5 ± 0.4 μm) (Figure 9D). Previous studies showed that the overexpression of GFP-mTalin (for expression of the actin binding domain of mouse talin fused to GFP) (Kost et al., 1998) induced MF bundling and suppressed the diurnal patterns of stomatal opening (Ketelaar et al., 2004a; Sheahan et al., 2004; Higaki et al., 2010). When we tested stomatal closure in OE-GFP-mTalin transgenic plants in response to abscisic acid, closure happened more slowly than in wild-type plants (Figure 9E), indicating that the retardation of stomatal closure in the SCAB1 overexpression lines was likely caused by excessive actin bundling.
To investigate whether the overexpression of SCAB1 alters stomatal closure-associated actin reorganization, we monitored the array changes of GFP-SCAB1-labeled F-actin in guard cells. Consistent with the pattern of actin reorganization during stomatal closure (Figures 8A to 8C), the pattern of GFP-SCAB1 also changed from a “radial array” (see Supplemental Figures 9A and 9D online) to a type 2 “random meshwork” (see Supplemental Figures 9B and 9E online), and finally to a type 3 “longitudinal array” (see Supplemental Figures 9C and 9F online). To investigate the effect of SCAB1 overexpression on MF reorganization in guard cells, we treated transgenic seedlings harboring np-GFP-SCAB1 and OE-GFP-SCAB1 with abscisic acid. As shown in Supplemental Figure 9G online, the switch from type 1 to type 3 took more time in the OE-GFP-SCAB1 plants than in np-GFP-SCAB1 plants. After 30 min of 20 μM abscisic acid treatment, 64% of the np-GFP-SCAB1 guard cells showed a change in actin organization to type 3 (see Supplemental Figure 9G online), which is similar to that of wild-type (Figure 8D). By contrast, less than 30% of the guard cells exhibited a type 3 actin organization in OE-GFP-SCAB1 guard cells (see Supplemental Figure 9G online), which is similar to that of scab1-1 (Figure 8D). These results demonstrate that overexpression of SCAB1 retards MF rearrangement during stomatal movement.
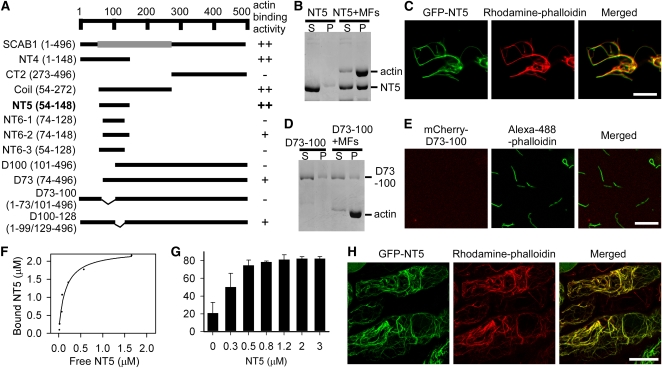
Identification of the Actin Binding Domain of SCAB1
The secondary structure of SCAB1 was predicted using the web-based program PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) (Jones, 1999). The predicted structure indicated the existence of three distinct regions in SCAB1: an N-terminal region (residues 1–53), a central α-helical region (residues 54–271), and a C-terminal region consisting of β-strands and α-helices (residues 272–496). SCAB1 is a new type of plant-specific actin binding protein and does not show significant sequence similarity to other known actin binding proteins. To identify the actin binding domain in SCAB1, various SCAB1 fragments were amplified by PCR based on the three predicted domains (Figure 10A). The resulting truncated proteins were purified and subjected to high-speed cosedimentation and fluorescence microscopic assays (Figures 10B to 10E; see Supplemental Figure 10 online). We found that fragment NT5 (residues 54–148) corresponding to the central α-helical region could bind F-actin (Figures 10B and 10C; see Supplemental Figure 10N online). Subsequent experiments revealed that NT6-2 (residues 74–148) possessed a functional actin binding domain with less activity compared with full-length SCAB1 (see Supplemental Figures 10D and 10O online). Deletion analyses revealed that residues 73 through 100 comprise the critical region for SCAB1-actin binding (Figures 10A to 10E; see Supplemental Figure 10 online). The deletion of residues 73 through 100 abolished SCAB1-actin binding activity (Figures 10D and 10E).
Figure 10.
Identification of the Actin Binding Domain of SCAB1.
(A) Diagram of the SCAB1-truncated proteins and their actin binding activity. The α-helical regions were predicted using PSIPRED and are represented by gray rectangles (54–272). Numbers indicate the positions of the first and last amino acids in each SCAB1 fragment. Actin binding activity was evaluated by a high-speed cosedimentation assay and fluorescence microscopy. ++, strong binding activity; +, weak binding activity; −, no binding activity.
(B) High-speed cosedimentation assay of NT5 in the presence or absence of MFs. After centrifugation at 150,000g, the proteins in the supernatant (S) and pellet (P) were resolved by SDS-PAGE.
(C) Rhodamine-phalloidin–stained actin filaments (middle) were incubated with GFP-tagged NT5 (left). Right panel: merged images.
(D) High-speed cosedimentation assay of SCAB1 with a deletion between 73 and 100 (D73-100) in the presence or absence of MFs.
(E) Alexa-488-phalloidin–stained actin filaments (middle) were incubated with mCherry-tagged D73-100 (left). Right panel: merged images.
