Abstract
AUXIN BINDING PROTEIN1 (ABP1) is one of the first characterized proteins that bind auxin and has been implied as a receptor for a number of auxin responses. Early studies characterized its auxin binding properties and focused on rapid electrophysiological and cell expansion responses, while subsequent work indicated a role in cell cycle and cell division control. Very recently, ABP1 has been ascribed a role in modulating endocytic events at the plasma membrane and RHO OF PLANTS-mediated cytoskeletal rearrangements during asymmetric cell expansion. The exact molecular function of ABP1 is still unresolved, but its main activity apparently lies in influencing events at the plasma membrane. This review aims to connect the novel findings with the more classical literature on ABP1 and to point out the many open questions that still separate us from a comprehensive model of ABP1 action, almost 40 years after the first reports of its existence.
INTRODUCTION
Given the variety of reports that implicate AUXIN BINDING PROTEIN1 (ABP1) in a collection of processes ranging from protoplast swelling to cell cycle control, a pessimist would state that almost 40 years after the first glimpse of ABP1, its function still remains largely unclear. Indeed, despite enormous research efforts, until today, ABP1 has kept most of its secrets. The reason might be partially that while plant biology advanced, the available experimental systems and approaches to elucidate ABP1 function also underwent major changes, creating a quite heterogeneous array of data points in which the connections are not always obvious. The main culprit, however, is the manifold nature of auxin responses that are the outcome of a highly complex network of auxin synthesis, transport, sensing, and signaling events in which many of the components themselves are regulated by auxin and form potential feedback loops. Hence, the specific biological role of an individual component is often difficult to isolate.
Auxin signaling as understood today is to a large extent governed by SKP-Cullin-F box (SCF)TIR1/AFB (for TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX)-dependent derepression of AUXIN RESPONSE FACTOR (ARF) transcription factors, and the resulting transcriptional regulation of response genes. In this pathway, the F box protein TIR1 and the closely related AFB proteins act as auxin receptors (Dharmasiri et al., 2005; Kepinski and Leyser, 2005), or more correctly, as coreceptors, because for high affinity binding, a complex with proteins of the AUXIN/INDOLE ACETIC ACID (Aux/IAA) family is needed (reviewed in Calderon-Villalobos et al., 2010). However, it has become clear that there are auxin responses that do not utilize this pathway, especially some very rapid responses at the plasma membrane. In this review, we will briefly outline some cornerstones of ABP1 research to provide the necessary context before we discuss the more recent findings that add some new, exciting twists to the story of an auxin receptor on the eve of the 40th anniversary of its discovery.
TIMELINE OF ABP1 RESEARCH
In the early phase of ABP1 research, the dominant model system was maize (Zea mays), as auxin binding activity was first reported in maize coleoptile membrane preparations (Hertel et al., 1972). The biochemical properties of this binding activity were characterized (Batt and Venis, 1976; Batt et al., 1976; Ray, 1977; Venis, 1977), but it was not until 1985 that Zm-ABP1 protein was purified (Löbler and Klämbt, 1985) and subsequently molecularly cloned (Hesse et al., 1989; Inohara et al., 1989; Tillmann et al., 1989). In the same year, it was also proven that ABP1 indeed binds auxin (Jones and Venis, 1989), a result that was later corroborated by the crystal structure determination of ABP1 cocrystallized with auxin (Woo et al. 2002). ABP1 antibodies and peptides generated in these biochemical studies were increasingly used for physiological studies in heterologous systems, showing auxin induction of cellular processes such as membrane hyperpolarization (Barbier-Brygoo et al., 1991; Venis et al., 1992), K+ fluxes (Thiel et al., 1993), or cell expansion, as evidenced by protoplast swelling assays (Steffens et al., 2001). The importance and function of ABP1 for the plant as a whole remained unclear because attempts to modulate ABP1 protein amounts (particularly, its' downregulation) proved difficult in planta. Data from ABP1 overexpression in tobacco (Nicotiana tabacum) suggested a role in cell expansion (Jones et al., 1998; Chen et al., 2001a), and heterologous expression of Zm-ABP1 in tobacco further corroborated a role in auxin-responsive K+ currents (Bauly et al., 2000).
With the advent of Arabidopsis thaliana as a model system, an abp1-null mutant was identified as embryo lethal and, thus, ABP1 was defined as an essential protein (Chen et al., 2001b). Unfortunately, this also limited its use for further functional studies. Attempts to downregulate ABP1 by antisense or clonal knock-out approaches met with little success, and attention was further driven away from ABP1 as an auxin receptor by the elucidation of the Aux/IAA-ARF-SCF auxin signaling pathway and the identification of the F box protein TIR1 (Dharmasiri et al., 2005; Kepinski and Leyser, 2005) as auxin receptors. A rapidly increasing body of data suggested that most macroscopic auxin responses could be attributed to a combination of genetic regulation via this pathway and local auxin accumulation caused by local synthesis and its polar, cell-to-cell transport by plasma membrane transport proteins, such as PIN-FORMED (PIN), AUXIN-RESISTANT1/LIKE AUX1 or MULTIDRUG-RESISTANT/P-GLYCOPROTEIN (reviewed in Vieten et al., 2007).
However, genetic regulation by Aux/IAA-ARF-SCFTIR1/AFB could not account for some auxin responses, namely, the fast reactions such as cell expansion and modification of ion fluxes at the plasma membrane that occur within minutes (Figure 1) and the rapid endocytosis inhibition of a number of plasma membrane proteins by auxin (Paciorek et al., 2005; Figure 2). Finally, attempts succeeded in downregulating ABP1 levels by the use of immunomodulation and antisense approaches, hinting at an involvement in the cell cycle and cell expansion (David et al., 2007; Braun et al., 2008). Recently, in planta studies have implicated ABP1 in two fundamental auxin-sensitive cell biological processes, cytoskeleton rearrangement via a Rho-GTPase-dependent (ROP) pathway (Xu et al., 2010; Figure 3) and clathrin-mediated endocytosis (Robert et al., 2010; Figure 2).
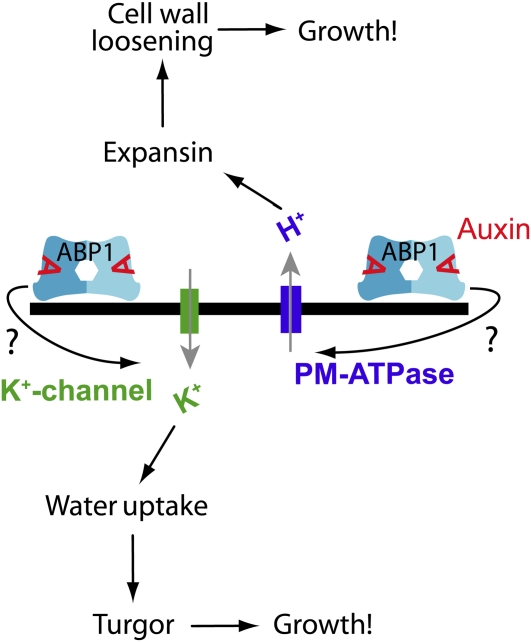
Figure 1.
ABP1 Regulates Cellular Growth.
