A rice male-sterile mutant, defective pollen wall (dpw), which has defective anthers and pollen grains, is isolated and characterized. DPW is found to encode a fatty acyl carrier protein reductase that is active during the synthesis of the anther cuticle and pollen sporopollenin.
Abstract
Aliphatic alcohols naturally exist in many organisms as important cellular components; however, their roles in extracellular polymer biosynthesis are poorly defined. We report here the isolation and characterization of a rice (Oryza sativa) male-sterile mutant, defective pollen wall (dpw), which displays defective anther development and degenerated pollen grains with an irregular exine. Chemical analysis revealed that dpw anthers had a dramatic reduction in cutin monomers and an altered composition of cuticular wax, as well as soluble fatty acids and alcohols. Using map-based cloning, we identified the DPW gene, which is expressed in both tapetal cells and microspores during anther development. Biochemical analysis of the recombinant DPW enzyme shows that it is a novel fatty acid reductase that produces 1-hexadecanol and exhibits >270-fold higher specificity for palmiltoyl-acyl carrier protein than for C16:0 CoA substrates. DPW was predominantly targeted to plastids mediated by its N-terminal transit peptide. Moreover, we demonstrate that the monocot DPW from rice complements the dicot Arabidopsis thaliana male sterile2 (ms2) mutant and is the probable ortholog of MS2. These data suggest that DPWs participate in a conserved step in primary fatty alcohol synthesis for anther cuticle and pollen sporopollenin biosynthesis in monocots and dicots.
INTRODUCTION
Fatty alcohols naturally exist in plants, animals, and algae in free forms (the component of cuticular lipids), but more frequently in esterified (wax esters) or etherified forms (glyceryl ethers). Fatty alcohols are important cellular components, including the fatty constituents of plant cuticle, insect surface coatings, animal skin, and others (Kunst and Samuels, 2003). It has been demonstrated that fatty alcohols are synthesized by fatty acyl-CoA reductase (FAR) through the four-electron reduction of fatty acyl-CoA, with NADPH as a cofactor (Kunst and Samuels, 2003; Samuels et al., 2008). Previous reports revealed that FARs in garden pea (Pisum sativum), jojoba (Simmondsia chinensis), Arabidopsis thaliana, wheat (Triticum aestivum), mouse (Mus musculus), and silkmoth (Bombyx mori) possibly perform alcohol formation from fatty acid precursors (Aarts et al., 1997; Metz et al., 2000; Moto et al., 2003; Wang et al., 2003; Cheng and Russell, 2004; Rowland et al., 2006; Doan et al., 2009; Domergue et al., 2010).
During pollen development in flowering plants, fatty alcohols and their derivatives are major components of the anther cuticle and pollen wall, which are rich in lipids (Ahlers et al., 1999; Meuter-Gerhards et al., 1999; Jung et al., 2006). The anther cuticle functions as a barrier enclosing the anther, which contains the meiotic cells (also called microsporocytes) at the center of each anther lobe, surrounded by four somatic layers, which from the surface to interior, are the epidermis, the endothecium, the middle layer, and the tapetum (Goldberg et al., 1993; Scott et al., 2004). Later, the pollen develops a protective wall, which has two layers, the outer exine and the inner intine. The exine is generally further divided into two sublayers, the sexine and nexine (Zinkl et al., 1999). The highly sculptured exine is mainly composed of a biopolymer sporopollenin, which occurs throughout flowering plants and is thought to have played a key role in land colonization by plants (Chaloner, 1976). The biochemical nature of the anther surface and pollen exine remains poorly understood due to difficulties in purifying and obtaining large quantity of materials for analysis. In addition, sporopollenin is highly insoluble, hardly degradable, and exceptionally stable (Brooks and Shaw, 1968; Bubert et al., 2002). Currently available evidence indicates that the major components of sporopollenin are straight-chain fatty acids and oxygenated aromatic monomers, such as p-coumaric and ferulic acids (Domínguez et al., 1999; Bubert et al., 2002; Blackmore et al., 2007).
Previous studies of male-sterile mutants with abnormal pollen wall formation have shown that both the diploid sporophytic tapetal cells and gametophytic microspores contribute to pollen wall synthesis (Bedinger, 1992; Li and Zhang, 2010; Ariizumi and Toriyama, 2011). The lipidic precursors of sporopollenin are considered to be mainly produced in the tapetum under the control of the sporophytic genome. Rice (Oryza sativa) and many other plants have a secretory type tapetum with Ubisch bodies or orbicules; these are specialized structures forming along the inner surface of the tapetum/anther locule, which transfer lipidic components from the tapetum to the microspores (Huysmans et al., 1998). Exine formation commences from the late tetrad stage, with the deposition of lipidic precursors onto the microspore surface, between the callose wall and the microspore plasma membrane (Paxson-Sowders et al., 1997). The primexine structure on the microspore plasma membrane functions as the exine template for the polymerization of these precursor components and exine patterning (Blackmore et al., 2007). DEFECTIVE IN EXINE PATTERN FORMATION (DEX1) is a putative membrane associated protein with predicted calcium binding domains. In Arabidopsis, the dex1 mutant has delayed development of the primexine, and the synthesized sporopollenin is therefore abnormally deposited on the mutant microspore surface (Paxson-Sowders et al., 1997), suggesting a crucial role for the microspore in pollen wall patterning. After the first pollen mitosis, the formation of the exine is almost complete; at later stages of pollen ontogeny, two other components (i.e., the pectocellulosic intine and the tryphine, or pollen coat) are deposited onto the pollen wall (Piffanelli et al., 1998).
Genetic analyses have revealed several genes critical for anther cuticle and pollen wall development, such as MALE STERILITY1 (MS1), MS2, ECERIFERUM1 (CER1), NO EXINE FORMATION1 (NEF1), FACELESS POLLEN1, CYP703A2, Acyl-CoA Synthetase 5 (ACOS5), CYP704B1, TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1), TKPR2, LAP6/POLYKETIDE SYNTHASE A (PKSA), and LAP5/POLYKETIDE SYNTHASE B (PKSB) in Arabidopsis (Aarts et al., 1995, 1997; Wilson et al., 2001; Ariizumi et al., 2003, 2004; Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009, 2010; Grienenberger et al., 2010; Kim et al., 2010), as well as Tapetum Degeneration Retardation (TDR), Wax-Deficient Anther1 (WDA1), CYP704B2, Os C6, Post-meiotic Deficient Anther1, and PERSISTENT TAPETAL CELL1 (PTC1) in rice (Jung et al., 2006; Zhang et al., 2008, 2010; Li et al., 2010, 2011; Hu et al., 2010; Li and Zhang, 2010). However, relatively little has been described on the biochemical activities of these gene products.
In heterotrophic eukaryotes, the usual pathway of fatty acid synthesis is found in the cytosol, whereas in plants, the generation of long-chain fatty acids up to C18 mainly occurs in semiautonomous plastids, such as the chloroplast (Ohlrogge et al., 1979; Li-Beisson et al., 2010), and some fatty acids are extended and used for lipid synthesis in the endoplasmic reticulum (ER; Kunst and Samuels, 2003). Plastids are assumed to play crucial roles for successful anther development in angiosperms, such as storing lipids in elaioplasts. In tapetal cells, proplastids undergo division during the early stages of microsporogenesis and subsequently develop into elaioplasts that are involved in the biosynthesis of tapetal lipids, which contribute to lipidic pollen wall formation (Clement and Pacini, 2001). However, the role of plastid-related lipid metabolism in plant development remains unknown.
In this study, we report the isolation of a rice recessive, nuclear male sterile mutation in the DEFECTIVE POLLEN WALL (DPW) gene, which encodes a fatty acyl reductase. We show that DPW expression is detectable in the tapetum and microspores and that its protein is mainly localized to the plastid by a DPW N-terminal transit peptide. Recombinant DPW enzyme produced in bacteria has the ability to convert palmiltoyl-acyl carrier protein (ACP), palmiltoyl-CoA, and palmitoleoyl-CoA to their corresponding alcohols. In addition, recombinant DPW has a high affinity for palmiltoyl-ACP. DPW is able to complement the Arabidopsis ms2 mutant, indicating that DPW is the putative rice ortholog of MS2. Our work therefore reveals a conserved pathway for primary fatty alcohol synthesis that is essential for pollen wall and anther cuticle development in both dicot and monocot plants.
RESULTS
Isolation and Genetic Analysis of the dpw Mutant
To identify rice genes that are important for normal anther and pollen development, we used 60Co γ-ray radiation to generate a rice mutant library in the 9522 background, which is a cultivar of O. sativa ssp japonica (Liu et al., 2005; Chen et al., 2006; Li et al., 2006; Wang et al., 2006). We isolated the dpw mutant by its complete male sterility and small anthers. The dpw mutant was backcrossed with wild-type plants (the 9522 cultivar) three times and used for genetic and phenotypic analyses.
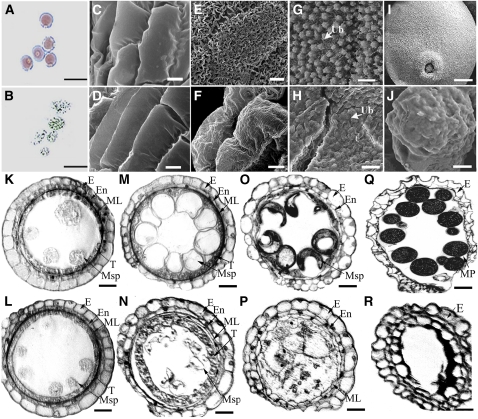
When the dpw plants were pollinated with wild-type pollen, all of the F1 progeny displayed the wide-type phenotype, indicating that dpw is a recessive mutant. F2 progeny tests yielded a segregation of 153 normal and 49 mutant plants (χ2 = 0.08, P > 0.05), indicating monofactorial recessive inheritance of the mutant characteristic. Vegetative and floral development seemed to be normal in the dpw mutant plant (Figures 1A to 1C); however, compared with the wild type, the mutant anthers were smaller (Figures 1D and 1E) and lacked normal mature pollen grains (Figures 1F and 1G).
Figure 1.
Phenotypic Comparison between the Wild Type and the dpw Mutant.
(A) A wild-type (WT) plant (left) and a dpw mutant plant (right) after bolting.
(B) A wild-type panicle (left) and a dpw mutant panicle (right) at the heading stage. The arrows indicate anthers.
(C) A wild-type spikelet (left) and a dpw mutant spikelet (right). gl, glume; le, lemma; pa, palea.
(D) A wild-type spikelet (left) and a dpw mutant spikelet (right) after removing half of the lemma and palea. st, stamen.
(E) A wild-type yellow anther (left) and a dpw mutant pale-yellow, smaller anther (right).
(F) Wild-type pollen grains stained with 1% I2-KI solution at stage 12 (Zhang and Wilson, 2009), showing mature pollen grains.
(G) Pollen grains of dpw stained with 1% I2-KI solution at stage 12; no mature pollen grains are present.
Bars = 2 mm in (C) to (E) and 5 μm in (F) and (G).
Phenotypic Analysis of the dpw Mutant Anther Development
To investigate the cellular defects of the dpw mutant during pollen development, we first examined the integrity of the pollen wall following chemical treatments. The abnormal pollen exine of pollen wall defective mutants is usually sensitive to chemical treatment, such as acetic anhydride (Aarts et al., 1997). We observed that wild-type pollen grains at stage 9 (Zhang and Wilson 2009) were insensitive to acetolysis and retained their integrity (Figure 2A), whereas the dpw mutant pollen grains at this stage were highly sensitive and severely damaged by this treatment (Figure 2B), indicating that the pollen exine in dpw was abnormal.
Figure 2.
Defects of dpw Anther Development and the Pollen Wall.
(A) and (B) Pollen grains of the wild type (A) and dpw (B) treated with an acetolysis mixture for 10 min.
(C) to (F) Scanning electron microscopy analysis of the anther surface of the wild type ([C] and [E]) and dpw ([D] and [F]) at stage 9 ([C] and [D]) and stage 12 ([E] and [F]).
(G) and (H) Scanning electron microscopy analysis of the inner surface of the anther wall layers of the wild type (G) and dpw (H), showing Ubisch bodies at stage 10.
(I) and (J) Scanning electron microscopy analysis of pollen grains of the wild type (I) and dpw (J) at stage 10.