(F) Increasing concentrations of NT5 (0.3–4 μM) were cosedimented with 2 μM F-actin. After gel quantification, the concentration of bound NT5 was plotted against the concentration of free NT5 and fitted with a hyperbolic function. In this representative experiment, the Kd value was 0.34 μM with a stoichiometry at saturation of 1.10 molecules of NT5 bound per actin subunit.
(G) A low-speed cosedimentation assay was used to assess the bundling activity of NT5. Increasing concentrations of NT5 were incubated with 2 μM F-actin and the reactions were centrifuged at 13,000g. The proteins in the supernatant and pellet were resolved by SDS-PAGE and examined by densitometry. The data given are the means ± sd (n = 3).
(H) GFP-NT5 (left) and rhodamine-phalloidin–stained actin filaments (middle) in hypocotyl epidermal cells. A merged image is shown on the right. Scale bars, 10 μm in (C) and (E), and 20 μm in (H).
To determine the binding affinity of NT5 for F-actin, increasing concentrations of NT5 were cosedimented with preformed F-actin. The concentration of F-actin–bound NT5 was plotted against the concentration of free NT5 and fitted to a hyperbolic function (Figure 10F). In this representative experiment, the calculated Kd was 0.34 μM with a stoichiometry at saturation of 1.17 mole of NT5 bound per mole of actin. From three independent experiments, a mean Kd value (±sd) of 0.35 ± 0.01 μM (n = 3) for NT5 binding to F-actin was calculated. At saturation, the stoichiometry of the interaction was 1.12 ± 0.06 mole of NT5 bound per mole of actin. As shown in Figure 10C, rhodamine-phalloidin–labeled F-actin formed bundles in the presence of GFP-NT5, indicating that NT5 is also capable of MF bundling. To confirm this, we performed a low-speed cosedimentation assay (Figure 10G). The amount of F-actin in the pellet fraction increased in proportion to the NT5 concentration, demonstrating that NT5 can bundle actin filaments in vitro. To determine whether NT5 binds F-actin in vivo, the plasmid 35S:GFP-NT5 was transformed into wild-type plants. GFP-NT5 colocalized with rhodamine-phalloidin–stained MFs in hypocotyl epidermal cells (Figure 10H). Moreover, this actin binding domain is conserved in SCAB1 homologs (see Supplemental Figure 11 online), suggesting that SCAB1 and its homologs comprise a group of plant-specific actin binding proteins.
DISCUSSION
SCAB1 Is a Previously Undiscovered Actin Binding Protein
Many F-actin binding proteins share conserved actin binding domains from other organisms. Plant villins contain a C-terminal villin headpiece domain that binds MFs (Vidali et al., 1999; Klahre et al., 2000; Yokota et al., 2003; Huang et al., 2005; Khurana et al., 2010; Zhang et al., 2010). Plant fimbrins contain two conserved actin binding domains (ABD1 and ABD2), each of which contains tandemly repeated calponin-homology domains (Kovar et al., 2000). Plant formins contain formin homology domain 1, which binds the barbed end or side of MFs (Cheung and Wu, 2004; Ingouff et al., 2005; Michelot et al., 2005, 2006; Li et al., 2010). Plant LIM domain–containing proteins contain two LIM domain–containing protein domains (Eliasson et al., 2000; Thomas et al., 2006; Thomas et al., 2007; Wang et al., 2008a; Papuga et al., 2010). SCAB1 shows sequence similarity only to plant proteins and contains a previously undiscovered actin binding domain; thus, SCAB1 and its homologs represent a new type of F-actin binding protein.
SCAB1-like proteins contain a conserved actin binding domain (see Supplemental Figure 11 online). Plants lacking or overexpressing SCAB1 displayed an obvious defect in stomatal closure but no other significant difference in plant growth or development, most likely because of functional redundancy with its homologs. Indeed, in Arabidopsis, there are two proteins with strong similarity to SCAB1 throughout their sequences, and the expression of these three genes overlaps in roots, stems, leaves, flowers, and siliques (see Supplemental Figure 12 online). Our results suggest that these three genes may have similar functions in bundling and stabilizing MFs during many biological processes, and that SCAB1 functions predominantly in the regulation of stomatal closure.
SCAB1 Regulates Stomatal Closure in Response to Drought Stress
Plant actin filaments play key roles in local expansion-based plant cell morphogenesis, including in pollen tubes, root hairs, trichomes, and guard cells (Mathur, 2006; Yang, 2008). The ability of vascular plants to close their stomata in aerial tissues is an important aspect of controlling water loss. Recent studies have shown that MF configurations undergo rapid dynamic changes during stomatal movement; specifically, MFs display a radial orientation in open stomata and a longitudinal orientation in closed stomata (Higaki et al., 2010).
How these actin dynamics trigger stomatal movement is not well understood. Under normal conditions, long actin filaments are organized at the cortex, radiating out from the stomatal pore. Upon abscisic acid treatment or drought stress, these filaments are rearranged to a longitudinal bundled orientation, which is associated with stomatal closure (Hwang and Lee, 2001; Higaki et al., 2010). The rearrangement of MFs must be temporally and spatially regulated so that the opening and closure of stomata can be accurately coordinated in response to environmental changes.