ABP1 rapidly stimulates the activity of the plasma membrane-ATPase and K+-channels by an as yet unknown pathway. The plasma membrane-ATPase will lead to the acidification of the extracellular space, which in turn activates pH-sensitive expansion, causing cell wall loosening. Simultaneously, K+ influx through inward rectifying-K+-channels will ultimately lead to enhanced cellular water uptake. Both processes jointly allow for turgor-induced growth.
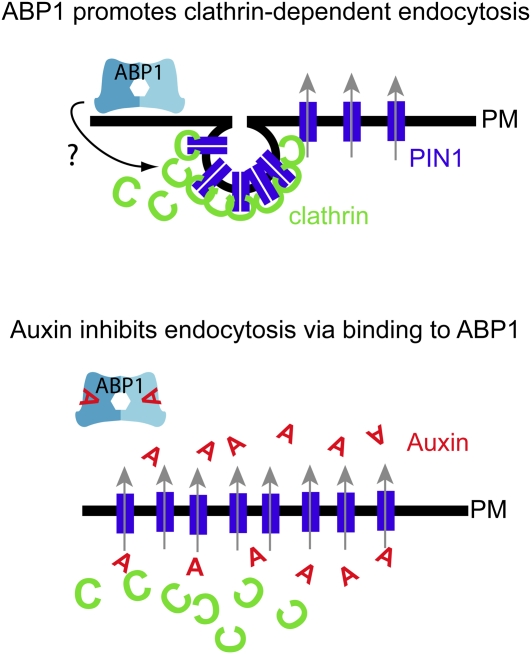
Figure 2.
ABP1 Regulates Clathrin-Dependent Endocytosis.
Unbound extracellular ABP1 appears to promote clathrin-dependent endocytosis (top scheme). Auxin binding to ABP1 interferes with this activity and hence reduces clathrin-dependent endocytosis, leading to higher retention of PIN proteins and, subsequently, higher auxin efflux rates (bottom scheme).
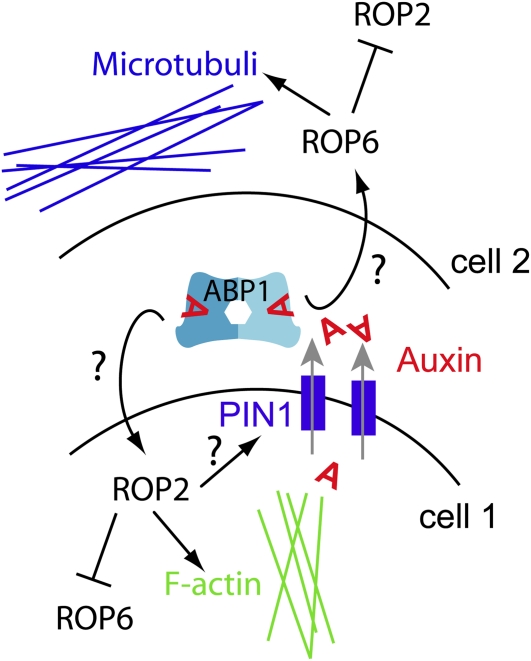
Figure 3.
ABP1 Regulates the Cytoskeleton.
Auxin binding to ABP1 in the cell wall appears to regulate the activity of the small GTPases ROP2 and ROP6 during interdigitative epidermal growth. ROP6 regulates the microtubuli cytoskeleton (top, indented cell 2), whereas ROP2 regulates the formation of actin filaments and promotes PIN1 localization at the plasma membrane (bottom, lobed cell 1). PIN polarization provides a positive feedback via increasing extracellular auxin concentration. ROP2 and ROP6 repress each other and are activated by auxin-bound ABP1 via distinct kinetics.
Although the data are insufficient to build a comprehensive model of ABP1 function at the molecular level, we now have some new concepts that need to be explored by further experimental work. At the same time, it is without doubt that ABP1 fulfills the criteria for an auxin receptor: It binds auxin specifically and saturably, it is required for specific physiological responses, and it has a rate-limiting function in these responses. In this review, we aim to connect the most recent data with the more classical literature on ABP1, and to highlight some of the many remaining unanswered questions.
IN AND OUT: ABP1 LOCALIZATION
There is ample evidence that in developing tissues, auxin serves as a short-range signal to coordinate cell/tissue polarity and patterning events (Sachs, 1969; Benková et al., 2003; Friml et al. 2003; Reinhardt et al., 2003; Sauer et al., 2006; Wabnik et al., 2011), somehow implying that cells are able to sense the positional information via an auxin signal. If this positional sensing were achieved by binding of auxin to a receptor, then this receptor should be located at the cell periphery. As TIR1/AFBs reside in the nucleus and thus are unlikely receptors for directional cues, the localization of ABP1 deserves special attention.
The canonical ABP1 of flowering plants contains an N-terminal signal peptide for entry into the secretory pathway and a C-terminal endoplasmic reticulum (ER) retention sequence of the KDEL type (Lys-Asp-Glu-Leu) (Hesse et al., 1989; Inohara et al., 1989; Tillmann et al., 1989). Other, less common types of ER retention signals exist in ABP1 of ferns and gymnosperms (Tromas et al., 2010), whereas ABP1 orthologs from several moss species do not contain a known ER retention motif (Panigrahi et al., 2009), and not all clades of green algae have bona fide ABP1 orthologs (Tromas et al., 2010). Although the functional importance and requirement of ER retention motifs in mosses and algae is unknown, numerous reports from seed plants indeed confirm that the bulk of ABP1 is in the ER lumen (Jones and Herman, 1993; Henderson et al., 1997). N-Glycosylation of ABP1, which contains one to several such motifs in its sequence (Massotte et al., 1995), most likely takes place here.
Despite the clear evidence for ABP1 as an ER-resident protein, there is little doubt from experimental data and theoretical considerations, that some ABP1 is secreted to the plasma membrane and/or the extracellular matrix. Small amounts of ABP1 have been visualized by immunocytochemistry close to the plasma membrane on the inner (cytoplasmic) side and on the outer side, extending into the cell wall (Jones and Herman, 1993; Diekmann et al., 1995). Henderson et al. (1997) failed to detect ABP1 in the cell wall, but they cautioned that ABP1 amounts might be at the detection limit of direct visualization. Secretion of ABP1 has been demonstrated using several different types of cell culture, and ABP1 could readily be found in the culture medium, albeit in small quantities (Jones and Herman, 1993; Henderson et al., 1997; Bauly et al., 2000). The mechanism for secretion is unclear, but the quantities are so low that they may be attributed to a simple escape from the ER retention machinery and further transport in the general secretory pathway. In line with this assumption, the vesicle transport inhibitor brefeldin A was found to inhibit ABP1 secretion into the culture medium (Jones and Herman, 1993).
For some time, auxin was suspected to affect secretion by binding to and masking the KDEL motif, but no experimental evidence for this hypothesis could be found (Tian et al., 1995; Henderson et al., 1997). The only effect of auxin on ABP1 localization reported so far is a clustering of ABP1 at the outer membrane of maize coleoptile protoplasts after auxin treatment (Diekmann et al., 1995). Interestingly, Jones and Herman (1993) reported that auxin starvation of cell cultures seemed to increase ABP1 secretion into the medium, but no other study followed up on this observation.