(K) to (R) Comparison of anther development in the wild type and dpw mutant. The images are of cross sections through a single locule. The wild-type anther is shown in (K), (M), (O), and (Q) and the dpw mutant anther in (L), (N), (P), and (R). Stage 9 ([K] and [L]); stage 10 ([M] and [N]); stage 11 ([O] and [P]); and stage 12 ([Q] and [R]).
E, epidermis; En, endothecium; ML, middle layer; MP, mature pollen. Msp, microspore; T, tapetum; Ub, Ubisch body. Bars = 10 μm in (A) and (B), 5 μm in (C) to (J), and 15 μm in (K) to (R).
[See online article for color version of this figure.]
The dpw anther was smaller than that of the wild type, and we analyzed its phenotype further by examining the anther surface structure using scanning electron microscopy. At stage 9 of anther development, the anther surfaces of the wild type and the dpw mutant exhibited no obvious differences (Figures 2C and 2D). At stage 12 (Zhang and Wilson, 2009), compared with the well-formed cuticle on the exterior of wild-type anthers (Figure 2E), the dpw anther outer surface was relatively smooth (Figure 2F). Intriguingly, unlike the wild-type anther, which at stage 9 had large numbers of granular Ubisch bodies on the inner locule surface (Figure 2G) (Zhang and Wilson 2009), the dpw mutant had empty and shrunken Ubisch bodies (Figure 2H). Moreover, at this stage, the wild-type pollen grains had a smooth and particulate exine patterning (Figure 2I), whereas the dpw pollen surface appeared severely shrunken (Figure 2J). This finding is in good agreement with the observed sensitivity to acetolysis treatment. Therefore, we named this mutant defective pollen wall (dpw).
To characterize the anther morphological defects in the dpw mutant, we examined wild-type and mutant anther development using light microscopy of transverse sections. No detectable differences were observed between wild-type and the dpw mutant anthers during the early stages of development, until stage 9 (Figures 2K and 2L); meiosis in the dpw mutant also seemed to be normal, as indicated using 4′,6-diamidino-2-phenylindole staining analysis (see Supplemental Figure 1 online). During stage 10, wild-type tapetal cells started to shrink and were deeply stained with toluidine blue. The middle layer became narrower and was barely visible, and the wild-type microspores were spherical with large central vacuoles (Figure 2M). By contrast, at this stage, the dpw mutant tapetal cells were larger than normal with reduced staining, and the microspores were less vacuolated, abnormally shaped, and starting to degenerate (Figure 2N). In the wild-type anthers, exine deposition was nearly completed by stage 11; at this stage, the uninucleate pollen had developed into trinucleate pollen through two mitotic divisions and the tapetal cells had differentiated and degenerated (Figure 2O) (Zhang and Wilson, 2009). However, at stage 11 in the dpw anthers, the microspores were abnormally developed with an irregular appearance, with most fragmenting and disintegrating. In addition, the dpw tapetal cells were lightly stained, collapsed into the anther locule, and degenerating, whereas the middle layer persisted (Figure 2P). At stage 12, wild-type pollen grains were full of starch and lipids, and the tapetum was completely degenerated (Figure 2Q); by contrast, in the dpw anther, the microspores had degenerated completely, leaving an empty anther locule (Figure 2R).
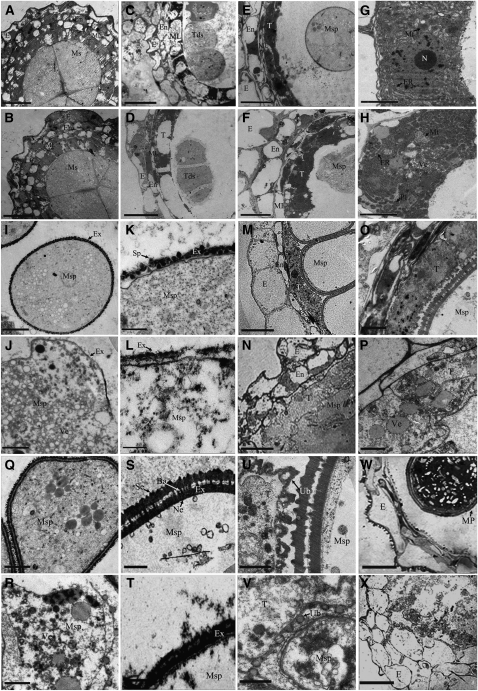
To obtain a more detailed understanding of the abnormalities of the dpw mutant anther development, transmission electron microscopy (TEM) was performed. In accordance with the light microscopy, from late stage 6 to stage 8, no significant differences were detected between the wild type and the dpw mutant in the anther cell layers and microspores (Figures 3A to 3D). In the wild type at stage 9, the tapetal cells contained numerous small vacuoles throughout the electron-dense cytoplasm; the tapetal cells then became deeply stained and started to degenerate (Figures 3E and 3G). However, more abnormal electron-dense structures were found in the dpw tapetal cells (Figures 3F and 3H). At this stage in the wild type, microspores were released from the tetrads into the locules and the pollen wall appeared thickened as more sporopollenin was incorporated into the exine (Figures 3I and 3K). By contrast, the dpw mutant pollen had irregularly distributed vacuoles and a surface with a coarse primexine matrix that lacked any obvious sporopollenin deposition (Figures 3J and 3L). At stage 10, wild-type tapetal cells appeared lightly stained, became thin, and continued to degenerate, and the middle layer degenerated and was no longer visible (Figures 3M and 3O) (Zhang and Wilson, 2009). However, in the dpw anthers, irregular cellular structures were apparent, the middle layer persisted, and the tapetal cells did not condense but rather became abnormally expanded with an increased number of vacuoles (Figures 3N and 3P). Furthermore, at this stage, wild-type microspores had nearly completed pollen exine deposition and exhibited distinctive sexine and nexine sublayers (Figures 3Q and 3S). However, the dpw mutant pollen failed to form separate sexine and nexine and had numerous abnormal vacuoles (Figures 3R and 3T). Most of the abnormal dpw pollen grains gradually degenerated, leaving only some distorted pollen with less exine in the locule. Consistent with the role of Ubisch bodies in the transport of nutrients and materials for pollen wall development, we observed in the wild-type anthers abundant Ubisch bodies with electron-dense sporopollenin precursors at the interface between the tapetal cells and microspores (Figure 3U, arrow). However, only a few abnormal Ubisch bodies were found in the dpw mutant (Figure 3V), implying that the dpw pollen wall defect is possibly due to a lack of Ubisch bodies and reduced secretion from the dpw tapetum. Consistently, unlike the mature pollen grains in the wild type at stage 12 (Figure 3W), we found nearly collapsed pollen in the dpw anther locule (Figure 3X). Meanwhile, the wild-type anther wall layers at stage 12 became more degenerated and formed electron-dense hair-like structures on the epidermis (Figure 3W). This suggests that lipophilic materials (cutin and wax) diffused to the surface of the anther cell wall. By contrast, the dpw anther wall layers appeared less degenerated and had obvious cellular structures and stained weakly (Figure 3X), suggesting that decreased amounts of lipophilic materials were deposited or transferred to the outer epidermal cell wall during these stages.
Figure 3.
TEMs of Anthers from the Wild Type and the dpw Mutant.
(A) and (B) Anthers of the wild type (A) and dpw (B) at late stage 6 showing meiocytes.
(C) and (D) Anthers of the wild type (C) and dpw (D) at stage 8 showing tetrads.
(E) The wild-type anther at stage 9 showing highly metabolically active cells and spherical microspores.
(F) A dpw mutant anther at stage 9 showing abnormal tapetal cells and microspores.
(G) Higher magnification of the wild-type tapetum in (E) showing mitochondrion and endoplasmic reticulum (arrows).
(H) Higher magnification of the dpw tapetum in (F) showing mitochondrion, ER, and large numbers of electron-dense structures (arrows).
(I) The microspore of the wild type at stage 9.
(J) The microspore of dpw at stage 9.
(K) Wild-type exine showing deposition of sporopollenin (arrows).
(L) The dpw mutant exine (arrows) showing abnormal deposition of sporopollenin.
(M) The wild-type anther at stage 10 showing degenerated tapetum and vacuolated microspores.
(N) A dpw mutant anther at stage 10 showing expanded tapetum and distorted microspores.
(O) Higher magnification of wild-type tapetum in (M).
(P) Higher magnification of dpw tapetum in (N).
(Q) Wild-type microspores at stage 10.
(R) dpw mutant microspores at stage 10.
(S) Higher magnification of wild-type exine in (Q) showing sexine, bacula, and nexine (arrows).
(T) Higher magnification of the dpw mutant abnormal exine in (R).
(U) Wild-type Ubisch bodies (arrows).
(V) dpw mutant Ubisch bodies (arrows).
(W) A wild-type anther at stage 12.
(X) A dpw mutant anther at stage 12.
Ba, bacula; E, epidermis; En, endothecium; Ex, exine; ML, middle layer; Ms, microsporocyte; Msp, microspore; Mt, mitochondrion; N, nucleus; Pl, plastid; Ne, nexine; Se, sexine; Sp, sporopollenin; T, tapetum; Tds, tetrads; Ub, Ubisch body; Ve, vacuole. Bars = 5 μm in (A) to (F), (M), (N), (W), and (X), 1 μm in (G) to (J) and (O) to (R), and 0.5 μm in (K), (L), and (S) to (V).
Altered Aliphatic Composition of the dpw Anther
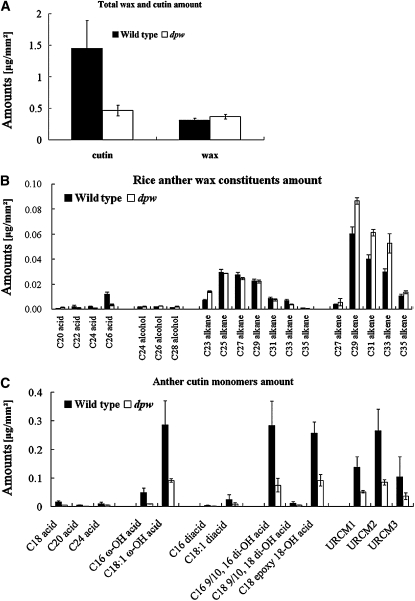
The defects in the anther cuticle and pollen wall, as well as the reduced number of lipidic Ubisch bodies in dpw, suggest abnormalities in the synthesis of aliphatic components. To test this hypothesis, we analyzed the composition of chloroform extractable cuticular waxes as well as aliphatic cutin monomers and total soluble lipids from wild-type and dpw anthers by gas chromatography–mass spectrometry (GC-MS) and gas chromatography–flame ionization detection (GC-FID) (Bonaventure et al., 2004; Franke et al., 2005). We employed an approach to measure the surface area of anthers, in which the calculated values of surface area of randomly selected samples were plotted against the weight of each corresponding sample (Li et al., 2010; see Supplemental Figure 2 online). Analytical results showed that the total wax in the dpw mutant anthers (0.37 μg/mm2) increased by 16.2% compared with the wild type (0.31 μg/mm2; P < 0.05) (Figure 4A). In particular, alkenes (C27, C29, C31, C33, and C35) were significantly increased in the mutant (P < 0.05 for C27; P < 0.01 for C29, C31, and C35) (Figure 4B; see Supplemental Table 1 online). In addition, a significant increase in the levels of C24 (P < 0.01), C26 (P < 0.01), and C28 (P < 0.01) alcohols and C23 alkane, as well as a decrease in C26 acid, were also observed (Figure 4B; see Supplemental Table 1 online).
Figure 4.
Analysis of Anther Wax and Cutin in the Wild Type and dpw.
(A) Total wax and cutin amount per unit surface area (μg mm−2) in the wild-type (black bars) and dpw (white bars) anthers. Error bars indicate sd (n = 5).
(B) Wax constituents, amount per unit surface area (μg mm−2) in the wild-type (black bars) and dpw anthers (white bars). Error bars indicate sd (n = 5).
(C) Cutin monomers, amount per unit surface area (μg mm−2) in the wild-type (black bars) and dpw anthers (white bars). Error bars indicate sd (n = 5).