In this study, we identified a previously unknown actin binding protein, SCAB1, which binds, bundles, and stabilizes actin filaments in vitro and in vivo. In contrast to other actin binding proteins, SCAB1 exists only in the plant kingdom and has strong affinity for F-actin. The lack of SCAB1 resulted in reduced MF stability, delayed MF reorganization, and reduced sensitivity to abscisic acid in guard cells. Overexpression of SCAB1 resulted in formation of MF bundles, enhanced MF stability, retarded MF reorganization, and impaired stomatal closure. When we overexpressed mTalin in wild-type plants, a similar phenotype was obtained in which stomatal closure in the transgenic plants was less sensitive to abscisic acid. These results suggest that excessive bundling of MFs is negatively correlated with stomatal closure.
Although the MFs disassembled more quickly in the scab1 knockout mutants and more slowly in the overexpression lines, we found that MF orientation was not altered in these mutant plants compared with wild type when we examined the change in MF configuration during stomatal closure. However, the time required for the transition in MF configuration was prolonged in the mutant plants, suggesting that SCAB1-regulated actin reorganization is important for the change in actin configuration. This result provides a potential explanation for the scab1 drought-sensitive phenotype. The role of MF reorganization in guard cells during stomatal closure is unclear. MFs play important roles in the localization, movement, and morphology of many organelles, including endosomes and vacuoles (Grebe et al., 2003; Higaki et al., 2006). Rapid changes in vacuole volume determine the turgor of guard cells, and are therefore a major factor in stomatal movement. When stomata are opened wide, the MFs in the cells radiate outward from the stomatal pore, the vacuolar membrane is fully spread, and the maximum volume of the vacuole emerges (Gao et al., 2005; Tanaka et al., 2007). When stomata are closed, the MFs are rearranged and bundled preferentially as long cables in the longitudinal direction and the vacuolar membrane folds, forming a luminal structure or small vacuole (Gao et al., 2005; Tanaka et al., 2007) with a minimum volume. Vacuolar structure is reportedly maintained by MFs in tobacco bright yellow-2 cells and Arabidopsis protoplasts (Higaki et al., 2006; Sheahan et al., 2007). SCAB1 participates in actin bundle formation in guard cells during stomatal closure, which might further regulate vacuolar structure and volume.
Additionally, homologs of SCAB1 have been identified in many plant species, including bryophytes and spermatophytes, suggesting that SCAB1-like proteins perform a general plant-specific function. However, except for the reported defects in stomatal movement, the scab1 mutants and overexpression lines showed no other significant defects in plant growth or development, suggesting that SCAB1 functions redundantly with its homologs in other biological processes.
METHODS
Plant Materials
Arabidopsis thaliana Col-0 was used as the wild type. Drought-sensitive scab1-1 mutant was isolated from a T-DNA insertion pool (pSKI015) in Col-0 background. The T-DNA insertion in scab1-1 was identified in exon seven of At2g26770 by plasmid rescue with PstI digestion and was confirmed by gene-specific primers scab1-1 F and scab1-1 R, and the T-DNA right border-specific primer pSKI015 RB (see Supplemental Table 2 online).
The scab1-2 mutant (SAIL_193_E09) was obtained from ABRC harboring a T-DNA insertion in the first intron of At2g26770. The insertion was confirmed by the gene-specific primers scab1-2 F and scab1-2 R, and the T-DNA (pCSA110) left border-specific primers pCSA110 LB1 and pCSA110 LB3 (see Supplemental Table 2 online).
Stomatal Aperture Measurement and Water Loss Analysis
Assays for stomatal responses to abscisic acid, H2O2, and CaCl2 were performed as described (Hugouvieux et al., 2001). Plants were grown in a growth chamber at 23°C under an 8-h light/16-h dark photoperiod and 70% relative humidity for 5 weeks. To study the promotion of stomatal closure by abscisic acid, H2O2, and CaCl2, rosette leaves were incubated in stomatal opening solution (10 mM MES, pH 6.15, 50 mM KCl, and 10 μM CaCl2) for 2 h in a growth chamber at 23°C under continuous white light. After incubation for 0.5 h in 20 μM abscisic acid, 2 h in 0.5 mM H2O2, or 2 h in 2 mM CaCl2, respectively, epidermal strips were peeled from the pretreated rosette leaves, and stomatal apertures were measured under the microscope (magnification ×20).
For water loss measurement, three to five rosette leaves at a similar developmental stage were detached from 5-week-old plants grown in the same conditions and weighed at the indicated times.
Expression Analyses
Total RNA was extracted by RNAVzol (Vigorous) from 10-d-old seedlings grown on Murashige and Skoog medium and treated with RNase-free DNase I (TAKARA). Reverse transcription was performed by M-MLV reverse transcriptase (Promega) using 3 μg of total RNA. The cDNAs were amplified with the following primers: SCAB1 F, SCAB1 R; and EF1α F, EF1α R (see Supplemental Table 2 online). EF1α (At5g60390) was used as an internal control.
Subcellular Localization of SCAB1 and Colocalization with MT and MF
The SCAB1 cDNA coding region was amplified by RT-PCR using the gene-specific primers SCAB1 CDSF and SCAB1 CDSR (see Supplemental Table 2 online), cloned into the BamHI and EcoRI sites of the binary vector pCAMBIA1205 (Zhao et al., 2007), and confirmed by sequencing. The resultant plasmid was transformed into scab1-1 mutant mediated by Agrobacterium tumefaciens GV3101.