Additional indirect evidence for ABP1 in the cell wall or at the outer side of the plasma membrane comes from physiological assays that have utilized the full protein, peptides, or antibodies against ABP1 to inhibit or mimic rapid auxin effects such as protoplast swelling or electrophysiological responses (Barbier-Brygoo et al., 1991; Venis et al., 1992; Thiel et al., 1993; Steffens et al., 2001). Neither antibody fragments nor peptides can pass the plasma membrane easily by diffusion, thus, if their mode of action is indeed by binding ABP1 or a supposed signaling component, this must take place in the extracellular matrix.
The fact that some of the studies were performed in protoplasts in which the cell wall is mostly degraded suggests that at least some fraction of ABP1 is attached to the outer side of the plasma membrane rather than freely diffusing among the cell wall material. A putative ABP1 interactor or docking protein was identified in maize as an extracellular, glycosylphosphotidylinositol-anchored plasma membrane protein with similarity to Arabidopsis SKEWED5 (SKU5) and SKU5 SIMILAR6 (SKS6) (Shimomura, 2006), but no follow-up study has confirmed such an interaction in Arabidopsis, nor an influence of these proteins on ABP1 abundance at the outer plasma membrane. The sku5 loss-of-function mutant phenotype of root twisting (Sedbrook et al., 2002) has little in common with the ABP1 downregulated lines, although analysis of heterozygous abp1/+ mutants also revealed a slight root slanting (Effendi et al., 2011). sks6 knock-out plants are defective in cotyledon vasculature (Jacobs and Roe, 2005), which might also be found also in ABP1 downregulated lines (unreferenced comment in Tromas et al., 2010). This evidence, although weak and circumstantial, deserves closer inspection. Undoubtedly, further approaches must be taken to identify ABP1 interactors at the plasma membrane, taking into account the special chemical properties of the extracellular matrix as an interaction environment. Recently, an interaction between ABP1 and the ER-localized E3 ubiquitin ligase RING MEMBRANE-ANCHOR2 (At-RMA2) has been reported (Son et al., 2010) (see below).
pH is a critical parameter for ABP1 auxin affinity. The pH in the ER lumen is estimated to be close to 7.0, and ABP1 affinity to auxin is nearly zero at this pH (Tian et al., 1995). The highest affinity was determined at approximately pH 5.5 (Löbler and Klämbt, 1985; Shimomura et al., 1986; Tian et al., 1995), which is a typical pH of the extracellular matrix. Purified Zm-ABP1 was found to bind specifically and saturatably to plasma membrane vesicles, independently of auxin concentrations (Schiebl et al., 1997). Again, the binding optimum occurred approximately at a pH of 5.0, which is typical of the extracellular space. Nevertheless, because the exact type of vesicles generated in this study was unclear (i.e., outside-in or inside-out), it cannot be concluded definitively that the observed binding was specific for a putative extracellular ABP1-docking protein.
With all the direct and indirect evidence, a proportion of ABP1—however small—is assumed generally to be secreted to the extracellular space. Whether the ER-resident ABP1 represents an inactive pool of proteins that awaits activation in the extracellular space or whether ER luminal and extracellular ABP1 have distinct functions remain unresolved.
AUXIN RESPONSES: ABP1 AMONG OTHER AUXIN RECEPTORS
Many proteins have the ability to bind auxins (for an overview, see Napier et al., 2002), but no auxin signaling responses have yet been attributed to these proteins. If we loosely define a receptor as a protein that binds its ligand and triggers a biological response, then, to date, three different auxin receptors can be discerned: ABP1, the TIR1/AFB family, and, very recently, the S-PHASE KINASE-ASSOCIATED PROTEIN 2A (SKP2A), an E3 ligase SCF complex constituting F box protein of which turnover and molecular interactions are altered upon auxin binding (Jurado et al., 2010). In the following sections, we will examine several auxin responses in which ABP1 has been implicated, as well as the potential overlap with other auxin receptor pathways.
Cell Cycle
SKP2A has been implicated directly in cell proliferation, as it was found to positively regulate degradation of the cell cycle transcription factors E2FC and DPB (del Pozo et al., 2006; Jurado et al., 2008). A binding site in SKP2A has been identified that is required for auxin binding and E2FC and DPB degradation (Jurado et al., 2010), and SKP2A with a mutated auxin binding site is no longer able to promote cell proliferation. Several questions still need to be addressed, such as partial functional overlap with the SCFTIR1/AFB pathway or the degradation of SKP2A itself in response to auxin by a yet uncharacterized mechanism. Nevertheless, the results are sufficiently clear to treat SKP2A as an auxin receptor with a main role in cell cycle control.
Several lines of evidence suggest the involvement of ABP1 in cell cycle control. Downregulation of ABP1 function by transient immunomodulation in tobacco Bright Yellow-2 (BY-2) cell cultures leads to cell cycle arrest, predominantly at the G1-to-S transition (David et al., 2007). Similar approaches in Arabidopsis and tobacco confirmed that several promotive cell cycle components, such as cyclins CYCD3 or CYCD6, are downregulated in response to ABP1 knock-down and with corresponding phenotypes (Braun et al., 2008; Tromas et al., 2009). ABP1 downregulation also led to an increase in RETINOBLASTOMA RELATED (RBR) expression, which negatively affects cell division and has recently been related to cell differentiation (Borghi et al., 2010). Overexpression of CYCD3;1 or downregulation of RBR in ABP1 knock-down plants partially restored the cell proliferation defects (Tromas et al., 2009), indicating that downregulation of ABP1 affects the cell cycle via the CYCD/RBR pathway.
What happens at the molecular level is yet unknown. A direct interaction of ABP1 with components of cell cycle control is improbable because these proteins are typically localized to the nucleus or cytosol, whereas ABP1 is a luminal endomembrane protein and, as such, is physically separated from the nucleus and cytosol. On the other hand, SKP2A and TIR1 are localized predominantly in the nucleus, and the direct interaction between SKP2A and several cell cycle components has been demonstrated (Jurado et al., 2010). No direct interaction of TIR1/AFBs with cell cycle components have been reported so far, and effects of the Aux/IAA-ARF-SCFTIR1/AFB pathway on cell proliferation may be more indirect via transcriptional control of cell cycle genes (Vanneste et al., 2005).
Although a direct regulation of cell cycle components by ABP1 seems unlikely, it is possible that ABP1 overlaps with or has indirect effects on the other two auxin response pathways. Intriguingly, ABP1 downregulation increases expression of E2FC (Braun et al., 2008), which is a substrate of SKP2A. It would be interesting to analyze SKP2 and E2FC protein levels in ABP1 knock-down plants to see if and to what extent this pathway is affected. The expression of many Aux/IAA proteins (components of the SCFTIR1/AFB pathway) was found to be downregulated in ABP1 knock-down plants (Braun et al., 2008), and the induction of several Aux/IAAs became insensitive to auxin application after ABP1 downregulation (Tromas et al., 2009) or in heterozygous abp1/+ plants (Effendi et al., 2011). Whether these effects are direct or are merely the consequence of preceding events on cell division or auxin transport, which might affect cell type or auxin levels, is still unknown.
To conclude, cell cycle/proliferation appears to be a highly complex, auxin-responsive system in which all three known auxin receptor systems (ABP1, TIR1/AFB, and SKP2A) presumably play a role. Further research is needed to differentiate these roles and to distinguish between direct and more indirect effects.