Compound names are abbreviated as follows: C18 acid, octadecanoic acid; C18(2) acid, linoleic acid; C18 (1/3) acids, oleic acid and linolenic acid; C22 acid, docosanoic acid; C24 acid: tetracosanoic acid; C16 ω-OH acid, 16-hydroxyhexadecanoic acid; C18 (1) ω-OH acid, 18-hydroxy-octadecanoic acid; C16 diacid, hexadecane-1,16-dioic acid; C18 (1) diacid, octadecene-1,18-dioic acid; C16 9/10,16-diOH acid, 9(10), 16-dihydroxy-hexadecanoic acid; C18 9/10, 18-diOH acid, 9(10), 18-dihydroxy-octadecanoic acid; C18 epoxy ω-OH acid, 9-epoxy-18-hydroxy-octadecanoic acid; URCM, unknown rice cutin monomer. Acids were analyzed as methyl esters, and hydroxyl groups were analyzed as trimethylsilyl esters.
To analyze anther cutin monomer composition, the cutin polyester from delipidated anthers was transesterified (Bonaventure et al., 2004; Franke et al., 2005). An earlier investigation by Jung et al. (2006) indicated that the applied transesterification conditions did not release cutin-like monomers out of isolated pollen grains; thus, the identified monomers were only released from the epidermal cutin of anthers by this method. In the wild type, the total amount of cutin was 1.45 μg/mm2. By contrast, in the dpw mutant, it was 0.46 μg/mm2, which corresponded to a reduction of 68.0% compared with the wild type (Figures 4A and 4C; see Supplemental Table 2 online). In the wild-type anthers, the major aliphatic cutin monomers were the 16-hydroxy-hexadecanoic acid, 9(10),16-dihydroxy-hexadecanoic acid, 18-hydroxy-octadecenoic acid, and 9-epoxy-18-hydroxy-octadecanoic acid and compounds reported as unidentified rice cutin monomers (Jung et al., 2006; Li et al., 2010). Compared with the wild type, all aliphatic cutin monomers observed in the dpw mutant were significantly reduced (Figure 4C).
To understand the change in internal fatty acid and alcohol content, the total soluble anther lipids were extracted for analysis. In the wild type, the total amount of fatty acids with carbon lengths from 12 to 24 was 34.9 μg/mg (Table 1). However, the dpw anthers had slightly more fatty acids in this size range (36.4 μg/mg) than those of the wild type. By contrast, the dpw anthers contained less C16 and C18 fatty alcohols (0.22 μg/mg) than the wild type (0.38 μg/mg) (Table 1), suggesting that fatty alcohol formation is altered in the mutant anthers.
Table 1.
Total Soluble Fatty Acids and Fatty Alcohols of Wild-Type and dpw Anthers
| Wild Type | dpw | ||
| Lipids | Mean ± sd (μg/mg Dry Weight) | Up | |
| Acids | |||
| C12 acid | 0.198 ± 0.060 | 20.233 ± 0.076 | 17.20% |
| C14 acid | 0.211 ± 0.021 | 0.243 ± 0.085 | 15.09% |
| C16 acid | 6.992 ± 1.019 | 6.612 ± 0.408 | −5.44% |
| C18(2/3) acid | 19.16 ± 1.182 | 20.76 ± 0.783 | 8.32% |
| C18(1) acid | 0.984 ± 0.077 | 1.346 ± 0.316 | 36.83% |
| C18 acid | 2.696 ± 0.201 | 2.500 ± 0.486 | −7.27% |
| C20(2) acid | 0.072 ± 0.057 | 0.209 ± 0.082 | 189.77% |
| C20(1) acid | 0.544 ± 0.145 | 0.795 ± 0.261 | 46.20% |
| C20 acid | 0.326 ± 0.032 | 0.882 ± 0.176 | 170.59% |
| C22(1) acid | 1.495 ± 0.160 | 0.704 ± 0.047 | −52.89% |
| C22 acid | 0.155 ± 0.042 | 0.626 ± 0.035 | 305.06% |
| C24(2) acid | 0.361 ± 0.051 | 0.357 ± 0.007 | −1.13% |
| C24(1) acid | 1.488 ± 0.161 | 0.616 ± 0.027 | −58.64% |
| C24 acid | 0.252 ± 0.075 | 0.464 ± 0.026 | 84.54% |
| Total acids | 34.943 ± 3.284 | 36.351 ± 2.814 | 4.03% |
| Alcohols | |||
| C16 alcohol | 0.110 ± 0.034 | 0.065 ± 0.014 | −40.86% |
| C18 alcohol | 0.268 ± 0.104 | 0.152 ± 0.031 | −43.39% |
| Total alcohols | 0.378 ± 0.138 | 0.217 ± 0.045 | -42.65% |
Isolation of DPW
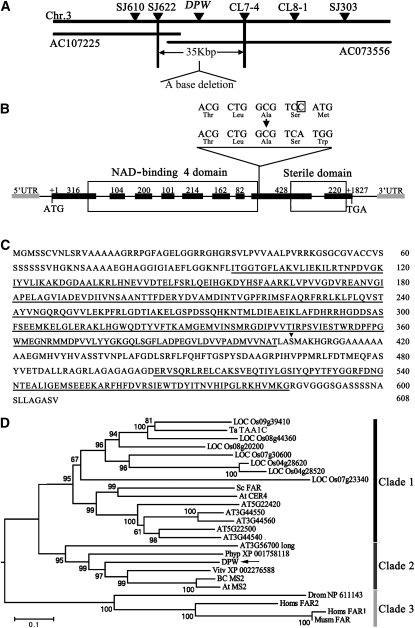
To identify DPW, we used a map-based cloning approach (see Methods). After fine mapping, we found that the dpw mutation had a single base deletion in the eighth exon of a candidate gene (LOC_Os03g07140) that consisted of nine predicted exons and eight introns (Figures 5A and 5B), resulting in frame shift and premature translational termination (Figure 5C). To confirm the annotation of DPW, we obtained a full-length cDNA (AK121254) from the Rice Genome Resource Center (RGRC-NIAS; http://www.rgrc.dna.affrc.go.jp/stock.html). The DPW transcribed region includes a 368-bp 5′ untranslated region (UTR), an 893-bp 3′ UTR, and an open reading frame of 2851 bp encoding 608 amino acids. This gene was confirmed to be DPW by functional complementation of dpw homozygous plants (see Supplemental Figure 3 online).
Figure 5.
Molecular Identification and Sequence Analysis of DPW.
(A) Fine mapping of the DPW gene on chromosome 3. The names and positions of the molecular markers are indicated. AC107225 and AC073556 are genomic DNA accession numbers. The DPW locus was mapped to a 35-kb region between two molecular markers (SJ622 and CL7-4).
(B) A schematic representation of the exon and intron organization of DPW. The mutant sequence has one base deletion in the eighth exon, which is boxed. +1 Indicates the starting nucleotide of translation (ATG), and +1827 indicates the stop codon (TAG). Black boxes indicate exons, intervening lines indicate introns, gray boxes indicate 5′-UTR and 3′-UTR, respectively, and the white boxes indicate the NAD binding 4 domain and male sterility domain.
(C) The DPW protein sequence. The NAD binding 4 domain and sterile domain are underlined. The black triangle shows the position of base deletion.
(D) A neighbor-joining phylogenetic tree summarizes the evolutionary relationships among the DPW-related proteins (E value of BLASTP result is <1E-11). The proteins are named according to their gene names or NCBI accession numbers: ABO14927 (BC MS2), AT3G11980 (At MS2), AAD38039 (Sc FAR), CAD30692 (Ta TAA1C), AT4G33790 (At CER4), NP_060569 (Homs FAR2), NP_115604 (Homs FAR1), and NP_081655 (Musm_FAR). The numbers under the branches refer to the bootstrap value of the neighbor-joining phylogenetic tree. The length of the branches is proportional to the amino acid variation rates. At, Arabidopsis thaliana; Ta, Triticum aestivum; Sc, Simmondsia chinensis; BC, Brassica rapa subsp chinensis; Vitv, Vitis vinifera; Phyp, Physcomitrella patens subsp patens; Homs, Homo sapiens; Musm, Mus musculus; Drom, Drosophila melanogaster. The bar indicates the estimated number of amino acid substitutions per site (for alignment, see Supplemental Figure 4 and Supplemental Data Set 1 online).
Functional Conservation of DPW in Flowering Plants
To test whether DPW function is rice specific or conserved in plants, we performed phylogenetic analysis of DPW and its close homologs. We searched public databases (National Center for Biotechnology Information [NCBI], Gramene, and The Arabidopsis Information Resource) using BLASTP with the DPW amino acid sequence as a query and obtained 24 closely related homologous sequences. Sequence comparison (see Supplemental Figure 4 online) indicated that all 25 proteins contained highly similar NAD binding 4 and a male sterile domain (Aarts et al., 1997), implying that these two functional domains are evolutionarily conserved between plants and animals.
Subsequently, we constructed a neighbor-joining phylogenetic tree of the 25 genes (Figure 5D), which were grouped into three clades. The anther-specific Ta TAA1 gene implicated in wheat pollen wall development (Wang et al., 2002) and seven rice genes were grouped into the first clade. Clade 1 also contained seven other genes from dicots: six from Arabidopsis and the FAR gene from jojoba (Metz et al., 2000). Moreover, Clade 2 contained DPW, Arabidopsis MS2 (Aarts et al., 1997), an Arabidopsis MS2-like gene (AT3G56700 long) (Doan et al., 2009), Physcomitrella patens XP 001758118, Vitis vinifera XP 002276588, and Bc MS2 from Brassica rapa subsp chinensis (Hu et al., 2006). A single putative paralog of this group seems to be present in the genome of land plants. The presence of the XP 001758118 gene from P. patens, a moss, suggests that DPW and its homologs represent a conserved group of genes with basic and important roles in the major lineages of land plants. Furthermore, Drom NP 611143, human FAR1 and FAR2, and mouse FAR1 from animals were grouped into Clade 3. These results suggest that DPW and its homologs play key roles in lipid biosynthesis among divergent plants and animals.
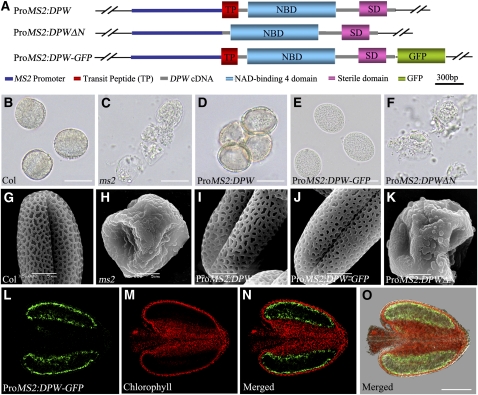
The phylogenetic analysis suggests that DPW and Arabidopsis MS2 are orthologs. In support of this hypothesis, ms2 also has abnormal pollen wall development, with a similar phenotype to the dpw mutant (Aarts et al., 1997; Dobritsa et al., 2009). However, detailed biochemical defects have not been described for the ms2 mutant. The DPW and MS2 proteins share only 59% identity overall and 65% identity within the two functional domains (Figure 5D; see Supplemental Data Set 1 and Supplemental Figure 5 online). To obtain direct evidence for the evolutionary relatedness of these two genes, we performed functional complementation of the ms2 mutant using full-length DPW cDNA and a DPW translationally fused construct, DPW-Green Fluorescent Protein (GFP), respectively (Figure 6A). Two binary plasmids, ProMS2:DPW and ProMS2:DPW-GFP, carrying the Arabidopsis MS2 promoter were individually introduced into MS2/ms2 heterozygous plants. The result showed that both DPW and DPW-GFP were able to rescue the pollen wall defects of the ms2 homozygous plants (Figures 6B to 6E and 6G to 6J), indicating that the function of DPW/MS2 is conserved in both dicots and monocots and possibly in all flowering plants.
Figure 6.
Functional Complementation of the Arabidopsis ms2 Mutant Using DPW.
(A) Diagrams of the constructs of the full-length DPW cDNA under the control of the MS2 promoter (ProMS2:DPW), DPW cDNA without the sequence encoding the putative targeting signal peptide under the control of the MS2 promoter (ProMS2:DPWΔN), and construct with the GFP sequence fused to the C terminus of DPW cDNA.
(B) to (F) Result of acetolysis treatment of microspores from wild-type and ms2 mutant transgenic lines at the mature stage.
(G) to (K) Scanning electron microscopy analysis of pollen wall surface of the wild-type and transgenic lines of the ms2 mutants at the mature pollen stage.
(B) and (G) The wild-type pollen grains.
(C) and (H) ms2 mutant pollen grains.
(D) and (I) Pollen grains of transgenic lines containing ProMS2:DPW in the ms2 background.
(E) and (J) Pollen grains of transgenic lines containing ProMS2:DPW-GFP in the ms2 background.