To generate ProSCAB1:SCAB1 construct, the 1.09-kb SCAB1 promoter fragment amplified from BAC plasmid F18A8 (ABRC) with the primers SCAB1Pro PstIF and SCAB1Pro BamHIR (see Supplemental Table 2 online) was cloned into the PstI and BamHI sites of the pCAMBIA1390 vector, and then the coding region of SCAB1 from pCAMBIA1205-SCAB1 was subcloned into the BamHI and EcoRI sites downstream of the SCAB1 promoter.
To generate ProSCAB1:GFP-SCAB1 construct, the GFP fragment amplified with the primers GFP BamHIF and GFP BamHIR (see Supplemental Table 2 online) was cloned into the BamHI site of pCAMBIA1390-ProSCAB1:SCAB1 between the SCAB1 promoter and SCAB1 coding sequence. The resultant plasmid was transformed into scab1-1 mutant. The T2 transgenic plants were used for the observation of subcellular localization of SCAB1.
To generate the ProSCAB1:mCherry-SCAB1 construct, the mCherry fragment amplified with the primers GFP BamHIF and GFP BamHIR (see Supplemental Table 2 online) was cloned into the BamHI site of pCAMBIA1390-ProSCAB1:SCAB1 between the SCAB1 promoter and SCAB1 coding sequence. The resultant plasmid was transformed into transgenic plants expressing GFP-fABD2-GFP (Wang et al., 2008b).
Pro35S:mCherry-SCAB1 was constructed by subcloning the coding region of SCAB1 from pCAMBIA1205-SCAB1 into the pCAMBIA1205-mCherry vector. The construct was transformed into transgenic plants expressing GFP-fABD2-GFP and GFP-tubulin (Ueda et al., 1999), respectively.
Pro35S:GFP-SCAB1 was constructed by subcloning the coding region of SCAB1 from pCAMBIA1205-SCAB1 into the pCAMBIA1205-GFP vector. The construct was transformed into Col-0 wild-type plants.
The images were taken from 6-d-old T2 transgenic seedlings with a Zeiss LSM510 Meta confocal microscope using a Plan-Apochromat 63×/1.4 oil immersion differential interference contrast lens in multitrack mode. GFP and mCherry were excited at 488 and 543 nm, respectively.
F-Actin Staining in Arabidopsis
F-actin staining was performed as described with slightly modification (Traas et al., 1987). In brief, suspension cells or 3-d-old seedlings were incubated in actin stabilizing buffer (100 mM PIPES, 10 mM EGTA, 5 mM MgSO4, 5% DMSO, and 0.05% Nonidet P-40, pH 6.8) with 0.18 μM rhodamine-phalloidin for 15 min or 3 h at room temperature, respectively.
Protein Expression and Purification
The SCAB1 cDNA coding region from pCAMBIA1205-SCAB1 was subcloned into the pGEX-6P-1 vector between BamHI and EcoRI sites. Escherichia coli strain BL21 (DE3.0) was used to express the glutathione S-transferase (GST)-SCAB1 recombinant protein. Protein expression was induced by 0.6 mM isopropyl β-d-thiogalactoside for 20 h at 16°C. The GST-SCAB1 was purified from the soluble fraction using Glutathione Sepharose 4B (Amersham) under native conditions according to the manufacturer’s instructions. The GST tag was removed by PreScission Protease at 4°C overnight in PBS (pH 7.3) with 1 mM DTT.
For His-tag fusion, SCAB1 from pCAMBIA1205-SCAB1 was subcloned into the pET28A vector between BamHI and EcoRI sites, and GFP-SCAB1 from pCAMBIA1205-GFP-SCAB1 was subcloned into the pET28A vector between SacI and HindIII sites. The plasmid was transformed into BL21 (DE3.0) cells. The His-SCAB1 and His-GFP-SCAB1 were purified from the soluble fraction using Ni-NTA agarose (QIAGEN) under native conditions according to the manufacturer’s protocol.
GST-Fim1 was purified according to the method for GST-SCAB1, and the GST tag was removed by PreScission Protease at 4°C for 3 h prior to use. Os-FH5 FH2 was purified according to the method of Yang et al. (2011).
To identify the actin binding domain in SCAB1, SCAB1 fragments were amplified by PCR. Fragments of Coil, NT4, NT5, NT6-1, NT6-2, NT6-3, CT1, and CT2, were cloned into the BamHI and EcoRI sites of pCAMBIA1205-GFP binary vector downstream of the GFP coding region. Fragments of D73, D100, D73-100, and D100-128 were cloned into the BamHI and EcoRI sites of pCAMBIA1205-mCherry binary vector downstream of the mCherry coding region. All primers and plasmid constructs used in these studies were listed in Supplemental Table 3 online. All fragments fused to GFP or mCherry at N termini were subcloned into the pET28A vector between SacI and HindIII sites. Fragments of NT5 and D73-100 were subcloned into the pGEX-6P-1 vector between BamHI and EcoRI sites. GST- or His-tagged proteins were purified according to the method for GST- or His-SCAB1.
Antibody Preparation and Immunoblotting
The recombinant SCAB1 was used as an antigen to raise polyclonal SCAB1 antibodies in mouse, which was digested from GST-SCAB1 recombinant protein with PreScission Protease.