Cell Expansion
Exit of proliferation and entry into differentiation typically involve an increase in cell size. Several lines of evidence support a role for ABP1 in control of cell expansion. Early protoplast swelling assays suggested a role for ABP1 in auxin-mediated cell expansion (Steffens et al., 2001), and overexpression of ABP1 in maize plants confirmed that ABP1 is a positive regulator of cell size (Jones et al., 1998). Accordingly, abp1-null mutant embryos have isodiametric cells that fail to elongate (Chen et al., 2001b), and downregulation of ABP1 in leaf tissues reduces the cell size (Braun et al., 2008). On the other hand, TIR1/AFBs do not seem to be involved in rapid cell expansion events, as multiple TIR1/AFB mutants still exhibit normal rapid elongation responses (Schenck et al. 2010). A potential mechanism is that extracellular ABP1 is required for auxin-mediated hyperpolarization of the plasma membrane, which has been proven experimentally in several, independent systems (Barbier-Brygoo et al., 1989; Rück et al., 1993; Thiel et al., 1993; Bauly et al., 2000; Dahlke et al., 2010). Activation of H-ATPases and the resulting acidification of the extracellular matrix actuates cell wall-modifying enzymes, such as expansins that, in turn, could loosen the cell wall and thus allow cell expansion (reviewed in Napier, 1995; Perrot-Rechenmann, 2010).
This relatively straightforward model (Figure 1) most certainly needs to be expanded to accommodate all observed effects of auxin with respect to cell expansion, especially the striking difference between shoot and root cells in which auxin promotes or inhibits cell expansion, respectively. Nevertheless, thus far, the experimental data seem to provide good support for the general hypothesis.
Clathrin-Dependent Endocytosis
Cell division and cell expansion are the building blocks of growth and patterning. Without doubt, auxin is an important phytohormone for patterning and organogenesis, and numerous reports have shown that local accumulation in developing tissue serves as a trigger and guide for organ development such as apical/basal patterning of the embryo (Friml et al., 2003), general organogenesis (Benková et al., 2003), phyllotaxis (Reinhardt et al., 2003), lateral root formation (Dubrovsky et al., 2008), and vasculature development (Sauer et al., 2006; Scarpella et al., 2006), but also directional growth responses such as gravitropism of the root or phototropism of the shoot (Friml et al., 2002a; Kleine-Vehn et al., 2010; Ding et al., 2011). Local auxin synthesis plays an important role in generating these auxin maxima (Cheng et al., 2006, 2007; Stepanova et al., 2008; Tao et al., 2008), but it is the directional (polar) cell-to-cell transport of auxin that offers a means not only to fine-tune the amount, but also to control the direction of auxin flux within a tissue.
High auxin flux rates have been proposed to enhance the cellular capacity for polar auxin transport and subsequently lead to the formation of conductive auxin channels, and ultimately, to vascular strands (Sachs, 1969). The feasibility of this hypothesis has been demonstrated by elegant experiments (Sachs, 1969) that inspired the formulation of mathematical feedback models for auxin transport-driven patterning in plant development. These theoretical approaches predicted the existence of polarly localized auxin efflux carriers that would determine the directionality of the polar auxin transport (Rubery and Sheldrake, 1974; Raven, 1975; Goldsmith et al., 1981; Mitchison et al., 1981; Wabnik et al., 2011).
Forward genetics later identified the PIN auxin efflux carriers that display the predicted polar localization at the plasma membrane (Okada et al., 1991; Chen et al., 1998; Gälweileret al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al. 1998; Berleth and Sachs, 2001; Friml et al., 2002a, 2002b). Their polar localization to a given cell side indeed determines the direction of intercellular auxin transport (Petrášek et al., 2006; Wiśniewska et al., 2006). PIN polarity and abundance at the plasma membrane are highly dynamic and rely on various vesicle transport-dependent processes such as the constitutive cycling of PIN proteins (Geldner et al., 2001; Paciorek et al., 2005; Dhonukshe et al., 2007; Kleine-Vehn et al., 2008a, 2009; Tanaka et al., 2009; Robert et al., 2010), de novo protein secretion (Heisler et al., 2005; Vieten et al., 2005; Dhonukshe et al., 2008), and protein degradation in lytic vacuoles (Abas et al., 2006; Kleine-Vehn et al., 2008b; Laxmi et al., 2008). Apparently, the dynamic nature of PIN polarization provides the needed flexibility to establish and maintain the auxin gradients within tissues during developmental stages, and, ultimately, shapes plant architecture (Benjamins and Scheres, 2008).
The concept of a positive auxin feedback mechanism for the regulation of polar auxin transport as initially suggested remains an important concept in auxin biology (Leyser, 2006). Because expression of PIN auxin carriers is directly controlled by auxin, presumably via the SCFTIR1/AFB pathway (Heisler et al., 2005; Vieten et al., 2005; Scarpella et al., 2006), an auxin-induced PIN expression mechanism could be part of the predicted positive feedback loop for auxin canalization. Additionally, in a subset of cells, PIN polarity has been shown to be regulated by auxin via a TIR1- and IAA17/AXR3-dependent mechanism that might substantially contribute to vascular patterning (Sauer et al., 2006).
Yet another, more direct, level of auxin feedback on PIN activity has been proposed, namely that auxin enhances its own efflux by modulating PIN protein trafficking. Auxin inhibits the internalization of PIN protein, thus leading to increased PIN abundance at the plasma membrane (Paciorek et al., 2005). Remarkably, this auxin-mediated inhibition of PIN protein internalization occurs within minutes (Paciorek et al., 2005; Robert et al., 2010). This auxin effect on endocytosis requires additional auxin binding sites distinct from TIR1/AFB and is, hence, independent of the SCFTIR1/AFB pathway (Robert et al., 2010). It should be noted that another report suggests TIR1 dependency of the auxin effect on endocytosis (Pan et al., 2009). These conflicting observations are possibly due to differences in experimental conditions and might rather reflect an auxin effect on PIN degradation (Pan et al., 2009; Robert et al., 2010).
The effect of auxin on endocytosis is related to the regulation of clathrin-dependent endocytosis and apparently requires ABP1 (Robert et al., 2010). Overexpression of an engineered secretable ABP1δKDEL mutant that lacks the KDEL ER retention signal-induced PIN internalization in transient tobacco BY-2 cell culture and stable Arabidopsis-overexpressing lines. In transient assays with tobacco BY-2, this action could be counterbalanced by pharmacological and genetic interference with clathrin activity (Robert et al., 2010), suggesting that extracellular ABP1 promotes clathrin-dependent endocytosis. Importantly, the promoting effect of ABP1 on clathrin-dependent endocytosis could be diminished by auxin application, indicating that auxin binding to ABP1 negatively regulates this activity (Robert et al., 2010). In line with this assumption, abp1-5 mutant seedlings that display a point mutation in the presumed auxin binding pocket (Xu et al., 2010) and are likely defective in auxin binding were partially resistant to the auxin effect on endocytosis (Robert et al., 2010). Transient overexpression of a putatively secretable ABP1-5δKDEL in tobacco BY-2 cell cultures fully induced PIN internalization similarly to the wild-type ABP1δKDEL (Robert et al., 2010). However, the mutation in the binding pocket affected its auxin sensitivity, leading to ABP1-induced PIN internalization even in the presence of auxin (Robert et al., 2010).