(F) and (K) Pollen grains of transgenic lines containing ProMS2:DPWΔN in the ms2 background.
(L) to (O) Fluorescence micrographs of an anther from a transgenic line expressing ProMS2:DPW-GFP in the ms2 background. (L) shows the green fluorescence of GFP; (M) shows the autofluorescence of chlorophyll; (N) is the merged image of (L) and (M); and (O) is a superimposed image from GFP (green), chlorophyll autofluorescence (red), and bright-field images.
Bars = 20 μm in (B) to (F), 5 μm in (G) to (K), and 100 μm in (L) to (O).
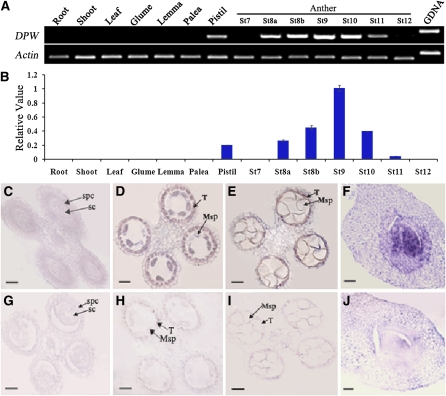
DPW Is Mainly Expressed in the Tapetum and Microspores
To test whether DPW acts within the anther, or in other tissues, we analyzed the DPW expression pattern. DPW expression analysis by RT-PCR using total RNA extracted from vegetative and reproductive organs indicated that the DPW transcripts were not detectable in roots, stems, leaves, and the nonreproductive floral organs, glume, lemma, and palea (Figures 7A and 7B). On the other hand, DPW expression was observed in the anthers at relatively early stages of anther development, starting at stage 8, peaking at stage 9, declining slightly at stage 10, and dissipating by stage 12. Meanwhile, a low level of DPW transcript was detected in the pistil. No obvious morphological abnormalities were observed in the dpw pistil; therefore, it is possible that there is gene redundancy or that DPW is not required for morphological development in the pistil.
Figure 7.
DPW Expression Pattern.
(A) Spatial and temporal expression analysis of DPW by RT-PCR. Each reaction had three biological repeats. Actin1 served as a control.
(B) qRT-PCR analysis of DPW. St8a, stage 8a; St8b, stage 8b; St9, stage 9; St10, stage 10; St11, stage 11; St12, stage 12. Error bars indicate sd, and each reaction had three biological repeats.
(C) to (J) In situ analyses of DPW in wild-type anthers and pistil.
(C) An anther at stage 7 showing no DPW expression.
(D) An anther at stage 8a showing DPW expression in tapetal cells and microspores.
(E) An anther at stage 10 showing reduced DPW expression in the tapetum and microspores.
(F) A pistil showing DPW expression.
(G) An anther at early stage 8 hybridized with the DPW sense probe.
(H) An anther at stage 8a hybridized with the DPW sense probe.
(I) An anther at stage 10 with the DPW sense probe.
(J) A pistil hybridized with the DPW sense probe.
Msp, microspore; SC, sporogenous cell; SPC, secondary parietal cell; T, tapetum. Bars = 25 μm.
To more precisely determine the spatial and temporal patterns of DPW expression, we performed RNA in situ hybridization with wild-type floral sections. At early stage 7, no DPW expression was detected (Figure 7C). From stage 8 to stage 10, strong expression of DPW was detected in the tapetum, but there was no detectable signal in the anther epidermis (Figures 7D and 7E). Also, the DPW transcripts could be detected in microspores at stage 8 (Figure 7D) but were reduced at stage 10 (Figure 7E). Consistent with RT-PCR analysis, we detected a weaker expression signal of DPW in the pistil, specifically in the ovule (Figure 7F). Only background levels of signal were detected with the sense probe (Figures 7G to 7J). Furthermore, the expression signal of DPW-GFP in the complemented lines was observed in the tapetal layer in the homozygous ms2/ms2 background (Figures 6L to 6O). These results demonstrate that DPW is indeed expressed in the tapetum and microspore, which confirms that it acts directly on anther development and most likely has a role in the production of an essential structural building block.
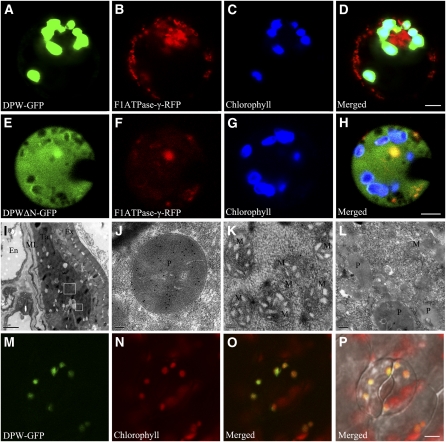
DPW Is Mainly Localized to Plastids
A putative chloroplast or mitochondrion targeting signal peptide was predicted at the N terminus of DPW from the first to 41st amino acid residues using the TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/) and SubLoc v1.0 (http://www.bioinfo.tsinghua.edu.cn/SubLoc/eu_predict.htm). To experimentally verify the subcellular localization of DPW, we constructed another fusion construct, DPWΔN-GFP, which contains a DPW fragment without the sequence encoding the putative N-terminal transit peptide. The constructs DPW-GFP, DPWΔN-GFP, and GFP alone were introduced into protoplasts isolated from etiolated rice stems. Plastid signals were observed by the autofluorescence of chlorophyll. To mark the mitochondrion, a translational fusion construct of the mitochondrial F1ATPase γ-subunit and the red fluorescent protein (RFP) (Kim et al., 2006) was cotransformed into the protoplasts. Our analysis indicated that the DPW-GFP signal colocalized with the chlorophyll autofluorescence signal in plastids but not obviously with the F1ATPase-γ-RFP signal in rice protoplasts (Figures 8A to 8D), suggesting that DPW is mainly localized in plastids. By contrast, the free GFP and DPWΔN-GFP signals were found in the cytoplasm and did not colocalize with the RFP signal or the chlorophyll autofluorescence signal (Figures 8E to 8H; see Supplemental Figure 6 online).
Figure 8.
Analysis of DPW Subcellular Localization.
(A) A rice protoplast expressing DPW-GFP showing green fluorescent signals.
(B) The same rice protoplast cell of (A) showing mitochondrion-localizing F1ATPase-γ-RFP red fluorescent signals as a control.
(C) The same protoplast cell of (A) showing the chlorophyll autofluorescence signal in the plastids.
(D) The merged signal of (A) to (C).
(E) A rice protoplast expressing DPW without its N-terminal targeting peptide fused with GFP showing the green fluorescent signal in the cytoplasm.
(F) The same rice protoplast of (E) expressing a mitochondrion-localizing F1ATPase-γ-RFP with red fluorescent signal.
(G) The same protoplast of (E) showing the chlorophyll autofluorescence signal in the plastids.
(H) The merged signal of (E) to (G).
(I) Section of wild-type anther at stage 9 probed with the purified DPW antibody.
(J) A higher magnification of the large box in (I) showing abundant DPW signals in plastids of the tapetum.
(K) A higher magnification of the small box in (I) showing labeling with a few gold particles in the mitochondrion of the tapetum.
(L) Negative control showing the plastid and the mitochondrion in wild-type tapetal cells probed with preimmune serum.
(M) to (P) The GFP signal (M) of the leaf of a 35s:DPW-GFP stable transgenic Arabidopsis seedling 2 weeks after germination.
(N) Chlorophyll autofluorescence showing chloroplasts.
(O) The merged signals of (M) and (N).
(P) Bright-field image of (O).
Bars = 4 μm in (A) to (H), 1 μm in (I), 0.1 μm in (J) and (K), 0.2 μm in (L), and 5 μm in (M) to (P). En, endothecium; ML, middle layer; Ta, tapetum; Ex, exine; M, mitochondrion; P, plastid.
Moreover, we produced rabbit polyclonal antibodies against recombinant DPW to perform immunological colloidal gold localization experiments in rice. The specificity of the antibodies was verified by immunoblot analysis. A specific signal corresponding to the expected size of DPW was observed using the DPW antibody in the wild-type anther but not in the dpw mutant (see Supplemental Figure 7 online). The molecular mass of the detected protein was estimated to be ~63 kD, as expected of DPW without its targeting signal peptide. Transverse sections of wild-type stage 9 anthers were incubated with the purified DPW antibody as the primary antibody. Subsequently, goat anti-rabbit IgG antibodies conjugated with 10-nm colloidal gold particles (Sigma-Aldrich) were used to recognize the primary antibody. The gold particles were observed in plastids and fewer in the mitochondria within the tapetal cells (Figures 8I to 8K), whereas no detectable signal was observed in the control sections when incubated with the preimmune antiserum as the primary antibody (Figure 8L). Furthermore, analysis of stable transgenic lines in a wild-type Arabidopsis background containing DPW-GFP driven by the cauliflower mosaic virus 35S promoter confirmed the presence of DPW-GFP signal in leaf chloroplasts (Figures 8M to 8P). These observations support the idea that DPW is mainly localized to plastids.
To confirm the biological importance of the N-terminal transit peptide of DPW, we made an additional fusion construct comprising of the Arabidopsis MS2 promoter and DPW without the targeting signal peptide (ProMS2:DPWΔN) (Figure 6A). Unlike the ms2 homozygous plants transformed with ProMS2:DPW, the ProMS2:DPWΔN transgenic plants displayed defective pollen wall development, similar to that of the ms2 mutants (Figures 6C, 6F, 6H, and 6K). These results indicate that the N-terminal transit peptide sequence of DPW is required for its functioning in pollen wall development.
Recombinant DPW Has the Fatty Acyl ACP Reductase Activity
As shown in Figure 5, DPW is predicted to be a FAR involved in the formation of fatty alcohols. To test this, we generated a bacterial expression vector by inserting the full-length DPW cDNA into the pET30a vector (Novagen) and introducing this construct into Escherichia coli BL21(DE3). The recombinant strain containing pET-DPW or the empty vector as a control was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 48 h and shaken slowly at 20°C to improve expression of the soluble, active form of DPW. Then, recombinant DPW was purified by affinity chromatography and eluted from the column using 10 mM imidazole. In agreement with the predicted size, the molecular mass of the affinity-purified protein was estimated to be 65 kD by SDS-PAGE analysis (see Supplemental Figure 8 online). The DPW sample contained nonspecific protein bands, which were also observed in the control bacteria containing the pET30a vector (see Supplemental Figure 8 online).
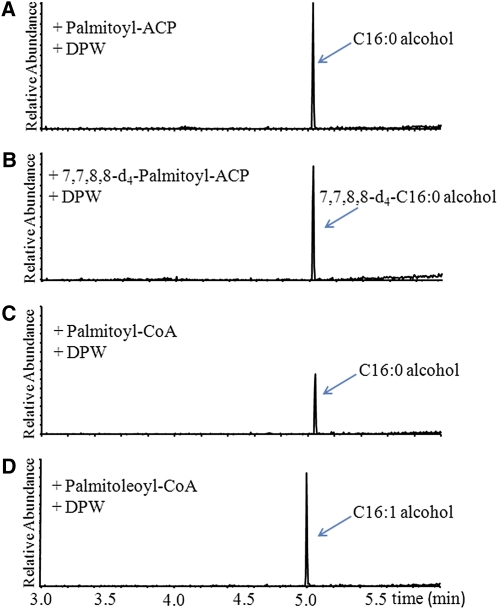
Long-chain fatty acids synthesized in plastids by fatty acid synthase are esterified to ACP, but long-chain acyl-ACPs are expected to be cleaved by an acyl-ACP thioesterase, exported from the plastid, and then linked to CoA (Li-Beisson et al., 2010). Since DPW localizes to the plastid, we tested the DPW activity against acyl-ACPs. A previous investigation suggested that jojoba FAR can use acyl-CoA as the substrate (Metz et al., 2000); therefore we also tested DPW activity against acyl-CoAs. Recombinant DPW converted palmiltoyl-ACP into hexadecanol (Figure 9A), which is confirmed using the deuterium-labeled 7,7,8,8-d4-palmitoyl-ACP as substrate, yielding a product at the same retention time as hexadecanol but with a mass ion 4 more than the d0hexadecanol standard (Figure 9B; see Supplemental Figure 9 online). Recombinant DPW could also reduce palmitoyl-CoA to produce C16:0 fatty alcohol (Figure 9C). DPW activity was not detectable in the absence of NADPH (see Supplemental Figure 10 online) or in the presence of NADH (see Supplemental Figure 11 online), suggesting the strict dependence of DPW activity on NADPH. In addition, C16:0 fatty alcohol was not formed when palmiltoyl-ACP, palmiltoyl-CoA, or palmiltoyl-ACP solution was incubated with control extracts obtained from cells containing only the empty pET30a vector. To investigate the substrate specificity of DPW, additional ACP and CoA compounds of saturated and unsaturated fatty acids with different chain lengths were tested (see Supplemental Figures 12 and 13 online). When palmitoleoyl-CoA was used as the substrate, C16:1 fatty alcohol was observed (Figure 9D). The identities of the generated fatty alcohols were confirmed by mass spectra (see Supplemental Figure 9 online). However, no derived alcohols were observed when ACP compounds, including myristoyl (14:0), palmitoleoyl (C16:1), and stearoyl (C18:0), or CoA compounds of decanoyl (C10:0), lauroyl (C12:0), myristoyl (14:0), stearoyl (C18:0), linoleoyl (C18:1), and oleoyl (C18:2) were tested individually (see Supplemental Figures 12 and 13 online). Moreover, the free palmitoyl and palmitoleoyl acids were not converted to fatty alcohol by the recombinant DPW enzyme (see Supplemental Figure 11 online). Table 2 shows the Km, Vmax, kcat, and kcat/Km values measured with affinity-purified recombinant DPW for palmiltoyl-ACP (C16:0), palmiltoyl-CoA (C16:0), and palmitoleoyl-CoA (C16:1) reduction. The specificity factor Km/kcat was >270-fold higher for palmitoyl-ACP versus palmitoyl-CoA, suggesting that DPW acts on acyl-ACPs rather than CoA esters in vivo. The large change in specificity factor is mostly accounted for by a much lower Km for the ACP ester, while the Vmax values are not significantly different. A similar discrimination between ACP esters and CoAs was reported for acyl-ACP desaturases (McKeon and Stumpf, 1982).