Total proteins were extracted from 1-d-old Arabidopsis seedlings in extraction buffer containing 10 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 0.5% (v/v) Nonidet P-40, and 2× protease inhibitor (Roche). Extracts were centrifuged at 16,200g for 15 min at 4°C and the resulting supernatants were subjected to immunoblot analyses. The blots were probed with primary mouse anti-SCAB1 (diluted 1:250), rabbit anti-MPK3 (diluted 1:1000; Sigma-Aldrich), or rabbit anti-actin (diluted 1:1000; Cytoskeleton) polyclonal antibodies. The chemiluminescence signals were detected by film with ECL Plus Western Blotting Detection reagents (GE Healthcare).
F-Actin Cosedimentation Assay
SCAB1 was dialyzed for 1 h against 1× KMEI buffer (10 mM imidazole, 100 mM KCl, 1 mM MgCl2, and 1 mM EGTA, pH 7.0). Protein concentration was determined using the BIO-RAD protein assay kit, with BSA as a standard.
Actin was purified from rabbit skeletal muscle acetone powder as described in Pardee and Spudich (1982) in G buffer (5 mM Tris–HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, and 0.01% NaN3).
Proteins were centrifuged at 400,000g for 1 h at 4°C prior to use, mixed with 2 μM preformed F-actin, and incubated in 50 μL volume of 1× KMEI buffer for 20 min at 23°C. The samples were centrifuged at 150,000g for 30 min at 4°C. Fim1 is a known F-actin binding protein (Kovar et al., 2000) and used as a positive control. BSA was used as a negative control. Proteins in supernatants and pellets were analyzed by SDS-PAGE, respectively.
For low-speed cosedimentation assays, increasing amounts (0, 0.3, 0.5, 0.8, 1.2, 2 and 3 μM) of SCAB1 were incubated with 2.0 μM preassembled F-actin for 60 min at 23°C. After centrifugation at 13,000g for 30 min at 4°C, the supernatants and pellets were separated and subjected to SDS-PAGE and visualized by Coomassie Blue staining. The amounts of actin in the pellets were quantified using ImageJ 1.38x (Wayne Rasband).
High- and low-speed cosedimentation assays were also performed in 1× KMEI with various concentrations of free Ca2+ (0.01, 0.1, 1, 10, 100, and 1000 μM) as described in Khurana et al. (2010). Free Ca2+ concentration was calculated with EGTA software by Petesmif (http://pcwww.liv.ac.uk/∼petesmif/petesmif/software/_webware06/EGTA/EGTA.htm). Experiments were performed with 0.5 μM SCAB1 and 2 μM F-actin.
Cosedimentation assays were also tested in 1× KME (100 mM KCl, 1 mM MgCl2, and 1 mM EGTA) buffered with 10 mM MES (pH 6.2), 10 mM PIPES (pH 6.8), 10 mM Tris (pH 7.4), and 10 mM Tris (pH 8.0) as described in Papuga et al. (2010). Experiments were performed with 0.5 μM SCAB1 and 2 μM F-actin.
To determine a Kd, increasing amounts (0.3, 0.5, 0.8, 1.2, 1.6, 2, 2.8, and 4 μM) of SCAB1 were incubated with 2.0 μM preassembled F-actin for 60 min at 23°C. After centrifugation at 150,000g for 40 min at 4°C, the supernatants and pellets were separated and subjected to SDS-PAGE. The amounts of SCAB1 in the pellets and supernatant were quantified using ImageJ 1.38x (Wayne Rasband). A Kd value for SCAB1 bound to actin was calculated by fitting the data of protein bound versus protein free to a hyperbolic function with SigmaPlot 2000 software (Systat Software).
F-Actin Bundling Assays
The actin filaments were visualized by rhodamine-phalloidin staining (Sigma). Preformed F-actin at 4 μM was mixed with or without 1 μM His-GFP-SCAB1, incubated for 30 min at room temperature, and then labeled with equimolar rhodamine-phalloidin for 15 min at 4°C. Actin filaments were observed by epifluorescence illumination with an IX71 microscope (Olympus) equipped with a 60×1.42–numerical aperture oil objective, and digital images were collected with a Retiga EXi Fast 1394 charge-coupled device camera (QImaging) using Image-Pro Express 6.3 software.
Skewness Analysis
Skewness analysis were performed as described in Higaki et al. (2010) and Khurana et al. (2010). Before experiments, proteins were centrifuged at 400,000g for 60 min at 4°C. Increasing amounts (0, 0.3, 0.5, 0.8, 1.2, and 2 μM) of SCAB1 were incubated with 2.0 μM preassembled F-actin for 60 min at 23°C. The actin filaments were labeled with 1.0 μM rhodamine-phalloidin. All reactions were diluted at a ratio of 3 μL in 100 μL of buffer and visualized as described previously. Micrographs were analyzed in ImageJ with plugins of Hig Skewness and KbiPlugins available at http://hasezawa.ib.k.u-tokyo.ac.jp/zp/Kbi/HigStomata.
F-Actin Depolymerization Assay
In vitro F-actin depolymerization was performed as described in Thomas et al. (2006). Various amounts of His-SCAB1 proteins (0, 0.5, 2, 4, 8, and 12 μM) were incubated with 7 μM preassembled F-actin for 40 min at 22°C. Then, 24 μM Lat A was added and incubated for 8 h. The F-actin alone, which was used to estimate the total amount of actin, was incubated with the same volume of DMSO as control. After centrifugation at 150,000g for 40 min at 4°C, the supernatants and pellets were separated and subjected to SDS-PAGE and visualized by Coomassie Blue staining. Gel scanning was performed on an AlphaImager 2200 (Alpha Innotech).