Intriguingly, these data indicate that auxin binding to ABP1 appears to inhibit the effect of ABP1 on the clathrin machinery (Figure 2). Therefore, ABP1 might not represent an auxin receptor in the classical sense, but rather it may modulate an intrinsic activity related to endocytosis. For further clarification, auxin binding assays with the abp1-5 mutant are needed to verify experimentally the assumed reduction in auxin binding affinity.
Furthermore, overexpression of ABP1 harboring the functional ER retention signal does not increase clathrin-dependent endocytosis (Robert et al., 2010), indicating that the ABP1 secretion mechanism, but not the gene expression level, is the rate-limiting step. Thus, the extracellular pool of ABP1 might be involved in regulation of endocytosis, but the underlying mechanism of how extracellular ABP1 could modulate the clathrin machinery (which resides at the opposite, cytosolic side of the plasma membrane) remains an open question.
ROP-Dependent Cytoskeleton Rearrangements
The direction of auxin flux is tightly connected with tissue polarization and the direction of cellular growth. At the tissue level, polarization of PIN proteins in individual cells is seemingly coordinated with the surrounding cells by a positive feedback between auxin and its directional transport (Mitchison, 1980; Sauer et al., 2006). Regulation of PIN internalization by extracellular ABP1 might guide tissue polarization by spatially defining PIN protein retention at the plasma membrane (Wabnik et al., 2010). However, such an ABP1-dependent PIN polarization mechanism remains to be experimentally assessed.
PIN1 polarization to the growing tip (lobes) of epidermal leaf pavement cells is required for interdigitated growth (Xu et al., 2010). In conditional abp1 knock-down and abp1-5 mutants, PIN1 polarization and pavement cell interdigitation are lost (Xu et al., 2010). In these pavement cells, auxin seemingly instructs interdigitated lobing and indentation by ROP2- and ROP6-dependent formation and rearrangement of the actin and microtubule cytoskeleton, respectively (Yang, 2008; Xu et al., 2010). ROP2/ROP6 are, as all RAC/ROP GTPases, plant-specific (Rho-related) molecular switches, of which the activity is regulated via shuttling between GDP- (inactive) and GTP-bound (active) states (Wu et al., 2011). Auxin rapidly activates ROP2 and ROP6 within minutes and requires an ABP1-dependent perception mechanism (Xu et al., 2010).
PIN1-mediated auxin efflux might lead to extracellular auxin accumulation, which will bind to extracellular ABP1 that subsequently coordinates cytosolic ROP2 and ROP6 activity with different kinetics (Xu et al., 2010). In turn, ROP2 activity appears to increase PIN1 abundance at the plasma membrane, leading to a positive feedback on ROP2-dependent formation of F-actin (Xu et al., 2010; Figure 3). However, whether ABP1-dependent PIN1 polarization in leaf pavement cells underlies the control of clathrin-dependent endocytosis (Robert et al., 2010), an actin-reliant secretion mechanism, or even the combination of both is currently not known.
Intriguingly, abp1 knock-down and the interference with auxin binding in abp1-5 mutants enhance intracellular PIN1 localization in leaf pavement cells, leading to reduced PIN1 labeling at the lobed cell side (Xu et al., 2010). By contrast, in root stele cells, PIN1 internalization is reduced in abp1 knock-down lines and is unchanged in untreated abp1-5 mutants (Robert et al., 2010). These findings indicate that the principal mechanisms of how auxin enhances PIN protein abundance might be distinct in root stele and leaf pavement cells. Such a difference could be on the level of ABP1-interacting proteins in roots versus shoots that may trigger distinct signaling pathways into the cytosol. Alternatively, ABP1 could act on ROP2/actin-dependent exocytosis of PIN1 in pavement cells, but on PIN endocytosis in root stele cells. In the latter scenario, it is tempting to speculate that it is the intracellular, rather than the extracellular, pool of ABP1 that contributes to a spatially defined PIN secretion mechanism for PIN polarization at the lobed pavement cell sides.
Other ROP-dependent processes, such as polar root hair positioning, might also require local auxin gradients (Fischer et al., 2006; Ikeda et al., 2009). However, the underlying mechanism is unknown and it would be interesting to see whether ABP1 regulates the polarized activity of ROPs during root hair positioning.
OPEN QUESTIONS
Even with the exciting new findings on the involvement of ABP1 in clathrin-dependent endocytosis and ROP-mediated cytoskeleton control, a comprehensive answer concerning the molecular function of ABP1 is still lacking. Thus, it seems fitting to conclude in the form of questions or musings to which only further experimental work can bring the answers.
Inside–Outside: Who Serves as Bridge?
Let us recapitulate: ABP1 is a luminal ER protein that is secreted (in small amounts) to the extracellular matrix. As a luminal ER protein, it is always inside the lumen of secretory vesicles and thus is unlikely to interact with cytosolic or nuclear components. Upon secretion, it becomes extracellular, where the acidic pH allows high-affinity binding to auxin. Whether intracellular or extracellular, ABP1 needs an interacting partner that bridges the membrane to affect events at the internal, cytosolic side of the ER or plasma membrane, such as clathrin or ROP activity. It is crucial to identify this (or these) interactor(s) to understand the function of ABP1 in events occurring at the cytosolic membrane side.
An interesting, but entirely speculative, idea in this context is that ABP1 might merely trigger a membrane hyperpolarization response, which, in turn, would serve as a signal for inhibition of clathrin-dependent endocytosis, ROP activation, cell wall loosening, and the whole range of downstream events related to these processes. It should not be overly difficult to devise experimental approaches to test this hypothesis.
ABP1 Turnover and Regulation of Transcription/Secretion?
Thus far, few studies systematically assessed the regulation of ABP1 at the transcriptional and posttranscriptional level, which is understandable given the technical difficulties of distinguishing between the large ER-resident pool and the minute secreted amounts of ABP1. Still, scattered evidence conveys the image of a stable protein that is continuously produced at low levels. Whereas in maize, a highly stable ABP1 protein was found at low levels irrespective of experimental conditions (Oliver et al., 1995), in Arabidopsis, ABP1 transcription was responsive to auxin (Effendi et al., 2011), but whether or not this results in higher protein levels has not been tested. In publicly available transcriptome databases, ABP1 expression seems low under most conditions. ABP1 might be degraded via the 26S proteasome after polyubiquitination in the ER by interaction with the E3 ubiquitin ligase At-RMA2 (Son et al., 2010), but further experiments will be needed to understand its biological importance.
The secretion of ABP1 seems to be limited by KDEL-mediated ER retention, as suggested by KDEL deletion constructs (Robert et al., 2010). Previously, in tobacco, mutation of KDEL to KEQL or KDELGL (which should render the motif inactive for ER retention) has been found to facilitate entry into the secretory pathway, although the ABP1 amounts in the extracellular space were not higher than those observed by overproduction of the wild-type protein (Bauly et al., 2000). This observation might hint that the amount of secreted ABP1 could be regulated by a more sophisticated mechanism than just ER escape and secretion by bulk flow. Whatever the mechanism, it seems unaffected by increased auxin concentrations, but as mentioned above, auxin starvation might induce ABP1 secretion (Jones and Herman, 1993).
What Is the Significance of ABP1 in Overall Auxin Signaling?