Figure 9.
Catalytic Activity Analysis of Recombinant DPW.
Gas chromatograms of metabolites generated from the incubation of palmitoyl-ACP (A), 7,7,8,8-d4-palmitoyl-ACP (B), palmitoyl-CoA (C), and palmitoleoyl-CoA (D) with affinity-purified DPW in the presence of NADPH.
[See online article for color version of this figure.]
Table 2.
Kinetic Properties of DPW
| Substrates | Km (μM) | Vmax (nmol min−1 mg−1) | kcat (s−1 10−3) | kcat/ Km (s−1 M−1) |
| Palmitoyl-ACP | 3.62 ± 0.79 | 4.54 ± 0.45 | 4.92 ± 0.49 | 1360 ± 136 |
| Palmitoyl-CoA | 1010.00 ± 310.00 | 4.32 ± 0.78 | 4.68 ± 1.1 | 4.67 ± 0.45 |
| Palmitoleoyl-CoA | 1150.00 ± 390.00 | 5.96 ± 1.1 | 5.72 ± 0.3 | 4.97 ± 1.2 |
Enzyme activity was measured by the assay with affinity-purified DPW and various concentrations of the listed substrates. All values represent the mean ± SD of three independent tests.
Expression of the Genes Involved in Anther and Pollen Wall Development in dpw
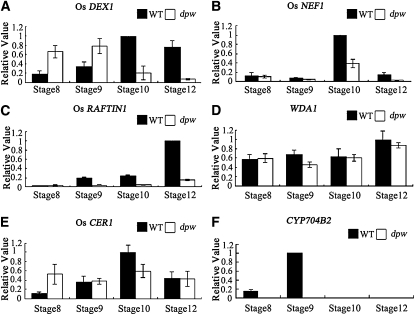
Because the dpw anther has an abnormal lipid composition and alteration in metabolites can modulate gene expression by feedback regulation, we postulated that the expression of genes critical for pollen wall and anther cuticle formation might be affected. To test this hypothesis, we examined the expression of the genes thought to be involved pollen wall and anther cuticle development in wild-type and mutant anthers. We first investigated the expression of the Os DEX1 gene, which is the putative homolog of Arabidopsis DEX1 with 64% amino acid sequence identity. DEX1 is a putative membrane-associated protein possibly functioning in primexine assembly (Paxson-Sowders et al., 2001). Quantitative RT-PCR (qRT-PCR) analysis revealed that Os DEX1 was highly expressed in wild-type rice anthers from stages 8 to 12, with maximal expression at stage 10, implying that Os DEX1 probably functions in rice pollen development. The Os DEX1 transcripts appeared to show earlier expression in the dpw mutant, with increased expression at stages 8 and 9 compared with the corresponding stages in the wild type, but greatly reduced expression at stages 10 and 12 (Figure 10A).
Figure 10.
Expression Analysis of Genes Related to Anther and Pollen Wall Synthesis Using qRT-PCR on Stage 8 to 12 Anthers.
Expression analysis of Os DEX1 (A), Os NEF1 (B), Os RAFTIN1 (C), WDA1 (D), Os CER1 (E), and CYP704B2 (F) in stages 8 to 12 anthers from the wild type (WT) and the dpw mutant using qRT-PCR. Rice Actin1 was used as a normalizer control. Error bars indicate sd, and each reaction had four biological repeats.
The Arabidopsis NEF1 gene is another regulator of primexine and exine development, which encodes a membrane-integrated protein in the tapetum plastid (Ariizumi et al., 2004). Os NEF1 shares 60% identity in amino acid sequence with NEF1; Os NEF1 expression was detected at high levels at stages 10 in the wild-type anther but was greatly decreased in the dpw mutant anther (Figure 10B). The sporophytic Os RAFTIN1 protein has been shown to accumulate in cereal Ubisch bodies and to be transported from the tapetum to the pollen exine (Wang et al., 2003), and transgenic plants with reduced Os RAFTIN1 expression due to RNA interference had aborted pollen development (Jung et al., 2006). Os RAFTIN1 is mainly expressed at late pollen developmental stage (stages 9 to 12) in the wild type, but its expression was virtually undetectable in dpw (Figure 10C). This result is consistent with the observation of abnormal Ubisch bodies in the dpw mutant at stage 10 (Figure 2H).
Compared with wild-type anthers, dpw anthers are smaller, similar in size to those of the wda1 mutant. The epicuticular structure is a mixture of long-chain fatty acid–derived substances (Kunst and Samuels, 2003). The rice WDA1 gene is mainly expressed in the anther epidermal cells and is thought to be involved in the decarbonylation pathway of very-long-chain fatty acid synthesis (Aarts et al., 1995; Jung et al., 2006; Bourdenx et al., 2011). We detected wild-type WDA1 expression from stage 8 to stage 12. The expression level of WDA1 was not greatly changed at the late pollen stage in the dpw mutant (Figure 10D). A putative WDA1 paralog, Os CER1 shares 54% amino acid sequence identity with Arabidopsis CER1 and is thought to be involved in the conversion of long-chain aldehydes to alkanes for wax biosynthesis (Aarts et al., 1995; Bourdenx et al., 2011). Expression of Os CER1 was detectable from stage 8 to stage 12 in the wild type; however, its expression level was increased only at stage 8 but decreased at stage 10 in the dpw mutant (Figure 10E).
CYP704B2 is a conserved cytochrome P450 family gene, which is critical for rice pollen wall and anther cuticle development (Li et al., 2010). CYP704B2 expression is specifically detected in the tapetum as well as in the microspores from stage 8 to stage 10. In dpw anthers, a dramatic reduction of CYP704B2 expression was observed (Figure 10F).
DISCUSSION
DPW Is Essential for Normal Anther Cuticle and Pollen Exine Development
Biosynthesis of fatty acid derivatives is required for reproductive development and fertility in higher plants, as supported by studies of wheat TAA1c (Wang et al., 2002), rice TDR (Li et al., 2006; Zhang et al., 2008), WDA1 (Jung et al., 2006), CYP703A3 (Aya et al., 2009), and CYP704B2 (Li et al., 2010; Li and Zhang, 2010), and Arabidopsis CER1 (Aarts et al., 1995), MS2 (Aarts et al., 1997), CYP703A2 (Morant et al., 2007), ACOS5 (de Azevedo Souza et al., 2009), CYP704B1 (Dobritsa et al., 2009; Yi et al., 2010), TKPR1/2 (Grienenberger et al., 2010), PKSA/LAP6, and PKSB/LAP5 (Dobritsa et al., 2010; Kim et al., 2010). Our results show that DPW is able to produce C16:0 alcohol, which is critical for pollen exine development. The spatio-temporal expression pattern of DPW in the tapetum from stage 8 to 10 is in good agreement with the role of the tapetum in synthesizing and secreting sporopollenin precursors. The dpw mutant has reduced sporopollenin deposition on the primexine of the pollen grain surface. The pollen wall patterning defect in dpw is probably due to abnormal tapetum development and function, with reduced numbers of Ubisch bodies generated from the tapetum. Recent evidence indicates that the wheat and rice tapetal RAFTIN protein is packaged into Ubisch bodies and incorporated into the pollen exine, and silencing of Os RAFTIN1 blocks rice pollen development (Wang et al., 2003). Os RAFTIN1 is expressed mainly during the late stages of anther development (Jung et al., 2006). Os RAFTIN1 expression was dramatically reduced in the dpw mutant. This is consistent with the observed accumulation of abnormal Ubisch bodies and less condensed tapetum with abnormally accumulated electron-dense structures that usually contain lipidic compounds (Yamamoto et al., 2003) in the dpw mutant.
Furthermore, mutation of DPW had significant effects on the gene expression patterns of other lipidic synthetic genes (e.g., CYP704B2 and Os CER1), which may account for the defect in the formation of the anther cuticle and pollen exine in dpw. The spatio-temporal expression pattern of CYP704B2 is similar to that of DPW, and its protein is putatively localized in the ER and catalyzes the hydroxylation of C16 and C18 fatty acids. cyp704B2 mutants display a defective pollen exine and an undeveloped anther cuticle (Li et al., 2010). Why and how the missing formation of C16:0 fatty alcohol by DPW in plastids affects CYP704B2 expression remains to be elucidated.
Even though the expression pattern of DPW is restricted to the tapetum and microspores, DPW is surprisingly critical for the development of the cuticle in anthers. It is interesting to observe that changes in lipid composition occurred in the dpw anther. There was a slight accumulation of soluble fatty acids and a significant reduction in the C16 fatty alcohols (P < 0.05) in the dpw mutant, which supports the biochemical role of DPW in the production of C16 primary alcohols from fatty acids during anther development. Residual C16 and C18 fatty alcohols in the dpw mutant may be explained by the expression of three other DPW putative paralogs (Os09g0567500, Os08g0557800, and Os08g0298700) in rice anthers (Hobo et al., 2008; Huang et al., 2009). Unexpectedly, upon cuticle analysis, we did not observe a concomitant accumulation of C16 monomers in the dpw anther but rather a reduction in cuticle C16 and C18 hydroxyacids, epoxyacids, and diacid monomers. The structural defect of the dpw anther cuticle is thus most likely explained by this significant reduction of all cutin monomers compared with the wild type (Figure 4C; see Supplemental Table 2 online). This same defect may also explain the major perturbation in sporopollenin formation and pollen development in the mutant. A slight increase in the levels of total wax monomers, C27, C29, C31, C33, and C35 alkenes, as well as C24, C26, and C18 alcohols is also detected in dpw. This may result from the feedback regulation in the mutant due to the defective synthesis of C16 fatty alcohols and/or to the rerouting of the precursors to an elongation/alkene pathway.
DPW Encodes a Fatty Acyl ACP Reductase
The framework of plant cuticle is determined by cutin, which is an insoluble polymer matrix containing the major constituents of hydroxylated and epoxy hydroxylated C16 and C18 fatty acids, as well as minor components such as fatty alcohols (Domínguez et al., 1999; Kolattukudy, 2001; Franke et al., 2005). Fatty alcohols with C16 and C18 account for 0 to 8% of cutin composition (Pollard et al., 2008). The alcohols naturally occur as free and esterified forms with various acyl groups, including aromatic and short (C2), long (C16 and C18), or very-long-chain aliphatic acids (Samuels et al., 2008). In plant reproductive organs, the surface of specialized anther cells accumulate abundant lipidic wax and cutin (Riederer and Schreiber, 2001). In this study, we demonstrate that DPW can reduce palmitoyl-ACP and CoA esters to 1-hexadecanol. It is perhaps surprising that DPW has very similar kinetic parameters for palmitoyl and palmitoleoyl CoAs, but that while palmitoyl-ACP is a better substrate than the CoAs, the enzyme shows no activity against palmitoleoyl-ACP. The palmitoleoyl-ACP in these experiments was 30 μM (i.e., an order of magnitude greater than the Km for palmitoyl-ACP). Lack of activity for palmitoleoyl-ACP likely reflects the fact that this substrate is present at low levels if at all in plastids.