Dilution-mediated actin depolymerization was performed as previously described in Huang et al. (2003). F-actin at 5 μM (50% pyrene labeled) was mixed with varying concentrations of His-SCAB1, incubated at room temperature for 5 min, and diluted 25-fold into Buffer G (2 mM Tris-HCl, pH 8.0, 0.01% NaN3, 0.2 mM CaCl2, 0.2 mM ATP, and 0.2 mM DTT) at room temperature. The decrease in pyrene fluorescence accompanying actin depolymerization was monitored for 1200 s after dilution.
Quantification of Filamentous Actin
Measurement of actin filament levels in Arabidopsis suspension cells was according to the method of Huang et al. (2006). Arabidopsis suspension cells of wild type, SCAB1 overexpression line, and scab1-1 mutant were incubated with 100 nM Lat A for various time period, and cells were then fixed by the addition of 300 μM maleimidobenzoyl-N-hydroxysuccinimide ester (Sigma) and 0.05% (v/v) NP-40. The cells were subsequently washed with 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.05% NP-40 three times and neutralized in this same solution by the addition of 1 mM DTT. The suspension cells were then incubated with 2 μM Alexa-488-phalloidin and 5 μM ethidium bromide (EB) overnight. After extensive washing, the level of actin filaments was determined by eluting bound phalloidin from cells with methanol, and the eluted solution was analyzed by a fluorimeter with excitation at 492 nm and emission at 514 nm. The fluorescence of eluted EB was determined with excitation at 513 nm and emission at 615 nm to estimate the cell number. The relative level of actin filament was defined by phalloidin fluorescence divided by EB fluorescence.
F-Actin Depolymerization Assay in Hypocotyl Epidermal Cells
F-actin depolymerization assay in hypocotyl epidermal cells were performed on hypocotyls of 6-d-old transgenic seedlings harboring Pro35S:fABD2-GFP. The seedlings were treated with 200 nM Lat A for 30 or 60 min. The status of actin filaments were observed on a LSM510 Meta confocal microscope (Zeiss) using a Plan-Apochromat 63×/1.4 oil objective. The cells with a random localized dot-like GFP fluorescence were considered as F-actin depolymerized. More than 100 cells were analyzed at each time point in each background.
Visualization of Actin in Guard Cells
Transgenic plants harboring Pro35S:GFP-fABD2-GFP (Wang et al., 2008b) in wild-type and scab1-1 background were used for F-actin visualization in guard cells of mature leaves. Plants with strong GFP fluorescence were grown in a greenhouse at 21 to 23°C under an 8-h light/16-h dark photoperiod and 70% relative humidity for 3 to 4 weeks. Rosette leaves were incubated in stomatal opening solution (10 mM MES, pH 6.15, 50 mM KCl, and 10 μM CaCl2) for 1 to 2 h under continuous white light. Fragments of leaves were cut off from the pretreated leaves with minimum damage because peeling off epidermal strips or other mechanical damage would induce the F-actin rapidly depolymerization. Actin organization was observed and captured on a LSM510 Meta confocal microscope (Zeiss) using a Plan-Apochromat 63×/1.4 oil objective. Stomatal aperture was measured with the LSM Image Browser software. To induce the stomatal closure, 20 μM abscisic acid was added and the images were randomly captured after abscisic acid treatment.
Promoter GUS Analysis
The SCAB1 promoter fragment containing 1090 base pairs upstream of the translation start site was digested from pCAMBIA1390-ProSCAB1:SCAB1 and was subcloned into the PstI and BamHI sites of the pCAMBIA1391 vector. The construct was introduced into A. tumefaciens GV3101 and was transformed into Arabidopsis. GUS staining of the T2 transgenic lines was performed as described in Zhao et al. (2007). Samples were incubated in reaction buffer containing 100 mM sodium phosphate, pH 7.0, 0.1% Triton X-100, 3 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 8 mM β-mercaptoethanol in the dark for 3 h at 37°C, and were then immersed in 75% ethanol at 37°C to extract chlorophyll.
Sequence Comparison
Protein sequences with similarity to At-SCAB1 (E value < 1e-90) were obtained using BLAST tools from Swiss-Prot databases (http://expasy.org/tools/blast/), JGI databases (http://www.jgi.doe.gov/), and ABRC (http://www.Arabidopsis.org/wublast/index2.jsp). To analyze the evolutionary relationships of SCAB1 and its homologs, protein sequences were aligned using ClustalX 2.0.5 (Larkin et al., 2007) with the default settings (see Supplemental Data Set 1 online), and a rooted phylogenetic tree was generated using MEGA 4.0 (Tamura et al., 2007) by the neighbor-joining method. The phylogeny test was processed with 1000 replicates of bootstrap analysis.
Accession Numbers
Sequence data from this article can be found in Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession number: SCAB1 (At2g26770, NM_128234). The accession number of the T-DNA insertion mutant is SAIL_193_E09 (scab1-2). Accession numbers of the proteins used in this study are shown in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of SCAB1 and Its Homologs in Arabidopsis.
Supplemental Figure 2. Analyses of Native SCAB1 Concentration.
Supplemental Figure 3. SCAB1 Lacks Nucleation or Capping Activity.