The most direct effects of auxin binding to ABP1 appear to occur at the cell periphery: Membrane hyperpolarization, cell expansion, inhibition of endocytosis, and activation of ROPs (Figures 1 to 3). These local, immediate effects on an individual cell will have an impact on temporally and spatially separated downstream events, such as changes of mechanical properties, inhibition of polar auxin transport, and cytoskeletal rearrangement. Among these, polar auxin transport has been best studied as a relay mechanism to orchestrate directional growth and patterning decisions by creating local auxin maxima, which, in turn, are read out by SCFTIR1/AFB- and SKP2-signaling networks. In this manner, ABP1 signaling at the plasma membrane could eventually lead to auxin responses that traditionally have been attributed to other pathways.
It will be a daunting but necessary task to assign the distinct observable auxin effects to the responsible signaling systems. A good example is the role of ABP1 in cell cycle/proliferation: It is difficult to determine to which extent the observed effects are directly related to downregulated ABP1 signaling or to secondary effects, such as changes in polar auxin transport. At least in roots, ABP1 downregulation has been reported not to affect auxin levels (Tromas et al., 2009). By contrast, long-term inactivation of ABP1 leads to roots with a terminated meristem where most cells have differentiated. This phenotype is remarkably similar to that of auxin starvation, as observed in weak gnom mutants in which auxin transport by the PIN efflux carriers is compromised (Geldner et al., 2004). Accordingly, slight auxin transport defects were reported in abp1/+ heterozygous plants (Effendi et al., 2011).
Then again, the consequences of slightly reduced auxin levels on cell proliferation are not understood and the altered transcriptional auxin responses of some Aux/IAA genes after ABP1 knock-down (Tromas et al., 2009) cannot easily be explained by a general auxin transport defect. With three different, but interconnected, receptor/signaling systems that can all affect cell proliferation, it will take some time before the role of ABP1 in this process is identified.
Why Is a Loss of ABP1 Function Lethal?
Whether full loss of function of the other auxin receptors, TIR1/AFB or SKP2, leads to embryo lethality is still unknown because most likely there is functional complementation by other members of the respective gene family. By contrast, ABP1 in Arabidopsis is a single-copy gene without homologs and full loss of function appears to be embryo lethal. However, although malformed, homozygous abp1-null embryos proliferate for a long time and form an undifferentiated mass of cells that are only aborted at a very late stage of seed development. A recent analysis has demonstrated that at very low frequencies, vegetative development can be observed to some extent even in full knock-out individuals (Effendi et al., 2011).
Loss-of-function abp1/abp1 embryos are characterized by severe patterning defects, over time increasingly random planes of cell division, and lack of cell elongation (Chen et al., 2001b). The novel findings discussed above suggest an involvement of ABP1 in clathrin-mediated endocytosis and ROP-mediated cytoskeleton rearrangements, which could explain the patterning defects as being related to defective polar auxin transport (as have been observed in gnom mutants; Geldner et al., 2003) and aberrant cell division and expansion as a consequence of a defective cytoskeleton. However, these results come from knock-down lines with reduced ABP1 function, and it is difficult to extrapolate the effect of complete absence of ABP1.
Hypothetically, if ABP1 were absent, auxin could not trigger membrane hyperpolarization, could not modulate clathrin-mediated endocytosis, and could not activate ROPs. The cell walls would be uniformly rigid, plasma membrane protein levels would be affected due to defects in endocytosis, and the cytoskeleton would likely be less organized. It will be interesting to see if these theoretical predictions can be verified experimentally.
Nevertheless, these defects alone might be not sufficient to explain the eventual arrest of cell proliferation and death of the embryo. Clearly, some genetic programs are not executed correctly that would ensure further development and survival, even as an unorganized callus-like mass of cells. It remains to be seen whether this lethality is due to communication problems with maternal tissues, auxin concentration-dependent effects on developmental programs, or a yet unknown direct function of ABP1 in cell maintenance.
What Is the Function of ABP1 in the ER Lumen?
Although no canonical ER retention signal has been found in nonvascular plant species, it has not been experimentally tested where ABP1 is localized in these clades. Too little is known about the secretory pathway in these plant species to predict whether a little or the majority of ABP1 would reside in the ER, or whether the bulk would be secreted. Until this is known, the idea of an evolutionarily recently acquired function in the ER of higher plants remains an interesting speculation.
The function (if any) of ABP1 in the ER lumen will probably remain an open question for some time. ABP1 affinity to auxin at ER luminal pH is assumed to be close to zero (Tian et al., 1995), thus a function related to auxin in the ER is unlikely. However, it must be said that the exact pH in the ER lumen of plant cells has not been directly measured. In addition, ABP1 binding partners or cofactors in the ER could affect auxin binding affinity. Therefore, one cannot completely rule out the possibility that auxin binds to ABP1 in the ER lumen.
Apart from this unresolved question, ER retention might be a means to reduce ABP1 secretion because overexpression of a KDEL deletion construct drastically affects PIN internalization (Robert et al., 2010). On the other hand, it seems rather inefficient to keep the majority of a protein stored in an inactive state when seemingly only very little is needed for the activity. It might be a means to guarantee a constant (small) amount of ABP1 secretion independent of transcriptional regulation. Also, the ER might possibly be important for ABP1 degradation, as suggested by the proposed interaction with the ER-resident E3 ligase At-RMA2 (Son et al., 2010).
To experimentally assess a function for ABP1 in the ER, KDEL deletion constructs may be a means to overcome the strict connection between ER-resident and ER-secreted ABP1. For example, it would be instructive to see whether this version could rescue the abp1-null mutant.
Bound or Free: Is This the Question?
The recent experiments with KDEL deletion constructs that presumably are secreted to the extracellular matrix more efficiently and the abp1-5 mutant allele that supposedly has reduced auxin affinity suggest an interesting idea: The ratio of auxin-free to auxin-bound ABP1 might be the deciding factor for biological responses. Overexpression of ABPδKDEL in BY-2 cell cultures strongly internalizes plasma membrane proteins. The addition of extracellular auxin restores normal plasma membrane protein abundance only for wild-type ABP1, but not for the abp1-5 mutant protein. Apparently, high amounts of auxin-free ABP1 destabilize plasma membrane proteins, potentially by an unrestricted clathrin-mediated endocytosis and/or by failure of ROP2/actin-dependent protein delivery to the membrane. This would mean that a certain amount of auxin-bound ABP1 is necessary at all times for proper plasma membrane protein abundance, and this might explain the drastic phenotypes of an ABP1 full loss-of-function mutant.
Once Bound: How Free?
Upon receiving a stimulus, a receptor triggers a downstream response. To avoid overreactions and to ensure a dynamic response, it is crucial that the receptor becomes inactive once the stimulus is over. The proposed stimulus for ABP1 is auxin binding, most likely in the extracellular matrix at an acidic pH. At this pH, ABP1 has a high auxin affinity and it is unlikely that auxin would spontaneously dissociate. Thus, there must be a mechanism that stops ABP1 from signaling once the auxin levels are low again. The nature of this mechanism has not been assessed experimentally thus far. One hypothesis would be that ABP1 binds to a membrane ligand and is endocytosed; another, that the stability of ABP1 would be reduced once bound to auxin, although all reports thus far indicate high protein stability regardless of auxin concentrations. Many more ways of deactivation can be imagined, but they remain pure speculation until experimental work provides further answers.
Can a Working Model Explain All Experimental Data?