The data presented here provide biochemical insight into the enzymes with high sequence similarity to the FARs from the green alga Euglena gracilis (Kolattukudy, 1970), jojoba (Metz et al., 2000), and Arabidopsis (Rowland et al., 2006; Doan et al., 2009; Domergue et al., 2010). The DPW protein may share a common catalytic function in lipid metabolism with other FARs described so far but differs from them in that it is mainly targeted to the plastids, and the purified DPW enzyme shows strong substrate preference for C16:0-ACP. The jojoba FAR is thought to contribute to the production of long-chain alcohols for wax accumulation in jojoba seeds, with recombinant enzyme producing C16:0 and C18:0 fatty alcohols (Metz et al., 2000). Arabidopsis CER4 is a key enzyme in the production of very-long-chain primary alcohols required for development of epidermal cells of aerial organs and roots, and yeast cells expressing Arabidopsis CER4 were able to generate C24:0 and C26:0 primary alcohols (Rowland et al., 2006). More recently, analysis of lipidic extracts of recombinant bacteria expressing five Arabidopsis FAR homologs revealed the formation of fatty alcohols with carbon lengths of C14, C16, and C18 from endogenous fatty acids. Production of C14:0, C16:0, and C18:1 alcohols was observed in bacteria expressing MS2 (Doan et al., 2009). Moreover, in yeast, recombinant FAR1, FAR4, and FAR5 are able to form alcohols using distinct but overlapping substrates with a chain length ranging from C18:0 to C24:0 (Domergue et al., 2010). In addition, yeast cells expressing silkmoth FAR could reduce fatty acids with 14 to 20 carbons in chain length to alcohols and preferentially to C15 and C16 fatty acids (Moto et al., 2003). Two mouse enzymes, FAR1 and FAR2, were also observed to convert fatty acyl-CoA ester substrates into fatty alcohols (Cheng and Russell, 2004).
DPW Defines Novel Pathways for Lipid Metabolism during Anther Development
Various types of plastids are found in different anther cells. Tapetal plastids are thought to play key roles in lipid synthesis and secretion for pollen wall synthesis during male gametophyte development (Clement and Pacini, 2001). Fatty acid biosynthesis starts from acetyl-CoA carboxylase, which catalyzes the conversion of acetyl-CoA to malonyl-CoA by carboxylation, an ATP-dependent reaction (Post-Beittenmiller et al., 1992). In plants, fatty acids with short carbon chain lengths of up to C18 are thought to be mainly synthesized in plastids (Ohlrogge et al., 1979; Li-Beisson et al., 2010). Subsequently, a portion of these fatty acids can be transported and elongated further in the ER. By sequence analysis and experimental evidence, we have shown that the DPW protein is localized predominantly in the plastids of tapetal cells during anther development. We also demonstrated that this plastidial localization relies on an N-terminal targeting signal peptide absolutely required for the DPW function in anther development as indicated by genetic complementation (Figure 6). A similar N-terminal transit peptide is observed in other DPW homologs, including Arabidopsis MS2, but not in the moss protein (see Supplemental Figure 4 online). Our findings thus seem to widen the metabolic functional range of plastids to include an important novel role in the conversion of fatty acids to fatty alcohols, which supports anther development in higher plants.
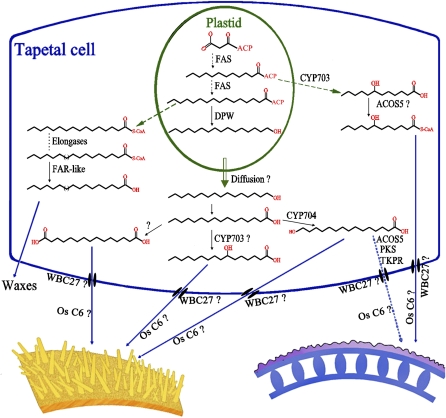
Here, we proposed a working model of how DPW regulates pollen exine and anther cuticle development (Figure 11). In plastids, fatty acids are synthesized by two enzyme systems: acetyl-CoA carboxylase and fatty acid synthase (Ohlrogge and Jaworski, 1997; Li-Beisson et al., 2010). Fatty acids are esterified to ACP as the main form in the plastid. Long-chain acyl-ACPs could be cleaved by an acyl-ACP thioesterase, transported from the plastid to ER, and then linked to CoA for direct elongation (Li-Beisson et al., 2010). The bulk of C16:0-ACP, however, seems to be converted to hexadecanol by DPW in plastids. This conversion into hexadecanol may be required for efficient export from the plastids, either via a transporter, membrane fusion, or possibly just via free diffusion through the lipidic phase into the cytoplasm (Spector and Soboroff, 1972). This model would imply subsequent oxygenation by P450 or other enzymes most likely in the ER to generate cytoplasmic fatty acids. Those enzymes could then be hydroxylated by fatty acid hydroxylases, such as CYP703As (Morant et al., 2007; Aya et al., 2009) and/or CYP704Bs (Dobritsa et al., 2009; Li et al., 2010). The derived hydroxylated fatty acids would then be available for conversion to acyl-CoAs by ACOS, such as ACOS5 (de Azevedo Souza et al., 2009), and the derived acyl-CoAs converted to polyketides by TKPR1/2 (Grienenberger et al., 2010), PKSA/LAP6 and PKSB/LAP5 (Kim et al., 2010). Fatty acids, alcohols, acyl-CoAs, polyketides, and other derived monomers may be exported from the tapetal cells by ATP binding cassette transporters, such as WBC27/ABCG26 (Choi et al., 2010; Quilichini et al., 2010; Xu et al., 2010; Dou et al., 2011), and/or lipid transport proteins, such as Os C6 (Zhang et al., 2010), into the locule and anther epidermis as sporopollenin and anther cuticle precursors.
Figure 11.
The Proposed Scheme of the Biosynthesis of Pollen Exine and Anther Cuticle.
In plastids, fatty acids are de novo synthesized and esterified to ACP. The long-chain acyl-ACPs could be cleaved, transported from the plastid to ER, and linked to CoA for direct elongation (Li-Beisson et al., 2010). C16:0-ACP is reduced to hexadecanol by DPW in plastids, and hexadecanol may freely diffuse or be transported into the lipidic phase in the cytoplasm (Spector and Soboroff, 1972). Hexadecanol may be further converted to fatty acid and oxygenated by P450 enzymes in the ER. The derived hydroxylated fatty acids may then be converted to acyl-CoAs by ACOS, such as ACOS5 (de Azevedo Souza et al., 2009), and the derived acyl-CoAs may be converted to polyketides by TKPR1/2 (Grienenberger et al., 2010), PKSA/LAP6, and PKSB/LAP5 (Kim et al., 2010). Fatty acids, alcohols, acyl-CoAs, polyketides, and other derived monomers may be translocated from the tapetal cells by ATP binding cassette transporters, such as WBC27/ABCG26 (Choi et al., 2010; Dou et al., 2011; Quilichini et al., 2010; Xu et al., 2010), and/or lipid transport proteins, such as Os C6 (Zhang et al., 2010), into the locule and anther epidermis as sporopollenin and anther cuticle precursors.
DPW Function Is Conserved among Plants
Production of fatty alcohols is a basic biological event for the generation of lipidic derivatives in organisms ranging from animals to plants. In this study, we identified and analyzed 24 putatively DPW-like proteins from lower plants, higher plants, insects, and mammals. Their phylogenetic relationship suggests that these genes probably share a common ancestor, highlighting the conserved role of DPW and their homologs in the production of fatty alcohols. The sequence conservation in Clade 2 also suggests that our analyses of DPW have uncovered a key step of fatty alcohol production common to land plants. In addition, the analysis of Arabidopsis transgenic plants carrying DPW indicates that it is a functionally equivalent ortholog of MS2.
In conclusion, we identified the rice DPW gene that encodes a fatty acyl ACP reductase, which we have shown to be principally localized to plastids. Loss of function of DPW caused abnormalities to the anther surface and pollen wall and reduction in the number of secretory, lipidic Ubisch bodies. Phylogenetic analyses and functional rescue experiments using DPW reveal a crucial pathway for fatty alcohol biosynthesis, which is essential for anther and pollen development and is conserved in both monocots and dicots.
METHODS
Mutant Material and Growth Conditions
All the plants were grown in the paddy field of Shanghai Jiao Tong University. The F2 mapping population was generated from a cross between dpw (Oryza sativa ssp japonica) and Guang Lu Ai (indica) for gene mapping.
Characterization of Mutant Phenotype
The photography of plant materials, nuclei staining using 4′,6-diamidino-2-phenylindole, and observation of anthers fixed in formaldehyde acetic acid using standard plastic sections and TEM were performed as described by Li et al. (2006). For scanning electron microscopy, fresh spikelets were coated with palladium-gold in a sputter coater (Hummer) and then observed in an autoscan scanning electron microscope (JEOL) with an acceleration voltage of 30 kV. Acetolysis was performed as described previously (Aarts et al., 1997).
Analysis of Anther Waxes, Cutin, and Total Soluble Lipids
Wax and cutin of anthers were analyzed as described previously (Jung et al., 2006; Li et al., 2010). To express amounts per unit surface area, we generated a ratio of anther dried weight to surface area (see Supplemental Figure 2 online). Area was determined from pixel numbers in microscopy images, assuming a cylindrical body for rice anthers. To extract waxes, ~10 mg of freeze-dried anther material corresponding to 125 to 350 mm2 of surface area were submersed in 1 mL chloroform for 1 min. The resulting chloroform extracts were spiked with 10 μg of tetracosane (Fluka) as internal standard and transferred to new vials. The solvents were evaporated under a gentle stream of nitrogen gas until a final volume of 100 μL was reached. Compounds containing free hydroxyl and carboxyl groups were converted to their trimethylsilyl ethers and esters by adding 20 μL of BSTFA (N,N-bis-trimethylsilyltrifluoroacitamide; Machery-Nagel) and 20 μL of pyridine to the extracts and incubating them for 40 min at 70°C. These derivatized samples were then analyzed by GC-FID (Agilent Technologies) and GC-MS (Agilent gas chromatograph coupled to an Agilent 5973N quadrupole mass selective detector). Results of anther wax analysis were related to unit surface area.
To extract all soluble lipids, anthers that had been used in the wax extraction were reextracted with 1 mL of chloroform/methanol (1:1 [v/v]). They were first incubated at 50°C for 30 min and then overnight with constant shaking at room temperature. This extraction was repeated three times to ensure that no soluble lipids were left in the anther samples. The combined lipid extracts were then evaporated under a gentle stream of nitrogen gas until they reached a final volume of 100 μL and used for analyzing total soluble lipids. The remaining delipidated anthers were dried over silica gel and used to analyze the monomer composition of cutin polyester as described by Franke et al. (2005). All soluble lipid samples and anther cutin samples were transesterified in 1 mL of 1 n methanolic HCl for 2 h at 80°C. After the addition of 2 mL of saturated NaCl/water and 20 μg of dotriacontane (Fluka) as internal standard, the hydrophobic monomers were subsequently extracted three times with 1 mL of hexane. The organic phases were combined, the solvent evaporated, and the remaining sample derivatized as described above. GC-MS and GC-FID analysis were performed as for the wax analysis. Results of anther cutin analysis were related to unit surface area, while soluble lipid analysis was related to unit dry weights of anthers.
Map-Based Cloning of the DPW Gene
For fine-mapping of the DPW locus, bulked segregant analysis was used to identify markers linked to DPW as described by Liu et al. (2005). The primer sequences for InDel markers are shown in Supplemental Table 3 online. The DPW locus was first mapped between two InDel molecular markers, OS302 and SJ302, on the short arm of chromosome 3. Then, 2160 F2 segregants from the mapping cross were generated, and three InDel markers (i.e., SJ622, CL7-4, and CL8-1) were used. DPW was finally defined between two of these InDel markers, SJ622 and CL7-4, within a 35-kb region (Figure 5). PCR was performed according to Chu et al. (2006). The PCR products were separated on 6% polyacrylamide denaturing gels, and bands were visualized by a silver-staining method (Liu et al., 2005).