Supplemental Figure 4. SCAB1 Delayed the Lat A-Induced Depolymerization of Actin Filaments in Guard Cells.
Supplemental Figure 5. Level of SCAB1 Protein in SCAB1-Overexpressing Plants.
Supplemental Figure 6. Histograms Showing the Stomatal Apertures with Actin Organization in Guard Cells from Wild-Type and scab1-1 Plants.
Supplemental Figure 7. SCAB1 Does Not Alter Skewness in Guard Cells.
Supplemental Figure 8. SCAB1 Bundles MFs in Vivo.
Supplemental Figure 9. Overexpression of SCAB1 Delays GFP-SCAB1-Labeled Actin Reorganization during Stomatal Closure.
Supplemental Figure 10. Identification of the Actin Binding Domain of SCAB1.
Supplemental Figure 11. Alignment of the Actin Binding Domains from SCAB1 Homologs.
Supplemental Figure 12. Expression of SCAB1 and Its Homologous Genes in Arabidopsis.
Supplemental Table 1. Accession Number of Proteins with Sequence Similarity to SCAB1 (E value < 1e-90).
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Primers Used for Plasmid Constructions.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 4.
Supplementary Material
Acknowledgments
We thank Xiaodong Wang, Xing Wang Deng, Jianmin Zhou, and Jianguo Chen for critical reading of the manuscript and stimulating discussions, Lei Wang and Jun Zhang for excellent technical assistance, and the ABRC for the T-DNA insertion lines. This work was supported by China National Funds for Distinguished Young Scientists (Grant 31025003 to Y.G.), by the 111 Project (Grant B06003 to M.Y.), and by the National Natural Science Foundation of China (Grants 30830058 and 30721062 to M.Y.). S.H. was supported by Chinese Academy of Sciences through its Hundred Talent program.
AUTHOR CONTRIBUTIONS
Y.G. and Y.Z. conceived the research and Y.G., Y.Z., M.Y., S.H., T.M., K.Y., and G.Z. designed the research. Y.Z., S.Z., T.M., X.Q., W.C., L.Z., W.Z., L.H., S.L., S.R., J.Z., and S.H. performed the experiments. Y.Z., S.Z., T.M., W.C., S.H., and Y.G. analyzed the data. Y.G. and Y.Z. wrote the article with input from all of the authors.
References
- Barrero R.A., Umeda M., Yamamura S., Uchimiya H. (2002). Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division. Plant Cell 14: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Lee Y., Jeon B.W., Staiger C.J., Lee Y. (2008). Phosphatidylinositol 3- and 4-phosphate modulate actin filament reorganization in guard cells of day flower. Plant Cell Environ. 31: 366–377 [DOI] [PubMed] [Google Scholar]
- Desikan R., Cheung M.K., Bright J., Henson D., Hancock J.T., Neill S.J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55: 205–212 [DOI] [PubMed] [Google Scholar]
- Djakovic S., Dyachok J., Burke M., Frank M.J., Smith L.G. (2006). BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133: 1091–1100 [DOI] [PubMed] [Google Scholar]
- Dong C.H., Xia G.X., Hong Y., Ramachandran S., Kost B., Chua N.H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson A., Gass N., Mundel C., Baltz R., Kräuter R., Evrard J.L., Steinmetz A. (2000). Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol. Gen. Genet. 264: 257–267 [DOI] [PubMed] [Google Scholar]
- Eun S.O., Lee Y. (1997). Actin filaments of guard cells are reorganized in response to light and abscisic acid. Plant Physiol. 115: 1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.Q., Chen J., Wei P.C., Ren F., Chen J., Wang X.C. (2008). Array and distribution of actin filaments in guard cells contribute to the determination of stomatal aperture. Plant Cell Rep. 27: 1655–1665 [DOI] [PubMed] [Google Scholar]
- Gao X.Q., Li C.G., Wei P.C., Zhang X.Y., Chen J., Wang X.C. (2005). The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol. 139: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Okubo E., Sano T., Hasezawa S. (2006). Actin microfilaments regulate vacuolar structures and dynamics: Dual observation of actin microfilaments and vacuolar membrane in living tobacco BY-2 Cells. Plant Cell Physiol. 47: 839–852 [DOI] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2010). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: Role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2007). Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Holweg C., Nick P. (2008). Retraction for Holweg and Nick: Arabidopsis myosin XI mutant is defective in organelle movement and polar auxin transport. Proc. Natl. Acad. Sci. USA 105: 3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E., et al. (2003). The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. USA 100: 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., Gao L., Blanchoin L., Staiger C.J. (2006). Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J.M., Schroeder J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Hwang J.U., Lee Y. (2001). Abscisic acid-induced actin reorganization in guard cells of dayflower is mediated by cytosolic calcium levels and by protein kinase and protein phosphatase activities. Plant Physiol. 125: 2120–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Suh S., Yi H., Kim J., Lee Y. (1997). Actin filaments modulate both stomatal opening and inward K+-channel activities in guard cells of Vicia faba L. Plant Physiol. 115: 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J.N., Guérin C., Robert H., Sørensen M.B., Van Damme D., Geelen D., Blanchoin L., Berger F. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7: 374–380 [DOI] [PubMed] [Google Scholar]
- Jedd G., Chua N.H. (2002). Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 43: 384–392 [DOI] [PubMed] [Google Scholar]