Thus far, the short answer is No. Given the enormously complex network of auxin signaling, and ABP1 being just one piece of it, it seems unlikely that all ABP1-related observations will be explained comprehensively in a simple model. Instead of risking failure in the attempt, we will conclude with some ideas that we believe are worth a second thought.
In all studies reported to date, except in full null alleles, the ABP1 and auxin amounts vary in the extracellular space, where ABP1 binds auxin with high affinity. Thus, the ratio of auxin-free to auxin-bound ABP1 fluctuates, depending on ABP1 and auxin levels. Is the ratio of auxin-free to auxin-bound ABP1 the decisive factor in the competition for a common docking protein, where only auxin-bound ABP1 triggers a response? Or does auxin-free ABP1 have a function as well, as indicated for ABP1-induced endocytosis? Could all observed effects at the membrane actually be secondary effects of a single, direct impact of ABP1 signaling exerted on the plasma membrane status? In conclusion, ABP1 retains much of the mystery that has surrounded it for nearly four decades, but recent evidence suggests that it may be a key player in auxin signaling and is deserving of further attention.
Acknowledgments
We thank Stéphanie Robert, Jiří Friml, and Juan Carlos del Pozo for helpful discussion and critical reading of the manuscript, and Martine De Cock for help in preparing it. M.S and J.K.-V. are supported by an European Molecular Biology Organisation long-term postdoctoral fellowship and by the Vienna Science and Technology Fund, respectively.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Barbier-Brygoo H., Ephritikhine G., Klämbt D., Ghislain M., Guern J. (1989). Functional evidence for an auxin receptor at the plasmalemma of tobacco mesophyll protoplasts. Proc. Natl. Acad. Sci. USA 86: 891–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H., Ephritikhine G., Klämbt D., Maurel C., Palme K., Schell J., Guern J. (1991). Perception of the auxin signal at the plasma membrane of tobacco mesophyll protoplasts. Plant J. 1: 83–93 [DOI] [PubMed] [Google Scholar]
- Batt S., Venis M.A. (1976). Separation and localization of two classes of auxin binding sites in corn coleoptile membranes. Planta 130: 15–21 [DOI] [PubMed] [Google Scholar]
- Batt S., Wilkins M.B., Venis M.A. (1976). Auxin binding to corn coleoptile membranes: Kinetics and specificity. Planta 130: 7–13 [DOI] [PubMed] [Google Scholar]
- Bauly J.M., Sealy I.M., Macdonald H., Brearley J., Dröge S., Hillmer S., Robinson D.G., Venis M.A., Blatt M.R., Lazarus C.M., Napier R.M. (2000). Overexpression of auxin-binding protein enhances the sensitivity of guard cells to auxin. Plant Physiol. 124: 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Berleth T., Sachs T. (2001). Plant morphogenesis: Long-distance coordination and local patterning. Curr. Opin. Plant Biol. 4: 57–62 [DOI] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Fütterer J., Laizet Y., Hennig L., Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N., Wyrzykowska J., Muller P., David K., Couch D., Perrot-Rechenmann C., Fleming A.J. (2008). Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Villalobos L.I., Tan X., Zheng N., Estelle M. (2010). Auxin perception: Structural insights. Cold Spring Harb. Perspect. Biol. 2: a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-G., Shimomura S., Sitbon F., Sandberg G., Jones A.M. (2001a). The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. Plant J. 28: 607–617 [DOI] [PubMed] [Google Scholar]
- Chen J.-G., Ullah H., Young J.C., Sussman M.R., Jones A.M. (2001b). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., Masson P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlke R.I., Luethen H., Steffens B. (2010). ABP1: An auxin receptor for fast responses at the plasma membrane. Plant Signal. Behav. 5: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David K.M., Couch D., Braun N., Brown S., Grosclaude J., Perrot-Rechenmann C. (2007). The auxin-binding protein 1 is essential for the control of cell cycle. Plant J. 50: 197–206 [DOI] [PubMed] [Google Scholar]
- del Pozo J.C., Diaz-Trivino S., Cisneros N., Gutierrez C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.-D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Diekmann W., Venis M.A., Robinson D.G. (1995). Auxins induce clustering of the auxin-binding protein at the surface of maize coleoptile protoplasts. Proc. Natl. Acad. Sci. USA 92: 3425–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski Ł., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benková E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi Y., Rietz S., Fischer U., Scherer G.F.E. (2011). The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin-regulated genes. Plant J. 65: 282–294 [DOI] [PubMed] [Google Scholar]
- Fischer U., Ikeda Y., Ljung K., Serralbo O., Singh M., Heidstra R., Palme K., Scheres B., Grebe M. (2006). Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr. Biol. 16: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002b). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002a). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.-D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Geldner N., Richter S., Vieten A., Marquardt S., Torres-Ruiz R.A., Mayer U., Jürgens G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400 [DOI] [PubMed] [Google Scholar]
- Goldsmith M.H.M., Goldsmith T.H., Martin M.H. (1981). Mathematical analysis of the chemosmotic polar diffusion of auxin through plant tissues. Proc. Natl. Acad. Sci. USA 78: 976–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Henderson J., Bauly J.M., Ashford D.A., Oliver S.C., Hawes C.R., Lazarus C.M., Venis M.A., Napier R.M. (1997). Retention of maize auxin-binding protein in the endoplasmic reticulum: Quantifying escape and the role of auxin. Planta 202: 313–323 [DOI] [PubMed] [Google Scholar]
- Hertel R., Thomson K.-S., Russo V.E.A. (1972). In-vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta 107: 325–340 [DOI] [PubMed] [Google Scholar]
- Hesse T., Feldwisch J., Balshüsemann D., Bauw G., Puype M., Vandekerckhove J., Löbler M., Klämbt D., Schell J., Palme K. (1989). Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. EMBO J. 8: 2453–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Men S., Fischer U., Stepanova A.N., Alonso J.M., Ljung K., Grebe M. (2009). Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat. Cell Biol. 11: 731–738 [DOI] [PubMed] [Google Scholar]
- Inohara N., Shimomura S., Fukui T., Futai M. (1989). Auxin-binding protein located in the endoplasmic reticulum of maize shoots: Molecular cloning and complete primary structure. Proc. Natl. Acad. Sci. USA 86: 3564–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J., Roe J.L. (2005). SKS6, a multicopper oxidase-like gene, participates in cotyledon vascular patterning during Arabidopsis thaliana development. Planta 222: 652–666 [DOI] [PubMed] [Google Scholar]
- Jones A.M., Herman E.M. (1993). KDEL-containing auxin-binding protein is secreted to the plasma membrane and cell wall. Plant Physiol. 101: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Venis M.A. (1989). Photoaffinity labeling of indole-3-acetic acid-binding proteins in maize. Proc. Natl. Acad. Sci. USA 86: 6153–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Im K.-H., Savka M.A., Wu M.-J., DeWitt N.G., Shillito R., Binns A.N. (1998). Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282: 1114–1117 [DOI] [PubMed] [Google Scholar]
- Jurado S., Abraham Z., Manzano C., López-Torrejón G., Pacios L.F., Del Pozo J.C. (2010). The Arabidopsis cell cycle F box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S., Díaz-Triviño S., Abraham Z., Manzano C., Gutierrez C., del Pozo C. (2008). SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J. 53: 828–841 [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Ding Z., Jones A.R., Tasaka M., Morita M.T., Friml J. (2010). Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. USA 107: 22344–22349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008b). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. (2006). Dynamic integration of auxin transport and signalling. Curr. Biol. 16: R424–R433 [DOI] [PubMed] [Google Scholar]