Complementation of the dpw Mutant
For functional complementation of the rice dpw mutant, a genomic fragment of ~6.0 kb containing the entire DPW coding region, the 2283-bp upstream sequence, and 890-bp downstream sequence was digested from the BAC clone AC073556 with EcoRI and KpnI and subcloned into the binary vector pCAMBIA1301 with a hygromycin resistance marker to generate the p1301-DPW construct. Calli that were induced from homogenous dpw young panicles were used for transformation with Agrobacterium tumefaciens EHA105 carrying the p1301-DPW plasmid.
qRT-PCR Assay
Total RNA was isolated using Trizol reagent (Invitrogen), as described by the supplier, from rice tissues, including root shoot, leaf, glume, lemma, palea, and anthers at different stages. The developing stages of anthers were classified into the categories according to spikelet length and anther morphology (Feng et al., 2001; Zhang and Wilson, 2009). RT-PCR procedures were performed as described by Li et al. (2006). All the primers for RT-PCR are listed in Supplemental Table 4 online. PCR was performed with TaKaRa Ex Taq DNA polymerase for 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 58°C, and extension for 90 s at 72°C, followed by a final extension for 10 min. qRT-PCR was performed on a Rotor-Gene RG3000A detection system (Corbett Research) using SYBR Green I master mix (Generay Biotech). All PCR experiments were conducted using 40 cycles of 94°C for 20 s, 58°C for 20 s, and 72°C for 20 s, in a reaction mixture containing 10 pmol of each primer and 3 mM magnesium chloride and a 1:10 dilution of each cDNA pool (per biological replicate) as a template; all reactions were performed in triplicate, with Actin1 as the normalizer reference gene for all comparisons.
In Situ Hybridization
To generate gene-specific and control sense probes, a 750-bp DPW cDNA fragment from a DPW cDNA clone, pCMVFL3 (RGRC-NIAS; http://www.rgrc.dna.affrc.go.jp/stock.html), was digested with BamHI and HindIII, respectively, and transcribed in vitro under the T7 and SP6 promoters with RNA polymerase using the DIG RNA labeling kit (Roche). In situ hybridization was performed according to the protocol of Kouchi and Hata (1993).
Complementation of the ms2 Mutant
For functional complementation of the ms2 mutant, we cloned an ~1.1-kb MS2 promoter sequence from ecotype Columbia Arabidopsis, and DPW cDNA fragments with 1.827 kb (DPW) or 1.602 kb (without the 222-bp fragment encoding the N-terminal transit peptide, DPWΔN) from the cDNA clone AK121254. The MS2 promoter and DPW,DPWΔN fragments and the DPW fragment fused with GFP were subcloned into the binary vector pCAMBIA1301 to yield ProMS2:DPW, ProMS2:DPWΔN, and ProMS2:DPW-GFP, respectively. All the primers used for vector construction are listed in Supplemental Table 5 online. These constructs were introduced into Agrobacterium GV3101 and transformed into MS2/ms2 heterozygous plants. The transformed seeds were selected using hygromycin (50 mg/L), and transgenic ms2 homozygous lines were identified.
Polyclonal Antibody Preparation and Immunoblotting
The DPW fragment was amplified from the cDNA clone pCMVFL3 (RGRC-NIAS) using the primers PET-F and PET-R. The PCR product was cloned into the KpnI and EcoRI sites of a pET30a vector (Novagen) to produce pET30a-DPW. The fusion protein purification and antibody preparation were performed as described by Huang et al. (2003). The preimmune antiserum was collected before the first injection, and the antiserum was collected after three injections. The antibody was purified according to the method described by Ritter (1991).
Using extraction buffer (0.1 mmol/L Tris-HCl, 0.1 mmol/L EDTA, 0.15 mol/L NaCl, and 0.5% butanol, pH 7.5-8.0), cellular protein was extracted from the wild type and dpw mutant using the protocol described by Huang et al. (2003) and separated by 12% SDS-PAGE. After electrophoresis, the proteins were electronically transferred onto nitrocellulose membranes. The protein blot was blocked with 5% casein in Tris-buffered saline (TBS) Tween-20 buffer (20 mM Tris-HCl, pH 7.6, 0.8% NaCl, and 0.1% Tween 20) and incubated with the purified antibodies (with 1:50 dilution) for 1 h at room temperature as described by Huang et al. (2003).
Subcellular Localization of DPW
For transient expression, full-length DPW cDNA and DPWΔN were cloned into the pCAMBIA1301-GFP plasmid. Then, the control mitochondrial F1ATPase-γ-RFP fusion construct (kindly provided by Hyun-Sook Paic, Yonsei University, Korea) (Kim et al., 2006), DPW-GFP and DPWΔN-GFP constructs, and the construct containing GFP alone were introduced into rice protoplasts isolated from partly etiolated stems, by polyethylene glycol–mediated transformation (Bart et al., 2006). The DPW-GFP construct was transformed into wild-type Arabidopsis plants (Columbia-0). Cells that exhibited GFP and RFP fluorescence were examined under a fluorescence confocal microscope (Leica TCS SP5).
For immunolocalization of DPW, rice anthers of wild type at stage 9 (Zhang and Wilson, 2009) were fixed in 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.2 n sodium phosphate buffer, pH 7.0, and dehydrated in an ethanol gradient. Then, the anthers were infiltrated with LR-white resin (London Resin) and polymerized in gelatin capsules at 60°C for 24 h. Microtome sections were hydrated in deionized water, etched with 0.56 M NaIO4 and 0.1 n HCL, and washed for 15 min in PBS, 0.1% (v/v) Triton X-100, and 0.2% (v/v) Gly at pH 7.2. Then, the sections were blocked two times for 30 min each in 3% BSA and 0.1% Tween 20 in PBS and incubated overnight with the purified primary antibody at a 1/5 dilution or with preimmune serum diluted similarly as a control. After washing the sections in PBS and placing them for 40 min on TBS, pH 8.2, 0.2% (v/v) Tween, 0.2% (v/v) Triton X-100, and 0.1% (w/v) BSA, the sections were treated with the secondary antibody (10-nm colloidal gold-conjugated anti-rabbit IgG developed in goat; Sigma-Aldrich), which was diluted 1:50 with 1% BSA for 2 h (Wang et al., 2003). The sections were washed in TBS and deionized water and incubated with saturated uranyl acetate (30 min) and lead citrate (10 min). The sections were examined with a JEM-1230 transmission electron microscope (JEOL).
Phylogenetic Analysis
Multiple sequences were aligned with the ClustalX tool using default parameters, and a phylogenetic tree was constructed with the alignment of DPW-like protein sequences of rice and other species using the MEGA program (version 3.0; http://www.megasoftware.net/index.html) (Kumar et al., 2004) and the neighbor-joining method with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed).
Purification and Activity Assay of Recombinant DPW
Full-length DPW cDNA was inserted into the bacterial expression vector pET30a (Novagen), and this construct was introduced into Escherichia coli BL21(DE3). DPW expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 48 h and shaken slowly at 20°C to improve the yield of soluble and active recombinant DPW in E. coli.
For DPW purification, the collected bacterial cells were resuspended in a buffer (135 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and 8 mM K2HPO4, pH 7.4) containing 0.1 mM phenylmethanesulfonyl fluoride and lysed by sonication or in a French press at 4°C. After removal of cell debris by centrifugation (13,000 rpm, 4°C, 10 min), the recombinant His-tagged DPW was purified by affinity chromatography using a Ni2+-NTA column, following the manufacturer’s instructions. Bound proteins were eluted with sodium phosphate buffer containing increasing concentrations of imidazole and detected by 12% SDS-PAGE. Nonspecific proteins purified from the bacteria with the pET30a vector were used as control.
C16:0-ACP, 7,7,8,8-d4-C16:0-ACP, C14:0-ACP, C18:0-ACP, C16:1-ACP, and [1-14C]16:0-ACP were synthesized enzymatically (Rock and Garwin,1979) using recombinant spinach ACP-I (Broadwater and Fox, 1999). The fatty acyl-ACP reductase activity of DPW and the FAR activity of DPW were analyzed as described by Metz et al. (2000). The reaction mixture for the DPW assay contained 15 mM NADPH, 25 mM HEPES-NaOH, pH 7.5, 1 mM DTT, 1 mM EDTA, 10% (w/v) glycerol, and 0.3% (w/v) CHAPS, plus the desired substrate. The reaction was initiated by the addition of the enzyme, and for negative control experiments, an equivalent volume of eluant from the Ni2+-NTA column that had been incubated with protein expressing the empty pET30a vector was added. Incubations were performed at 30°C for 30 min and terminated by the addition of an equal volume of cyclohexane. The products were extracted into 1.2 volumes of hexane, and 2 μL of this extract was subjected to GC-MS analysis.
The Agilent GC was coupled with either an Agilent 5975C inert XL MSD with triple axis dector or an Agilent 5975C quadrupole mass selective detector. The detector and injector temperature were maintained at 280°C. The oven temperature of 75°C was increased to 320°C at a rate of 40°C min−1 and held at 320°C for 2 min. Alternatively, the initial oven temperature was programmed at 50°C for 2 min, increased to 180°C at a rate of 5°C min−1 for 2 min, and held at 180°C for 20 min.
[1-14C]16:0-ACP was used for kinetics analysis of DPW. The reaction was incubated at 30°C for 10 min and extracted twice with the use of 2 volumes of hexane, and the radioactivity of the hexane fraction was measured by liquid scintillation counting. As described above, extracts from bacteria expressing pET30a minus insert were used as the negative control.
Accession Numbers
Sequence data from this article for the mRNA and genomic DNA of DPW can be found in the GenBank/EMBL data libraries under accession numbers NM_001055618 and AC073556, respectively. GenBank accession numbers of all genes used in this study are NM_001058293 (Os DEX1), NM_001074533 (Os NEF1), NM_001068672 (Os RAFTIN1), NM_001053994 (Os CER1), NM_001071361 (WDA1), and NM_001055627 (CYP704B2). Accession numbers for the sequences used in the phylogenetic analysis are listed on the tree and the legend of Figure 5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DAPI Staining of the dpw Microspores.
Supplemental Figure 2. Weight/Surface Area Ratios of Wild-Type and dpw Anthers.
Supplemental Figure 3. Complementation of the dpw Mutant by DPW Genomic DNA.
Supplemental Figure 4. Sequence Alignment of DPW and 24 DPW-Related Proteins.
Supplemental Figure 5. Sequence Alignment of DPW and MS2 Proteins.
Supplemental Figure 6. The Subcellular Localization of Free GFP in Rice Protoplasts.
Supplemental Figure 7. Immunobloting Analysis of DPW.
Supplemental Figure 8. SDS-PAGE Analysis of Affinity-Purified DPW.
Supplemental Figure 9. The Mass Spectra of Fatty Alcohols.
Supplemental Figure 10. Catalytic Activity Analysis of Recombinant DPW Dependent on NADPH.
Supplemental Figure 11. Evaluation of the Recombinant DPW Activity Using Free Fatty Acids and NADH.
Supplemental Figure 12. Substrate Specificity Analysis of Recombinant DPW Using Acyl-ACPs.
Supplemental Figure 13. Substrate Specificity Analysis of Recombinant DPW.
Supplemental Table 1. Wax Composition of the Wild-Type and dpw Anthers.
Supplemental Table 2. Cutin Composition of the Wild-Type and dpw Anthers.
Supplemental Table 3. Primers Used for DPW Mapping.
Supplemental Table 4. Primers Used for RT-PCR Analyses.
Supplemental Table 5. Primers Used for Vector Construction.
Supplemental Data Set 1. Sequences Used to Generate the Phylogenetic Tree Presented in Figure 5D.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers, D. Werck-Reichhart, Z. Wilson, and Y. Zhang for helpful comments, H.S. Paic for providing F1ATPase-γ:RFP fusion construct, and B. Han and the RGRC for providing the BAC clone and the cDNA clone, respectively. We thank Z.J. Luo and M.J. Chen for mutant screening and generation of F2 populations for the mapping, C.M. Zhang for rice transformation, X.Y. Gao for plastic sections, scanning electron microscopy and TEM, W.W. Li and W. Kong for vector construction and transformation, C.S. Yin and D.S. Zhang for in situ and immunological assay, X. Yao for bioinformatics analysis, Y.M. Liu and W. Jia for lipidic testing, H. Li for figure preparation, and Z. Yuan for discussion. This work was supported by funds from the National Key Basic Research Developments Program of the Ministry of Science and Technology, the People’s Republic of China (2009CB941500 and 2007CB108700), the National “863” High-Tech Project (2006AA10A102), the National Natural Science Foundation of China (30725022 and 90717109), the Shanghai Leading Academic Discipline Project (B205), a Deutsche Forschungsgemeinshaft grant to L.S., the National Science Foundation (Grant DBI 0701919) for support of X.-H.Y., and the Office of Basic Energy Sciences of the U.S. Department of Energy to J.S.