- Jones D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Karpova T.S., Tatchell K., Cooper J.A. (1995). Actin filaments in yeast are unstable in the absence of capping protein or fimbrin. J. Cell Biol. 131: 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004b). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Anthony R.G., Hussey P.J. (2004a). Green fluorescent protein-mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor’s actin depolymerizing activity in vitro. Plant Physiol. 136: 3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Hepler P.K., Eun S.O., Ha K.S., Lee Y. (1995). Actin filaments in mature guard cells are radially distributed and involved in stomatal movement. Plant Physiol. 109: 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Friederich E., Kost B., Louvard D., Chua N.H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 122: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Staiger C.J., Weaver E.A., McCurdy D.W. (2000). AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lemichez E., Wu Y., Sanchez J.P., Mettouchi A., Mathur J., Chua N.H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15: 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shen Y., Cai C., Zhong C., Zhu L., Yuan M., Ren H. (2010). The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22: 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Luan S. (1998). Voltage-dependent K+ channels as targets of osmosensing in guard cells. Plant Cell 10: 1957–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie E.A., Kurup S. (2007). Signalling mechanisms in the regulation of vacuolar ion release in guard cells. New Phytol. 175: 630–640 [DOI] [PubMed] [Google Scholar]
- Mathur J. (2006). Local interactions shape plant cells. Curr. Opin. Cell Biol. 18: 40–46 [DOI] [PubMed] [Google Scholar]
- Michelot A., Derivery E., Paterski-Boujemaa R., Guérin C., Huang S., Parcy F., Staiger C.J., Blanchoin L. (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr. Biol. 16: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003). Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papuga J., Hoffmann C., Dieterle M., Moes D., Moreau F., Tholl S., Steinmetz A., Thomas C. (2010). Arabidopsis LIM proteins: A family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22: 3034–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee J.D., Spudich J.A. (1982). Purification of muscle actin. Methods Cell Biol. 24: 271–289 [DOI] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Dolja V.V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Avisar D., Dolja V.V. (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky A.I., Peremyslov V.V., Dolja V.V. (2008). Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. USA 105: 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sheahan M.B., Rose R.J., McCurdy D.W. (2007). Actin-filament-dependent remodeling of the vacuole in cultured mesophyll protoplasts. Protoplasma 230: 141–152 [DOI] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Teanby N.A., Hawes C. (2008). Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: A genetic tool for the next generation. J. Exp. Bot. 59: 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Yamada N., Kagawa T., Yonekura H., Uyeda T.Q., Kadota A., Wada M. (2010). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 8860–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D.B. (2005). Breaking the WAVE complex: The point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8: 103–112 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Kutsuna N., Kanazawa Y., Kondo N., Hasezawa S., Sano T. (2007). Intra-vacuolar reserves of membranes during stomatal closure: The possible role of guard cell vacuoles estimated by 3-D reconstruction. Plant Cell Physiol. 48: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18: 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Moreau F., Dieterle M., Hoffmann C., Gatti S., Hofmann C., Van Troys M., Ampe C., Steinmetz A. (2007). The LIM domains of WLIM1 define a new class of actin bundling modules. J. Biol. Chem. 282: 33599–33608 [DOI] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton 66: 940–957 [DOI] [PubMed] [Google Scholar]
- Tian M., Chaudhry F., Ruzicka D.R., Meagher R.B., Staiger C.J., Day B. (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 150: 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J.A., Doonan J.H., Rawlins D.J., Shaw P.J., Watts J., Lloyd C.W. (1987). An actin network is present in the cytoplasm throughout the cell cycle of carrot cells and associates with the dividing nucleus. J. Cell Biol. 105: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitschke S., Doehlemann G., Schuster M., Steinberg G. (2010). The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell 22: 2476–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., Hasezawa S., Dolja V.V., Hara-Nishimura I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Matsuyama T., Hashimoto T. (1999). Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tüzel E., Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Yokota E., Cheung A.Y., Shimmen T., Hepler P.K. (1999). The 135 kDa actin-bundling protein from Lilium longiflorum pollen is the plant homologue of villin. Protoplasma 209: 283–291 [Google Scholar]
- Wang H.J., Wan A.R., Jauh G.Y. (2008a). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147: 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu Y.M., Li Y. (2005). Comparison of F-actin fluorescent labeling methods in pollen tubes of Lilium davidii. Plant Cell Rep. 24: 266–270 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Motes C.M., Mohamalawari D.R., Blancaflor E.B. (2004). Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil. Cytoskeleton 59: 79–93 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Yoo C.M., Blancaflor E.B. (2008b). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Ren S., Zhang X., Gao M., Ye S., Qi Y., Zheng Y., Wang J., Zeng L., Li Q., Huang S., He Z. (2011). BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Vidali L., Tominaga M., Tahara H., Orii H., Morizane Y., Hepler P.K., Shimmen T. (2003). Plant 115-kDa actin-filament bundling protein, P-115-ABP, is a homologue of plant villin and is widely distributed in cells. Plant Cell Physiol. 44: 1088–1099 [DOI] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Fan L.M., Wu W.H. (2007). Osmo-sensitive and stretch-activated calcium-permeable channels in Vicia faba guard cells are regulated by actin dynamics. Plant Physiol. 143: 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., Yuan Z., Hu J., Ren H., Zhang D. (2011). RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23: 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang W., Zhao Y., Gong X., Guo L., Zhu G., Wang X., Gong Z., Schumaker K.S., Guo Y. (2007). SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.