- Löbler M., Klämbt D. (1985). Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). I. Purification by immunological methods and characterization. J. Biol. Chem. 260: 9848–9853 [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R.A., Grisafi P., Fink G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massotte D., Fleig U., Palme K. (1995). Purification and characterization of an auxin-binding protein from Arabidopsis thaliana expressed in baculovirus-infected insect cells. Protein Expr. Purif. 6: 220–227 [DOI] [PubMed] [Google Scholar]
- Mitchison G.J. (1980). The dynamics of auxin transport. Proc. R. Soc. Lond. B Biol. Sci. 209: 489–511 [Google Scholar]
- Mitchison G.J., Hanke D.E., Sheldrake A.R. (1981). The polar transport of auxin and vein patterns in plants. Philos. Trans. R. Soc. B Biol. Sci. 295: 461–471 [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier R.M. (1995). Towards an understanding of ABP1. J. Exp. Bot. 46: 1787–1795 [Google Scholar]
- Napier R.M., David K.M., Perrot-Rechenmann C. (2002). A short history of auxin-binding proteins. Plant Mol. Biol. 49: 339–348 [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.C., Venis M.A., Freedman R.B., Napier R.M. (1995). Regulation of synthesis and turnover of maize auxin-binding protein and observations on its passage to the plasma membrane: Comparisons to maize immunoglobulin-binding protein cognate. Planta 197: 465–474 [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.-D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Pan J., Fujioka S., Peng J., Chen J., Li G., Chen R. (2009). The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi K.C.S., Panigrahy M., Vervliet-Scheebaum M., Lang D., Reski R., Johri M.M. (2009). Auxin-binding proteins without KDEL sequence in the moss Funaria hygrometrica. Plant Cell Rep. 28: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010). Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Raven J.A. (1975). Transport of indoleacetic acid in plant cells in relation to pH and electrical potential gradients, and its significance for polar IAA transport. New Phytol. 74: 163–172 [Google Scholar]
- Ray P.M. (1977). Auxin-binding sites of maize coleoptiles are localized on membranes of the endoplasmic reticulum. Plant Physiol. 59: 594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.-R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Robert S., et al. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubery P.H., Sheldrake A.R. (1974). Carrier-mediated auxin transport. Planta 118: 101–121 [DOI] [PubMed] [Google Scholar]
- Rück A., Palme K., Venis M.A., Napier R.M., Felle H.H. (1993). Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J. 4: 41–46 [Google Scholar]
- Sachs T. (1969). Polarity and the induction of organized vascular tissues. Ann. Bot. (Lond.) 33: 263–275 [Google Scholar]
- Sauer M., Balla J., Luschnig C., Wisniewska J., Reinöhl V., Friml J., Benková E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20: 2902–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck D., Christian M., Jones A., Lüthen H. (2010). Rapid auxin-induced cell expansion and gene expression: A four-decade-old question revisited. Plant Physiol. 152: 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebl C., Walther A., Rescher U., Klämbt D. (1997). Interaction of auxin-binding protein 1 with maize coleoptile plasma membranes in vitro. Planta 201: 470–476 [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Carroll K.L., Hung K.F., Masson P.H., Somerville C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura S. (2006). Identification of a glycosylphosphatidylinositol-anchored plasma membrane protein interacting with the C-terminus of auxin-binding protein 1: A photoaffinity crosslinking study. Plant Mol. Biol. 60: 663–677 [DOI] [PubMed] [Google Scholar]
- Shimomura S., Sotobayashi T., Futai M., Fukui T. (1986). Purification and properties of an auxin-binding protein from maize shoot membranes. J. Biochem. 99: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Son O., Cho S.K., Kim S.J., Kim W.T. (2010). In vitro and in vivo interaction of AtRma2 E3 ubiquitin ligase and auxin binding protein 1. Biochem. Biophys. Res. Commun. 393: 492–497 [DOI] [PubMed] [Google Scholar]
- Steffens B., Feckler C., Palme K., Christian M., Böttger M., Lüthen H. (2001). The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J. 27: 591–599 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.-Y., Doležal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397 [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G., Blatt M.R., Fricker M.D., White I.R., Millner P. (1993). Modulation of K+ channels in Vicia stomatal guard cells by peptide homologs to the auxin-binding protein C terminus. Proc. Natl. Acad. Sci. USA 90: 11493–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Klämbt D., Jones A.M. (1995). Auxin-binding protein 1 does not bind auxin within the endoplasmic reticulum despite this being the predominant subcellular location for this hormone receptor. J. Biol. Chem. 270: 26962–26969 [DOI] [PubMed] [Google Scholar]
- Tillmann U., Viola G., Kayser B., Siemeister G., Hesse T., Palme K., Löbler M., Klämbt D. (1989). cDNA clones of the auxin-binding protein from corn coleoptiles (Zea mays L.): Isolation and characterization by immunological methods. EMBO J. 8: 2463–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A., Braun N., Muller P., Khodus T., Paponov I.A., Palme K., Ljung K., Lee J.-Y., Benfey P., Murray J.A.H., Scheres B., Perrot-Rechenmann C. (2009). The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS ONE 4: e6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A., Paponov I., Perrot-Rechenmann C. (2010). AUXIN BINDING PROTEIN 1: Functional and evolutionary aspects. Trends Plant Sci. 15: 436–446 [DOI] [PubMed] [Google Scholar]
- Utsuno K., Shikanai T., Yamada Y., Hashimoto T. (1998). Agr, an Agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 39: 1111–1118 [DOI] [PubMed] [Google Scholar]
- Vanneste S., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venis M.A. (1977). Solubilisation and partial purification of auxin-binding sites of corn membranes. Nature 266: 268–269 [Google Scholar]
- Venis M.A., Napier R.M., Barbier-Brygoo H., Maurel C., Perrot-Rechenmann C., Guern J. (1992). Antibodies to a peptide from the maize auxin-binding protein have auxin agonist activity. Proc. Natl. Acad. Sci. USA 89: 7208–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A., Sauer M., Brewer P.B., Friml J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12: 160–168 [DOI] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wiśniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C., Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Wabnik K., Govaerts W., Friml J., Kleine-Vehn J. (June 10, 2011). Feedback models for polarized auxin transport: An emerging trend. Mol. Biosyst. http://dx.doi.org/10.1039/C1MB05109A [DOI] [PubMed] [Google Scholar]
- Wabnik K., Kleine-Vehn J., Balla J., Sauer M., Naramoto S., Reinöhl V., Merks R.M., Govaerts W., Friml J. (2010). Emergence of tissue polarization from synergy of intracellular and extracellular auxin signaling. Mol. Syst. Biol. 6: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P.B., Růžička K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Woo E.J., Marshall J., Bauly J., Chen J.G., Venis M., Napier R.M., Pickersgill R.W. (2002). Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 21: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.M., Hazak O., Cheung A.Y., Yalovsky S. (2011). RAC/ROP GTPases and auxin signaling. Plant Cell 23: 1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J.-G., Wu M.-J., Perrot-Rechenmann C., Friml J., Jones A.M., Yang Z. (2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]