References
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Aarts M.G., Keijzer C.J., Stiekema W.J., Pereira A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7: 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlers F., Thom I., Lambert J., Kuckuk R., Rolf W. (1999). 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 50: 1095–1098 [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Sato S., Kato T., Tabata S., Toriyama K. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Aya K., Ueguchi-Tanaka M., Kondo M., Hamada K., Yano K., Nishimura M., Matsuoka M. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P. (1992). The remarkable biology of pollen. Plant Cell 4: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S., Wortley A.H., Skvarla J.J., Rowley J.R. (2007). Pollen wall development in flowering plants. New Phytol. 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Bonaventure G., Beisson F., Ohlrogge J., Pollard M. (2004). Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 40: 920–930 [DOI] [PubMed] [Google Scholar]
- Broadwater J.A., Fox B.G. (1999). Spinach holo-acyl carrier protein: Overproduction and phosphopantetheinylation in Escherichia coli BL21(DE3), in vitro acylation, and enzymatic desaturation of histidine-tagged isoform I. Protein Expr. Purif. 15: 314–326 [DOI] [PubMed] [Google Scholar]
- Bourdenx B., Bernard A., Domergue F., Pascal S., Léger A., Roby D., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Joubes J. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J., Shaw G. (1968). Chemical structure of the exine of pollen walls and a new function for carotenoids in nature. Nature 219: 532–533 [DOI] [PubMed] [Google Scholar]
- Bubert H., Lambert J., Steuernagel S., Ahlers F., Wiermann R. (2002). Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Z. Naturforsch. C. 57: 1035–1071 [DOI] [PubMed] [Google Scholar]
- Chaloner W. (1976). The evolution of adaptive features in fossil exines. In Evolutionary Significance of the Exine, Ferguson I.K, Muller J., (London: Academic Press; ), 1–14 [Google Scholar]
- Chen L., Chu H.W., Yuan Z., Pan A.H., Liang W.Q., Huang H., Shen M.S., Zhang D.B., Chen L. (2006). Isolation and genetic analysis for rice mutants treated with 60 Co γ-ray. J. Xiamen Univ. 45: 82–85 [Google Scholar]
- Cheng J.B., Russell D.W. (2004). Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 279: 37789–37797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Jin J.Y., Choi S., Hwang J.U., Kim Y.Y., Suh M.C., Lee Y. (2010). An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 65: 181–193 [DOI] [PubMed] [Google Scholar]
- Chu H.W., Qian Q.Q., Liang W.Q., Yin C.S., Tan H.X., Yao X., Yuan Z., Yang J., Huang H., Luo D., Ma H., Zhang D.B. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142: 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C., Pacini E. (2001). Anther plastids in angiosperms. Bot. Rev. 67: 54–73 [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Lei Z.T., Nishikawa S.I., Urbanczyk-Wochniak E., Huhman D.V., Preuss D., Sumner L.W. (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Møller B.L., Preuss D. (2009). CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T.T., Carlsson A.S., Hamberg M., Bülow L., Stymne S., Olsson P. (2009). Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 166: 787–796 [DOI] [PubMed] [Google Scholar]
- Domergue F., Vishwanath S.J., Joubès J., Ono J., Lee J.A., Bourdon M., Alhattab R., Lowe C., Pascal S., Lessire R., Rowland O. (2010). Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 153: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez E., Mercado J.A., Quesada M.A., Heredia A. (1999). Pollen sporopollenin: Degradation and structural elucidation. Sex. Plant Reprod. 12: 171–178 [Google Scholar]
- Dou X.Y., Zhang K.Z., Zhang Y., Wang W., Liu X.L., Chen L.Q., Zhang X.Q., Ye D. (2011). WBC27, an ATP-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J. Integrative Plant Biol. 53: 74–88 [DOI] [PubMed] [Google Scholar]
- Feng J.H., Lu Y.G., Liu X.D., Xu X.B. (2001). Pollen development and its stages in rice (Oryza sativa L.). Chinese J. Rice Sci. 15: 21–28 [Google Scholar]
- Franke R., Briesen I., Wojciechowski T., Faust A., Yephremov A., Nawrath C., Schreiber L. (2005). Apoplastic polyesters in Arabidopsis surface tissues—A typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Grienenberger E., Kim S.S., Lallemand B., Geoffroy P., Heintz D., Souza Cde.A., Heitz T., Douglas C.J., Legrand M. (2010). Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R.B., Beals T.P., Sanders P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T., et al. (2008). Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol. 49: 1417–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.F., Tan H.X., Liang W.Q., Zhang D.B. (2010). The Post-meiotic Deficicent Anther1 (PDA1) gene is required for post-meiotic anther development in rice. J. Genet. Genomics 37: 37–46 [DOI] [PubMed] [Google Scholar]
- Hu S.W., Fan Y.F., Zhao H.X., Guo X.L., Yu C.Y., Sun G.L., Dong C.H., Liu S.Y., Wang H.Z. (2006). Analysis of MS2Bnap genomic DNA homologous to MS2 gene from Arabidopsis thaliana in two dominant digenic male sterile accessions of oilseed rape (Brassica napus L.). Theor. Appl. Genet. 113: 397–406 [DOI] [PubMed] [Google Scholar]
- Huang M.D., Wei F.J., Wu C.C., Hsing Y.I., Huang A.H. (2009). Analyses of advanced rice anther transcriptomes reveal global tapetum secretory functions and potential proteins for lipid exine formation. Plant Physiol. 149: 694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.H., Liang W.Q., Pan A.H., Zhou Z.A., Huang C., Chen J.Q., Zhang D.B. (2003). Production of FaeG, the major subunit of K88 fimbriae, in transgenic tobacco plants and its immunogenicity in mice. Infect. Immun. 71: 5436–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans S., El-Ghazaly G., Smets E. (1998). Orbicules in angiosperms: Morphology, function, distribution, and relation with tapetum types. Bot. Rev. 64: 240–272 [Google Scholar]
- Jung K.H., Han M.J., Lee D.Y., Lee Y.S., Schreiber L., Franke R., Faust A., Yephremov A., Saedler H., Kim Y.W., Hwang I., An G. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Lim J.H., Ahn C.S., Park K., Kim G.T., Kim W.T., Pai H.S. (2006). Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.S., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy P.E. (1970). Reduction of fatty acids to alcohols by cell-free preparations of Euglena gracilis. Biochemistry 9: 1095–1102 [DOI] [PubMed] [Google Scholar]
- Kolattukudy P.E. (2001). Polyesters in higher plants. Adv. Biochem. Eng. Biotechnol. 71: 1–49 [DOI] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Kunst L., Samuels A.L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Li H., Pinot F., Sauveplane V., Werck-Reichhart D., Diehl P., Schreiber L., Franke R., Zhang P., Chen L., Gao Y.W., Liang W.Q., Zhang D.B. (2010). Cytochrome P450 family member CYP704B2 catalyzes the omega-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yuan Z., Vizcay-Barrena G., Yang C.Y., Liang W.Q., Zong J., Wilson Z.A., Zhang D.B. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhang D.B. (2010). Biosynthesis of anther cuticle and pollen exine in rice. Plant Signal. Behav. 5: 1121–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y.H., et al. (2010). Acyl-lipid metabolism. In The Arabidopsis Book 8: e0133, doi/10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.S., et al. (2005). Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (Osms-l) mutant. Chin. Sci. Bull. 50: 122–125 [Google Scholar]
- McKeon T.A., Stumpf P.K. (1982). Purification and characterization of the stearoyl-acyl carrier protein desaturase and the acyl-acyl carrier protein thioesterase from maturing seeds of safflower. J. Biol. Chem. 257: 12141–12147 [PubMed] [Google Scholar]
- Metz J.G., Pollard M.R., Anderson L., Hayes T.R., Lassner M.W. (2000). Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 122: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuter-Gerhards A., Riegart S., Wiermann R. (1999). Studies on sporopollenin biosynthesis in Curcurbita maxima (DUCH)-II: The involvement of aliphatic metabolism. J. Plant Physiol. 154: 431–436 [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B.L., Werck-Reichhart D., Bak S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moto K., Yoshiga T., Yamamoto M., Takahashi S., Okano K., Ando T., Nakata T., Matsumoto S. (2003). Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori. Proc. Natl. Acad. Sci. USA 100: 9156–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J.B., Jaworski J.G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J.B., Kuhn D.N., Stumpf P.K. (1979). Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc. Natl. Acad. Sci. USA 76: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders D.M., Dodrill C.H., Owen H.A., Makaroff C.A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders D.M., Owen H.A., Makaroff C.A. (1997). A comparative ultrastructural analysis of exine pattern development in wild-type Arabidopsis and a mutant defective in pattern formation. Protoplasma 198: 53–65 [Google Scholar]
- Piffanelli P., Ross J.H., Murphy D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11: 65–80 [Google Scholar]
- Pollard M., Beisson F., Li Y., Ohlrogge J.B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D., Roughan G., Ohlrogge J.B. (1992). Regulation of plant fatty acid biosynthesis: Analysis of acyl-coenzyme A and acyl-acyl carrier protein substrate pools in spinach and pea chloroplasts. Plant Physiol. 100: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Friedmann M.C., Samuels A.L., Douglas C.J. (2010). ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M., Schreiber L. (2001). Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 52: 2023–2032 [DOI] [PubMed] [Google Scholar]
- Ritter K. (1991). Affinity purification of antibodies from sera using polyvinylidenedifluoride (PVDF) membranes as coupling matrices for antigens presented by autoantibodies to triosephosphate isomerase. J. Immunol. Methods 137: 209–215 [DOI] [PubMed] [Google Scholar]
- Rock C.O., Garwin J.L. (1979). Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J. Biol. Chem. 254: 7123–7128 [PubMed] [Google Scholar]
- Rowland O., Zheng H., Hepworth S.R., Lam P., Jetter R., Kunst L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Dickinson H.G. (2004). Stamen structure and function. Plant Cell 16 (suppl): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector A.A., Soboroff J.M. (1972). Studies on the cellular mechanism of free fatty acid uptake using an analog, hexadecanol. J. Lipid Res. 13: 790–796 [PubMed] [Google Scholar]
- Wang A., Xia Q., Xie W., Datla R., Selvaraj G. (2003). The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. USA 100: 14487–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Xia Q., Xie W., Dumonceaux T., Zou J., Datla R., Selvaraj G. (2002). Male gametophyte development in bread wheat (Triticum aestivum L.): Molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. Plant J. 30: 613–623 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y.F., Zhang D.B. (2006). [Identification of the rice (Oryza sativa L.) mutant msp1-4 and expression analysis of its UDT1 and GAMYB genes]. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 32: 527–534 [PubMed] [Google Scholar]
- Wilson Z.A., Morroll S.M., Dawson J., Swarup R., Tighe P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Xu J., Yang C.Y, Yuan Z., Zhang D.S., Gondwe M.Y., Ding Z.W., Liang W.Q., Zhang D.B., Wilson Z.A. (2010). The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Nishimura M., Hara-Nishimura I., Noguchi T. (2003). Behavior of vacuoles during microspore and pollen development in Arabidopsis thaliana. Plant Cell Physiol. 44: 1192–1201 [DOI] [PubMed] [Google Scholar]
- Yi B., Zeng F.Q., Lei S., Chen Y., Yao X., Zhu Y., Wen J., Shen J., Ma C., Tu J., Fu T. (2010). Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 63: 925–938 [DOI] [PubMed] [Google Scholar]
- Zhang D.B., Wilson Z.A. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 2342–2353 [Google Scholar]
- Zhang D.S., Liang W., Yin C., Zong J., Gu F., Zhang D. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.S., Liang W.Q., Yuan Z., Li N., Shi J., Wang J., Liu Y.M., Yu W.J., Zhang D.B. (2008). Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zinkl G.M., Zwiebel B.I., Grier D.G., Preuss D. (1999). Pollen-stigma adhesion in Arabidopsis: A species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.